Lead-In
Posttranslational modifications (PTMs) on histone proteins have emerged as a central theme in the regulation of gene expression and other chromatin-associated processes. The discovery that certain protein domains can recognize acetylated and methylated lysine residues of histones has spurred efforts to uncover and characterize histone PTM-binding proteins. In this task, chromatin biology has strongly benefited from synthetic approaches stemming from chemical biology. Peptide-based techniques have been instrumental in identifying histone mark binding proteins and analyzing their binding specificities. To explore how histone PTMs carry out their function in the context of chromatin, reconstituted systems based on recombinant histones carrying defined modifications are increasingly being used. They constitute promising tools to analyze mechanistic aspects of histone PTMs, including their role in transcription and their transmission in replication. In this review, we present strategies that have been used successfully to investigate the role of histone modifications, concepts that have emerged from their application, and their potential to contribute to current developments in the field.
Introduction
The DNA of eukaryotes is packed into the cell nucleus as chromatin, an assembly of DNA and DNA-binding proteins. Most prominent among these proteins are the histones, highly basic proteins that form an octamer structure as the fundamental building block of chromatin. Histones H3 and H4 form a central tetramer as a dimer of H3-H4 complexes, which is accompanied by two dimers of histones H2A and H2B. 147 base pairs (bp) of DNA bind to the basic surface provided by the histone octamer in 1.65 superhelical turns.[1, 2] DNA between two such nucleosomes can further be complexed by the linker histone H1, which is involved in establishing higher-order chromatin structure.
Instead of representing static entities that merely bind DNA to form a structural scaffold, nucleosomes are now widely recognized as being key players in the dynamic regulation of gene expression, DNA repair, and other cellular processes that rely on access to or sequestration of DNA. After the early discovery of the histone component of chromatin by Albrecht Kossel in 1884 and the identification of individual histone proteins from 1950 on,[3] covalent modifications of histone polypeptide side chains soon took center stage with the demonstration of methylated and acetylated lysine residues in histones.[4, 5] In a seminal study, Allfrey and coworkers showed that acetylation of histones substantially relieves the repression of mRNA synthesis associated with the addition of histones to naked DNA[6], leading them to hypothesize that relatively minor modifications in histone structure could provide means to facilitate RNA synthesis from chromatin in a locus-specific way and without histone removal.[7] Although the function of covalent histone modifications remains far from being deciphered, this insightful hypothesis has proven to be essentially correct.
Besides acetylation and methylation, lysine residues in histones are also subject to ubiquitylation and sumoylation. Other histone posttranslational modifications (PTMs) include methylation and deimination of arginines, phosphorylation at serine, threonine, and tyrosine residues as well as ADP ribosylation of glutamate residues, among others.[8] With the increasing sensitivity of mass spectrometry-based methods, more modifications will likely be added to this collection in the future. Most of these modifications are found at the N- and C-terminal histone tails that protrude from the nucleosome, rendering them accessible to the modifying machinery, interactions with neighboring nucleosomes in higher-order chromatin structures, and recognition by effector proteins. Genome-wide studies based on chromatin immunoprecipitation (ChIP) and subsequent microarray (ChIP-chip) or, more recently, deep sequencing (ChIP-seq) analyses provided us with a detailed view of how histone marks are distributed across the genome in various cell types and how histone PTMs and their combinations correlate with transcriptional activation or repression.[9-11] These static snapshots of PTM distributions are highly informative, but provide little insight into their actual mode of action. Despite tremendous advances, the mechanistic intricacies of histone PTMs remain largely elusive. It is beyond doubt that these modifications play important roles in the regulation of gene expression and in maintaining the integrity of the genome. Understanding the homeostasis as well as the function of histone PTMs, their impact on chromatin structure, and their recognition by effector proteins is an essential part of chromatin biology.
In this review we attempt to provide an overview of chemical biology-inspired strategies that are available and have been successfully employed in the field of epigenetics to unravel the function of histone PTMs, while highlighting general concepts and insights that have emerged from these studies. The first section illustrates the use of peptide-based strategies for the identification of histone PTM binding proteins and for the determination of the binding specificities of these proteins. Elucidating the underlying structural features of histone mark-binding proteins has significantly contributed to understanding the function of these proteins. Exploring exactly how histone PTMs exert their functions through either effector proteins or direct influences on chromatin structure is an ongoing quest. Techniques that allow for the incorporation of defined histone marks into nucleosome particles and chromatinized fragments have greatly facilitated the analysis of PTM function in reconstituted systems. These techniques and their applications will be dealt with in the second section of this text. Lastly, we briefly discuss how these and other techniques might help to clarify whether histone PTMs function as inheritable epigenetic signals.
Probing histone mark recognition
The introduction of synthetic peptide approaches in the study of histone PTMs
The structure of the histones and the localization of their PTMs make them ideal candidates for synthetic peptide-based approaches. Most of the modifications on histones H3 and H4 are present within the first 30 amino acids (aa) comprising their N-terminal tails. These tails are devoid of appreciable tertiary structure and any local structure is likely independent of the histone globular domain. Although substantially oversimplified relative to chromatin, many aspects of histones and of their interactions with other proteins can be recapitulated with the use of comparatively short peptides. Pioneering studies in the 1970s on histone deacetylase (HDAC) activity from calf thymus and histone acetylase (HAT) activity from rat liver have demonstrated the feasibility of such approaches.[12-16] Peptides were either derived from purified histones by proteolysis with chymotrypsin or acetic acid.[14, 16] or generated by solid-phase peptide synthesis[12-15] As a significant advantage, the synthetic approach afforded the possibility of differentially radio-labeling acetyl-lysine precursors to test for the preferred target sites of particular HDACs.[15] Short peptides spanning aa 14–21 of H4 were sufficient to allow deacetylation of K16, provided that the N and C termini of the peptide were acetylated and amidated, respectively.[12] Given that interactions between the active site of an enzyme and its substrate are spatially restricted and that histone tails in particular presumably lack defined folding, it is not surprising that peptide-based assays are still the most common approach to assess enzymatic properties of histone-modifying enzymes.
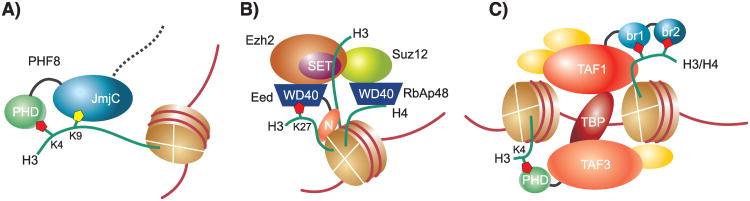
Of note, however, recent studies indicate that often neighboring modifications and even more distant features within nucleosomes can alter affinities towards substrate sites and reaction kinetics. For example, the histone demethylase PHF8 contains a H3K4me3-binding PHD finger adjacent to its catalytic jumonji domain, which demethylates H3K9me2 and H3K27me2. Peptides that carry both H3K4me3 and H3K9me2 are markedly better substrates for the enzymes, because the PHD domain significantly contributes to substrate binding (Figure 1A).[17] Within the polycomb repressive complex 2 (PRC2), whose catalytic Ezh2 subunit di- and trimethylates H3K27, multiple subunits are engaged in binding to nucleosomes. While RbAp48 binds H4,[18] Eed binds to H3 with its N-terminal region[19] and to H3K27me3 with its C-terminal WD40 domain (Figure 1B).[20] All these interactions likely modulate Ezh2-dependent methylation of H3K27 in vivo, but cannot be recapitulated on a peptide substrate. Thus, while the peptide-based assay results are often a valid approximation, the information may be limited in scope when considering the chromatin context. The same caveat also affects the peptide- and peptide array-based identification and characterization of histone PTM binding proteins. Still, significant insight has been gained based on carefully designed synthetic histone peptides, and peptides will remain an extremely helpful tool in chromatin biology even as more sophisticated models become available.
Figure 1. Multiple binding determinants cooperate to recruit histone binding proteins to chromatin.
A) Demethylation of H3K9me2 at the catalytic JmjC domain of PHF8 is enhanced through the presence of H3K4me3 on the same H3 peptide (illustrated here as part of a nucleosome). The PHD finger located adjacent to the JmjC domain recognizes H3K4me3. B) The PRC2 complex interacts with several surfaces of a nucleosome. The WD40 domain of RbAp48 binds to H4, whereas the N-terminal domain and WD40 domain of Eed binds to the globular domain of H3 and to H3K27me3, respectively. Moreover, the SET domain of Ezh2 interacts with H3 tails during catalysis. These determinants might be present on the same, as illustrated here, or neighboring nucleosomes. C) Subunits of the TFIID complex contain multiple domains capable of recognizing histone PTMs, which are implicated in recruiting the complex to promoter regions. The PHD domain of TAF3 binds to H3K4me3, while acetylated residues in the H3 or H4 tail are recognized by the tandem bromodomain of TAF1. TBP and other components (not shown) of the TFIID complex interact with DNA elements at the promoter.
Identifying histone PTM binding proteins
Synthetic peptides have also been instrumental in the identification of histone PTM binding proteins. The first histone effector proteins and their prototypical recognition domains were uncovered through structural studies designed to test the hypothesis that a candidate protein might interact with a histone modification, based on functional clues. Peptide pull-down assays were soon employed as well to identify novel proteins that bind to histone marks.
