SUMMARY
Mouse p202 containing two HIN domains antagonizes AIM2 inflammasome signaling and potentially modifies lupus susceptibility. We found only HIN1 of p202 binds dsDNA, while HIN2 forms a homo-tetramer. Crystal structures of HIN1 revealed that dsDNA is bound on the opposite face to the site used in AIM2 and IFI16. The structure of HIN2 revealed a dimer of dimers, with the face analogous to the HIN1 dsDNA binding site being a dimerization interface. Electron microscopy imaging showed that HIN1 is flexibly linked to HIN2 in p202, and tetramerization provided enhanced avidity for dsDNA. Surprisingly, HIN2 of p202 interacts with AIM HIN domain. We propose this results in spatial separation of AIM2 pyrin domains, and indeed p202 prevented dsDNA-dependent clustering of ASC and AIM2 inflammasome activation. We hypothesize that while p202 was evolutionarily selected to limit AIM2-mediated inflammation in some mouse strains, the same mechanism contributes to increased interferon production and lupus susceptibility.
INTRODUCTION
The innate immune system directly recognizes pathogen molecules to alert the host of infection. Viral and bacterial nucleic acids can elicit innate immune responses due to unique structural features not found in the host, or due to an abnormal location in the cell. Within the endosomal system, Toll-like receptors 3, 7, 8 and 9 recognize dsRNA, ssRNA, G-rich oligonucleotides, and DNA containing unmethylated CpG dinucleotides, respectively [reviewed in (Kawai and Akira, 2010)]. In the cytosol, while the helicase domain-containing RIG-I like receptors sense cytosolic RNAs and induce type I interferon [reviewed in (Loo and Gale, 2011)], cytosolic DNA recognition leads to both inflammasome activation and interferon induction. Although a number of candidates have been proposed (Ablasser and Hornung, 2013), recent work suggested a novel protein, cyclic GMP-AMP synthase (cGAS), to be the DNA sensor leading to interferon-β induction via production of a cyclic dinucleotide second messenger (Sun et al., 2013; Wu et al., 2013). On the other hand, activation of inflammasome function by cytosolic DNA depends on recognition by absent in melanoma 2 (AIM2), a member of the pyrin and HIN200 domain-containing protein family (PYHIN, also known as p200 or HIN200 proteins) (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009)
Most PYHIN proteins contain an N-terminal pyrin domain (PYD), and the C-terminal region is comprised of one or two HIN domains (Cridland et al., 2012). The PYD belongs to the death domain fold superfamily (Park et al., 2007) that mediates homotypic interactions in assembly of oligomeric complexes important in both cell death and innate immune signalling pathways (Ferrao and Wu, 2012). HIN domains are unique to mammals (Cridland et al., 2012) and consist of tandem oligonucleotide /oligosaccharide binding-folds (OB-folds) known to interact with nucleic acids (Albrecht et al., 2005). The AIM2 HIN domain binds to cytosolic DNA, and the PYD mediates nucleation of an inflammasome by recruitment of ASC through a PYD/PYD interaction. ASC in turn recruits and activates procaspase-1 through a CARD/CARD interaction (Hornung and Latz, 2010; Rathinam et al., 2012; Schroder and Tschopp, 2010). Activated caspase-1 cleaves prointerleukin-1β (pro-IL-1β) and pro-IL-18 to generate mature proinflammatory cytokines IL-1β and IL-18. Caspase-1 activation can also induce rapid lytic cell death known as pyroptosis (Miao et al., 2011). Recent work showed that, in addition, ASC can recruit and activate procaspase-8, leading to apoptotic responses (Pierini et al., 2012; Sagulenko et al., 2013).
The mammalian PYHIN gene family is highly variable between species, with four members in human, one in cow and at least fourteen in mice, all clustered on chromosome 1 (Cridland et al., 2012). Apart from AIM2, the role of most other PYHIN proteins is not clearly defined. Human IFI16 and mouse p204 were suggested as DNA receptors leading to the production of type I interferon (Unterholzner et al., 2010). Since cGAS provides a good candidate for this role, it may be that the PYHIN proteins, which certainly bind to introduced foreign DNA, can modify or prolong the interferon response. The other PYHIN protein for which functional data has been obtained is mouse p202, which is an antagonist of the AIM2 inflammasome (Roberts et al., 2009).
Mouse p202 was the first member of the PYHIN family characterized, and was discovered as an interferon-inducible protein (Kingsmore et al., 1989). Unlike AIM2 and IFI16, p202 lacks a PYD but consists of two HIN domains. p202 binds rapidly to DNA introduced into the cytosol, and RNAi-mediated knockdown of p202 showed that it inhibits AIM2 inflammasome signaling (Roberts et al., 2009). Intriguingly, high p202 expression is seen in three strains of mice (NZB, BXSB and MRL) used as models of systemic lupus erythematosus (SLE or lupus), and is proposed to potentiate the disease (Choubey and Panchanathan, 2008; Haywood et al., 2006; Ichii et al., 2010; Rozzo et al., 2001). SLE is an autoimmune disease characterized by auto-antibodies directed against DNA and nucleic acid-associated proteins, and by elevated type I interferon. One of the few characterized monogenic causes of lupus is mutation in the gene for the DNase, Trex1, which is known to degrade retroviral cDNA (Crow and Rehwinkel, 2009; Yan et al., 2010). The trex1 knockout mouse accumulates non-chromosomal DNA, some derived from retroelements, which drives type I interferon production and autoimmunity (Stetson et al., 2008). Since misplaced DNAs in the cytosol can be sensed by PYHIN family members, whether p202 plays a role in lupus via inhibition of AIM2 response to DNA, requires further investigation.
Because of their biological importance, extensive efforts have been directed towards understanding the molecular mechanisms of DNA recognition by PYHIN proteins. Initial structure examination of both HIN domains from human IFI16 confirmed the OB-fold architecture of HIN domain (Liao et al., 2011), as proposed previously based on sequence analysis (Albrecht et al., 2005). Both OB folds in the HIN domain conform to the canonical OB-fold framework, with five twisted β-strands forming a closed β barrel and an α-helix connecting β3 and β4 strands (Theobald et al., 2003). The two OB folds are arranged in a roughly parallel manner and connected to each other by a bridging α-helix. Further crystallographic studies on AIM2 HIN and IFI16 HIN2 in complex with dsDNA showed that both OB folds and the linker in between engage in DNA phosphate backbone binding (Jin et al., 2012). Crystal structure and mutagenesis of mouse AIM2 suggests it binds to dsDNA similarly (Ru et al., 2013; Sung et al., 2012).
Regulation of cytosolic DNA sensing may be crucial for the balance between physiological host defense and pathological DNA-induced inflammation. To elucidate the role of p202 in this function, we performed extensive structural, biochemical and cellular studies. Unexpectedly, our crystal structures at 2.1Å, 1.9Å, 3.0Å and 3.4Å resolutions revealed that p202 HIN1 binds to dsDNA using the opposite surface relative to the site used for AIM2 and IFI16, and that HIN2 not only lacks DNA binding capability but also forms a tetrameric core for full-length p202. While the current manuscript was under revision, a crystal structure of p202 HIN1 in complex with a 14-mer dsDNA at 3.0Å resolution was also reported (Ru et al., 2013) and confirmed our observation on HIN1. We found surprisingly that there is a direct interaction between p202 HIN2 and AIM2 HIN both in vitro and in cells. Our data support that p202 specifically inhibits AIM2 signaling through direct binding to AIM2 and disruption of AIM2 clustering induced by recognition of double-stranded DNA, which may lead to enhanced interferon production and correlate with the role of p202 in lupus.
