Abstract
The circadian system orchestrates the temporal organization of many aspects of physiology, including metabolism, in synchrony with the 24 hr rotation of the Earth. Like the metabolic system, the circadian system is a complex feedback network that involves interactions between the central nervous system and peripheral tissues. Emerging evidence suggests that circadian regulation is intimately linked to metabolic homeostasis, and that dysregulation of circadian rhythms can contribute to disease. Conversely, metabolic signals also feed back into the circadian system, modulating circadian gene expression and behavior. Here, we review the relationship between the circadian and metabolic systems, and the implications for cardiovascular disease, obesity and diabetes.
“It don’t mean a thing if it ain’t got that swing”
Edward Kennedy “Duke” Ellington (1899–1974)
What would music be without rhythm? So much in music depends upon timing, as do many aspects of our daily lives, including the rhythms of eating, fasting, sleep and wakefulness. These behaviors, although seemingly second nature, are now recognized to be governed by an intricate system of internal molecular clocks. These clocks coordinate biological processes to maintain synchrony with the environmental cycles of light and nutrients. It has been known for many years that numerous aspects of metabolism exhibit daily rhythmicity, including many types of circulating and intracellular metabolites, feeding-related hormones, and ingestive behaviors. Many of these rhythms are driven, at least in part, by circadian clocks. However, it has become clear that this is not a simple, linear relationship. Rather, the metabolism and circadian clocks are tightly interlocked: clocks drive metabolic processes and various metabolic parameters affect clocks, producing complex feedback relationships (Figure 1).
Figure 1. The sleep/wake and fasting/feeding cycles.
Genetic and molecular evidence suggest that within both the central nervous system (CNS) and peripheral tissues, co-regulation of the daily alteration between the sleep/wake and fasting/feeding cycles reflects coupling of both behavioral and metabolic pathways. Because the availability of food and the risk of predators are tied to the environmental cycle of light and darkness, these interlinked cycles may have provided selective advantages. Although this review focuses primarily on the interdependence of circadian and metabolic systems, it is important to note that the circadian system also impacts sleep, the restriction of which is sufficient to lead to abnormalities in metabolic homeostasis.
The mechanism underlying circadian rhythmicity is composed of a set of interlocking transcription/translation feedback loops that result in cascades of gene expression with 24-hour periodicity (for a more detailed review of this mechanism, see Lowrey and Takahashi, 2004) (Figure 2). At the core of this mechanism in mammals is the heterodimeric transcription factor complex of CLOCK and BMAL1, which activates transcription of the Period (Per) genes and Cryptochrome (Cry) genes via E-box enhancer elements in their promoters. The products of these genes interact to form complexes with each other. They also interact with other proteins such as casein kinase Iɛ, a protein that eventually translocates into the nucleus to inhibit CLOCK/BMAL1 transactivation activity, resulting in the repression of the Per and Cry genes. Removal of the Iɛ repressor complex, at least in part through ubiquitination and degradation by the proteasome (Siepka et al., 2007), eventually relieves repression, thereby allowing the negative feedback loop to start again.
Figure 2. Building blocks of the molecular clock.
The discovery of the core components of the circadian clock was first revealed through forward genetics showing that the clock is encoded by a transcription-translation feedback loop that oscillates with a periodicity of 24 hr in pacemaker neurons and peripheral cells. Subsequent analyses have identified robust outputs of the clock on metabolic pathways in liver, fat and muscle, suggesting convergence of circadian and metabolic pathways at the transcriptional level. The core mammalian clock is comprised of the heterodimeric activators CLOCK and BMAL1 that activate transcription of the genes encoding the repressors PERIOD (PER) and CRYPTOCHROME (CRY). An interlocked regulatory loop directs alternating activation and repression of BMAL1 expression by the nuclear receptors RORα and REV-ERBα, respectively, via binding at the ROR enhancer elements (ROREs) in the BMAL1 promoter. Several other metabolically active nuclear receptors have been identified as modulators of BMAL and CLOCK, including PPARγ and PGC1α. DBP, Albumin D-binding protein; TEF, thyrotroph embryonic factor; HLF, hepatocyte leukemia factor; HSF-1, heat shock factor 1.
This core negative feedback loop is modulated by other interlocking feedback loops. The best characterized of these loops involves the orphan nuclear receptors REV-ERBα and RORα, which drive rhythmic Bmal1 expression. The Bmal1 promoter contains ROR enhancer elements (ROREs) that can be bound by RORα and REV-ERBα:RORα activates transcription whereas REV-ERBα represses transcription. The Bmal1 rhythmicity is driven by a rhythmic change in RORE occupancy by RORα and REV-ERBα. This alternating promoter occupancy occurs because REV-ERBα levels are robustly rhythmic, a result of direct transcriptional activation of the Rev-erbα gene by CLOCK/BMAL1 (Preitner et al., 2002).