Even before the groundbreaking structural reports, individual studies probed for interaction with histone tails by pull-down-based approaches. Using bacterially expressed glutathione S-transferase (GST)-fused N-terminal peptides representative of the four core histones, Michael Grunstein's laboratory reported that the yeast silent information regulator proteins Sir3 and Sir4 interact with the N- terminal tails of histones H3 and H4.[21] This assay was next applied to the yeast HAT Gcn5 yielding a first clue to the function of the bromodomain, as Gcn5 was shown to interact with the N termini of H3 and H4 in a bromodomain-dependent manner.[22] The bromodomain was initially described for the protein brahma, a Drosophila orthologue of the yeast chromatin remodeler Swi2/Snf2, and subsequently identified in many nuclear proteins, especially HATs and chromatin remodeling complexes.[23-26] Shortly after the pull-down experiments on Gcn5, the NMR solution structure of the bromodomain of the p300/CBP coactivator P/CAF was reported.[27] As bromodomains are found in virtually all nuclear HATs the authors chose P/CAF as a candidate protein and probed for binding to an H4(1-12) peptide acetylated at K8. NMR titration revealed an interaction with a dissociation constant (KD) of 346 μM.[27] More recent studies have confirmed that this bromodomain along with others exhibit comparatively low affinities towards their targets.[28, 29] A notable exception is the tandem bromodomain of the TAF1 subunit of TFIID. In this case, the multi-site recognition of adjacent acetyl-lysines in an H4(1-36)K5ac/K12ac peptide gives rise to a dissociation constant of 1.4 μM, in comparison to 39 μM for a single acetyl-lysine peptide H4(1-36)K16ac, and >250 μM for unmodified peptide H4(1-36).[30]
This successful identification of the bromodomain–acetyl-lysine interaction led the Jenuwein and Kouzarides groups to employ peptide pull-down-based candidate approaches to probe for binders of methylated H3K9.[31, 32] Earlier studies had shown that the histone methyltransferase (HMT) Suv39H1 in humans (Clr4 in S. pombe) targets K9 on H3 in the context of constitutive heterochromatin[33] and that heterochromatin protein 1 (HP1) localization to these regions is dependent on its chromodomain[34] and on Suv39H1[35]. Indeed, in vitro pull-down assays revealed interaction of the yeast HP1 orthologue Swi6[31] and the murine HP1[32], with H3K9me3- and H3K9me2-containing peptides, respectively. These assays scored for the retention of radiolabeled proteins on affinity matrixes that were generated by maleimid[32] or iodoacetyl[31] coupling of synthetic, specifically modified peptides via their C-terminal cysteines (Figure 2A). The interaction of HP1 with H3K9me3-containing peptides exhibits a KD of about 70 nM, as determined by surface plasmon resonance measurements with the full-length protein.[31] For the isolated chromodomain, however, affinities of 0.7 μM and 2.5 μM were found by NMR titration with H3K9me2 peptide[36] and by isothermal calorimetry (ITC) with H3K9me3 peptide[37], respectively. These findings suggest that HP1 dimerization via its chromo-shadow domain leads to an increase in binding affinity through synergistic recognition of two H3 tails by the chromodomains. In support of this notion, efficient HP1 targeting to heterochromatin entails its binding to two methylated H3 molecules.[38]
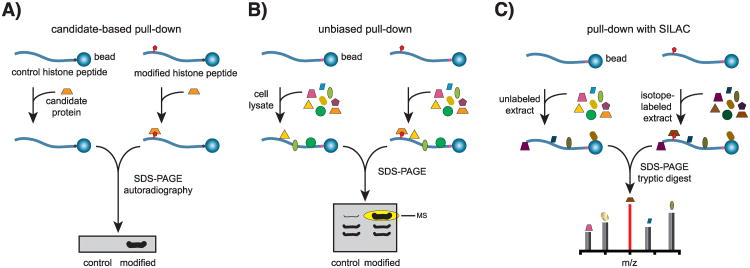
Figure 2. Peptide pull-down approaches for the identification of histone PTM binding proteins.
A) Candidate-based peptide pull-downs. Affinity matrixes are generated by coupling either modified or control peptides to beads. The beads are then incubated with a radiolabeled or affinity-tag-labeled candidate protein. Retention of the candidate protein on the affinity matrix is indicative of an interaction. B) To search for interacting proteins in an unbiased manner, cell lysates are employed instead of a purified candidate protein. Modification-specific interacting proteins, as well as proteins that bind nonspecifically to the matrix, and the peptide are collected and resolved by SDS polyacrylamide gel electrophoresis (SDS-PAGE). Proteins enriched in the pull-down with the modified peptide are identified by MS analysis. C) In pull-downs combined with SILAC, control and modified peptide are incubated with lysates from unlabeled and isotope-labeled cells, respectively. In subsequent MS analysis of recovered material, proteins binding to both affinity matrixes appear as peak doublets, while peptides corresponding to specific binders are strongly enriched in their labeled form versus unlabeled form. Therefore, unspecific binding can be dealt with in an effective way.
Polycomb (Pc), a component of the polycomb repressive complex (PRC) 1, and chromodomain helicase DNA binding protein 1 (CHD1) feature chromodomains similar to that of HP1. Not surprisingly, analogous peptide pull-down-based candidate approaches uncovered their methyl-lysine binding properties soon after the reports on HP1. A direct mechanistic link between PRC1 and PRC2 was recognized based on their respective connection with H3K27me3. On the one hand, the H3K27-methylating activity of the Ezh2 subunit of PRC2 was identified and characterized.[39-41] The same studies also demonstrated binding of the PRC1 component Pc to K27-methylated peptides that were immobilized to beads via covalent attachment or through biotin/streptavidin interaction.[39-41] The affinity of Pc for H3K27me3 is 5 μM, compared to 125 μM for H3K9me3.[42] Biotin/streptavidin-immobilized peptides also gave evidence of an interaction between the second chromodomain of yeast CHD1 and H3K4me2. This interaction is believed to enhance the activity of the SAGA-like CHD1-containing acetyltransferase complex SLIK.[43] Although yeast CHD1 binding to H3K4me2 could not be reproduced and remains somewhat controversial[44, 45], the double chromodomains of human CHD1 bind H3K4me3 with a KD of about 5 μM.[44, 46]
Examples of similar biased approaches based on functional similarities and/or homology of candidate proteins to known binders of the so-called ‘royal superfamily’[47] of chromo-, tudor-, malignant brain tumor (MBT), and PWWP domains can be found throughout the literature and continue to be instrumental in furthering our knowledge on histone PTM binding proteins. However, to uncover families of possible effectors whose structural binding modules do not share homology with those previously identified, unbiased methodologies must be incorporated. The advent of high-sensitivity mass spectrometry (MS) analysis greatly facilitated such endeavors and made it possible to identify even low abundance histone PTM binding proteins in an unbiased manner.
A common approach combines peptide pull-downs with subsequent MS-based identification of the recovered proteins (Figure 2B). The zinc-binding plant homeodomain (PHD) had been identified as a common structural motif in many nuclear proteins and was postulated to function as a protein-interaction domain.[48] Independent studies by several groups demonstrated that a subset of these proteins actually binds to histone methyl marks, most prominently H3K4me2/3. The first to be identified were inhibitor of growth (ING) family of tumor suppressor proteins, and the bromodomain and PHD finger transcription factor (BPTF) subunit of the chromatin remodeling complex NURF.[49-52] The identification of BPTF came about through the use of affinity pull-downs with methylated histone peptides as baits, and subsequent MS identification.[49, 52] Accompanying crystallographic studies revealed that PHD fingers exhibit a completely different overall structural fold, yet bind methylated lysines in a manner similar to that of the tudor domain family of proteins.[49, 50] Studies from our laboratory uncovered a number of indirect effectors that operate downstream of CHD1 using a histone peptide pull-down-MS approach.[53] Among other activities, complexes involved in transcript elongation (such as PAF and FACT) and components of the spliceosome were affinity-purified with H3K4me3-peptide-coupled resin.[53] CHD1 was shown to interact with these complexes and to bridge them to H3K4me[53], thereby extending the range of H3K4me-stimulated activities to include those that do not themselves contain a H3K4me3-binding domain.
A serious drawback of pull-down assays is the inherently high background of proteins that bind non-specifically to the affinity matrix or to the histone peptide regardless of its modification state. Vermeulen et al. devised a technique based on stable isotope labeling by amino acids in cell culture (SILAC) that elegantly eliminates background binding from subsequent MS fingerprinting.[54] In SILAC, stable heavy isotope-labeled variants of select aa replace their unlabeled forms in the cell culture medium, thus generating labeled “heavy” proteins. After digestion, the resultant labeled peptides can be distinguished from unlabeled counterparts by a mass shift. Pull-downs with unmethylated and methylated histone peptide are performed with unlabeled and labeled cell extracts, respectively (Figure 2C). Elutions from both assays are pooled and MS analysis is performed. All proteins binding to both affinity matrixes show up with two sets of peaks, one shifted with respect to the other by the isotope label. The vast majority of proteins (above 99%) bound to both matrixes equally well, while specific interactors showed enrichment in isotope-labeled peptides.[54] With such effective means to identify specific binders, the approach confirmed binding of H3K4me3 by ING proteins, BPTF, and CHD1, and also revealed new binders, the PHD finger proteins (PHF) 2 and 8, which were later shown to be H3K9-directed histone demethylases recruited and stimulated by H3K4me3.[17, 55] Moreover, the TAF3 subunit of the basal transcription factor TFIID was shown to bind H3K4me3, augmenting its recruitment to transcriptional start sites.[54] The bromodomains of its TAF1 subunit further facilitate TFIID recruitment through interaction with promoter-associated acetylation marks H3K9ac and H3K14ac.[30] The application of this exhaustive and unbiased technique to other histone PTMs, ideally in the context of fully reconstituted nucleosomes (see below), would most definitely prove fruitful in detailing the complement of histone PTM binding proteins. Indeed, in a recent comprehensive study the Mann laboratory extended the SILAC peptide pull-down approach to other trimethyl lysine modification sites. Among other interesting observations, a range of PWWP domain-containing proteins were identified to bind the H3K36me3 mark.[56] The origin recognition complex was found to be recruited to the repressive histone marks H3K9me3, H3K27me3 and H4K20me3.[56] As had been observed in the case of CHD1,[53] many proteins retrieved in the peptide pull-downs were recruited indirectly as part of macromolecular complexes. Genome-wide ChIP profiling of the identified binding proteins demonstrated good correlation with their respective histone marks, showing e.g. accumulation of H3K4me3-binding proteins at gene promoters.[56]
Array-based approaches for identifying PTM binding proteins
Peptide arrays have proven useful in the characterization of protein-protein interactions that involve recognition domains for linear peptide motifs, such as SH2, SH3, and PDZ domains.[57] Although less widely employed, array-based techniques also added to the exploration and characterization of histone PTM-interacting proteins.
