Abstract
Circadian clocks orchestrate 24-h oscillations of essential physiological and behavioral processes in response to daily environmental changes. These clocks are remarkably precise under constant conditions yet highly responsive to resetting signals. With the molecular composition of the core oscillator largely established, recent research has increasingly focused on clock-modifying mechanisms/molecules. In particular, small molecule modifiers, intrinsic or extrinsic, are emerging as powerful tools for understanding basic clock biology as well as developing putative therapeutic agents for clock-associated diseases. In this review, we will focus on synthetic compounds capable of modifying the period, phase, or amplitude of circadian clocks, with particular emphasis on the mammalian clock. We will discuss the potential of exploiting these small molecule modifiers in both basic and translational research.
Keywords: Metabolites, Synthetic compounds, Period, Phase, Amplitude, Clock-associated diseases, Chronotherapy
Introduction
To cope with daily environmental changes due to the earth’s rotation, virtually all living organisms have evolved an intrinsic time-keeping mechanism called the circadian clock [1–8]. The fundamental unit of animal clocks is a cell-autonomous oscillator consisting of transcriptional-translational feedback loops [9, 10]. In the primary feedback loop of the mammalian oscillator, heterodimeric transcription factors CLOCK/BMAL1 and NPAS2/BMAL1 activate expression of the Period1/2 and Cryptochrome1/2 genes. The resulting protein products, PER1/2 and CRY1/2, translocate to the nucleus where they inhibit CLOCK/BMAL1 and NPAS2/BMAL1 and repress their own expression. Various transcriptional and post-transcriptional mechanisms impinge on this primary loop to generate the ~24-h rhythms [11–16]. In particular, nuclear hormone receptors REV-ERBs and RORs act antagonistically to regulate transcription of several core clock genes, including Bmal1, through the shared RORE promoter element [17]. PER and CRY protein turnover is also tightly regulated via coupled phosphorylation/ubiquitination pathways. For example, CRYs have been shown to be phosphorylated by AMP-activated kinase AMPK [18] prior to ubiquitination by the F-box E3 ligase FBXL3 [19–21]. Likewise, sequential phosphorylation by NEMO/NEMO-like kinase and Casein kinase I primes PER proteins for ubiquitination by SLIMB/β-TRCP E3 ligase and proteosomal degradation [13, 22–25]. At the organismal level, the molecular oscillators throughout the body that perform tissue-specific physiological functions are coordinated by the central pacemaker in the hypothalamic SCN [26–29].
Given their key roles in coordinating cellular and physiological processes in anticipation of environmental rhythms, clocks are critical for the well-being and even survival of organisms. Desynchrony between intrinsic and environmental rhythms has been found to render growth disadvantage for cyanobacteria and plants and shortened lifespan in mice [30–33]. Ablation of the SCN central clock in chipmunks adversely affected their survival in the wild, likely attributable to impairments in foraging and predator avoidance [34]. Whereas genetic disruption of clock genes does not lead to acute lethality in laboratory settings, circadian mutant mice show a wide spectrum of physiological deficits [6], including metabolic syndromes in Clock∆19 mutant mice and premature ageing in Bmal1 knockout mice [35–39]. In humans, epidemiological and laboratory studies have also demonstrated increased risks of metabolic and cardiovascular diseases and cancer as a result of circadian disruption [40–43]. For example, within 10 days of living on an enforced 28-h rhythm, human subjects were found to suffer impaired glucose tolerance and hyperinsulinemia [41], similar to that seen with dysregulated pyruvate tolerance in a 2-week mouse model of shift work [44].
It is now well accepted that clocks play a fundamental role in metabolic regulation [45]. For photosynthetic organisms, nitrogen fixation is highly sensitive to oxygen and thus temporally sequestered from daytime photosynthesis. In mammals, hepatic gluconeogenesis takes place in the resting phase to maintain blood glucose homeostasis [46]. Concordantly, genomic and metabolomic studies have found tissue-specific oscillation of mRNA and metabolite accumulation in metabolically active tissues [47–53]. On the other hand, the clock is also highly amenable to reciprocal regulation by metabolites [45, 54, 55]. A number of metabolites can activate upstream signaling pathways that feed into the core oscillator, thereby altering cellular and physiological rhythms [55]. For example, cAMP levels were found to oscillate in the SCN, and the cAMP signaling pathway reciprocally resets the clock by inducing immediate early genes such as Per1 [56–58]. Importantly, certain metabolites can directly modulate clock protein functions by serving as endogenous ligands, including adenosine dinucleotides (NAD and FAD), heme and diatomic gases (NO and CO), and cholesterols [59–69]. For example, NAD levels oscillate in cells, as Nampt, the gene encoding nicotinamide phosphoribosyltransferase that catalyzes the rate-limiting step of NAD biosynthesis, is subject to direct transcriptional control by CLOCK/BMAL1 via its E-box promoter element [70, 71]. Oscillatory NAD levels in turn modulate the activities of NAD-dependent protein modifying enzymes SIRT1 and PARP1 that respectively deacetylate and poly(ADP-ribosyl)ate clock proteins [59, 60, 66], closing the NAD-centric feedback loop imposed on the transcriptional loop.
The revelation that circadian clocks are susceptible to manipulation by small molecule metabolites ushers in an exciting era to develop synthetic small molecule clock modifiers [72, 73]. A number of promising chemical modifiers have been uncovered in recent years, through either phenotypic functional screens or targeted ligand development. In this review, we discuss these small molecule modifiers of the circadian clock and their potential therapeutic application in clock-associated diseases.
Overview of synthetic compounds as clock modifiers
Whereas classical genetics produces inherited changes in the sequence and/or abundance of the target protein, most synthetic small molecule modifiers allosterically alter the protein in a reversible, time-controlled and dose-dependent manner. Small molecules may also bind to a particular domain and consequently modulate the cognate function of a multi-domain protein, leaving the other parts of the protein and associated functions intact. If the binding surface is conserved among multiple paralogous proteins, small molecules can concurrently regulate their activities to circumvent functional redundancy commonly observed in classical genetic studies. Thus, the small molecule-based chemical genetic approach is a powerful tool to perturb the system of interest [72, 74, 75].
Two complementary methods have been utilized to identify small molecule modifiers of the clock. The first approach, based on phenotypic functional assays, interrogates broad chemical space via screening of diverse chemical libraries. In published studies, the reporter assays involved stable cell lines expressing either luciferase alone from an exogenous Bmal1 promoter [76–80] or PER2::luciferase fusion proteins from the endogenous Per2 promoter [81], corresponding to mRNA or protein rhythm, respectively. Bioluminescence is monitored over several days in the so-called kinetic, as opposed to end-point, assay to visualize circadian reporter rhythms. Changes in key clock parameters, including period, phase, and amplitude, can then be measured to identify small molecule modifiers. In these screens, small molecule modifiers may act on an intracellular target in the upstream input pathway, the core oscillator, or any output pathways with feedback regulatory functions, such as metabolism (Fig. 1). Furthermore, novel screening assays targeting additional clock regulatory pathways will likely lead to an enriched repertoire of clock modifiers.