RESULTS
p202 HIN1 Binds to dsDNA Differently From Other Known HIN Domains
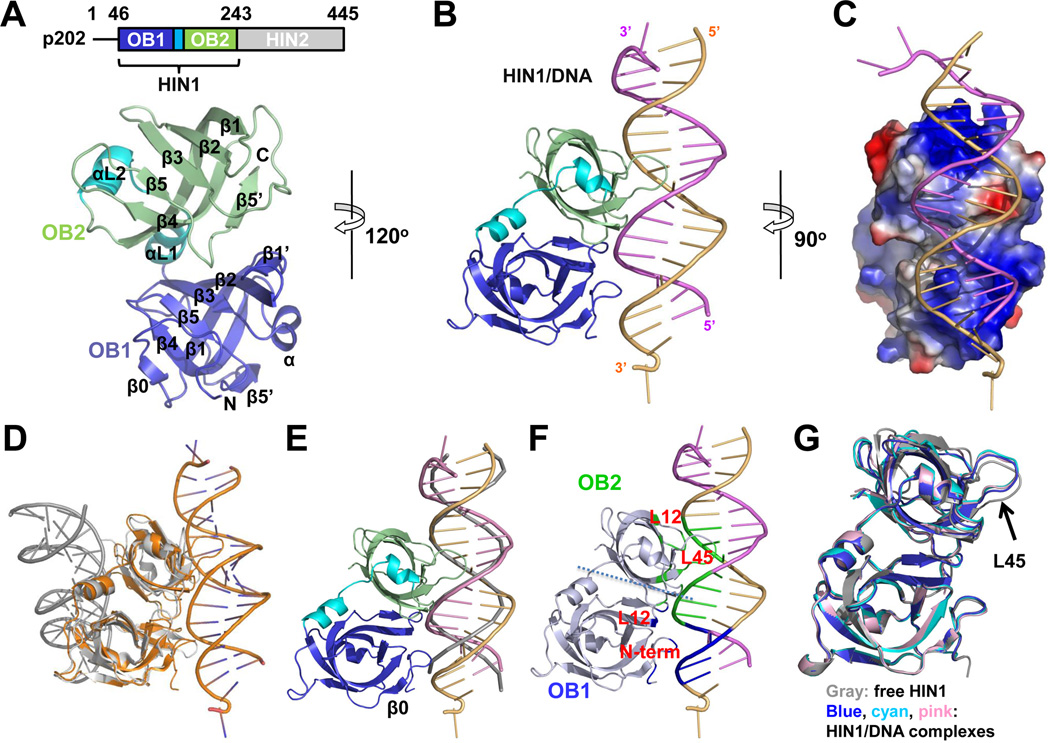
To elucidate the molecular basis of p202 function, we determined four crystal structures of p202: HIN1 at 2.1 Å resolution, HIN1 in complex with a 20-mer dsDNA and a 12-mer dsDNA at 1.9 Å and 3.0 Å resolutions, respectively, and HIN2 at 3.4 Å resolution (Figures 1A–1C, 2A, 3A, Table 1 and Figure S1). Although p202 HIN2 enhances dsDNA binding for full-length p202 (Roberts et al., 2009), HIN2 alone does not have measurable DNA-binding activity (Figure S1F). Surprisingly, despite the similar structure of p202 HIN1 to other HIN domains (Jin et al., 2012; Liao et al., 2011) (Table S1), the dsDNA binding surface is opposite to those in AIM2 HIN and IFI16 HIN2 (Jin et al., 2012) (Figure 1D).
Figure 1. p202 HIN1 Binds dsDNA in a Different Mode From Known HIN Domains.
(A) Above, domain diagram of p202. Below, free HIN1 structure with secondary structures labeled.
(B) Cartoon representation of the HIN1/20-mer structure. The two DNA chains are colored violet and light orange.
(C) HIN1 is shown as surface electrostatics and rotated 90° vertically from the orientation illustrated in (B).
(D) Superposition of the HIN1/20-mer (gold) and AIM2/DNA (gray) complexes. The HIN domains are superimposed, showing the opposite locations of the DNA relative to the HIN domains.
(E) Superposition of HIN1/DNA structure with ideal B-form DNA. HIN1 and 20-mer dsDNA are colored as in (B). Ideal B-form DNA is shown as a gray ribbon.
(F) Mapping of three interaction regions onto the HIN1/20-mer structure. N-terminal residues and the L12 loop of OB1 contact the minor groove while the L12 loop and the L45 region contact the major groove.
(G) Superposition of free HIN1 and HIN1 in complex with 20-mer or 12-mer dsDNA. DNA strands are omitted for clarity. OB1 is used for superposition to accentuate the movement of OB2 L45 (arrow).
See also Figure S1.
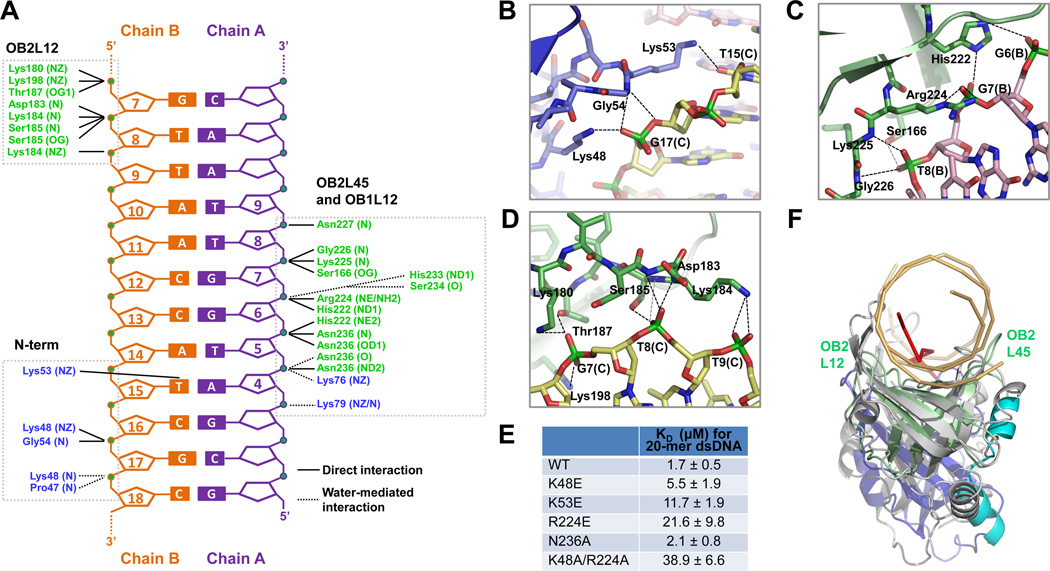
Figure 2. Detailed Interaction Between p202 HIN1 and dsDNA.
(A) Diagram summarizing HIN1/20-mer interactions. Non-interacting nucleotides are omitted. Three clusters of interaction (N-term, OB2L45 and OB1L12, and OB2L12) are boxed. Residues from OB1 and OB2 are shown in blue and green, respectively.
(B, C, D) Interaction details at N-terminus (B), OB2L45 and OB1L12 (C) and OB2L12 (D). Hydrogen bonds are represented by dashed lines.
(E) Summary of KD values of wild-type and mutant HIN1 with 20-mer dsDNA, determined by fluorescence polarization with fluorescein labeled 20-mer.
(F) Structural superposition of the HIN1/20-mer complex with the RPA/ssDNA complex. HIN1 and RPA (PDB ID: 1JMC) are superimposed using secondary structure (SSM) methods (Krissinel and Henrick, 2004). HIN1 is colored as in Figure 1A and dsDNA is in light orange. RPA is colored in gray and ssDNA in red. Dotted magenta arrows indicate the different conformations of L12 and L45 loops in HIN1 OB2 and RPA.
See also Figure S2.