The Gozani laboratory extensively used peptide arrays generated by spotting conventionally synthesized biotinylated histone peptides onto streptavidin-coated matrixes. An initial study employed arrays containing 12 H3-derived peptides with various methylation states to screen the complement of PHD finger proteins found in S. cerevisiae for their ability to bind to methyl-lysine; the majority of these domains recognized H3K4me3 or H3K36me3.[58] In subsequent applications with increasingly complex arrays, H3K4me3-binding PHD fingers were identified in the RAG2 subunit of the V(D)J recombinase[59] and the ING4 subunit of the HBO1 acetyltransferase complex[60], among others. Recently, efforts culminated in the generation of an array dubbed ‘human epigenome peptide microarray platform’ that comprises 63 different methylated and acetylated peptides.[61] These arrays were used to screen 67 members of the royal superfamily, confirming known interactors and uncovering three novel effectors. It was estimated that interactions with KD values ≤ 150-300 μM could be detected, based on a comparison with the known values of previously identified binders that were also detected on these arrays. These arrays have also proven useful for testing antibody specificity, as demonstrated for a selection of commercially available antibodies.[61] Antibodies against histone PTMs are invaluable tools for a plethora of approaches, e.g. genome-wide ChIP analysis. Yet cross-reactivity with other marks or with different degrees of methylation at the same residue can severely compromise the interpretation of results and must be guarded against with stringent controls.
A similar peptide array approach was taken to analyze the binding specificity of isolated bromodomains from 14 yeast proteins for 32 acetylated histone peptides, covering known acetylation sites in the four core histones. Combined analysis of binding data from the arrays, NMR titration, and structural models of the bromodomains underscored the importance of their ZA loop and their negatively charged residues, which anchor the histone peptides, for their affinity and selectivity in histone peptide binding.[62]
The peptide SPOT technique, introduced by Ronald Frank in 1992[63], allows parallel synthesis of peptides directly onto a solid-phase support.[63] Compared to arrays generated by spotting peptides post-synthesis, peptide SPOT arrays are less time-consuming and more cost-effective. They are, however, hampered by at times less efficient synthesis rates and more cumbersome quality controls of the synthesized, array-bound peptides. Surprisingly, histone peptide SPOT arrays have so far been used to only a limited extent. A pilot study describes their usability for assessing antibody and effector protein binding specificities, providing data on a limited set of interactions.[64] Jeltsch and coworkers employed peptide SPOT arrays in a number of studies. They analyzed the target specificity of the H3K9-specific HMT, Dim-5, from N. crassa by permuting all residues within the N-terminal 21 residues of H3 and performing HMT assays on the resulting arrays.[65] Application of an analogous approach to the SET domain of G9a led to the identification of novel non-histone targets for this HMT. So-called ‘CelluSPOTs’ arrays contain higher densities of spots on a smaller surface area than SPOT arrays, reducing reagent requirements. To generate such peptide ‘microarrays’, SPOT-synthesized peptides and their cellulose support are dissolved by e.g. TFA, and the resulting cellulose-peptide conjugates are respotted on glass slides.[66] The Jeltsch laboratory used CelluSPOTs arrays of N-terminal core histone peptides featuring 59 PTMs in various combinations to analyze the PTM specificity of the ADD and PWWP domain of DNMT3a, revealing that they bind to unmodified H3(1-19) and H3K36me3 peptides, respectively.[67, 68]
An even greater diversity of peptides can be generated with so-called ‘split-pool’ synthesis.[69] In this technique, the resin beads, on which the peptides are synthesized, are distributed to reaction vessels that each contain a single aa. After the coupling reaction is complete, the beads are collected and pooled. For the next round of synthesis, the beads are again split into separate pools and distributed to the reaction vessels. Alternatively, by reacting all beads in the same reaction vessel, common residues can be introduced at desired positions to yield a shared backbone sequence. By iterating this process, ‘one-bead, one-peptide’ libraries are generated that consist of fully or partially randomized peptides. Interactions are detected through ‘on-bead western’ assays using either fluorescently labeled antibodies or chromogenic reactions via alkaline phosphatase. Reactive beads are isolated and the corresponding peptides identified by sequencing or MS. A 512-peptide library containing randomized PTM combinations within the N-terminal 21 aa of H4 was screened for the preferred targets of interaction in the case of the JMJD2A double tudor domain. Verified by ITC measurements, the study confirmed that the double tudor domain binds not only to H3K4me3 but also to H4K20me3, with neighboring PTM sites modulating its binding affinity in a rheostat- rather than switch-like fashion.[70] Recently, the approach was applied to the N-terminal 10 aa of H3, generating a 5,000-peptide library with randomized PTMs.[71] A selection of H3K4me3 binders was characterized, revealing that PTMs at H3 sites that neighbor K4, i.e. R2, T3, and T6, can strongly influence binding to H3K4me3. R2 and T6 are differentially recognized by the domains assayed, indicating a possible regulatory mechanism that targets a subset of binders to specific sites.[71] Care must be taken, however, in the interpretation of binding data from such combinatorial libraries that likely contain some PTM combinations not present in vivo. In this study, evidence was provided that phosphorylation of H3T6 does occur in vivo, buttressing the claim based on the library screening.[71]
Protein arrays have been used to uncover histone interaction partners as well. In a pioneering study, Kim et al. generated a protein domain microarray featuring GST fusions of 109 domains of the ‘royal’ superfamily.[72] With the use of fluorescently labeled H3 or H4 peptides methylated to various degrees at H3K4, H3K9, or H4K20, the authors not only confirmed known interactions, but also uncovered a range of novel interactions, including the binding of MBT domains found in L(3)MBTL1 and CGI-72 to monomethylated H3K4 and mono/di-methylated H4K20.[72] It is still a matter of debate as to which histone PTM is recognized by L(3)MBTL1 in vivo.[73]In vitro and structural studies have demonstrated binding to mono- and di-methylated peptides with virtually no preference for the site of methylation.[74, 75] However, both L3(MBT)L1 association with chromatin and its role in transcriptional repression of the cyclin E gene in K562 cells have been shown to depend on the presence of PR-SET7, the H4K20 monomethylase, but not on G9a that catalyzes H3K9me2 in vivo, even though both marks are similarly recognized by L3(MBT)L1 in vitro.[76] These data point to a predominant role for H4K20me1 in vivo.
Histone PTM binding proteins – lessons learned
Since the groundbreaking discovery of the acetyl-lysine binding properties of the bromodomain, tremendous effort has been directed towards identifying and matching binders to the growing set of histone PTMs. As outlined above, most of these efforts were based on synthetic peptides in one form or another. These approaches have proven extremely successful, and a wealth of information has been obtained on how effector proteins recognize histone PTMs. Further intricacies are surely in store given the large and growing number of modifications described in the literature, as well as the range of protein families that contain potential histone binding domains. Unbiased approaches and techniques amenable to high-throughput screening will surely play a major role in this quest for histone effectors.
The use of peptides, however, is not without drawbacks. Histone tails represent only a fraction of the histone polypeptides. Moreover, within chromatin, histones are present as a DNA-bound octamer. In this context, many additional structural features will undoubtedly influence the recruitment of effector proteins in both positive and negative ways. As will be described in the next section, tools are now widely available to generate histones with specific PTMs and to reconstitute them into nucleosomes and model chromatin particles. With such reconstituted complexes as baits in pull-down assays, physiologically relevant, PTM-dependent interacting proteins could be revealed that require additional binding determinants, which are only present in a nucleosome or chromatin context, to generate sufficient affinity. As these highly complex systems offer a myriad of potential interaction surfaces with high charge density, unspecific binding might also become an even greater issue. As already demonstrated with peptide pull-downs,[54, 56] SILAC-based MS techniques could provide the means to overcome these obstacles and present us with hitherto unknown histone PTM effectors.
Even well established histone mark binding proteins often exhibit comparatively low affinities for their targets. Regulated protein-protein interactions in signal transduction cascades span a wide range of affinities with KD values often well below 1 μM (e.g. SH2 domains)[77] but also in the order of 100 μM (e.g. PDZ domains)[78]. Yet the absolute value of interaction affinities might be misleading outside of the physiological context.[79] It is conceivable for example that on a chromatin template within the cell nucleus, the local concentration of a histone PTM might be so high that low affinity binding would suffice to tether an effector to its binding site, even if the process might involve frequent binding-unbinding events. Stable compartments would still be maintained, even if individual molecules exchange. Such a scenario might facilitate rapid adaptation to external stimuli that require changes in gene expression and chromatin architecture. Indeed, evidence for such a dynamic behavior on the scale of individual molecules within stable chromatin domains has been furnished, e.g. for HP1.[80, 81]
Readily reversible binding can be preserved even under circumstances where higher binding affinity is required. It appears likely that binding to a histone PTM is often only one part of a complex net of interactions between a protein complex and chromatin. This phenomenon of recognition of multiple determinants on the same substrate via distinct binding interfaces is often referred to as multivalency. Hints at such cooperative binding in the context of histones can be found in multiple instances, some of which have been described in excellent reviews.[82, 83] Nucleosomes provide ample features that can be simultaneously engaged by an effector complex or histone-modifying activity. Additional affinity might stem from interactions with DNA, the histone octamer surface, or further PTMs on the same or adjacent histone tails (see examples in Figure 1). One can speculate that sequence-specific binding through transcription factors that are part of a recruited complex might contribute as well and provide further specificity for gene loci.