This model of the molecular mechanism of the mammalian circadian clock has been developed relatively recently – the first mammalian clock genes were not cloned until 1997 (Antoch et al., 1997; King et al., 1997; Sun et al., 1997; Tei et al., 1997). Thus, much of the early work in circadian rhythms in mammals focused on behavioral assays such as locomotor activity. These studies found that the clock that controlled circadian rhythms was localized in the suprachiasmatic nucleus (SCN) in the hypothalamus (Ralph et al., 1990). It took the identification of core clock components that cycled at the mRNA level to provide “universal” markers for examination of rhythmicity in many tissues. The development of transgenic animals containing luciferase reporter constructs driven by the promoters of cycling clock genes were particularly instrumental in showing that circadian clocks existed in tissues throughout the body (Yamazaki et al., 2000; Yoo et al., 2004). The function of these peripheral clocks in most cases remains to be defined, but as discussed below, it is likely that these clocks are important for driving local rhythms that are physiologically relevant for each specific tissue. The system appears to be arranged in a hierarchical manner with the SCN acting as the “master pacemaker” in mammals. The SCN drives rhythmic behavior and coordinates the many peripheral clocks through as yet poorly defined humoral and neural signals as well as indirectly by modulating activity and food intake Figure 3). This complex interplay between the CNS and periphery in driving circadian rhythms is reminiscent in many ways of the metabolic system. The peripheral metabolic tissues respond in various ways to metabolic signals, and the CNS coordinates these peripheral events through direct control and indirect means such as regulation of food intake and energy expenditure.
Figure 3. Central pacemaker and peripheral clocks.
The master pacemaker encoding the mammalian clock resides within the suprachiasmatic nucleus (SCN), although clock genes are also expressed in other regions of the brain and in most peripheral tissues. Emerging evidence suggests that peripheral tissue clocks are synchronized through humoral, nutrient, and autonomic wiring, and that the cell autonomous function of the clock is important in pathways involved in fuel storage and consumption. A hierarchical model indicates that all peripheral clocks are subordinate to the SCN. However, more recent work suggests that peripheral clocks play a broader role than previously realized in health and disease.
Linking circadian rhythms to metabolic control
The pervasive circadian control of metabolism is exemplified by microarray studies that have examined gene expression profiles throughout the circadian cycle in the mammalian liver, skeletal muscle, and brown and white adipose tissue (Akhtar et al., 2002; Kita et al., 2002; McCarthy et al., 2007; Panda et al., 2002; Reddy et al., 2006; Storch et al., 2002; Ueda et al., 2002; Zvonic et al., 2006). The numbers of genes judged to be rhythmic ranged from 3–20% in the different microarray studies of gene expression profiles, suggesting that a large proportion of the transcriptomes in these tissues are under circadian control. Among the rhythmic genes identified, many have roles in biosynthetic and metabolic processes, including cholesterol and lipid metabolism, glycolysis and gluconeogenesis, oxidative phosphorylation, and detoxification pathways. Importantly, the rate-limiting enzymes in many of these pathways are under circadian control (Panda et al., 2002), suggesting that the clock’s influence on these processes may be even broader than indicated by the numbers of rhythmic genes. The idea that these circadian oscillations in gene expression are physiologically meaningful is also supported by data showing that rhythmic genes in different tissues have varying degrees of overlap, with different rhythmic gene patterns in the different tissues (McCarthy et al., 2007; Panda et al., 2002; Storch et al., 2002; Ueda et al., 2002; Zvonic et al., 2006). This suggests that rhythmic gene expression is regulated in a tissue-specific manner to enable each cell/tissue to appropriately carry out its unique function.
Similar results were also found in a comprehensive analysis of nuclear receptor mRNA profiles measured throughout the daily cycle in liver, skeletal muscle, and brown and white adipose tissues (Yang et al., 2006). Of the 49 nuclear receptor mRNAs analyzed, more than half were rhythmic, with some variation between the profiles in the different tissues. The broad rhythmicity of this class of transcription factors, which interact with dietary lipids and fat-soluble hormones as ligands, suggests a rather direct link between nutrient sensing pathways and the circadian control of gene expression.
Considering these circadian gene expression patterns, it is difficult to infer whether the rhythmic patterns are driven directly by the core circadian clock mechanism or instead are driven indirectly by the rhythmic feeding profiles in the animals being tested (Figure 4). Because many metabolic genes are affected by the availability of nutrients, rhythmic feeding is likely to generate many such rhythms. In addition, rhythmic activity of animals affects body temperature, which again may contribute to driving rhythms in gene expression. This issue becomes even more complicated when one considers that the clocks located in many peripheral tissues such as the liver are rapidly entrained to food availability. Therefore, many possible signals may control rhythmic gene expression in the liver, including the central SCN clock, the mysterious “food entrainable oscillator” (FEO; see below) in the brain (through some type of systemic signal), the local endogenous liver clock (which is coupled to the SCN clock), or by food availability (either through signals from the food or through food entrainment of the liver clock).