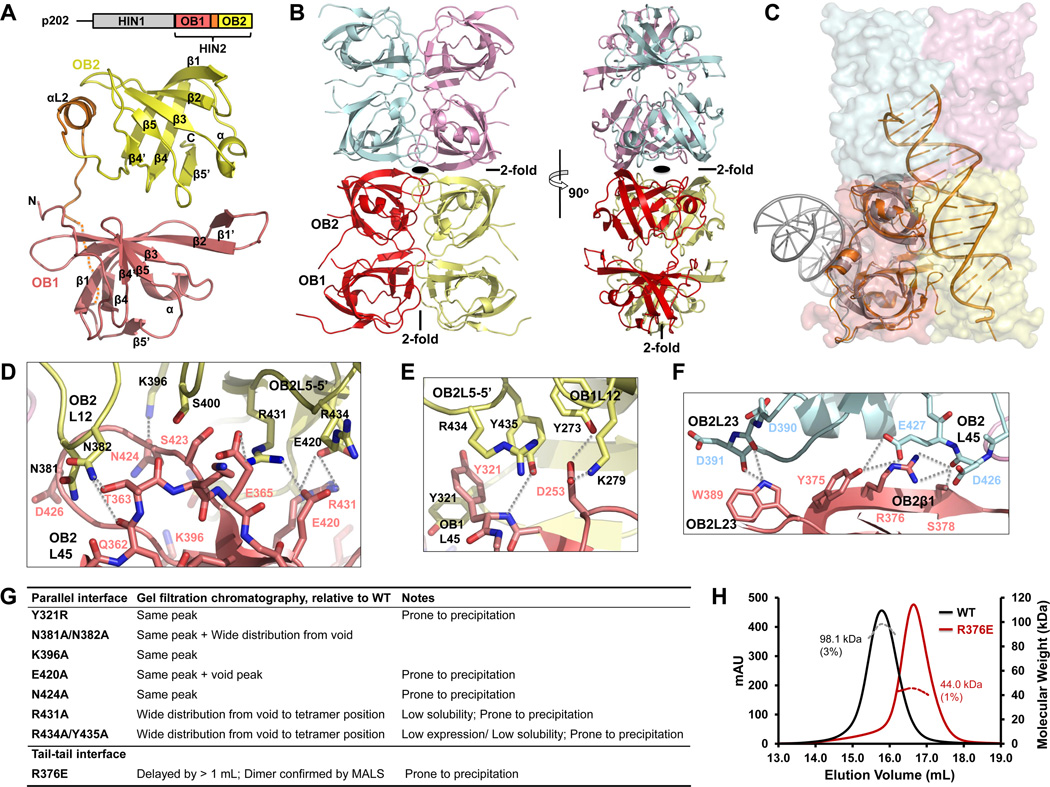
Figure 3. Tetramer Structure of p202 HIN2.
(A) Above, domain diagram of p202. Below, HIN2 monomer structure with secondary structures labeled. Residues without defined density are represented by the dotted line.
(B) HIN2 tetramer (dimer of dimers) structure in two orthogonal views with each monomer shown in light pink (A), yellow (B), pale cyan (C) and violet (D) , respectively. Locations of 2-old axes are either labeled or shown with a symbol.
(C) Monomer A (in light pink) in HIN2 tetramer is superimposed with the p202 HIN1/DNA structure (orange) and the AIM2 HIN/DNA structure (gray). While p202 HIN1 uses the similar interface for DNA interaction as in HIN2 for dimerization, AIM2 HIN uses the opposite surface for DNA interaction.
(D) Detailed interactions at the OB2-OB2 interface between parallel HIN dimers. Molecules A and B are colored in light pink or yellow as in (B). Residues mediating OB2-OB2 interaction are shown as sticks and labeled in light pink or black, respectively and secondary structures are labeled similarly. Hydrogen bonds are represented by dotted lines.
(E) Detailed interactions at the OB1-OB1 and OB2-OB1 interface between parallel HIN2 dimers. Residues and secondary structure motifs are colored and labeled as in (D). Hydrogen bonds are represented by dotted lines.
(F) Detailed interactions at the tail-to-tail interface of HIN2. Molecules A and C are colored in light pink or pale cyan as in (B).
(G) A summary of solution behaviors of HIN2 dimeric interface mutants.
(H) Molecular masses (MW) of wild-type HIN2 (black) and R376E mutant (dark red) were measured by multi-angle light scattering (MALS) coupled with size-exclusion chromatography.
See also Figure S3.
Table 1.
Crystallographic Statistics of p202 HIN1 and HIN2
| HIN1 | HIN1/20-mer | HIN1/12-mer | HIN2 | |
|---|---|---|---|---|
| Constructs | T46-E243 | T46-E243 | T46-E243 | K244-K445 |
| Structure determination | SAD | MR | MR | MR |
| Data collection | ||||
| Beamlines | X25 of NSLS | X29 of NSLS | X29 of NSLS | 24-ID-C of APS |
| Space group | P21212 | P21 | C2221 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 71.5, 76.2, 45.8 | 46.4, 86.4, 51.4 | 111.3, 156.1, 117.6 | 61.9, 71.9, 95.5 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 113.3, 90.0 | 90.0, 90.0, 90.0 | 90.0, 102.4, 90.0 |
| Resolution | 50–2.1 Å | 50–1.9Å | 50–3.0 Å | 50–3.4 Å |
| Rsym | 13.7% (70.9%) | 9.4% (67.0%) | 14.6% (73.9%) | 21.0% (87.2%) |
| I/σI | 9.3 (1.1) | 34.0 (1.9) | 22.4 (2.4) | 14.5 (3.5) |
| Completeness | 99.7% (97.9%) | 98.7% (91.4%) | 100% (100%) | 99.7% (97.9%) |
| Redundancy | 3.3 (2.7) | 7.1 (5.8) | 14.4 (14.7) | 6.7 (6.8) |
| Refinement | ||||
| Resolution | 50–2.1 Å | 25–1.9 Å | 50–3.0 Å | 50–3.4 Å |
| No. Protein | 1 | 1 | 2 | 4 |
| No. DNA | - | 2 | 7* | - |
| No. reflections | 21,940 | 30,294 | 22,039 | 21,483 |
| Rwork / Rfree | 19.7% / 24.0% | 19.1% / 22.8% | 20.4% / 25.5% | 21.5% / 27.3% |
| No. atoms | ||||
| Protein | 1604 | 1590 | 3180 | 5922 |
| DNA | - | 816 | 1504 | - |
| Water and ion | 66 | 179 | 24 | - |
| Average B-factors | ||||
| Protein | 33.9 Å2 | 36.9 Å2 | 66.9 Å2 | 108.0 Å2 |
| DNA | - | 54.8 Å2 | 74.7 Å2 | - |
| Water and ion | 36.2 Å2 | 46.7 Å2 | 90.9 Å2 | - |
| R.m.s deviations | ||||
| Bond lengths/angles | 0.008 Å / 1.22° | 0.006 Å / 1.16° | 0.008 Å / 1.49° | 0.01 Å / 1.31° |
| Ramachandran Plot | ||||
| Most favored/allowed | 95.4% / 100.0% | 96.5% / 100.0% | 88.7% / 99.2% | 91.3% / 100.0% |
Highest resolution shell is shown in parenthesis.
5 of the 7 DNA molecules form 5 dsDNA chains through crystallographic symmetry. One HIN1 and its symmetry mate interact with 3 DNA duplexes while the other HIN1 and its symmetry mate interact with 2 DNA duplexes. The remaining 2 DNA molecules form a duplex that does not interact significantly with HIN1.
Because the HIN1/20-mer dsDNA structure is of much higher resolution, our discussion will be based on this structure. The p202 HIN1 contacts a footprint of about 12 base pairs (bp) on nearly ideal B-form dsDNA (Figures 1E and 2A). In OB1, the N-terminal residues and the loop between β1 and β2 (L12) interact with edges of the dsDNA minor groove. The N-terminal residues provide key contacts using positively charged side chains and main chain amides (Figures 1F, 2A and 2B). The loop preceding β0 inserts itself into the minor groove and slightly pries it open (Figure 1E). In L12, two Lys residues interact with the other edge of the minor groove (Figure 2A). In OB2, the topologically equivalent L12 loop and the region around the loop between β4 and β5 (L45) protrude out to clasp the DNA duplex between them at the major groove. The most extensive interactions are from the L45 region, with intimate and extensive contacts to five consecutive backbone phosphates at phosphoryl oxygen atoms, utilizing both side chain atoms and main chain amides (Figures 1F, 2A and 2C). In fact, L45 region of OB2 and L12 of OB1 form an almost continuous surface (Figures 1F and 2A). The L12 loop interacts with three consecutive phosphate groups at the opposing edge of the major groove (Figures 1F, 2A and 2D). Overall, the interactions mediated by N-terminal residues and OB2 L12 are more electrostatic than those mediated by OB2 L45 and OB1 L12 (Figures 1C and 1F). Only minor structural perturbations in the OB2 L45 loop are observed in the HIN domain upon DNA binding (Figure 1G).