The nuclear proteome is replete with factors containing multiple interaction domains that may simultaneously engage different histone PTMs. As mentioned above, TFIID contains an H3K4me3-binding PHD finger in its TAF3 subunit as well as a double bromodomain in TAF1. Interaction of TFIID with H3K4me3 is enhanced in the presence of K9 and K14 acetylation.[54] Moreover, TFIID subunits can recognize core promoter elements, which is likely to significantly contribute to TFIID promoter recruitment (Figure 1C). To meaningfully assess combinatorial readout of histone marks, it is crucial to clarify which combinations of marks occur within the same nucleosome or on the same histone tail in vivo. Top and middle down MS has provided a framework for tremendous advances in the elucidation of histone mark co-occurrence within histone tails, demonstrating the presence of a multitude of histone PTM combinations in vivo.[84, 85] Beyond a single nucleosome, the formation of complex chromatin geometries/structures might foster the binding of effectors to surfaces composed of distinct nucleosome particles, including those that would seem quite distant from the perspective of the linear genome.
Multivalent binding might also resolve issues regarding the specificity of histone effector proteins. As discussed above, the binding specificities of L(3)MBTL1, and MBT domains in general, are defined rather loosely in vitro. Yet, L(3)MBTL1 binding to H4K20me1 leads to specific functional outcomes in vivo, suggesting that the in vitro binding is not fully reflective of the case in vivo. This may be a consequence of additional sites on L(3)MBTL1 that may recognize other signals or perhaps particular chromatin geometries that in summation contribute to its binding and increase its binding specificity. A surface groove on the MBT domains might comprise such an additional binding site.[73] It is also worthwhile to consider the degree of stringency required to couple binding of a histone effector to a specific outcome. So far, this question has not been addressed in a comprehensive fashion, and the solution will most likely be case-specific. For some effectors, strict discrimination of a specific mark might be determinant to the functional outcome, while for others distinguishing between so-called ‘active’ and ‘repressive’ marks might suffice.
Functional analysis of histone modifications in the context of nucleosomes and chromatin models
Histone marks are thought to elicit their functions both in direct and effector-mediated ways.[83, 86] A PTM may directly influence intrinsic properties of the nucleosomes by altering DNA–histone contacts, leading to increased nucleosome mobility and changes in local conformation. Extrinsic effects, in contrast, affect inter-nucleosomal contacts and thus higher-order chromatin structure. Interactions with effector proteins may lead to altered nucleosomal properties as a result of the effector's enzymatic activity and/or the establishment of higher-order structures via the recruitment of scaffolding proteins. To study the functional consequences of a histone PTM, it is preferential to investigate its role within its native context, that being nucleosomes and chromatin. While the in vivo case is mostly too complex and heterogeneous for mechanistic studies, reconstituted in vitro systems of defined composition and reduced complexity can be faithful models for cellular processes and are ideally suited to biochemically dissect their mechanistic aspects. For the chromatin field, the methodology to reconstitute such recombinant species containing defined histone PTMs has become increasingly accessible in recent years. We will introduce both chemical ligation-based and cysteine modification-based strategies and provide examples for their successful application to current topics in chromatin biology.
Assembly of recombinant chromatin
Studies describing the reconstitution of nucleosomes and chromatin-like fragments from separate DNA and histone preparations date back to at least the 1970s. In these early studies, however, the spacing of nucleosomes within reconstituted material did not resemble that observed in native chromatin. Crucial advances rested on the observation that chicken erythrocyte core histones reside at a distinct position when reconstituted on a 260-bp DNA fragment containing the 5S rRNA gene of the sea urchin Lytechinus variegatus.[87] In a seminal 1985 study, correctly spaced nucleosomes, as gauged relative to the in vivo case, were reconstituted from DNA constructs containing up to 50 copies of truncated 5S rRNA fragments and chicken core histones upon gradient or step-wise dialysis starting from 2 M NaCl to salt-free conditions.[88] Micrococcal nuclease digestion gave evidence of distinct bands indicative of the repeat length of the DNA, with regular spacing of nuclease-inhibiting core particles on the DNA. Electron microscopy (EM) confirmed the presence of regularly spaced histone octamers on these DNA fragments. These early techniques devised to obtain nucleosomes spaced in a manner reflective of the in vivo case, are employed to this date, with few changes. A selection of nucleosome positioning sequences beyond the original 5S rRNA are now commonly used, most notably the so-called ‘601′ fragment. 601 was obtained in a screen of random synthetic DNA fragments that selected for high histone octamer affinity and strong nucleosome positioning.[89] The cloning of histone genes from Xenopus laevis[90] and other organisms led to the isolation of homogeneous unmodified core histones, that in turn provided the resources to develop fully reconstituted in vitro systems used for many studies including those that tackled the structural determinants of the core nucleosome particle.[2] Besides salt dialysis-based methods, ATP-dependent chromatin remodelers such as ACF and RSF are also widely employed to assemble regularly spaced chromatin.[91-93] Although technically more demanding, these systems offer the advantage of utilizing a broader range of DNA sequences that would not give rise to regular spacing by salt dialysis.
Generating histones with site-specific marks: Native chemical and expressed protein ligation
Native chemical ligation (NCL)[94] and its close relative, expressed protein ligation (EPL)[95], have been developed by the Kent and Cole labs, respectively. These methods greatly facilitated the generation of proteins with defined site-specific modifications representative of in vivo post-translational events. In NCL, two peptides, derived from conventional solid-state peptide synthesis, are ligated via a C-terminal thioester on one peptide to an N-terminal cysteine on the other (Scheme 1). The initial thioester ligation product spontaneously rearranges to yield a native peptide bond at the ligation site. The size of constructs that can be generated by NCL is set by the limits of peptide synthesis. To generate larger proteins by NCL, the C-terminal peptide can easily be replaced by a recombinant protein with an N-terminal cysteine liberated, e.g. by proteolysis. This approach is ideally suited to histone proteins as most modifications are confined to their N termini. A more elaborate strategy is required to introduce specific modifications at the C terminus of a protein. In EPL, the recombinant part is fused to a mutated protein splice site (intein) at its C terminus.[95] The mutation of the intein traps the cleavage reaction, usually leading to an amide bond in equilibrium with the thioester intermediate. Thiolysis produces the recombinant protein in the thioester form, which can then be readily ligated to a peptide with an N-terminal cysteine (Scheme 2A). Combined with the methods for nucleosome reconstitution described above, NCL and ECL paved the way to study histone PTMs in a defined manner, reflective of native chromatin, and thus superior to peptide models.
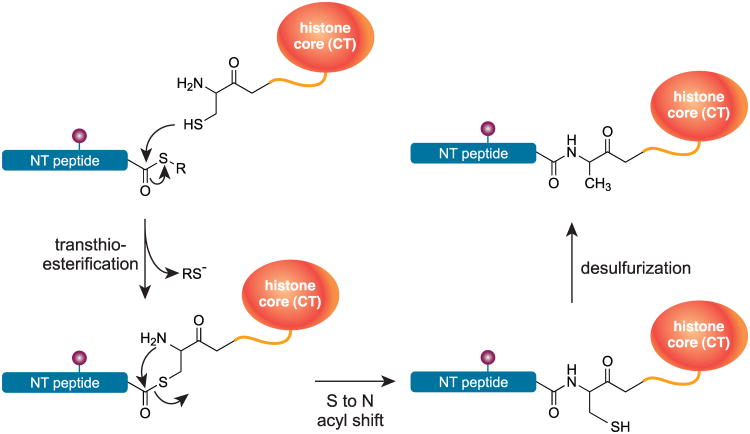
Scheme 1.
Native chemical ligation[94] and subsequent desulfurization[98] exemplified by a histone core domain and a peptide with a C-terminal thioester (SR).
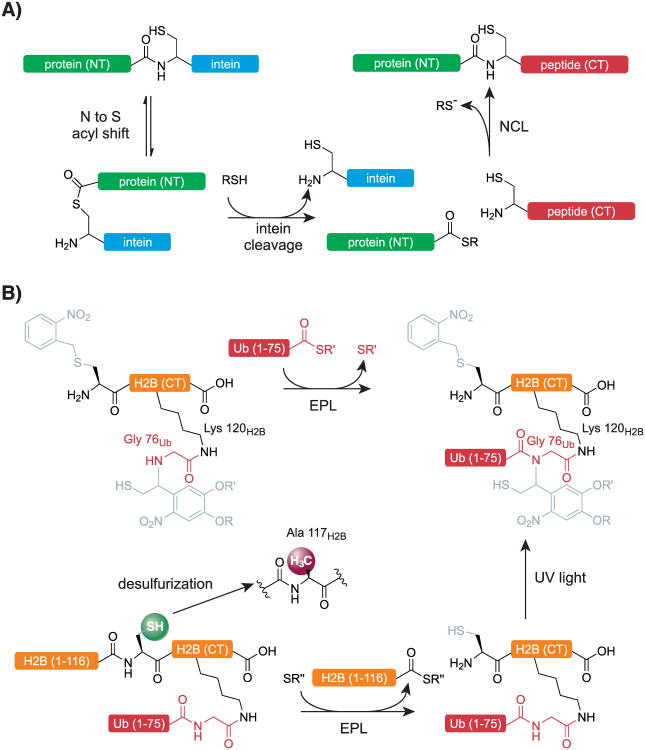
Scheme 2.
A) Expressed protein ligation of an intein-fused protein to a C-terminal peptide via intein cleavage with a thioester (RSH).[95] B) Generation of ubiquitinated H2B from a synthetic peptide corresponding to the C terminus of H2B and intein-fused ubiquitin and the H2B N terminus.[107] Residues of H2B are shown in black, those of ubiquitin in red. Protective groups of the ligation auxiliary are in light gray.