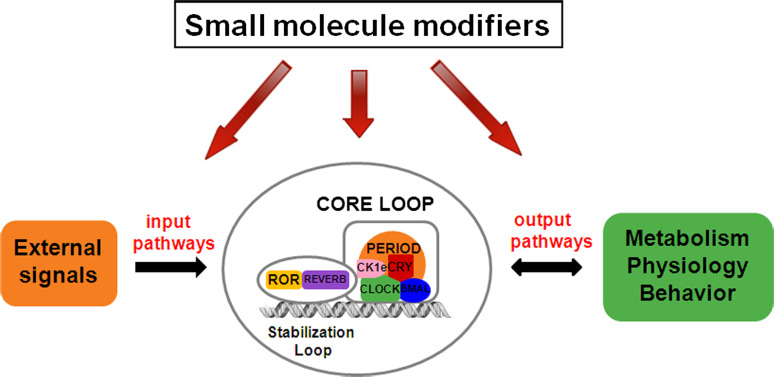
Fig. 1.
Small molecule modifiers of circadian clocks. In the mammalian circadian clock system, external signals are transmitted via input pathways to the molecular oscillator consisted of interlocked feedback loops. The molecular oscillator in turn orchestrates output functions which may reciprocally regulate the clock via feedback regulation. Small molecule modifiers of the clocks may target the input pathways, the core clock, or output pathways with feedback regulatory functions
Small molecule modifiers can also be identified based on direct interaction with particular clock proteins or regulatory factors. For example, IC261 and CKI-7 have been shown to lengthen the clock period as expected from their known CKI inhibitory activities [22, 82] (see also Tables 1, 2). On the other hand, to generate novel and/or improved ligands for a particular target, it is often necessary to conduct deliberate chemical derivatization of small molecule analogs based on prior knowledge of known ligands and/or binding cavity structures [83]. An interesting example is the development of a selective inhibitor of casein kinase Iε, PF-4800567 which confers >20-fold selective inhibition over CKIδ [84–86]. More recently, this approach has been successfully applied to the nuclear hormone receptors REV-ERBs and RORs, which constitute the stabilization loop of the core oscillator [17]. Whereas the endogenous ligands are known for these proteins (heme and cholesterols respectively) [63–65], small molecule ligands are highly desirable to circumvent intracellular complications that altering metabolites commonly incurs, including nonspecific actions, cytotoxicity and redox imbalance [87]. Starting with privileged scaffolds known to target ligand binding domains of nuclear hormone receptors, investigators were able to identify tertiary amines with three lipophilic substituents as agonists of REV-ERBα [87–90]. Novel RORα/γ ligands, most of them sulfonamide derivatives, have also been shown to modulate hepatic metabolism [91, 92] or to attenuate expression of downstream cytokines and alleviate autoimmune disease symptoms [93, 94]; however, the role of the clock in these settings is currently unknown. In an attempt to correlate bona fide clock effects of small molecules with physiological consequences, we describe below known small molecule clock modifiers based on their activities in modifying the three major clock characteristics, namely period, phase, and amplitude. The classification is based on their primary, most pronounced phenotype since many small molecules are able to co-regulate more than one clock parameter.
Table 1.
Summary of small molecules capable of altering the circadian period
| Name | CAS # | Molecular targets | Period effects | References |
|---|---|---|---|---|
| IC261 | 186611-52-9 | CKIδ/ε | Lengthening | [22] |
| CKI-7 | 1177141-67-1 | CKIδ/ε | Lengthening | [82] |
| CK01 | N/A | CKIδ/ε | Lengthening | [152] |
| D4476 | 301836-43-1 | CKIδ/ε | Lengthening | [95] |
| DMAT | 749234-11-5 | CK2, CKI? | Lengthening | [25, 97] |
| PF-4800567 | 1188296-52-7 | CKIε | Lengthening | [84] |
| PF-670462 | 950912-80-8 | CKIδ/ε | Lengthening | [96] |
| Roscovitine | 186692-46-6 | CDK, CKIδ/ε | Lengthening | [77] |
| TG003 | 300801-52-9 | CLK, CKIδ/ε | Lengthening | [77] |
| SB202190 | 152121-30-7 | P38, CKIδ/ε | Lengthening | [77] |
| PD169316 | 152121-53-4 | P38, CKIδ/ε | Lengthening | [77] |
| SU5416 | 204005-46-9 | VEGFR PTK, CKIδ/ε | Lengthening | [77] |
| DRB | 53-85-0 | CK2, CKIδ/ε | Lengthening | [77] |
| SP600125 | 129-56-6 | JNK, CKIδ/ε | Lengthening | [77] |
| CGS-15943 | 104615-18-1 | AR agonist, CKIδ/ε | Lengthening | [77] |
| PPT | 263717-53-9 | ERα agonist, CKIδ/ε | Lengthening | [77] |
| Calyculin A | 101932-71-2 | PP2A | Lengthening | [22] |
| 17-OHP | 68-96-2 | Progesterone receptor | Lengthening | [77] |
| Vincristine | 57-22-7 | Tubulin/microtubule | Shortening | [76] |
| Etoposide | 33419-42-0 | DNA topoisomerase II | Shortening | [76] |
| Mitoxantrone | 65271-80-9 | DNA topoisomerase II | Shortening | [76] |
| Amsacrine | 51264-14-3 | DNA topoisomerase II | Shortening | [77, 103] |
| PMA | 16561-29-8 | PKC agonist | Shortening | [76] |
| SKF-96365 | 130495-35-1 | Ca channel | Shortening | [76] |
| Indirubin-3’-oxime | 160807-49-8 | CDK, GSK-3β | Shortening | [76, 83] |
| Kenpaulone | 142273-20-9 | CDK, GSK-3β | Shortening | [76] |
| SB216763 | 280744-09-4 | GSK-3β | Shortening | [76, 77] |
| Longdaysin | 1353867-91-0 | CKIδ/α, ERK2, CDK7 | Lengthening | [76] |
| LH846 | 639052-78-1 | CKIδ | Lengthening | [79] |
| KL-001 | 309928-48-1 | CRY | Lengthening | [80] |
| Cmpd-1 | 683807-31-0 | CKIε, CKIδ? | Lengthening | [81] |
| Cmpd-2 | 892293-00-4 | CKIε, CKIδ? | Lengthening | [81] |
| Cmpd-3 | 422279-51-4 | CKIε, CKIδ? | Lengthening | [81] |
| Cmpd-4 | 533873-00-6 | GABAAR agonist and ? | Lengthening | [81] |
| Cmpd-7 | 416879-98-6 | Unknown | Shortening | [81] |
Except as otherwise indicated, the small molecules herein negatively regulate their respective targets. For analog series, representative compounds are listed. The clock modifying activities of IC261, CKI-7, D4476, DMAT, and Calyculin A were specifically tested based on their known enzymatic targets. These small molecules were not identified via circadian-based screening or chemical derivatization approaches
Table 2.