The HIN1 of p202 mainly interacts with backbone phosphates of the dsDNA (Figures 2A–D), explaining its indiscriminate binding to dsDNA (Roberts et al., 2009). We did not observe any clashes if the 2’ hydrogen atoms in the deoxyribose groups are replaced by hydroxyls as in dsRNA. However, dsRNA usually assumes an A-form double helix conformation, which has a helical pitch much shorter than the B-form dsDNA. Since HIN1 recognizes both major and minor grooves on dsDNA, the change in pitch will preclude simultaneous binding of both sites. Similarly, ssDNA does not provide a double helical structure for HIN1 recognition, explaining the lack of observed binding to either dsRNA or ssDNA (Roberts et al., 2009).
Structure-based mutagenesis of OB1 N-terminal residues (Lys48 and Lys53) for minor groove interaction and of OB2 residues (Arg224) for major groove recognition showed that these positively charged residues are crucial for HIN1/dsDNA interaction (Figure 2E, Figures S2A, S2B and S2D). In contrast, mutation of conserved positive residues on the opposite side of the molecule that are involved in AIM2 DNA binding (Jin et al., 2012) did not significantly affect DNA binding (Figures S2C and S2D). Mutation of Asn236 had minimal effects, suggesting the major contribution to the HIN1/DNA interaction may be electrostatic, consistent with the DNA sequence-independent nature of the interaction (Roberts et al., 2009). Interestingly, like the OB2 of p202 HIN1, L12 and L45 loops of an OB-fold are used for recognition of ssDNA (Raghunathan et al., 2000) such as by RPA and BRCA2 (Bochkarev et al., 1997; Yang et al., 2002) (Figure 2F). However, in p202, the L12 and the L45 loops are positioned remarkably more outward to accommodate the wider dsDNA (Figure 2F).
p202 HIN2 Forms a Tetramer in Solution and Crystal
Unlike IFI16, in which both HIN domains bind to dsDNA, albeit with different affinities (Unterholzner et al., 2010), p202 HIN2 not only lacks the ability to bind dsDNA (Figure S1F) but also unexpectedly forms a tetramer in solution (Figure S3A). Multi-angle light scattering measurement (MALS) determined the experimental molecular mass to be 98.1 kDa (3% error), in good agreement with a tetramer when compared to the calculated molecular mass of 23.2 kDa for monomeric HIN2. In the crystal structure of p202 HIN2 (Figure 3A), there are four molecules in each asymmetric unit (A, B, C and D), forming a dimer of dimers (Figure 3B). A nine-residue stretch connecting the two OB folds, 340KEDSSSSDE348, only has poorly defined main chain density and is not included in the structure. Two HIN2 molecules form a parallel dimer (A and B, or C and D), with a buried surface area of 1,683 Å2, and two such dimers further dimerize tail-to-tail. All four N-termini point outwards, consistent with their connection to HIN1 in full-length p202. Strikingly, the interface within the parallel dimer overlaps almost completely with the dsDNA binding surface in p202 HIN1, but is opposite of the dsDNA binding surface of the AIM2 HIN domain (Figure 3C).
The symmetrical parallel dimer interface contains a mixture of hydrophobic, hydrogen bonding and salt bridge interactions. It can be further divided into OB2-OB2 interface (Figure 3D) and OB1-OB1 interface which also contains contributions from OB2-OB1 “cross” interaction (Figure 3E). OB2-OB2 interface is the larger of the two. It consists of regions around loop L45 and residues preceding β1 from one protomer and loop L12 and residues connecting β5 and β5’ (L5-5’) of the other. L45 loop of one protomer is positioned between L12 and L45 loops of the neighboring protomer. An intermolecular hydrogen bond between Asn424 in L45 and Lys396 likely stabilizes the conformation of L45 loop. The stretch of residues preceding β1 in OB2, Gln362 and Thr363, form hydrogen bonds with Asn382 in L12 in the neighboring protomer. Glu420 on the short β4’ strand and Arg431 on L5-5’ protrude out, forming intermolecular salt bridges with Arg431 and Glu420 (Figure 3D). The OB1-OB1 interface is smaller and involves interaction between loops L45 in both protomers and the loop preceding β1 in one protomer and loop L12 in the other. Carboxylate group of Asp253 can form hydrogen bonds simultaneously with Tyr273 and Lys279 of the other molecule, while Tyr321 in L45 contacts its equivalent in the other molecule via hydrophobic and possibly aromatic interaction. Arg434 and Tyr435 from OB2 L5-5’ loop point towards OB1, forming hydrogen bonds and salt bridges with Asp253 and Tyr321 as well (Figure 3E). The tail-to-tail dimer interface is composed of OB2 exclusively, involving L23 and L45 loops in one protomer and the C-terminal half of β1 and the L23 loop in the other (Figure 3F). L23 loop interaction is more hydrophobic while the interaction between the C-terminal β1 and L45 loop contains more hydrogen bonds.
We have generated a number of mutations on the tetramerization interface of p202 HIN2. Most of the mutants at the extensive parallel dimer interface were compromised in protein structural integrity as shown by the tendency to precipitate, wide distribution in gel filtration chromatography, lower solubility and aggregation (Figure 3G). These data are consistent with the largely hydrophobic nature of this interface. A mutation at the smaller tail-to-tail interface, R376E, however, was well behaved and shown to be a dimer by MALS (Figure 3H)
Full-length p202 is Tetrameric in Solution and in Cells
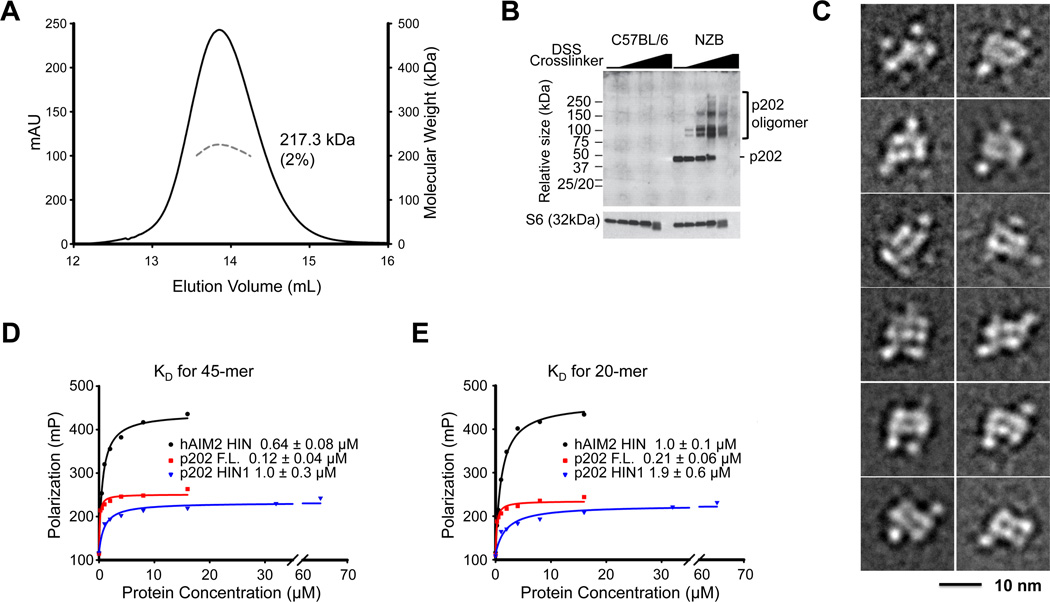
Full-length p202 is also a tetramer in solution, measured by gel filtration and MALS (Figure 4A). Results from protein cross-linking within cells are consistent with a tetramer form of p202 existing in vivo (Figure 4B). To obtain structural information on full-length p202, we carried out electron microscopy (EM) studies on HIN2 and full-length p202. Class averages of EM images on negatively stained HIN2 showed the similar elongated molecule as the crystal structure (Figure S3B). However, EM images of full-length p202 were heterogeneous (Figure S4A), and in the class averages, only the central HIN2 structures are consistently visible despite the very large number of particles (>34,000) and classes (400) (Figure 4C, S4B). The data suggest HIN1 domains are flexibly attached to the HIN2 core, which accounts for additional surrounding densities in the class averages. There is a linker of 14 residues in the primary sequence of p202 between the HIN1 and the HIN2 structures (Figure S1A). As expected from deletion of HIN2 in reducing p202/DNA interaction (Roberts et al., 2009), we found that full-length p202 exhibits ~10-fold and ~5-fold enhanced affinity for DNA relative to p202 HIN1 and AIM2 HIN respectively (Figures 4D and 4E), suggesting that full-length p202 may compete favorably with AIM2 for limited dsDNA in the cytosol.