Application of NCL and EPL to study histone PTMs
The Peterson group introduced NCL to the field of chromatin biology. In an initial study exploring the feasibility of the approach, they generated H3 specifically phosphorylated at S10 by ligation of an H3(1-31) peptide to an N-terminally truncated Xenopus laevis H3 with a T32C mutation to allow ligation.[96] This ligation product assembled well into histone octamers, which could further be incorporated into 11-repeat nucleosomal arrays. Remodeling of these arrays by yeast SWI/SNF was unaltered, compared to wild-type. In partial contrast to previous findings that Gcn5 activity as part of the SAGA complex was stimulated by S10 phosphorylated-peptides, only Gcn5 in isolation was stimulated by S10 phosphorylation on these arrays.[96] In the peptide-based experiments, the stimulatory effect was attributed to an increase in catalytic activity or to a higher substrate affinity.[97] On reconstituted chromatin however, the phosphorylation-mediated increase in affinity might be masked by additional interactions between other SAGA components and the nucleosomal array. This finding underscores that multi-site binding on a substrate as complex as a nucleosomal array might play important roles that cannot be recapitulated by, and thus go unnoticed in, simple peptide models.
Shortly after this initial report, the NCL strategy was further developed in the McCafferty and Reinberg laboratories. Refining the strategy employed by the Peterson group, the ligation was rendered traceless by adding a desulfurization step after the ligation that converts cysteine to alanine at the junction (Scheme 1).[98] Thus, an alanine such as A25 in H3 or A15 in H4 was chosen as the border. H3 and H4 versions with up to 5 specific acetylation sites per tail were generated, along with the first example of a methylated version of a synthetic histone, H3K9me3. All these semi-synthetic histones could be incorporated into H3-H4 tetramers and subsequently assembled along with H2A/H2B dimers onto DNA templates through the RSF nucleosome deposition complex.[98] The H3K4ac/K9ac/K14ac/K18ac/K23ac-containing tetramers were substrates for the HDAC Sir1 and, after the deacetylation step, also for the H3K9-specific HMT G9a.[98] The Muir laboratory likewise demonstrated traceless ligation for H2B carrying phosphorylation at S14 and acetylation at K5, K11, K12, and K15.[99] These studies further illustrate the potential of semi-synthetic histones for the study of chromatin-associated processes.
As of today, the most recognized contribution of NCL to chromatin biology came about from the Peterson group's analyses of the effects of H4K16ac on chromatin structure.[100] K16-acetylated H4 derived from NCL could readily be assembled into histone octamers, nucleosomes and 12-repeat nucleosomal arrays. In the context of mononucleosomes, H4K16ac exerted a slight inhibitory effect on ACF-mediated nucleosomal sliding, which was attributed to reduced interaction between ACF and its nucleosome substrate.[100] However, striking effects were observed on nucleosomal arrays. While compaction of such arrays can usually be induced by divalent cations in vitro, analytical ultracentrifugation revealed that MgCl2-mediated compaction into 30-nm-like chromatin fibers is severely impaired in the case of H4K16ac-containing arrays.[100] Moreover, the self-association of these arrays, believed to mimick fiber-fiber interactions in higher-order structures, was diminished.[100] Taken together, these data suggested that H4K16ac may promote the formation of a decondensed chromatin structure by modulating nucleosome-nucleosome interactions, possibly through diminishing electrostatic interactions with an acidic patch on H2A,[101] while also interfering with recruitment of histone-binding proteins.
Although the introduction of methyl marks by NCL is feasible as well, subsequent applications focused on the analysis of histone acetylation and chromatin remodeling. The introduction of 4 acetyl-lysines in each of the H3 and H4 N-terminal tails led to differential, complex-specific effects on the yeast Chd1, Isw2, and RSC remodeling complexes in nucleosome sliding assays and for octamer transfer between DNA fragments.[102] Expression of a C-terminally truncated H3 as an intein fusion protein and subsequent EPL enabled the introduction of specific acetylation at K115 and K122.[103] These sites are located close to the dyad axis of the nucleosome, where essential contacts are established at the H3/H4 tetramer interface and histone-DNA interactions are formed. Acetylation at these sites reduces affinity for DNA, possibly facilitating nucleosome repositioning or disassembly.[103] These histones were further employed in a study that revealed the nucleosome disassembly properties of the mismatch recognition proteins hMSH2-hMSH6.[104]
The Muir laboratory, responsible for many advances in EPL, devised strategies to link ubiquitin and other large peptidic modifiers to histones in a site-specific fashion. Nucleosomes can carry ubiquitin moieties at H2AK119 and H2BK120. Ubiquitination of H2A is catalyzed by the Ring1A/B components of PRC1 and is associated with silenced chromatin, whereas ubiquitination of H2B is brought about by the RNF20 (Bre1 in S. cerevisiae) E3 ligase in the context of active transcription.[86, 105, 106] The initial strategy for synthetic ubiquitination of H2B utilized a short synthetic, modified C-terminal peptide of H2B(117-125) and recombinant, intein-fused C-terminally truncated H2B(1-116) and ubiquitin (1-75).[107] First, the ubiquitin thioester was ligated to the peptide fragment with the help of a ligation auxiliary (Scheme 2B). In the next step, UV irradiation was used to remove the ligation auxiliary yielding G76 of ubiquitin, and the protective group on the cysteine that was subsequently ligated to H2B and desulfurized. H2BK120ub was sufficient to stimulate DOT1L HMT activity towards H3K79 in reconstituted mono- and dinucleosomes.[107] Use of synthetically modified H2BK120ub in in vitro transcription assays showed that this modification does not influence transcription per se.[108] However, a stimulatory effect was observed on SET1-dependent H3K4 di- and trimethylation, potentially explaining the presence of H2BK120ub at actively transcribed genes.[108] An alternative strategy that links ubiquitin to H2B via a disulfide linkage was developed to simplify the generation of H2BK120ub.[109] Here, K120 is replaced by a cysteine, which is activated by DTNP to an asymmetric disulfide, whereas full-length ubiquitin is expressed as a C-terminal intein fusion protein. After intein cleavage with cysteamine, both recombinant proteins are fused via a disulfide linkage mimicking the native isopeptide linkage. With this strategy, ubiquitin was attached to variable sites within the C terminus of H2B and replaced by ubiquitin-like proteins to assess the structural requirements for H2Bub–DOT1L crosstalk.[109] Analysis of H2A ubiquitination with these tools has yet to be reported.
Amber suppression and orthogonal ribosomes
Amber suppression and related approaches allow for the generation of modification-bearing histones that, like those generated by NCL, are indistinguishable from their in vivo counterparts. Unnatural or modified aa can be incorporated co-translationally into proteins in E. coli or yeast by making use of the unique amber codon and a corresponding tRNA–aminoacyl tRNA synthetase pair.[110] Although amber suppression strategies have been applied to histone modifications as well, their use has been reported in only very few studies.[111] This is mainly due to the limited accessibility and distribution of the required tools and the comparatively low efficiency in the synthesis of modified proteins. To incorporate two distinct modified aa, the Schultz laboratory evolved an orthogonal pair that recognizes a quadruplet codon coding for homoglutamine.[112] To circumvent interference with the synthesis of the host cell proteome, the Chin group developed a fully orthogonal ribosome system that decodes orthogonal mRNAs not recognized by the endogenous E. coli ribosomes, thereby increasing the amount of available codons and significantly enhancing yields.[113] By synthetic evolution they adapted the orthogonal ribosome to efficiently decode quadruplet codons, increasing the potential number of unnatural amino acids that can be incorporated to about 200.[114] In principle, with the directed evolution of orthogonal, quadruplet-decoding tRNA–aminoacyl-tRNA synthetase pairs for all histone PTMs, the efficient generation of recombinant histones with extensive PTM combinations appears feasible. However, as differentially methylated lysine residues are structurally very similar, evolution of specific synthetases for these residues is an especially daunting task. At this stage, application of orthogonal ribosome strategies to histones with complex modification patterns remains a promising, but as yet a far away prospect.
Generating methylation marks by cysteine modification strategies
Despite their proven advantage, there is an enormous gap between the plethora of questions that can be addressed with NCL and EPL and the number of studies published so far. First and foremost, this is due to the relatively high requirements of these approaches. Both the instrumentation for peptide synthesis and the requisite chemical skills might not be readily available to biology-focused labs. A method recently developed by Kevan Shokat and coworkers has the potential to significantly widen the study of histones with site-specific modifications due to its relative ease and affordability.
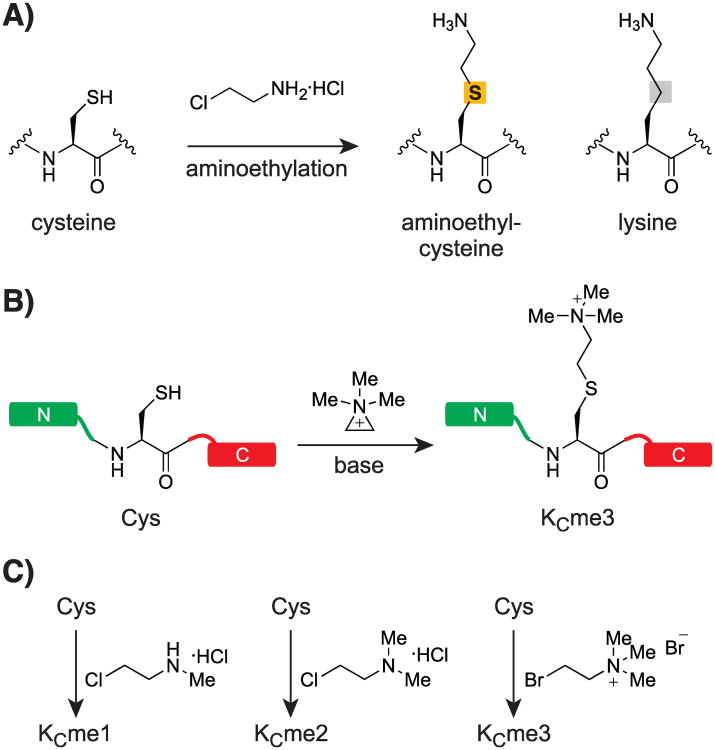
The reactivity of cysteine as a nucleophile has been exploited for various site-directed labeling and coupling strategies in proteins. It has long been known that cysteine can be alkylated to aminoethylcysteine (designated as Kc), an analog of lysine distinguished by sulfur replacing the γ-carbon (Scheme 3A). Simon et al. extend this reaction by employing N-methyl aminoethylhalides to obtain methyl-lysine analogs (MLAs, Scheme 3B,C).[115] As Xenopus laevis H3 and human H3.3 contain only a single cysteine residue (C110) that can be mutated to alanine without noticeable effects on nucleosome structure and function, any lysine is amenable to conversion into an MLA by site-directed replacement with cysteine in the C110A background. The alkylating reagents are commercially available, and the reactions have been optimized to go to completion and do not require special instrumentation. Detailed protocols have been published that render the procedure readily available to biology- and biochemistry-focused laboratories.[116]
Scheme 3.