Small molecules capable of altering the circadian phase and/or amplitude
| Name | CAS # | Molecular targets | Circadian effects | References |
|---|---|---|---|---|
| U0126 | 109511-58-2 | ERK | Attenuated phase shift | [119–121] |
| KN-62 | 127191-97-3 | CaMKII | Attenuated phase shift | [122] |
| KT5823 | 126643-37-6 | PKG | Attenuated phase advance | [118] |
| SB431542 | 301836-41-9 | ALK | Attenuated phase delay | [124] |
| Cmpd-5 | 361469-09-2 | cAMP inducer | Phase delay | [81] |
| Cmpd-6 | 443097-13-0 | cAMP inducer | Phase delay | [81] |
| Rolipram | 61413-54-5 | PDE | Phase delay | [81] |
| GSK4112 | 1216744-19-2 | REV-ERBα | Amplitude reduction | [87, 89] |
| SR9011/SR9009 | 1379686-29-9/1379686-30-2 | REV-ERBs | Amplitude reduction | [90] |
| T0901317 | 293754-55-9 | RORα/γ | N/A | [93] |
| SR1001 | 1335106-03-0 | RORα/γ | N/A | [94] |
| SR1078 | 1246525-60-9 | RORα/γ agonist | N/A | [91] |
| SR3335 | 293753-05-6 | RORα | N/A | [92] |
| CEM1/Cmpd-8 | 329903-11-9 | Unknown | Amplitude enhancement, period shortening | [81] |
| CEM2/Cmpd-9 | 687581-48-2 | Unknown | Amplitude enhancement, period shortening | [81] |
| CEM3/Cmpd-10 | 305334-67-2 | Unknown | Amplitude enhancement, period shortening | [81] |
| CEM4/Cmpd-11 | 892267-62-8 | Unknown | Amplitude enhancement, period shortening | [81] |
Except as otherwise indicated, the small molecules herein negatively regulate their respective targets. For analog series, representative compounds are listed. The clock modifying activities of U0126, KN-62, KT5823, and SB431542 were specifically tested based on their known enzymatic targets. These small molecules were not identified via circadian-based screening or chemical derivatization approaches
Period-altering modifiers
Circadian period has been a reliable assay parameter traditionally in rodent genetic studies in rodents and more recently in high-throughput chemical screens [6, 81]. In several independent chemical screens, small molecules showing the most significant period-lengthening activities were found to be predominantly CKI inhibitors (Table 1) [76–82, 95]. These compounds show diverse scaffold structures and are able to prolong the period of luciferase reporter rhythms to 48 h at 25 μM [77]. Inhibition of CKI slows down PER protein turnover, thus decelerating clock progression and lengthening the circadian period [6, 13]. The mechanistic convergence of these potent period-lengthening molecules highlights the central role of PER degradation cycles in setting the clock speed.
Kinase inhibitors are known to be promiscuous in target selectivity, and most CKI inhibitors appear to target paralogous CKI enzymes [58, 77, 78, 84]. In contrast, the selective CKIε inhibitor PF-4800567 caused insignificant period lengthening in cells and mice [84, 86], consistent with genetic evidence supportive of a predominant role of CKIδ in determining circadian speed [96]. Recently, three period-lengthening compounds (Cmpd-1, -2, -3) were shown to inhibit CKIε in vitro [81]; given their robust period-lengthening effects, it is possible that they also target CKIδ. Unlike Cmpd-1 and Cmpd-2, Cmpd-3 significantly increased the levels of Per2 mRNAs, suggesting a divergent mechanism for this CKI inhibitor in addition to PER protein stabilization. In addition to CKIδ and CKIε, casein kinase 2 (CK2) has also been shown to directly phosphorylate PER and regulate its nuclear localization and turnover in Drosophila and mammalian cells; in agreement, inhibitors of CK2 were also found to lengthen the circadian period [25, 97, 98]. Furthermore, a number of period-lengthening small molecules are known to inhibit CKIα, ERK, CDKs, p38, or c-JNK [25, 77, 78, 97] (Table 1). Since most of these kinase inhibitors also acts on CKIδ, the exact contribution of inhibiting these other kinases to period lengthening of these compounds requires further study.
Apart from the above kinase inhibitors, several carbazole derivatives also lengthened the circadian period, but appeared to function via potentiating the transcriptional repression by CRY proteins [80]. In this study, purified CRY proteins were found to directly bind to affinity resins conjugated with an active derivative KL001; furthermore, point mutation in the FAD binding pocket of CRY1 strongly attenuated its binding to the conjugated resin. Previously, hypomorphic mutations in Fbxl3, encoding the F-box E3 ligase FBXL3 required for CRY degradation, were found to lengthen the circadian period [19–21]. These studies thus indicate that activation of the primary repressor CRYs in the mammalian clock, by either binding small molecule agonists or blocking its turnover, lengthens the circadian period. Together with the above studies on CKI inhibitors, identification of CRY agonist molecules highlights the importance of the clock proteins in the negative arm of the feedback loop (PERs and CRYs) in setting the speed of the clock.
The target and mechanism of a period-lengthening benzodiazepine derivative, Cmpd-4, is currently unclear [81]. In central neurons, it appears to act as a canonical agonist for GABAA receptors, contributing moderately to period lengthening. Its predominant activity, however, is likely mediated by a novel target, leading to significant period lengthening in peripheral, non-neuronal cells where GABAA receptors are not abundantly expressed. This dual action by Cmpd-4 highlights the complexity of circadian regulation, and also reveals unexpected versatility of small molecules.
In comparison, period-shortening small molecules are less common. In contrast to early studies showing inhibition of GSK-3β activity by lithium or via genetic manipulation caused period lengthening [99, 100], several selective inhibitors of GSK-3β, including Indirubin-3′-oxime, Chir99021, Kenpaullone, and SB216763, were recently found to shorten the circadian period in reporter cell assays [72, 77, 83]. GSK-3β has been shown to phosphorylate both PER2 and CRY2 proteins [101, 102], modulating their nuclear localization and proteosomal degradation, respectively. Given the important roles of PER and CRY proteins in period regulation as mentioned above, it will be of interest to determine the specific mechanism by which GSK-3β inhibitors modulate circadian progression.
Three DNA topoisomerase II inhibitors and chemotherapeutic agents, namely etoposide, mitoxantrone, and Amsacrine, have also been shown to cause period shortening and phase advance [76, 77, 103]. It has been proposed that DNA damage constitutes a circadian resetting cue, or zeitgeber, capable of altering circadian progression [104]. For example, γ-irradiation and the radiomimetic agent methylmethane sulphonate (MMS) were shown to cause phase advance in mouse and Neurospora, respectively, when administered during the subjective day [105, 106]. On the other hand, circadian clock genes have been implicated in mediating the DNA damage response and cell cycle gating [107–112]. For example, the clock has been shown to transcriptionally regulate a key nucleotide excision repair factor XPA, conferring robust defense against cisplatin-induced DNA damage in the late afternoon [110]. Likewise, a yeast metabolic clock also gates cell cycle progression as a means of minimizing oxidative DNA damage [8, 113, 114]. Further investigation of the circadian function of the above DNA damage and chemotherapy agents may reveal important insight into the detailed mechanism underlying the reciprocal relationship between the clock and the DNA damage response/cell cycle progression.