Figure 4. Tetramer Organization of Full-length p202.
(A) Molecular mass (MW) of full-length p202 was measured by multi-angle light scattering (MALS) coupled with size-exclusion chromatography. Elution peak is shown as the solid line while molecular mass distribution is shown as the dashed line. Calculated molecular mass with error is labeled on p202 elution peak.
(B) p202 forms oligomers in cells. NZB and C57BL/6 bone marrow-derived macrophages (BMM) were left untreated, or treated with 4-fold serial dilutions of DSS amine-reactive crosslinker from 5mM to 26µM, and p202 (monomer = 50 kDa) was analyzed by western blotting.
(C) Selected class averages of negatively stained full-length p202 revealing the conformational heterogeneity of the particles. Although the averages show the tetrameric HIN2 core, none of them show clear density for all four HIN1 domains. The side length of the individual panels is 21.8 nm.
(D, E) Fluorescence polarization measurement of DNA affinities between AIM2 HIN, p202 fulllength or HIN1 and 45-mer (D) or 20-mer (E) dsDNA.
See also Figure S4.
Direct Interaction Between p202 and AIM2 Underlies Specific Inhibition of AIM2 Activation by p202
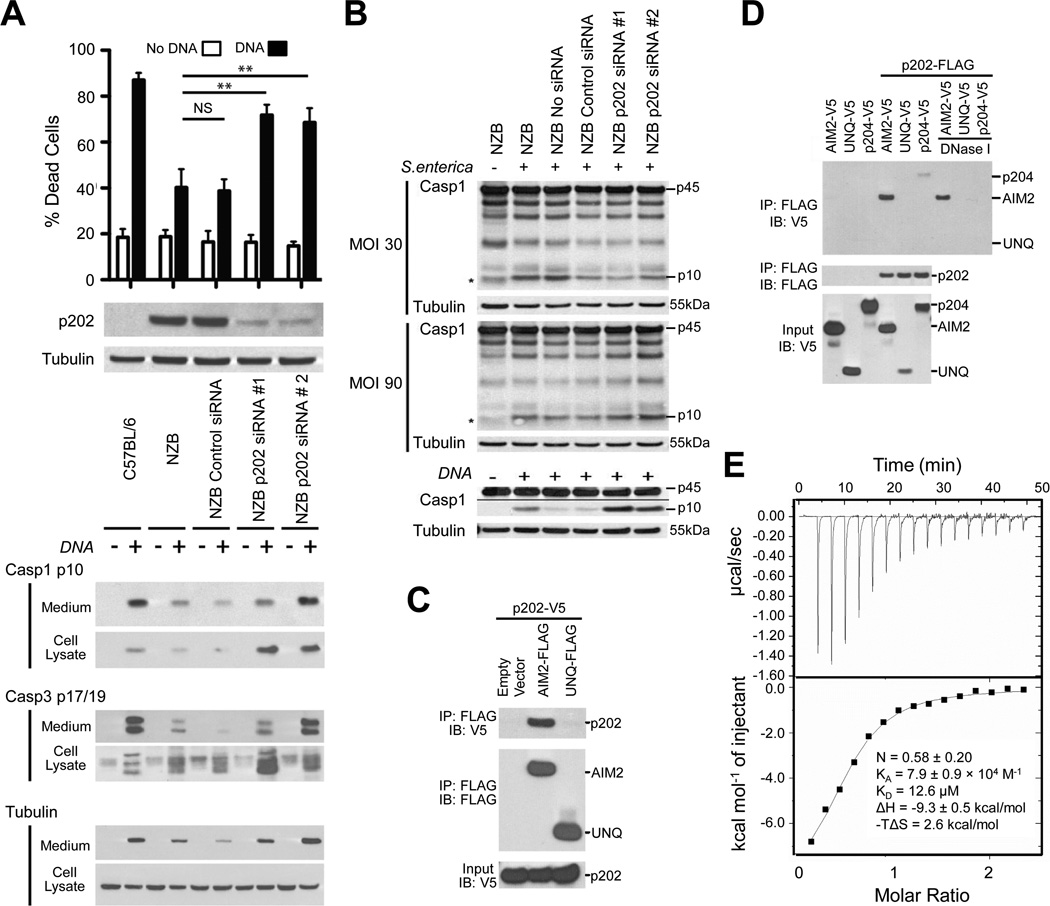
The activity of the AIM2 inflammasome can be observed following transfection of bone marrow macrophages (BMM) with dsDNA, by appearance of cleaved forms of caspase-1 and caspase-3, and cell death (Figure 5A). The processed caspases are split between the cell lysate and medium due to rapid caspase-1-dependent pyroptotic death. p202 expression is elevated in lupus-susceptible mouse strains, including NZB, in comparison with the C57BL/6 strain (Choubey and Panchanathan, 2008; Roberts et al., 2009; Rozzo et al., 2001), and AIM2 inflammasome function is low in NZB macrophages (Roberts et al., 2009). Knockdown of p202 in NZB BMM significantly enhanced DNA-dependent caspase-1 and caspase-3 cleavage and cell death (Figure 5A, Figures S5A and S5B). Under these conditions, cell death is entirely dependent on AIM2 and ASC (Figure S5C), and consequently we conclude that the knockdown of p202 alleviates inhibition of the AIM2 response to DNA in NZB cells. Results for C57BL/6 cells are shown to illustrate the level of response in a strain fully competent for AIM2 inflammasome function. Due to the undoubted genetic variability at many loci, we do not infer that the difference between these strains is necessarily solely due to p202. However the effect of p202 within the NZB strain is clearly seen from the effect of knockdown. To eliminate the possibility of some broad effect of p202 on general inflammasome function, we demonstrated that there was little or no effect of p202 knockdown on the NLRC4-mediated response to infection of BMM with Salmonella enterica serovar Typhimurium (Figure 5B). A small increase in caspase-1 cleavage with p202siRNA#2 was not corroborated by the other siRNA; any effect is likely insignificant compared to that seen on the AIM2-mediated response to DNA in the same experiment (Figure 5B). While the higher dsDNA-binding affinity of p202 suggests it could act to sequester cytosolic DNA from AIM2, such a model does not explain why p202 selectively inhibits AIM2-mediated caspase-1 activation, but not the induction of interferon-β (Figures S5D and S5E), which also requires receptor recognition of DNA.
Figure 5. p202 Binds Directly to AIM2 and Inhibits its Function.
(A) Knockdown of p202 enhances caspase activation and cell death in response to electroporated DNA. Degree of knockdown is shown by western blot in the middle panel. The upper panel shows cell death assessed by propidium iodide staining one hour after electroporation of BMM with or without 20µg calf thymus (CT) DNA. Bars show mean and SEM for 4 independent experiments (** p<0.01, paired one tailed t test). The lower panel shows a western blot for cleavage of caspases-1 and −3, at 30 minutes after electroporation with or without 20µg CT DNA. Proteins are found both in cell extracts and medium, due to cell death.
(B) Knockdown of p202 has no effect on NLRC4 inflammasome responses to Salmonella. siRNA-transfected BMM were infected with Salmonella enterica Typhimurium SL1334 at multiplicity of infection (MOI) 30 and 90 for 5 hours. Combined culture medium and cell lysate were immunoblotted for caspase-1. Procaspase-1 (p45) and cleaved caspase-1 (p10) are shown. The asterisk denotes a nonspecific band. Under these conditions we find caspase cleavage is almost entirely dependent on NLRC4 (unpublished data). The AIM2-mediated DNA response measured within the same experiment is shown in the lower panels.
(C) p202 associates with AIM2 in cells. Epitope-tagged proteins were expressed in HEK293 cells, immunoprecipitated (IP) from cell extracts with anti-FLAG and immunoblotted (IB) as indicated.