A) Generation of aminoethyl-cysteine and comparison to lysine. B) Reaction of a cysteine residue with (2-bromoethyl)-trimethylammonium bromide via an aziridinium intermediate. C) Conversion of cysteine to mono-, di-, and trimethylated aminoethyl-cysteine as described by Simon et al.[115].
MLAs incorporate into reconstituted octamers and nucleosomes and are recognized by antibodies against the corresponding methylated lysine residues, although with about 5-fold reduced affinity in the case of an H3K9me2 antibody.[115] Using biotinylated MLA-containing mononucleosomes as baits in pull-down assays, HP1α but not the PRC2 component Suz12 was specifically enriched for H3Kc9me2 compared to wild-type mononucleosomes. Moreover, the H3K9 HMT Suv39H1 was able to utilize Kc9-containing peptides as substrates, although with moderately reduced efficiency. Lastly, ACF-mediated remodeling was found to be unaffected by the introduction of MLAs into mononucleosomes.[115]
Taken together, these initial studies demonstrated that MLA-containing histones are reasonable models for methylated histones. Compared to NCL and EPL strategies, the cysteine modification approach is easy to perform and also applicable to sites that are beyond the reach of ligation techniques. Incorporation of methylated K36 and especially K79 into H3 is currently not feasible by semisynthetic strategies due to limitations in peptide synthesis. Synthetic peptides, however, permit the simultaneous incorporation of multiple distinct modifications. With the MLA approach, multiple lysines can potentially be modified within the same histone, but only with the same degree of methylation. Moreover, one should bear in mind that aminoethylcysteine is chemically distinct from lysine, potentially diminishing its recognition and modification by binding proteins and HMTs or demethylases. The replacement of the γ-carbon by sulfur changes the geometry of the side chain, extending the bonds to neighboring carbons to 1.82 Å from 1.54 Å for C-C bonds and reducing the bond angle by about 9°. Moreover, the presence of the sulfur reduces the pKa of the amine group by 1.1 units. Still, these changes seem to be generally well tolerated, but this may not hold true for all sites and applications. In addition to the proof-of-concept experiments performed in the initial paper from the Shokat lab, the Zhu lab performed extensive characterization to confirm the compatibility of MLAs with a variety of assays. They recapitulated mark-dependent binding of HP1α, 53BP1, and the ankyrin repeats of G9a to H3KC9me2/3, H4KC20me1/2, and H3KC9me1/2, respectively.[117] The HMTs HYPB, PR-Set7, Suv4-20h1, Suv4-20h2, and Dot1L were all able to methylate MLAs in a nucleosomal context although for some, a slight reduction in efficiency was observed with the unmethylated pseudolysine.[117] Moreover, JMJD3 and JHDM1a were shown to demethylate H3KC27me3 and H3KC36me1 peptides.[117] These results hint at a broad range of possible applications for MLA-containing histones.
Applications of MLA-containing histones
H3K79 is located in a solvent-exposed region within the structured core domain of H3. Site-directed modification of this residue with a MLA has been used to obtain crystal structures of mononucleosomes containing H3K79me2.[118] Whereas overall nucleosome structure is unaffected by H3K79me2 or H4K20me3, which was analyzed in parallel, methylation of H3K79 leads to local changes in surface electrostatic potential close to the DNA superhelix.[118] H4K20me3-containing nucleosomal arrays showed enhanced propensity for MgCl2-mediated compaction, potentially antagonizing the role of H4K16ac in the same region.[118]
Xu et al. reported an NMR structure of an N-terminal fragment of the Eaf3 subunit of an HDAC complex denoted reduced potassium dependency 3 (Rpd3S) from S. cerevisiae.[119] It adopts a chromo barrel-related fold and employs four aromatic residues in a cage structure to bind H3K36me2. The affinity is in the range of 1–3 mM, so that stable peptide binding could only be achieved by expressing the Eaf3 fragment directly fused to the H3 peptide. The methylation was introduced in the form of a MLA, showing that MLAs might also be helpful in the elucidation of histone – effector complex structures. Binding of the full Rpd3S complex to nucleosomes was further studied with histones containing KC36 methylated to different degrees. In electrophoretic mobility shift assays with reconstituted, MLA-containing mononucleosomes, Rpd3S was shown to bind both H3KC36me2 and H3KC36me3, whereas monomethylation of K36 was not sufficient.[120]
ING4 is a subunit of the HBO1 HAT complex and carries a PHD finger that recognizes H3K4me3. Implications of this binding event for the function of this HAT complex were assessed in HAT assays on wild-type vs. H3KC4me3 mononucleosomes.[60] Whereas acetylation of H4 was unaffected, acetylation of H3 was enhanced in the presence of H3KC4me3, indicating a possible H3K4me3-dependent switch in substrate specificity for this HAT complex.[60]
The substrate of the nuclear receptor SET domain-containing (NSD) family of HMTs has been a matter of debate since initial reports pointed to a broad range of targeted residues in H3 and H4. Two recent reports, however, indicated that, within a nucleosomal context, H3K36 is the target of the NSD HMTs.[121, 122] The SET domain of NSD2 was able to utilize nucleosomes carrying H3KC36 in either un- or monomethylated, but not di- or trimethylated form, indicating that NSD2 is a H3K36 dimethylase.[121] In the characterization of dKDM4A, the Drosophila orthologue of the histone demethylase Rph1, MLAs were used to confirm its substrate specificity, which was initially established on HeLa cell core histones with the use of antibodies. While H3KC36me3 is a substrate of dKDM4A, it cannot demethylate H3KC36me1.[123] In a study primarily reporting the inhibition of the HDAC Rpd3 upon complex formation with the histone demethylase Lid3 in Drosophila, concurrent effects on Lid3 were assessed.[124] H3KC4me3-modified H3 was readily demethylated by Lid3, irrespective of its interaction with Rpd3. Despite the presence of an H3K36me3-binding chromodomain protein Mrg15 in the complex, Lid3-dependent demethylation was unaltered in the presence of KC36me3 on the same histone.[124] This study is one of the very few examples of the use of multiply modified MLA histones.
Our laboratory has recently shown that the Eed subunit of the PRC2 complex binds to H3K27me3 and other repressive histone marks through an aromatic cage located at the top of the WD40 β-propeller, leading to allosteric activation of the HMT activity of the Ezh2 subunit.[20] These findings suggest a potential mechanism for the propagation of H3K27me3 and the maintenance of repressive chromatin domains. Binding to repressive marks was initially observed in a co-crystal structure of Eed with modified histone peptides and subsequently analyzed on reconstituted nucleosomal arrays containing H3KC9, H3KC27, or H3KC36 MLA histones with different methylation degrees.[20] Binding of PRC2 complexes was strongly enhanced by H3K27me3 and H3K9me3, whereas H3K36me3 was ineffectual.[20]
Recent studies from several groups have revealed that Jarid2 is associated with the PRC2 complex.[125-128] Jarid2 (also known as Jumonji or Jmj) is a member of the jumonji family of histone demethylases, but lacks demethylation activity. In genome-wide ChIP-seq analysis, binding of Jarid2 significantly overlapped with PRC2 target sites and consequently sites of H3K27me3 enrichment.[125-128] Recruitment of PRC2 to these sites depended strongly on Jarid2 and vice versa.[125-128] To assess the impact of Jarid2 on the catalytic activity of PRC2 in vitro, the Orkin laboratory employed MLA-containing histones as substrates for recombinant PRC2 complexes. As expected, these assays showed that PRC2 could utilize un-, mono-, di-, but not trimethylated H3KC27-containing octamers and nucleosomes as substrates.[128] In these assays, Jarid2 inhibited PRC2 activity in a dose-dependent manner.[128] In contrast, stimulation of PRC2 by Jarid2 was observed on recombinant nucleosome substrates containing unmodified H3.[125] It appears likely that differences in the composition of PRC2 complexes used and/or other assay conditions gave rise to these discrepancies, rather than the properties of the MLA-containing histones used as substrate.
Francis and coworkers employed MLA histones to investigate the behavior of polycomb proteins in an in vitro replication assay.[129] Their study was aimed at clarifying whether chromatin-bound factors remain associated with DNA during the process of replication. Although PRC1 complex inhibits in vitro replication in a dose-dependent manner, replication at sub-inhibitory concentrations of PRC1 does not fully dissociate the prebound PRC1 complex from the recombinant chromatin templates.[129] Although the PRC1 subunit Pc has been shown to bind H3K27me3, use of H3KC27me3-containing chromatin templates neither enhanced the inhibitory effect of PRC1 on replication nor altered the association of PRC1 to templates during replication.[129] This unexpected observation might be explained by the high concentration of the factors involved that greatly exceed the KD values for the interactions involved, thereby masking potential effects.[129]
Reconstituted transcription systems containing modified histones
Many histone PTMs have been implicated in the activation or repression of transcription. This notion is based on their correlation with active and repressed gene loci and is supported by functional data on their influence on chromatin structure and on their recognition by factors involved in either the formation of heterochromatin or various steps of transcription. However, direct evidence for stimulation or inhibition of transcription through histone PTMs is sparse. Partially, and later also fully, reconstituted in vitro transcription systems have been an integral part of research on transcription for many years and have been instrumental in the identification of the general transcription factors and beyond. They provide an ideal resource to directly analyze how histone PTMs affect the transcription process.