Phase-altering modifiers
Phase-resetting mechanisms allow the clocks to respond to environmental changes, conferring crucial adaptability in physiology and behavior. Whereas period changes can cause chronic phase delay or advance, acute phase shifts independent of sustained period changes play a predominant role in entrainment of the clock in response to the environment. Compounds that perturb the input pathways or downstream processes with feedback functions may transiently alter the circadian phase of the core oscillator. In mammals, acute phase resetting, or entrainment, of SCN clocks involves immediate early induction of Per1, and more weakly Per2, by the cAMP/CREB signaling pathway [10, 26]. Following the initial discovery of serum-induced synchronization of circadian gene oscillation in Rat-1 cells [57], many chemicals, including a number of kinase inhibitors (Table 2), have been shown to synchronize peripheral clocks and induce phase shifts in vitro [58, 72, 115–118]. Many such compounds also converge on the cAMP/CREB pathway and induce Per expression, such as U0126 (ERK inhibitor) and KN-62 (CamKII inhibitor) [119–123]. In fibroblast cells, two cAMP-inducing compounds, Cmpd-5 and Cmpd-6, were found to cause acute induction of Per1 mRNA levels and PER2::Luc reporter bioluminescence, followed by significant phase delay and amplitude damping of reporter rhythms [81]. These observations are reminiscent of the effects seen previously in SCN slices treated with the adenylyl cyclase activator Forskolin [56]. Furthermore, an inhibitor of the cAMP-catabolizing enzyme phosphodiesterase 4 (PDE4), Rolipram, also caused acute bioluminescence induction and subsequent phase delay [81]. In the SCN, the guanine exchange factors Epac1/2, but not the hyperpolarizing cyclic nucleotide-gated ion channels (HCN), were previously found to be involved in mediating cAMP-induced clock resetting [56]. Elucidation of the direct targets and downstream effectors for Cmpd-5 and Cmpd-6 requires further investigation.
Phase resetting independent of acute Per induction has also been reported. SB431542, an inhibitor of activin receptor-like kinase (ALK), was found to attenuate alkaline shock-induced phase delays via SMAD3-dependent acute induction (within 20 min to 1 h) of the circadian transcriptional regulator Dec1, but not Per1 [124]. DEC1 and its paralog DEC2 were initially found to play a role in suppressing Per1 transcription [125]. On the other hand, double knockout of Dec1 and Dec2 severely attenuated photic induction of Per1, also suggesting a potential positive role of DECs in Per1 transcription [126]. In a photic phase resetting experiment, a 30-min light pulse administered at night was able to acutely induce Dec1 in the SCN [125], mimicking the well-established light induction of Per1. In contrast, a light pulse showed no effects on Dec2. These observations together suggest that different cues may differentially cause acute induction of Per1 and/or Dec1 to reset the circadian phase. In the case of SB431542, whether Per1 contributes to its overall phase resetting effects requires further studies.
Amplitude-altering compounds
Amplitude represents the robustness of oscillation, clearly an important characteristic of any rhythmic process. The amplitude of mouse free-running activity rhythms can be measured as the relative power of the circadian component via fast-Fourier transformation (FTT) algorithms [127]. More recently, circadian reporter assays based upon cycling core clock elements allow convenient measurement of rhythm amplitude as the difference between the peak and the trough [10, 128]. In our previous chemical screen, 4 compounds (Cmpd-8, -9, -10, -11; Table 2) were identified to dose-dependently enhance the amplitude of PER2:luciferase reporter rhythm in fibroblast cells and pituitary explants [81], hereafter renamed as clock-enhancing small molecules (CEMs). These CEMs showed only modest stimulatory effects on Per2 transcript levels, suggesting post-transcriptional mechanisms required for the induction of PER2::Luc reporter bioluminescence. In addition, CEMs showed distinct effects on Bmal1-luc reporter rhythms in U2OS cells as well as transcript oscillation of Bmal1 target genes Dbp and Rev-erbα. For example, whereas CEM1 appeared to strongly induce Bmal1-luc oscillatory amplitude, CEM4 appeared to increase the magnitude (absolute value) of both trough and peak reporter expression, leading to only minor enhancement in amplitude (the difference between peak and trough). These observations underscore the complexity of the clock feedback regulatory circuit, particularly with regard to clock amplitude.
Apart from amplitude effects, CEMs also caused period shortening [81]; for example, at the concentration of 5 μM, CEMs were able to shorten the circadian period by 1–3 h in fibroblast cells. Whereas reciprocal regulation between amplitude and phase shifts has been demonstrated in rodents and humans [129–131], the relationship between amplitude and period is not well understood. Previously, classical mouse genetic studies have shown that overexpression of a bacterial artificial chromosome (BAC) transgene of Clock shortened the circadian period by approximately 1 h [132]. In flies, attaching a strong transcriptional activator VP-16 to CYCLE, the equivalent of mammalian BMAL1, or increasing the copy number of dClock, has been shown to enhance circadian transcription and reporter oscillatory amplitude [133]. Interestingly, enhanced transcription under these conditions correlated with shorter periods, likely attributable to accelerated dPER accumulation and subsequent transcriptional repression. These studies suggest that potentiating the activities or levels of the positive factors can both enhance the amplitude and shorten the period, primarily by accelerating the circadian phase when these factors are active. On the other hand, in a detailed biochemical study of mouse embryonic fibroblast (MEF) cells [134], CLOCK and BMAL1 were found to be enriched relative to PERs and CRYs. Ectopic expression of CLOCK and BMAL1 by adenoviral expression specifically increased the basal levels and thus dampened the overall rhythm of the PER2::Luc reporter. In contrast, overexpression of the less abundant PERs in MEFs enhanced the reporter rhythms. Therefore, maximum circadian amplitude appears to depend on stoichiometic levels of the positive and negative factors in the core clock feedback loop [134]. Observations from these studies can be unified if CLOCK or CYCLE are limiting in flies. Regardless, future mechanistic studies using CEMs will reveal key insights into the regulatory mechanisms of clock amplitude and period.
Elucidation of the direct signaling pathways or proteins targeted by CEMs is of significant interest. Previously, in an siRNA functional genomic screen using a U2OS cell line containing a Bmal1-driven destabilized luciferase reporter, over 50 genes were identified whose knockdown increased the circadian amplitude [128]. Examination of the gene list reveals highly divergent intracellular processes, suggesting that clock amplitude regulation is subjected to broad network control. Furthermore, the central SCN clock in the mammalian clock system has been shown to be particularly robust due to intercellular coupling [135] and resistant to genetic perturbation [136]. Among the identified CEMs showing general efficacy in peripheral clocks, only CEM3 appeared to enhance the reporter rhythm in SCN explants. At the cellular level, the failure of other CEMs to activate SCN clocks could result from the lack of expression of the protein target in the SCN (assuming they are not core clock proteins), or the inability to overcome the strong coupling among the SCN neurons. Future studies will investigate the effects of these CEMs on single-cell bioluminescence [137] to distinguish between these possibilities. Such studies will also shed new light onto the effects of CEMs on rhythm damping in cultured cells, generally considered to be a consequence of loss of synchrony.