(D) DNaseI treatment does not prevent interaction of AIM2 and p202. Epitope-tagged proteins were expressed in HEK293 cells, immunoprecipitated (IP) from cell extracts with anti-FLAG, and beads washed in the presence or absence of DNaseI.
(E) p202 HIN2 binds to mouse AIM2 HIN as measured by isothermal titration calorimetry (ITC).
See also Figure S5.
To address the specific inhibition of AIM2 responses by p202, we examined by immunoprecipitation whether p202 could directly interact with AIM2. Following co-expression in HEK293 cells, V5-tagged p202 co-immunoprecipitated with FLAG-tagged AIM2, but not with the control protein FLAG-UNQ (Figure 5C). Reversal of the tags confirmed the specific association, and this interaction was not destroyed by treatment of the bead-bound complex with DNaseI (Figure 5D). This indicates that the two proteins are not merely co-precipitating due to binding to the same piece of DNA. Coexpression of FLAG-p202 with the V5-tagged mouse p204 showed a small degree of coimmunoprecipitation, but this was lost with DNaseI treatment (Figure 5D). Using purified recombinant mouse AIM2 HIN and p202 HIN2, we further mapped and measured the interaction by isothermal titration calorimetry (ITC) as exhibiting a dissociation constant of 12.6 µM and a stoichiometry of 2 AIM2: 4 p202 (Figure 5E). p202 HIN2 interacted with human AIM2 HIN as well, but did not bind IFI16 HIN1 or HIN2 (Figure S5F). These results show a specific interaction between AIM2 and p202 rather than a general affinity between HIN domains. Although an AIM2/p202 interaction has been proposed previously (Choubey et al., 2000), the interaction was thought to be mediated by a motif MFHATVAT (Choubey et al., 2000; Koul et al., 1998), which is actually buried in the core of HIN domain structures (Figure S5G).
p202 Prevents AIM2-mediated ASC Clustering
We then investigated whether HIN2 binding to AIM2 prevents its interaction with dsDNA. DNA binding affinity of mouse AIM2 HIN remained very similar in the absence or presence of excess p202 HIN2 (Figure S6A), suggesting that the DNA binding surface on AIM2 is not blocked by HIN2. Disruption of the tail-to-tail dimer interaction in the R376E mutant did not significantly change binding affinity and stoichiometry between HIN2 and AIM2 HIN (Figure S6B), suggesting that each HIN2 dimer mediated by the parallel interface binds to one AIM2 HIN molecule. Computational docking supported this observation in which AIM2 HIN domains bind at both ends of HIN2 tetramer (Figure S6C).
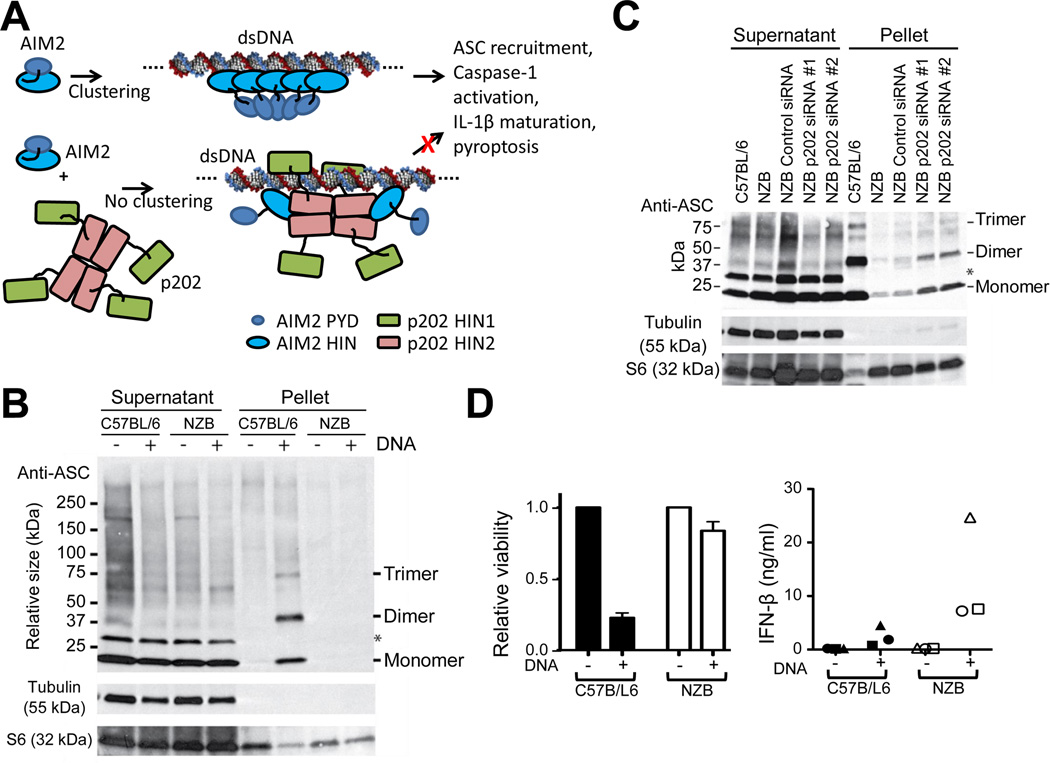
The generation of the DNA inflammasome response relies on the recruitment and clustering of ASC mediated by homotypic PYD interactions between AIM2 and ASC. ASC then forms an oligomeric structure, recruiting procaspase-1 via homotypic caspase recruitment domain (CARD) interactions. We propose that the interaction of AIM2 with p202 leads to spatial separation of AIM2 PYDs, preventing ASC clusters from forming (Figure 6A).
Figure 6. p202 Prevents Clustering of ASC and Interferes with AIM2 Signaling.
(A) Schematic model of p202 interference with AIM2 signaling. Upon dsDNA stimulation, multiple AIM2 bind to the same strand of dsDNA, resulting in ASC clustering, inflammasome formation and signaling. In contrast, the interaction between p202 and AIM2 dilutes AIM2 clustering to inhibit inflammasome formation.
(B) DNA transfection strongly induces ASC clustering in inflammasome structures in C57BL/6 but not NZB BMM. Cells were electroporated with or without 10µg CT DNA and at 10 minutes post electroporation, were treated with DSS cross-linker for a further 30 minutes. Cell extracts were pelleted at 6000g, and pellet and supernatant fractions were assessed by western blot for ASC. Transfer of ASC into the pellet fraction and formation of multimers are indications of inflammasome formation. ASC monomer, dimer and trimer bands are marked to the right. The asterisk denotes an unidentified protein which does not form part of the inflammasome ASC speck.
(C) Knockdown of p202 increases ASC clustering in DNA-transfected NZB cells. Cells were treated with or without the indicated siRNAs for 48 hours, then electroporated with 10µg CT DNA. Formation of ASC multimers was assessed as per (B).
(D) Rapid death of C57BL/6 BMM correlates with lower production of IFN-β. BMM were electroporated with or without 20 µg CT DNA, and viability was assessed by MTT cleavage after 3 hours (left panel). Results are the mean of three independent experiments +/− SEM. IFN-β was measured by ELISA of culture medium after 6 hours (right panel). Results of the three experiments are shown as different shaped symbols, with each data point the average of triplicate cell treatments.
See also Figure S6.
ASC recruitment into an inflammasome structure was assessed in C57BL/6 and NZB macrophages treated with a protein crosslinking reagent after electroporation with DNA. In control (no DNA) cells, ASC was a monomer in the soluble cellular fraction (Figure 6B). Following DNA transfection of C57BL/6 cells ASC was found in the 6000g pellet fraction of cell extracts, however was absent from the similarly treated NZB pellet fraction (Figure 6B). This, together with cross-linking of ASC into dimers and trimers indicated inflammasome formation in the C57BL/6 cells. Knockdown of p202 led to an increase in total ASC and cross-linked ASC oligomers in the pellet fraction of DNA-treated NZB cells (Figure 6C). These results are consistent with the model presented above, where p202 spatially separates AIM2 molecules, and therefore prevents recruitment of ASC into a dense inflammasome structure.