The effects of H3K4 trimethylation and H2B ubiquitination have been assessed in reconstituted transcription systems. These marks are interrelated, as it has been shown that ubiquitination of H2B at lysine 120 is a prerequisite for methylation of H3K4 and H3K79 by Set1 and Dot1, respectively.[86, 106] In a fully reconstituted system that supports ligand-dependent transcription initiation and elongation on a chromatinized template containing the retinoic acid–RAR/RXR-responsive RARβ2 promoter, enzymatic ubiquitination of H2B directly stimulated transcription at the post-initiation stage.[130] As has been shown before in various instances (see e.g. [131, 132]), histone acetylation, in this case mediated by p300, was required for transcription initiation on chromatin in vitro.[130] This report further analyzed the effect of H3K4me3 on transcription. In contrast to its presence at high levels at actively transcribed loci, enzymatic introduction of H3K4me3 did not enhance transcription in this system.[130] This may either be due to the absence of H3K4me3-binding proteins besides TFIID in this highly purified system, or indicate that H3K4me3 is not affecting transcription per se but mainly stimulates events downstream of transcription, such as splicing (see e.g. [53]). Although this study does not employ chemically, but instead enzymatically modified histones, it illustrates the great potential of combining fully reconstituted systems with synthetically modified histones to uncover the role of specific histone PTMs in the context of transcription.
The Roeder and Muir groups investigated the role of H2B monoubiquitination in a similar reconstituted transcription system. To analyze the influence of H2B ubiquitination separately from the ubiquitination machinery, they reconstituted a p53-dependent template on octamers containing semi-synthetic ubiquitinated H2B[107] (see above) and monitored transcription.[108] In this setting, H2B ubiquitination did not stimulate transcription.[108] Both the components and the chromatin template employed in this fully reconstituted transcription system differ from the scenario described above and may lack downstream factors dependent on H2B ubiquitination, one of which might be FACT. The chemically ubiquitinated chromatin was effective in stimulating H3K4 trimethylation by the human SET1 complex, confirming earlier studies.[108]
In the future we will surely see many more studies combining reconstituted transcription systems and recombinant histones with defined PTMs. However, both establishing a robust transcription assay and generating meaningful chromatin templates are by no means trivial tasks, as reflected by the small number of such studies published thus far. Reconstituted systems employing either nuclear extract or partially purified fractions as well as fully reconstituted systems have specific advantages for the study of histone PTMs. Although fully reconstituted systems are ideally suited to pinpointing the function of candidate, purified factors with respect to histone PTMs, the effects of such histone marks might also be missed due to the absence of other pertinent factors. Such effects of histone PTMs might be evident using nuclear extracts, but the identification of hitherto unknown factors that stimulate or inhibit transcription in a PTM-dependent fashion would necessitate fractionation of the extract. Conventional purification approaches have been employed successfully in the identification of the general transcription factors[133-135] for example, and might prove useful again to unravel the function of histone PTMs in transcription.
Fate of histone PTMs during DNA replication
The potential role of histones and their PTMs in transmitting epigenetic information is intimately connected to their fate during DNA replication. Epigenetic phenomena are commonly considered changes in gene expression and function that are not caused by alterations in the DNA sequence, but nonetheless are stably inherited during cell divisions.[136] The specific requirement for inheritability, however, is often neglected in dubbing processes or factors epigenetic, leading to a broad inclusion of numerous phenomena that establish and sustain chromatin states, resulting in altered gene activity.[137, 138] DNA methylation has been shown to be faithfully duplicated during replication, and passed on to daughter cells (reviewed in [139, 140]). It represents to date the sole chromatin-associated modification that can be considered a bona fide epigenetic mark. It is generally assumed that histone PTMs are key players in the regulation of gene expression and other chromatin-associated processes; however, solid proof of their transmission through cell division has not been furnished yet. Clarifying this issue of hereditability is pivotal to our understanding of their function in the establishment, maintenance, and transmission of chromatin states.
Due to its relatively high stability, lysine methylation, restricted to H3 and H4, has the highest potential for transmission of epigenetic signals. Over the past four decades, several studies have investigated the segregation of parental histones to the DNA daughter strands in replication (reviewed in [86, 141, 142]). These studies have shown that parental histones are re-deposited onto both daughter strands along with newly synthesized histones in what appears to be a random fashion. The overwhelming majority of studies further indicated that parental H3-H4 remain as intact tetramers during this process. However, the histone chaperone Asf1, besides its well-established role in supplying newly synthesized histones during replication,[143] has the ability to split H3-H4 tetramers into dimers[144] and has been implicated in handling parental histones at the replication fork as well.[145] This was seen as support for a splitting model that argues for semi-conservative deposition of parental histones, with one dimer being deposited onto each daughter strand paired with a novel H3-H4 dimer. A recent report provided strong evidence against the general validity of this model, showing that the overwhelming majority of the canonical H3.1-H4 tetramers remain intact over at least two rounds of replication.[146]
Given that parental H3-H4 tetramers are re-deposited onto both daughter strands, possibly retaining their PTM state, it is conceivable that histone PTM patterns on naïve H3-H4 tetramers are re-established by using the neighboring parental nucleosomes as templates. Support for such a model comes from reports demonstrating that certain HMTs themselves contain or interact with factors that recognize the PTM they catalyze.[147] A notable example is the PRC2 complex, whose Eed subunit binds H3K27me3 and thereby allosterically stimulates catalytic activity.[20]
The complexity of the machinery orchestrating chromatin replication renders its reconstitution in vitro a challenging task. A commonly used in vitro replication system is based on the single origin of replication of the simian virus (SV) 40 genome and depends on the SV40 large tumor (T) antigen.[148] Full reconstitution of the machinery required for SV40 replication in vitro from defined, purified proteins has been achieved,[149, 150] providing a system for characterizing the pertinent factors. Replication of chromatinized templates involves histone clearance and re-deposition via a network of histone chaperones, whose intricacies are still not completely resolved. Although histone chaperones have been employed in replication systems, a fully reconstituted system capable of proper histone handling has not been developed yet. Such systems could prove invaluable in elucidating the segregation of histones and their associated PTMs. Histones with defined modifications or chemical labels to trace their fate might be just one of many tools chemical biology has to offer to shed light on these intriguing processes.
Summary and Outlook
From the early stages of the discovery of histone PTMs to the present, chemical biology-derived tools have significantly advanced studies of chromatin biology. Histone tails were one of the first peptides to be synthesized by automated solid-phase synthesis and are being used extensively to this day to study histone-modifying enzymes and histone binding proteins. Peptide pull-down assays were instrumental in the discovery of a large number of histone PTM binding proteins; others were uncovered through peptide and protein arrays. Unbiased approaches such as the combination of pull-downs with SILAC-based MS will continue to add factors to the ever-growing list of histone PTM effectors. Peptide systems, however, are not without drawbacks as the soundness of the information obtained rests on how closely these systems mimic the case in vivo. Analysis of binding properties in more complex systems might resolve issues regarding the specificity of binding domains and further reveal tighter binding due to multi-site recognition, possible only with more physiological, nucleosomal targets.
Tools to generate recombinant systems carrying defined histone PTMs have become available in recent years and are beginning to be employed on a wider scale. Whereas NCL-based techniques enable the generation of PTM-carrying histones indistinguishable from their native counterparts, they are hampered by the limits of peptide synthesis and their relatively high technical requirements. MLAs on the other hand are comparatively easy to generate, restricted to methylated lysines, and they differ from native histones by a carbon-to-sulfur exchange. Initial reports have demonstrated that this change is well tolerated, but MLAs might exhibit limitations in certain applications. Although there are great challenges to developing orthogonal ribosome systems that will undoubtedly take many years to overcome, they are a highly promising tool for generating histones with complex modification patterns, without unnatural substitutions, and in a manner that should prove to be eventually less technically demanding, relative to semisynthetic techniques.
Initial applications have mostly focused on nucleosome and chromatin structure and chromatin remodeling activities, but properties of HMTs and demethylases have been analyzed on chromatin templates as well. To directly assess the function of histone PTMs on transcription, reconstituted transcription systems have been combined with semi-synthetic histones in pioneering studies. Histone marks have been correlated with activation or repression of transcription, but little is known as to how they exert their function in this setting. The combination of in vitro transcription systems with templates containing histones carrying one or more defined modifications provides a powerful resource to further our knowledge on the mechanistic role of histone PTMs, and we should expect more studies utilizing these techniques. The fate of histone marks in replication can be studied in a similar manner in reconstituted replication systems, helping to clarify their potential role as epigenetic signals. This combination of classical biochemical techniques and novel approaches stemming from chemical biology will open new avenues to the understanding of histone mark function.
Acknowledgments
PV is supported by a fellowship from the Deutsche Akademie der Naturforscher Leopoldina (LPDS 2009-5). Work in the Reinberg laboratory is funded by the Howard Hughes Medical Institute and the National Institute of Health (grants GM064844 and R37GM037120). We thank Drs. Lynne Vales, Eric Campos, and David Beck for helpful discussions and critical reading of the manuscript. We apologize to authors whose work could not be cited due to space limitations.