As opposed to CEMs, an inverse agonist of REV-ERBs, SR9011, was recently found to significantly repress oscillation amplitude without altering the clock period [90], consistent with the notion that the secondary feedback loop, consisting of REV-ERBs and RORs, functions to confer robustness and stability of the clock [138]. SR9011 also disrupted wheel-running activity immediately after administration. Notably, SR9011 appeared to promote energy expenditure and reduce weight gain in a diet-induced obesity model, providing an interesting example of beneficial effects of repressing the clock amplitude on energy metabolism. Whether this represents a general strategy or a specific case involving a derivatized nuclear receptor ligand remains to be seen. In the above chemical screen [81], a significant number of small molecules, estimated to be 1–3 % of the total compounds screened, strongly reduced the amplitude of reporter rhythms (data not shown). Visual examination at the end of the experiments revealed widespread cytotoxicity in these samples. Therefore, identification and utilization of amplitude-repressing compounds will require careful selection of secondary screens to eliminate cytotoxic and other complicating factors.
Therapeutic potentials in clock-related diseases
Circadian disruption is well known to contribute to pathologies with a strong temporal basis such as jetlag, sleep disorders, and seasonal affective disorders [6]. In recent years, a host of exciting studies have provided key mechanistic insights into circadian control of other physiological processes, and thus greatly expanded the spectrum of clock-related diseases. For example, the dominant negative Clock∆19 mutation or Bmal1 knockout led to impaired pancreatic insulin secretion and caused diabetic glucose intolerance in mice [37, 139, 140]. The Clock∆19 mutant mice are also prone to diet-induced obesity [141], perhaps in part due to the increased intestinal absorption of monosaccharides and lipids in these mice [142]. In a recent study, the Krüppel-like transcription factor 15 (KLF15) was found to be directly activated by CLOCK/BMAL1 via the E-box promoter element, and KLF15 in turn regulated the transcription of the gene encoding KvCHIP2, an important component of the cardiac ion channel required for myocardial repolarization [143]. Disruption of this transcriptional cascade was shown to render increased susceptibility to ventricular arrhythmias, thus providing a mechanistic explanation for the high incidence rates of myocardial infarction in the early morning. Recent studies have also revealed circadian controls of key regulators of immune responses in both mice and plants [144, 145]. These advances in circadian biology lay the foundation for applying clock-based therapies to a wide variety of diseases.
There are two general strategies in exploiting circadian rhythms to combat clock-related diseases. Traditional chronotherapy entails optimizing the circadian timing for existing therapies, such as cancer chemotherapy, to improve efficacy and/or reduce toxicity [73, 146]. On the other hand, small molecule modifiers with desirable pharmacokinetic and pharmacodynamic characteristics afford a novel strategy involving direct manipulation of the clock to improve output pathophysiology intrinsic to disease etiology. The small molecule modifiers may be administered by themselves or in conjunction with complementary therapies. The general rationale is to match the phenotypic or molecular function of small molecules with corresponding diseases with known clock dysfunctions. Jetlag is fundamentally a phase misalignment and therefore can be targeted by phase-resetting molecules. Given the reciprocal relationship between phase shift and amplitude, an amplitude repressor could be co-administered to augment or accelerate a phase shift. Another clock-related disorder is the familial advanced sleep phase syndrome (FASPS), characterized by short circadian periods, and in one family linked to a T44A missense mutation in human CK1δ located within the N-terminal ATP-binding motif [147, 148]. Paradoxically, this FASPS mutation repressed CK1δ kinase activity, suggesting distinct effects on PER proteins and circadian period compared with the aforementioned CKI inhibitors. It would be interesting to investigate whether known CKI inhibitors can act on this mutant CK1δ to prolong the period and alleviate the sleep syndrome. One good candidate is the selective CK1δ inhibitor PF-670462. Previously, daily dosing of PF-670462 has been shown to induce behavioral rhythms in mice that were arrhythmic due to either constant light exposure or disruption in the Vipr2 gene encoding a G protein-coupled receptor required for SCN pacemaker functions [86], indicating in vivo activity in a circadian mouse mutant.
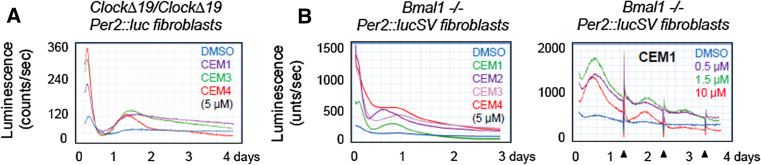
Several studies have revealed a strong correlation between clock dampening (reduced amplitude) and various pathological conditions [37, 129]. In particular, the ClockΔ19 mutant mice are known to exhibit damped amplitude and lengthened period of circadian rhythms, accompanied by various physiological and behavioral deficiencies [129, 141, 142, 149–151]. Using this mouse line as a disease model, a recent study showed that a CKIδ/ε inhibitor CK01, similar to PF-670462, was able to alleviate the manic-like behaviors in these mice [152]. In cell culture, ClockΔ19/+ heterozygous cells displayed approximately threefold reduction in reporter rhythm amplitude relative to wild-type cells [81], and CEM treatment largely restored the normal amplitude in ClockΔ19/+ cells. Moreover, CEM3 also enhanced reporter amplitude in ClockΔ19/+ SCN explants. Certain CEMs were able to acutely induce reporter expression followed by a descending phase in ClockΔ19/ClockΔ19 or even Bmal1-deficient cells using a daily dosing protocol (Fig. 2). Detailed circadian gene analysis will help elucidate whether and how such CEM-induced reporter oscillations resemble canonical circadian cycles. It is possible that even slight amplitude enhancement of individual cellular oscillators can combine to elicit significant physiological improvement in patients with partially impaired clocks.
Fig. 2.
Stimulatory effects of clock-enhancing small molecules (CEMs) on reporter rhythms in homozygous Clock∆19/Clock∆19 (a) and Bmal1 −/− (b) fibroblast cells. Luminescence recording was carried out as previously described [81]. Compared with the Per2::luc reporter, Per2::lucSV contains an exogenous SV40 polyA element which increases reporter luminescence [81]. The X- and Y-axes indicates time and luminescence, respectively. In (b) right panel, daily dosing times of CEM1 are indicated by the arrowheads corresponding to the spikes in bioluminescence recordings
Future directions
It is useful to expand the ensemble of small molecules capable of manipulating the clock by chemical screening or targeted ligand development. For example, new screens can utilize neuronal (or SCN derived) stable reporter cells [153], additional clock promoters (e.g., Dec2) [154], nuclear localization via high content screening [155], or simply exploring new chemical space. For ligand development, one particularly interesting target is the PER-ARNT-SIM (PAS) domains present in PER proteins and the bHLH-PAS family of transcription factors including CLOCK/NPAS2, BMAL1, HIF, and ARNT. In microorganisms and plants, PAS domain proteins are required for photic and two-component signaling pathways [156, 157]. In mammals, PAS domains mainly function in protein–protein interaction and recruitment [158, 159]. However, recent crystal structures of HIF2α-ARNT PAS domains revealed a buried internal pocket in the PAS-B domain of HIF2α, and artificial bicyclic ligands were capable of allosterically modulating heterodimer formation [160, 161]. More recently, the crystal structure of the full-length CLOCK/BMAL1 heterodimer also showed highly conserved structural features in the asymmetrically positioned CLOCK/BMAL1 PAS-B domains [158]. For example, Trp427 on the BMAL1 PAS-B domain inserts into a binding pocket on the CLOCK PAS-B domain that resembles cofactor-binding motifs in other PAS proteins. Interestingly, the corresponding Tryptophan residue on the CLOCK PAS-B domain protrudes away from the CLOCK:BMAL1 dimer and may interact with the PAS domains of CRY [158, 162]. These findings suggest a central role of the binding pockets on PAS-B domains during dynamic circadian complex formation. It will be of strong interest to derive ligands capable of binding PAS domains of clock proteins.