Given the suggested role of p202 as a murine lupus susceptibility modifier, it is relevant to ask whether interference with AIM2 inflammasome function by p202 could have a causal role in lupus. p202 could hamper the clearance of microorganisms which are detected via the AIM2 inflammasome, by reducing the production of IL-1β and IL-18. An inflammatory environment promoted by the resulting prolonged infections could promote lupus. Alternatively, p202 could prevent the appropriate cell death of macrophages when they encounter cytosolic DNA. In the latter scenario, surviving cells would synthesize elevated levels of cytokines such as type I interferon, which is induced by cytosolic DNA, and increased serum levels of which are associated with the development of lupus (Agrawal et al., 2009; Elkon and Stone, 2011). Consistent with this proposal, NZB BMMs were more viable than C57BL/6, and secreted greater levels of interferon-β following DNA transfection (Figure 6D). This is likely due to the increased cell survival, as NZB and C57BL/6 cells produce similar levels of interferon-β mRNA in response to cytosolic DNA (Figures S6D and S6E).
DISCUSSION
Here we have shown that the p202 HIN1 interacts with dsDNA in a manner that is completely different from those of AIM2 HIN and IFI16 HIN2 (Jin et al., 2012). In fact, the DNA binding surface is on the opposite side of these previously reported HIN/dsDNA structures. This finding is quite unexpected as the HIN domain structures themselves superimpose well (Table S1). While the current manuscript was under revision, a crystal structure of p202 HIN1 in complex with a 14-mer dsDNA at 3.0Å resolution was reported (Ru et al., 2013), confirming our observation.
HIN domains fall into three sequence subtypes, HIN-A, -B and -C, although no functional difference has been ascribed to these (Cridland et al., 2012; Ludlow et al., 2005). While the previous structures of AIM2 HIN/dsDNA and IFI16 HIN2/dsDNA are from HIN-C and -B family, respectively, p202 HIN1 belongs to the HIN-A family. Although some DNA binding residues in AIM2 HIN-C or IFI16 HIN-B are conserved in p202 HIN-A, mutations on these residues did not affect p202/dsDNA interaction. Conversely, p202 residues that contact DNA backbone are not all conserved in AIM2 HIN-C or IFI16 HIN-B (Cridland et al., 2012). The interactions between these three HIN domains and dsDNA are mostly electrostatic in nature, consistent with sequence-independent recognition. It appears then that the dsDNA-binding abilities of HIN-A in p202 must have evolved separately from the HIN-B and -C domains in AIM2 and IFI16. HIN-C domains are the most basal in phylogenetic analysis, with HIN-A and -B evolving later (Cridland et al., 2012). There is thus no simple explanation for the similarity of the DNA binding face used in p202 HIN-A and related OB-fold proteins such as RPA and BRCA2. It also remains to be determined whether other HIN-A domains bind similarly, or whether p202 alone uses this DNA binding mode.
On the other hand, HIN2 of p202 not only lacks dsDNA binding ability, but exists as a tetramer in solution, which was shown to be a dimer of dimers in the crystal. HIN2 first dimerizes in a parallel way with both OB1 and OB2 participating. Two such dimers then organize tail-to-tail, using only OB2 to complete tetramerization. The parallel dimer interface largely overlaps with the dsDNA binding interface in p202 HIN1. The full-length p202 is also a tetramer both in cells and as the purified protein in vitro. The tetrameric HIN2 serves as the central platform for HIN1 to append to and increases the avidity of dsDNA binding of HIN1. Although p202 HIN2 belongs to the HIN-B subtype, there is a remarkable divergence of its sequence from other HIN-B domains observed in a phylogenetic analysis (Cridland et al., 2012), consistent with evolution of a new function for the p202 HIN-B domain. Homo-oligomerization may be a desired attribute, since p204 HIN2 displayed a tendency to dimerize in solution (unpublished data).
Recent work speculated that p202 inhibits the function of AIM2 by effective competition for DNA (Ru et al., 2013). We have previously shown that upon introduction of DNA, all the p202 in a cell colocalizes with the DNA (Roberts et al., 2009). This suggests that p202 is not present in excess and is therefore unlikely to completely coat the DNA. In addition, p202 does not inhibit the induction of interferon-β, showing that it is not inhibiting access of all proteins to DNA. We showed instead that AIM2 HIN and p202 HIN2 directly interact. This interaction cannot be mediated by the highly conserved MFHATVAT motif proposed previously to mediate HIN domain homo- or hetero-dimerization (Choubey et al., 2000) because the motif is buried in the core of the structure. Regardless of the site of contact, p202 reduced AIM2-mediated ASC clustering in cells. This supports the hypothesis that p202 binds and spatially separates AIM2 such that its pyrin domains cannot nucleate the ASC speck formation.
The apparent rapid evolution of p202 suggests a significant selective pressure to limit AIM2-mediated functions. Indeed during mammalian evolution the AIM2 gene has been independently lost from several lineages (Cridland et al., 2012), suggesting that its function is not always advantageous. In addition to evolution of the gene sequence encoding p202, high expression of p202 in the NZB mouse strain may be due to gene amplification (unpublished data) and promoter variation (Choubey and Panchanathan, 2008). A relevant selective pressure driving increased expression could have been the need to limit AIM2-mediated IL-1β production and macrophage cell death in response to endogenous retroviruses. Lupus-prone mouse strains NZB, BXSB, and MRL, but not control strains, all express full length mRNA for an endogenous modified polytropic retrovirus in the thymus from birth (Krieg and Steinberg, 1990), and the same strains have elevated gene expression for p202 (Haywood et al., 2006; Ichii et al., 2010; Rozzo et al., 2001). A link between endogenous retroviruses and autoimmunity, particularly murine and human lupus, has long been suggested (Balada et al., 2010; Baudino et al., 2010; Perl et al., 2010). Trex1, which participates in degradation of retroviral cDNA, is mutated in a subset of lupus patients, and this suggests a causal role for aberrant cytosolic DNA in this disease through induction of type I interferon (Crow and Rehwinkel, 2009; Stetson et al., 2008; Yan et al., 2010).
With either endogenous retroviral expression or infection, p202 could specifically inhibit AIM2-mediated cell death, allowing increased production of type I interferon in response to cytosolic DNA, promoting murine lupus. Low AIM2 function would also reduce DNA-dependent release of IL-1β and IL-18. This could reduce clearance of certain pathogens or alter interaction with commensals, possibly contributing to lupus triggering. However, we do not infer that these cytokines are never elevated in established lupus, as there may be chronic inflammasome stimuli. In addition, IL-1β promoted disease in an induced model of murine lupus (Voronov et al., 2006). Although there is no human ortholog to p202, there are other ways in which the AIM2 inflammasome could be inhibited. Consequently, limited AIM2 function should be considered as a potential causal correlate in human SLE as well.
EXPERIMENTAL PROCEDURES
Cloning and Protein Purification
Mouse p202 cDNA was purchased from OpenBiosystems and was found to be consistent with BC018233.1. Sequences encoding T46-E243 (HIN1) and K244-K445 (HIN2) were cloned into pSMT3 vector using standard PCR protocols. Proteins were expressed in BL21 DE3 Codon-Plus RIPL cells, and purified by affinity column, heparin column and size-exclusion chromatography.
Crystallization, Data Collection, Structure Determination and Refinement
All crystals were crystallized using the hanging drop vapor diffusion method. Free HIN1 and HIN1/dsDNA diffraction data were collected at NSLS beamlines X25 and X29, while HIN2 diffraction data were collected at APS beamline 24-ID-C. HIN1 structure was determined by single-wavelength anomalous dispersion and the initial model was built using PHENIX (Adams et al., 2010). Both HIN1/dsDNA complexes were solved by molecular replacement using free HIN1 structure as search model using PHENIX (Adams et al., 2010). HIN2 structure was determined by molecular replacement. The templates used for molecular replacement are themselves generated by modeling using Swiss-Model (http://swissmodel.expasy.org/) with IFI16 HIN1 (PDB ID 2OQ0) and HIN2 (PDB ID 3B6Y) as templates. Iterative model building and refinement were carried out using Coot (Emsley and Cowtan, 2004), CNS (Brunger et al., 1998) and PHENIX (Adams et al., 2010).