Biographies
Biographical sketch D.R. Danny Reinberg received his B.S. in Biochemistry from the Catholic University in Valaparaiso, Chile. He then moved to New York to work on phage DNA replication in Jerard Hurwitz's laboratory at the Albert Einstein College of Medicine. He obtained his Ph.D. in Molecular Biology in 1982. For his postdoctoral work he joined Robert Roeder's group at the Rockefeller University to purify and study general transcription factors. In 1986, he set up his own laboratory at the University of Medicine and Dentistry of New Jersey, where he stayed until 2006 before relocating to the New York University School of Medicine. Research in the Reinberg laboratory is focused on mechanisms of transcription and the regulation of gene expression through chromatin and chromatin-modifying enzymes. Danny Reinberg has been a Howard Hughes Medical Institute Investigator since 1994.
Biographical sketch P.V. Philipp Voigt was trained at the Free University of Berlin. As part of his undergraduate studies, he worked with Ernst-Walter Knapp on electrostatic calculations on photosynthetic reaction centers. He received his PhD in Biochemistry from the Free University of Berlin, studying phosphoinositide signaling pathways in the laboratory of Michael Schaefer. In 2008 he joined the Reinberg lab for his postdoctoral training, working on mechanisms of polycomb gene silencing.
References
- 1.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Stedman E, Stedman E. Nature. 1950;166:780–781. doi: 10.1038/166780a0. [DOI] [PubMed] [Google Scholar]
- 4.Murray K. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 5.Phillips DM. Biochem J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang RC, Bonner J. Proc Natl Acad Sci USA. 1962;48:1216–1222. doi: 10.1073/pnas.48.7.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allfrey VG, Faulkner R, Mirsky AE. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kervabon A, Mery J, Parello J. FEBS Lett. 1979;106:93–96. doi: 10.1016/0014-5793(79)80702-4. [DOI] [PubMed] [Google Scholar]
- 13.Kervabon A, Parello J, Mery J. FEBS Lett. 1979;98:152–156. doi: 10.1016/0014-5793(79)80172-6. [DOI] [PubMed] [Google Scholar]
- 14.Krieger DE, Levine R, Merrifield RB, Vidali G, Allfrey VG. J Biol Chem. 1974;249:332–334. [PubMed] [Google Scholar]
- 15.Vidali G, Erickson B, Allfrey V, Merrifield R. Bioorg Chem. 1979 [Google Scholar]
- 16.Waterborg JH, Matthews HR. Anal Biochem. 1982;122:313–318. doi: 10.1016/0003-2697(82)90288-3. [DOI] [PubMed] [Google Scholar]
- 17.Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tie F, Stratton CA, Kurzhals RL, Harte PJ. Mol Cell Biol. 2007;27:2014–2026. doi: 10.1128/MCB.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 22.Ornaghi P, Ballario P, Lena AM, González A, Filetici P. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 23.Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 25.Mujtaba S, Zeng L, Zhou MM. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 26.Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 27.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 28.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson RH, Ladurner AG, King DS, Tjian R. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 31.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 32.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 33.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 34.Platero JS, Hartnett T, Eissenberg JC. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekwall K, Nimmo ER, Javerzat JP, Borgstrøm B, Egel R, Cranston G, Allshire R. J Cell Sci. 1996;109(Pt 11):2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- 36.Jacobs SA, Khorasanizadeh S. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 38.Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, Hirshberg M, Verreault A, Murzina NV, Laue ED. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 40.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 41.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, Grant PA. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 44.Sims RJ, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okuda M, Horikoshi M, Nishimura Y. J Mol Biol. 2007;365:1047–1062. doi: 10.1016/j.jmb.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 46.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 47.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 48.Bienz M. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Côté J, Chua KF, Gozani O. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 53.Sims RJ, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers HTM. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 55.Wen H, Li J, Song T, Lu M, Kan PY, Lee MG, Sha B, Shi X. J Biol Chem. 2010;285:9322–9326. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 57.Li SSC, Wu C. Methods Mol Biol. 2009;570:67–76. doi: 10.1007/978-1-60327-394-7_3. [DOI] [PubMed] [Google Scholar]
- 58.Shi X, Kachirskaia I, Walter KL, Kuo JHA, Lake A, Davrazou F, Chan SM, Martin DGE, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matthews AGW, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. Mol Cell. 2009;33:248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, Guccione E, Gozani O. PLoS ONE. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Q, Chakravarty S, Ghersi D, Zeng L, Plotnikov AN, Sanchez R, Zhou MM. PLoS ONE. 2010;5:e8903. doi: 10.1371/journal.pone.0008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frank R. Tetrahedron. 1992;48:9217–9232. [Google Scholar]
- 64.Nady N, Min J, Kareta MS, Chédin F, Arrowsmith CH. Trends Biochem Sci. 2008;33:305–313. doi: 10.1016/j.tibs.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 65.Rathert P, Zhang X, Freund C, Cheng X, Jeltsch A. Chem Biol. 2008;15:5–11. doi: 10.1016/j.chembiol.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkler DFH, Hilpert K, Brandt O, Hancock REW. Methods Mol Biol. 2009;570:157–174. doi: 10.1007/978-1-60327-394-7_5. [DOI] [PubMed] [Google Scholar]
- 67.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, Jeltsch A. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 70.Garske AL, Craciun G, Denu JM. Biochemistry. 2008;47:8094–8102. doi: 10.1021/bi800766k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garske AL, Oliver SS, Wagner EK, Musselman CA, Leroy G, Garcia BA, Kutateladze TG, Denu JM. Nat Chem Biol. 2010;6:283–290. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bonasio R, Lecona E, Reinberg D. Semin Cell Dev Biol. 2010;21:221–230. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. Mol Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. Nat Struct Mol Biol. 2007;14:1229–1230. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- 76.Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, Rice JC, Nimer SD. Oncogene. 2008;27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones RB, Gordus A, Krall JA, MacBeath G. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 78.Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pawson T. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 80.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 81.Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, Kioussis D. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 82.Ruthenburg AJ, Allis CD, Wysocka J. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 83.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Nat Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 85.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. Mol Cell Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campos EI, Reinberg D. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 87.Simpson RT, Stafford DW. Proc Natl Acad Sci USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Simpson RT, Thoma F, Brubaker JM. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 89.Lowary PT, Widom J. Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 90.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 91.LeRoy G, Orphanides G, Lane WS, Reinberg D. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 92.Lusser A, Kadonaga JT. Nat Methods. 2004;1:19–26. doi: 10.1038/nmeth709. [DOI] [PubMed] [Google Scholar]
- 93.Loyola A, Reinberg D. Methods. 2003;31:96–103. doi: 10.1016/s1046-2023(03)00093-8. [DOI] [PubMed] [Google Scholar]
- 94.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 95.Muir TW, Sondhi D, Cole PA. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shogren-Knaak MA, Fry CJ, Peterson CL. J Biol Chem. 2003;278:15744–15748. doi: 10.1074/jbc.M301445200. [DOI] [PubMed] [Google Scholar]
- 97.Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 98.He S, Bauman D, Davis JS, Loyola A, Nishioka K, Gronlund JL, Reinberg D, Meng F, Kelleher N, McCafferty DG. Proc Natl Acad Sci USA. 2003;100:12033–12038. doi: 10.1073/pnas.2035256100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chiang KP, Jensen MS, McGinty RK, Muir TW. ChemBioChem. 2009;10:2182–2187. doi: 10.1002/cbic.200900238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 101.Shogren-Knaak M, Peterson CL. Cell Cycle. 2006;5:1361–1365. doi: 10.4161/cc.5.13.2891. [DOI] [PubMed] [Google Scholar]
- 102.Ferreira H, Flaus A, Owen-Hughes T. J Mol Biol. 2007;374:563–579. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ. J Biol Chem. 2009;284:23312–23321. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Javaid S, Manohar M, Punja N, Mooney A, Ottesen JJ, Poirier MG, Fishel R. Mol Cell. 2009;36:1086–1094. doi: 10.1016/j.molcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Simon JA, Kingston RE. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 106.Weake VM, Workman JL. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 107.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chatterjee C, McGinty RK, Fierz B, Muir TW. Nat Chem Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 110.Xie J, Schultz PG. Nat Rev Mol Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 111.Chatterjee C, Muir TW. J Biol Chem. 2010;285:11045–11050. doi: 10.1074/jbc.R109.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. Proc Natl Acad Sci USA. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rackham O, Chin JW. Nat Chem Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 114.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 115.Simon M, Chu F, Racki L, de la Cruz C, Burlingame A, Panning B, Narlikar G, Shokat K. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Simon MD. Curr Protoc Mol Biol. 2010;Chapter 21:21–10. doi: 10.1002/0471142727.mb2125s101. Unit 21.18. [DOI] [PubMed] [Google Scholar]
- 117.Jia G, Wang W, Li H, Mao Z, Cai G, Sun J, Wu H, Xu M, Yang P, Yuan W, Chen S, Zhu B. Cell Research. 2009;19:1217–1220. doi: 10.1038/cr.2009.110. [DOI] [PubMed] [Google Scholar]
- 118.Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. Nat Struct Mol Biol. 2008;15:1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu C, Cui G, Botuyan MV, Mer G. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. J Biol Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, Qiao Q, Neubert TA, Xu RM, Gozani O, Reinberg D. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 123.Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL. Mol Cell. 2008;32:696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee N, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Mol Cell Biol. 2009;29:1401–1410. doi: 10.1128/MCB.01643-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 127.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 131.Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Mol Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 132.Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 133.Matsui T, Segall J, Weil PA, Roeder RG. J Biol Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- 134.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 135.Thomas MC, Chiang CM. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 136.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bird A. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 138.Ptashne M. Curr Biol. 2007;17:R233–236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 139.Law JA, Jacobsen SE. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jaenisch R, Bird A. Nat Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 141.Annunziato AT. J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 142.Ransom M, Dennehey BK, Tyler JK. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 144.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 145.Groth A, Corpet A, Cook AJL, Roche D, Bartek J, Lukas J, Almouzni G. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 146.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 147.Margueron R, Reinberg D. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li JJ, Kelly TJ. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Waga S, Bauer G, Stillman B. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 150.Waga S, Stillman B. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]