Small molecule modifiers are useful probes to understand basic circadian biology. As mentioned above, detailed characterization of how the modifiers regulate the core oscillator will reveal important insight into the regulatory mechanism of clock amplitude and its relationship with circadian period. Ultimately, the holy grail of small molecule studies is to identify the cellular pathways or proteins that are directly targeted by small molecules. Both functional genomic screens (siRNA, shRNA libraries) [74] as well as chemical proteomics [80] have been successfully utilized to identify small molecule targets. On the other hand, we can now envision using circadian mouse mutants as disease models to investigate whether restoring normal clock functions by small molecules will improve clock output physiology. The next step will be to directly applying small molecule modifiers including CEMs to canonical disease models, e.g., ob/ob mice in obesity and diabetes. A rational approach is to characterize the circadian features, at both molecular and physiological levels, of these disease models [163] in order to select small molecules with the best chance of therapeutic efficacy, either alone or in combination. Humans display a wide range of circadian phenotypes [164, 165]; of note, the period lengths of fibroblast cells from human subjects have been shown to correlate well with behavioral chronotypes. Therefore, application of small molecules in human fibroblast cells constitutes an in vitro experimental system toward the ultimate goal of pharmacologically manipulating human circadian rhythms.
In conclusion, small molecule modifiers have taught us much about how clocks are intricately constructed and broadly regulated. Identification of their underlying mechanisms will continue to unravel key regulatory nodes in the clock network. As we increasingly appreciate the importance of timing in biology and disease, the timing is also opportune to fully exploit small molecule modifiers for exciting advances in both basic research and therapeutic development.
Acknowledgments
We thank J.A. Mohawk and C.C. Lee for critical reading of the manuscript, M.R. Blackburn, B. He and Y. Chelliah for helpful discussions, and N. Koike for help with literature search. Small molecules research in Z.C.’s laboratory is supported by grants from the Robert A. Welch Foundation (AU-1731), American Heart Association (11SDG7600045) and Texas Medical Center Digestive Diseases Center funded by NIH Center Grant DK56338. J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Abbreviations
- ARNT
Aryl hydrocarbon receptor nuclear translocator
- bHLH PAS
Basic helix–loop–helix PER-ARNT-SIM
- BMAL1
Brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like
- CEM
Clock-enhancing molecule
- CLOCK
Circadian locomotor output cycles kaput
- CREB
cAMP response element-binding protein
- CRY
Cryptochrome
- FASPS
Familial advanced sleep phase syndrome
- HIF
Hypoxia-inducible factor
- NPAS2
Neuronal PAS domain protein 2
- PER
Period
- ROR
Retinoid acid receptor-related orphan receptor
- SCN
Suprachiasmatic nuclei
Contributor Information
Zheng Chen, Email: Zheng.chen.1@uth.tmc.edu.
Joseph S. Takahashi, Email: Joseph.Takahashi@UTSouthwestern.edu
References
- 1.Dong G, Golden SS. How a cyanobacterium tells time. Curr Opin Microbiol. 2008;11:541–546. doi: 10.1016/j.mib.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys. 2011;40:143–167. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7:e62. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eelderink-Chen Z, Mazzotta G, Sturre M, Bosman J, Roenneberg T, Merrow M. A circadian clock in Saccharomyces cerevisiae . Proc Natl Acad Sci USA. 2010;107:2043–2047. doi: 10.1073/pnas.0907902107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O’Neill JS, Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 10.Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- 11.Zheng X, Sehgal A. Speed control: cogs and gears that drive the circadian clock. Trends Neurosci. 2012;35(9):574–585. doi: 10.1016/j.tins.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124:311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 14.Schibler U. The daily timing of gene expression and physiology in mammals. Dialogues Clin Neurosci. 2007;9:257–272. doi: 10.31887/DCNS.2007.9.3/uschibler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337(6094): 599– 602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 16.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26:490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solt LA, Kojetin DJ, Burris TP. The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med Chem. 2011;3:623–638. doi: 10.4155/fmc.11.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 21.Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O’Neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 22.Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu JC, Ko HW, Edery I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145:357–370. doi: 10.1016/j.cell.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu W, Houl JH, Hardin PE. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol. 2011;21:756–761. doi: 10.1016/j.cub.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, Schlosser A, Kramer A. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 28.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boothroyd CE, Young MW. The in(put)s and out(put)s of the Drosophila circadian clock. Ann NY Acad Sci. 2008;1129:350–357. doi: 10.1196/annals.1417.006. [DOI] [PubMed] [Google Scholar]
- 30.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 32.Libert S, Bonkowski MS, Pointer K, Pletcher SD, Guarente L. Deviation of innate circadian period from 24 h reduces longevity in mice. Aging Cell. 2012;11(5):794–800. doi: 10.1111/j.1474-9726.2012.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16:R914–R916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeCoursey PJ, Krulas JR. Behavior of SCN-lesioned chipmunks in natural habitat: a pilot study. J Biol Rhythms. 1998;13:229–244. doi: 10.1177/074873098129000075. [DOI] [PubMed] [Google Scholar]
- 35.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/S0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, Takahashi JS. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westgate EJ, Cheng Y, Reilly DF, Price TS, Walisser JA, Bradfield CA, FitzGerald GA. Genetic components of the circadian clock regulate thrombogenesis in vivo. Circulation. 2008;117:2087–2095. doi: 10.1161/CIRCULATIONAHA.107.739227. [DOI] [PubMed] [Google Scholar]
- 40.Roenneberg T, Allebrandt KV, Merrow M, Vetter C. Social jetlag and obesity. Curr Biol. 2012;22:939–943. doi: 10.1016/j.cub.2012.03.038. [DOI] [PubMed] [Google Scholar]
- 41.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheer FA, Hu K, Evoniuk H, Kelly EE, Malhotra A, Hilton MF, Shea SA. Impact of the human circadian system, exercise, and their interaction on cardiovascular function. Proc Natl Acad Sci USA. 2010;107:20541–20546. doi: 10.1073/pnas.1006749107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arendt J. Shift work: coping with the biological clock. Occup Med (Lond) 2010;60:10–20. doi: 10.1093/occmed/kqp162. [DOI] [PubMed] [Google Scholar]
- 44.Barclay JL, Husse J, Bode B, Naujokat N, Meyer-Kovac J, Schmid SM, Lehnert H, Oster H. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One. 2012;7:e37150. doi: 10.1371/journal.pone.0037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 46.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 48.Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, Yasui A, van der Horst GT, Soga T, Ueda HR. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109:2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, Gillette MU. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337(6096):839–842. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasukawa T, Sugimoto M, Hida A, Minami Y, Mori M, Honma S, Honma K, Mishima K, Soga T, Ueda HR. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci USA. 2012;109:15036–15041. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 55.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/S0092-8674(00)81199-X. [DOI] [PubMed] [Google Scholar]