Salmonella Infection
siRNA transfected BMM were primed with 10ng/ml LPS for 4 hours, then infected with Salmonella enterica Typhimurium SL1334 by centrifugation at 700g for 10min. After incubation for 30 min to facilitate invasion, medium was replaced with gentamycincontaining serum-free medium.
Immunoprecipitation
HEK293 cells were chemically transfected with expression constructs for p202, p204, AIM2, or UNQ tagged with either FLAG or V5 epitope tags using Lipofectamine2K as per manufacturer’s instructions. After 24 hours cells were lysed in RIPA buffer and immunoprecipitated with anti-FLAG(M2) (F3165, Sigma) and protein G dynabeads. Beads were washed three times with 10mM HEPES pH7.4, 1.5mM MgCl2, 10mM KCl, 150mM NaCl. Where DNaseI was used, it was added to wash solutions. Control reactions showed the DNaseI used in these conditions was sufficient to completely degrade 6 µg plasmid within 3 minutes. Pulldown of proteins was assessed by western blot using anti-V5 tag-HRP (ab1325, AbD Serotec) or anti-FLAG(M2)-HRP (A8592, Sigma).
Cellular Protein Cross-linking
Unless otherwise stated cells were incubated for 30 minutes with 0.1mM cell-permeable amine-reactive cross-linker DSS (Thermo-Fisher Scientific). Following lysis in 20mM HEPES, 150mM KCl, 1% IGEPAL CA-630 (Sigma), supplemented with complete-mini protease inhibitors (Roche Applied Science, Indianapolis, IN), samples were pelleted at 6000g. Protein was precipitated from the supernatant by addition of 4 volumes of acetone, and both pellet and supernatant proteins were analyzed by western blot.
Supplementary Material
Highlights.
p202 HIN1 uses a surface for DNA binding distinct from that used by AIM2 and IFI16
p202 HIN2 mediates tetramerization of p202 and interaction with AIM2 HIN
p202 inhibits DNA/AIM2-dependent clustering of ASC and inflammasome activity
Reduced AIM2 inflammasome activity enhances Type-I IFN and may contribute to lupus
ACKNOWLEDGMENTS
We thank the staff scientists at beamlines X25 and X29 of National Synchrotron Light Source and beamline 24-ID-C of Advance Photon Source for assistance with data collection and Dr. Yang Li in Wu lab for help with structure refinement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Coordinates of p202 HIN1, p202 HIN1 with 20-mer and 12-mer dsDNA, and p202 HIN2 have been deposited in the RCSB Protein Data Bank under accession numbers 4L5Q, 4L5R, 4L5S, and 4L5T.
REFERENCES
- Ablasser A, Hornung V. DNA sensing unchained. Cell Res. 2013 doi: 10.1038/cr.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal H, Jacob N, Carreras E, Bajana S, Putterman C, Turner S, Neas B, Mathian A, Koss MN, Stohl W, et al. Deficiency of type I IFN receptor in lupus-prone New Zealand mixed 2328 mice decreases dendritic cell numbers and activation and protects from disease. J Immunol. 2009;183:6021–6029. doi: 10.4049/jimmunol.0803872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–687. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- Balada E, Vilardell-Tarres M, Ordi-Ros J. Implication of human endogenous retroviruses in the development of autoimmune diseases. International reviews of immunology. 2010;29:351–370. doi: 10.3109/08830185.2010.485333. [DOI] [PubMed] [Google Scholar]
- Baudino L, Yoshinobu K, Morito N, Santiago-Raber ML, Izui S. Role of endogenous retroviruses in murine SLE. Autoimmunity reviews. 2010;10:27–34. doi: 10.1016/j.autrev.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Choubey D, Panchanathan R. Interferon-inducible Ifi200-family genes in systemic lupus erythematosus. Immunol Lett. 2008;119:32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D, Walter S, Geng Y, Xin H. Cytoplasmic localization of the interferon-inducible protein that is encoded by the AIM2 (absent in melanoma) gene from the 200-gene family. FEBS Lett. 2000;474:38–42. doi: 10.1016/s0014-5793(00)01571-4. [DOI] [PubMed] [Google Scholar]
- Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC evolutionary biology. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon KB, Stone VV. Type I interferon and systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31:803–812. doi: 10.1089/jir.2011.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Haywood ME, Rose SJ, Horswell S, Lees MJ, Fu G, Walport MJ, Morley BJ. Overlapping BXSB congenic intervals, in combination with microarray gene expression, reveal novel lupus candidate genes. Genes and immunity. 2006;7:250–263. doi: 10.1038/sj.gene.6364294. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- Ichii O, Kamikawa A, Otsuka S, Hashimoto Y, Sasaki N, Endoh D, Kon Y. Overexpression of interferonactivated gene 202 (Ifi202) correlates with the progression of autoimmune glomerulonephritis associated with the MRL chromosome 1. Lupus. 2010;19:897–905. doi: 10.1177/0961203310362534. [DOI] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, et al. Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kingsmore SF, Snoddy J, Choubey D, Lengyel P, Seldin MF. Physical mapping of a family of interferonactivated genes, serum amyloid P-component, and alpha-spectrin on mouse chromosome 1. Immunogenetics. 1989;30:169–174. doi: 10.1007/BF02421202. [DOI] [PubMed] [Google Scholar]
- Koul D, Obeyesekere NU, Gutterman JU, Mills GB, Choubey D. p202 self-associates through a sequence conserved among the members of the 200-family proteins. FEBS Lett. 1998;438:21–24. doi: 10.1016/s0014-5793(98)01263-0. [DOI] [PubMed] [Google Scholar]
- Krieg AM, Steinberg AD. Analysis of thymic endogenous retroviral expression in murine lupus. Genetic and immune studies. The Journal of clinical investigation. 1990;86:809–816. doi: 10.1172/JCI114778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Liao JC, Lam R, Brazda V, Duan S, Ravichandran M, Ma J, Xiao T, Tempel W, Zuo X, Wang YX, et al. Interferon-Inducible Protein 16: Insight into the Interaction with Tumor Suppressor p53. Structure. 2011;19:418–429. doi: 10.1016/j.str.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev. 2011;243:206–214. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A, Fernandez D, Telarico T, Phillips PE. Endogenous retroviral pathogenesis in lupus. Current opinion in rheumatology. 2010;22:483–492. doi: 10.1097/BOR.0b013e32833c6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierini R, Juruj C, Perret M, Jones CL, Mangeot P, Weiss DS, Henry T. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 2012;19:1709–1721. doi: 10.1038/cdd.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15:435–443. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- Ru H, Ni X, Zhao L, Crowley C, Ding W, Hung LW, Shaw N, Cheng G, Liu ZJ. Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 2013 doi: 10.1038/cr.2013.52. Published online April 9, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko V, Thygesen SJ, Sester DP, Idris A, Cridland JA, Vajjhala PR, Roberts TL, Schroder K, Vince JE, Hill JM, et al. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 2013 doi: 10.1038/cdd.2013.37. Published online May 3, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Sester DP, Brion K, Trieu A, Goodridge HS, Roberts TL, Dunn J, Hume DA, Stacey KJ, Sweet MJ. CpG DNA activates survival in murine macrophages through TLR9 and the phosphatidylinositol 3-kinase-Akt pathway. J Immunol. 2006;177:4473–4480. doi: 10.4049/jimmunol.177.7.4473. [DOI] [PubMed] [Google Scholar]
- Stacey KJ, Ross IL, Hume DA. Electroporation and DNA-dependent cell death in murine macrophages. Immunology and cell biology. 1993;71(Pt 2):75–85. doi: 10.1038/icb.1993.8. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell- intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MW, Watts T, Li P. Crystallographic characterization of mouse AIM2 HIN-200 domain bound to a 15 bp and an 18 bp double-stranded DNA. Acta crystallographica Section F, Structural biology and crystallization communications. 2012;68:1081–1084. doi: 10.1107/S174430911203103X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronov E, Dayan M, Zinger H, Gayvoronsky L, Lin JP, Iwakura Y, Apte RN, Mozes E. IL-1 betadeficient mice are resistant to induction of experimental SLE. European cytokine network. 2006;17:109–116. [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.