- 58.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 59.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 60.Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 61.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 62.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 63.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 65.Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- 66.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+ -dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sancar A. Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem. 2004;279:34079–34082. doi: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- 68.Zoltowski BD, Vaidya AT, Top D, Widom J, Young MW, Crane BR. Structure of full-length Drosophila cryptochrome. Nature. 2011;480:396–399. doi: 10.1038/nature10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 70.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirota T, Kay SA. High-throughput screening and chemical biology: new approaches for understanding circadian clock mechanisms. Chem Biol. 2009;16:921–927. doi: 10.1016/j.chembiol.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farrow SN, Solari R, Willson TM. The importance of chronobiology to drug discovery. Expert Opin Drug Discov. 2012;7:535–541. doi: 10.1517/17460441.2012.689283. [DOI] [PubMed] [Google Scholar]
- 74.Lehar J, Stockwell BR, Giaever G, Nislow C. Combination chemical genetics. Nat Chem Biol. 2008;4:674–681. doi: 10.1038/nchembio.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frye SV. The art of the chemical probe. Nat Chem Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 76.Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci USA. 2008;105:20746–20751. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, Kito R, Nakao K, Kishimoto W, Yoo SH, Shimomura K, Takao T, Takano A, Kojima T, Nagai K, Sakaki Y, Takahashi JS, Ueda HR. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, Garcia M, Peters EC, Etchegaray JP, Traver D, Schultz PG, Kay SA. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol. 2010;8:e1000559. doi: 10.1371/journal.pbio.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee JW, Hirota T, Peters EC, Garcia M, Gonzalez R, Cho CY, Wu X, Schultz PG, Kay SA. A small molecule modulates circadian rhythms through phosphorylation of the period protein. Angew Chem Int Ed Engl. 2011;50:10608–10611. doi: 10.1002/anie.201103915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, Doyle FJ, Schultz PG, Kay SA. Identification of small molecule activators of cryptochrome. Science. 2012;337(6098):1094–1097. doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, Ye ZY, Pan HL, Takahashi JS. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci USA. 2012;109:101–106. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vougogiannopoulou K, Ferandin Y, Bettayeb K, Myrianthopoulos V, Lozach O, Fan Y, Johnson CH, Magiatis P, Skaltsounis AL, Mikros E, Meijer L. Soluble 3′,6-substituted indirubins with enhanced selectivity toward glycogen synthase kinase-3 alter circadian period. J Med Chem. 2008;51:6421–6431. doi: 10.1021/jm800648y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, Sladek M, Adams J, Bass M, Chandrasekaran R, Butler T, Griffor M, Rajamohan F, Serpa M, Chen Y, Claffey M, Hastings M, Loudon A, Maywood E, Ohren J, Doran A, Wager TT. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther. 2009;330:430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 85.Badura L, Swanson T, Adamowicz W, Adams J, Cianfrogna J, Fisher K, Holland J, Kleiman R, Nelson F, Reynolds L, St Germain K, Schaeffer E, Tate B, Sprouse J. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J Pharmacol Exp Ther. 2007;322:730–738. doi: 10.1124/jpet.107.122846. [DOI] [PubMed] [Google Scholar]
- 86.Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, Gibbs JE, Dupre SM, Chesham JE, Rajamohan F, Knafels J, Sneed B, Zawadzke LE, Ohren JF, Walton KM, Wager TT, Hastings MH, Loudon AS. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci USA. 2010;107:15240–15245. doi: 10.1073/pnas.1005101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grant D, Yin L, Collins JL, Parks DJ, Orband-Miller LA, Wisely GB, Joshi S, Lazar MA, Willson TM, Zuercher WJ. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbalpha. ACS Chem Biol. 2010;5:925–932. doi: 10.1021/cb100141y. [DOI] [PubMed] [Google Scholar]
- 88.Meng QJ, McMaster A, Beesley S, Lu WQ, Gibbs J, Parks D, Collins J, Farrow S, Donn R, Ray D, Loudon A. Ligand modulation of REV-ERBalpha function resets the peripheral circadian clock in a phasic manner. J Cell Sci. 2008;121:3629–3635. doi: 10.1242/jcs.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, Loudon AS. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, Takahashi JS, Butler AA, Kamenecka TM, Burris TP. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y, Kumar N, Nuhant P, Cameron MD, Istrate MA, Roush WR, Griffin PR, Burris TP. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORalpha and RORgamma. ACS Chem Biol. 2010;5:1029–1034. doi: 10.1021/cb100223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kumar N, Kojetin DJ, Solt LA, Kumar KG, Nuhant P, Duckett DR, Cameron MD, Butler AA, Roush WR, Griffin PR, Burris TP. Identification of SR3335 (ML-176): a synthetic RORalpha selective inverse agonist. ACS Chem Biol. 2011;6:218–222. doi: 10.1021/cb1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, Busby SA, Garcia-Ordonez RD, Burris TP, Griffin PR. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethy l]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol Pharmacol. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, Istrate MA, Kamenecka TM, Roush WR, Vidovic D, Schurer SC, Xu J, Wagoner G, Drew PD, Griffin PR, Burris TP. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 96.Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NR, Piggins HD, Chesham JE, Bechtold DA, Yoo SH, Takahashi JS, Virshup DM, Boot-Handford RP, Hastings MH, Loudon AS. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsuchiya Y, Akashi M, Matsuda M, Goto K, Miyata Y, Node K, Nishida E. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal. 2009;2:ra26. doi: 10.1126/scisignal.2000305. [DOI] [PubMed] [Google Scholar]
- 98.Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- 99.Quiroz JA, Gould TD, Manji HK. Molecular effects of lithium. Mol Interv. 2004;4:259–272. doi: 10.1124/mi.4.5.6. [DOI] [PubMed] [Google Scholar]
- 100.Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/S0092-8674(01)00383-X. [DOI] [PubMed] [Google Scholar]
- 101.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- 102.Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- 103.O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Z, McKnight SL. A conserved DNA damage response pathway responsible for coupling the cell division cycle to the circadian and metabolic cycles. Cell Cycle. 2007;6:2906–2912. doi: 10.4161/cc.6.23.5041. [DOI] [PubMed] [Google Scholar]
- 105.Pregueiro AM, Liu Q, Baker CL, Dunlap JC, Loros JJ. The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles. Science. 2006;313:644–649. doi: 10.1126/science.1121716. [DOI] [PubMed] [Google Scholar]
- 106.Oklejewicz M, Destici E, Tamanini F, Hut RA, Janssens R, van der Horst GT. Phase resetting of the mammalian circadian clock by DNA damage. Curr Biol. 2008;18:286–291. doi: 10.1016/j.cub.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 107.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 108.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/S0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 109.Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci USA. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci USA. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 112.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 113.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 114.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 115.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/S0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 116.Yagita K, Okamura H. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 2000;465:79–82. doi: 10.1016/S0014-5793(99)01724-X. [DOI] [PubMed] [Google Scholar]
- 117.Izumo M, Sato TR, Straume M, Johnson CH. Quantitative analyses of circadian gene expression in mammalian cell cultures. PLoS Comput Biol. 2006;2:e136. doi: 10.1371/journal.pcbi.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ding JM, Buchanan GF, Tischkau SA, Chen D, Kuriashkina L, Faiman LE, Alster JM, McPherson PS, Campbell KP, Gillette MU. A neuronal ryanodine receptor mediates light-induced phase delays of the circadian clock. Nature. 1998;394:381–384. doi: 10.1038/28639. [DOI] [PubMed] [Google Scholar]
- 119.Butcher GQ, Doner J, Dziema H, Collamore M, Burgoon PW, Obrietan K. The p42/44 mitogen-activated protein kinase pathway couples photic input to circadian clock entrainment. J Biol Chem. 2002;277:29519–29525. doi: 10.1074/jbc.M203301200. [DOI] [PubMed] [Google Scholar]
- 120.Coogan AN, Piggins HD. Circadian and photic regulation of phosphorylation of ERK1/2 and Elk-1 in the suprachiasmatic nuclei of the Syrian hamster. J Neurosci. 2003;23:3085–3093. doi: 10.1523/JNEUROSCI.23-07-03085.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Akashi M, Nishida E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 2000;14:645–649. [PMC free article] [PubMed] [Google Scholar]
- 122.Golombek DA, Ralph MR. KN-62, an inhibitor of Ca2+/calmodulin kinase II, attenuates circadian responses to light. Neuroreport. 1994;5:1638–1640. doi: 10.1097/00001756-199408150-00024. [DOI] [PubMed] [Google Scholar]
- 123.Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- 124.Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y. Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat Cell Biol. 2008;10:1463–1469. doi: 10.1038/ncb1806. [DOI] [PubMed] [Google Scholar]
- 125.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 126.Rossner MJ, Oster H, Wichert SP, Reinecke L, Wehr MC, Reinecke J, Eichele G, Taneja R, Nave KA. Disturbed clockwork resetting in Sharp-1 and Sharp-2 single and double mutant mice. PLoS One. 2008;3:e2762. doi: 10.1371/journal.pone.0002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kolker DE, Vitaterna MH, Fruechte EM, Takahashi JS, Turek FW. Effects of age on circadian rhythms are similar in wild-type and heterozygous Clock mutant mice. Neurobiol Aging. 2004;25:517–523. doi: 10.1016/j.neurobiolaging.2003.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, Su AI, Hogenesch JB, Kay SA. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, Schook A, Antoch MP, Turek FW, Takahashi JS. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci USA. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pulivarthy SR, Tanaka N, Welsh DK, De Haro L, Verma IM, Panda S. Reciprocity between phase shifts and amplitude changes in the mammalian circadian clock. Proc Natl Acad Sci USA. 2007;104:20356–20361. doi: 10.1073/pnas.0708877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jewett ME, Kronauer RE, Czeisler CA. Light-induced suppression of endogenous circadian amplitude in humans. Nature. 1991;350:59–62. doi: 10.1038/350059a0. [DOI] [PubMed] [Google Scholar]
- 132.Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/S0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 139.Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54:120–124. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee J, Kim MS, Li R, Liu VY, Fu L, Moore DD, Ma K, Yechoor VK. Loss of Bmal1 leads to uncoupling and impaired glucose-stimulated insulin secretion in beta-cells. Islets. 2011;3:381–388. doi: 10.4161/isl.3.6.18157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res. 2009;50:1800–1813. doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, Cutler MJ, Gulick J, Sanbe A, Robbins J, Demolombe S, Kondratov RV, Shea SA, Albrecht U, Wehrens XH, Rosenbaum DS, Jain MK. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483:96–99. doi: 10.1038/nature10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36:251–261. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang W, Barnaby JY, Tada Y, Li H, Tor M, Caldelari D, Lee DU, Fu XD, Dong X. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 147.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 148.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Roybal K, Theobold D, Graham A, DiNieri JA, Russo SJ, Krishnan V, Chakravarty S, Peevey J, Oehrlein N, Birnbaum S, Vitaterna MH, Orsulak P, Takahashi JS, Nestler EJ, Carlezon WA, Jr, McClung CA. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci USA. 2007;104:6406–6411. doi: 10.1073/pnas.0609625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, Jeong WJ, Tsai JY, Bugger H, Zhang D, Rohrwasser A, Rennison JH, Dyck JR, Litwin SE, Hardin PE, Chow CW, Chandler MP, Abel ED, Young ME. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol. 2008;294:H1036–H1047. doi: 10.1152/ajpheart.01291.2007. [DOI] [PubMed] [Google Scholar]
- 151.Anea CB, Zhang M, Stepp DW, Simkins GB, Reed G, Fulton DJ, Rudic RD. Vascular disease in mice with a dysfunctional circadian clock. Circulation. 2009;119:1510–1517. doi: 10.1161/CIRCULATIONAHA.108.827477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Arey R, McClung CA. An inhibitor of casein kinase 1 epsilon/delta partially normalizes the manic-like behaviors of the ClockDelta19 mouse. Behav Pharmacol. 2012;23:392–396. doi: 10.1097/FBP.0b013e32835651fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kawaguchi S, Shinozaki A, Obinata M, Saigo K, Sakaki Y, Tei H. Establishment of cell lines derived from the rat suprachiasmatic nucleus. Biochem Biophys Res Commun. 2007;355:555–561. doi: 10.1016/j.bbrc.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 154.He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL, Jr, Rossner MJ, Nishino S, Fu YH. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee S, Howell BJ. High-content screening: emerging hardware and software technologies. Methods Enzymol. 2006;414:468–483. doi: 10.1016/S0076-6879(06)14025-2. [DOI] [PubMed] [Google Scholar]
- 156.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science. 2012;337:189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Partch CL, Gardner KH. Coactivator recruitment: a new role for PAS domains in transcriptional regulation by the bHLH-PAS family. J Cell Physiol. 2010;223:553–557. doi: 10.1002/jcp.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B. J Am Chem Soc. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci USA. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Crane BR. Biochemistry. Nature’s intricate clockwork, Science. 2012;337:165–166. doi: 10.1126/science.1224611. [DOI] [PubMed] [Google Scholar]
- 163.Kudo T, Schroeder A, Loh DH, Kuljis D, Jordan MC, Roos KP, Colwell CS. Dysfunctions in circadian behavior and physiology in mouse models of Huntington’s disease. Exp Neurol. 2011;228:80–90. doi: 10.1016/j.expneurol.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pagani L, Semenova EA, Moriggi E, Revell VL, Hack LM, Lockley SW, Arendt J, Skene DJ, Meier F, Izakovic J, Wirz-Justice A, Cajochen C, Sergeeva OJ, Cheresiz SV, Danilenko KV, Eckert A, Brown SA. The physiological period length of the human circadian clock in vivo is directly proportional to period in human fibroblasts. PLoS One. 2010;5:e13376. doi: 10.1371/journal.pone.0013376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Roenneberg T, Merrow M. Entrainment of the human circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:293–299. doi: 10.1101/sqb.2007.72.043. [DOI] [PubMed] [Google Scholar]