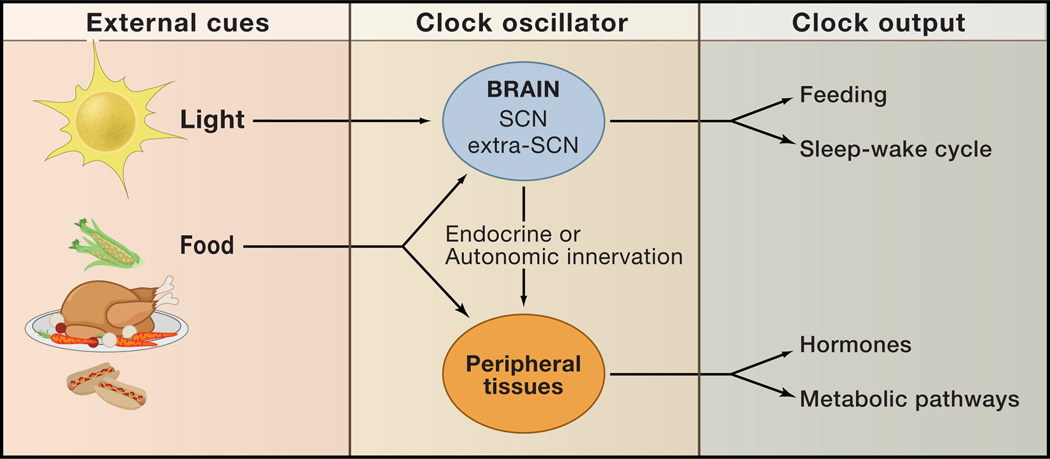
Figure 4. External cues and clock outputs.
The predominant external cue (zeitgeber) of the SCN clock is light. Clocks in peripheral tissues such as the liver also can be entrained by food. Nutrient and hormonal cues may also affect the period and phase characteristics of the master clock neurons, although little is known about how metabolic signals are communicated to the SCN. Outputs of both the SCN and peripheral clocks impact behavioral and metabolic responses such as feeding, sleep-wakefulness, hormone secretion, and metabolic homeostasis.
To address such issues, a conditional transgenic mouse has been generated in which the circadian clock could be reversibly and specifically disabled only in the liver, keeping the rest of the circadian system intact (Kornmann et al., 2007). Tetracycline-responsive, hepatocyte-specific over-expression of Rev-erbα in these mice causes constitutive repression of Bmal1 transcription when the tetracycline analog doxycycline is absent, resulting in the loss of clock function. Normal Bmal1 expression and normal hepatic circadian clock function is restored by feeding the mice doxycycline. Microarray analysis of the livers of the doxycycline-fed mice revealed 351 circadian transcripts, with significant overlap with the previous liver microarray datasets. However, following removal of doxycycline from the food, the great majority of these rhythmic genes no longer cycled in the liver, indicating that these genes are normally driven by the local liver clock and not by rhythmic systemic signals. Interestingly, 31 genes were still observed to have robust circadian patterns of expression in the absence of doxycycline, suggesting that this small subset of rhythmic liver genes are driven by some systemic signal independent of the liver clock. Thus, the control of metabolically-relevant hepatic gene expression is complex, most likely responding to both external and cell-autonomous signals to maintain appropriate temporal coordination of these metabolic pathways.
Food can entrain circadian clocks
The circadian/metabolic relationship is complicated by the fact that many tissues and cell types contribute to the metabolic state. These tissues and cell types all contain endogenous circadian oscillators that communicate with each other in ways that are not well understood. With regard to the circadian system, the clock in the hypothalamic SCN is thought to be the “master clock” and is entrained by an external cue (zeitgeber): light from the eyes via the retinohypothalamic tract. This clock drives behavioral rhythms such as that of locomotor activity and coordinates the many peripheral clocks so that they maintain proper phase-relationships with each other. It is now known that peripheral clocks can themselves also be entrained by various stimuli, with feeding being a dominant zeitgeber for many. In rodents that are given a restricted food (RF) paradigm where food is only available for a few hours during the day (a time when nocturnal rodents do not normally eat), the phase of many peripheral clocks shift by as much as 180° within about a week (Damiola et al., 2000; Stokkan et al., 2001). The liver clock entrains particularly rapidly to food availability with large phase shifts within two days of an altered feeding schedule.
An interesting example of food’s effect on the liver peripheral clock comes from studies of the common vole Microtus arvalis, which normally has an ultradian pattern of activity with short bouts of feeding every 2–3 hours (van der Veen et al., 2006). These animals have circadian rhythms of gene expression in their SCN, but not in their livers. However, if the voles are given food on a circadian time schedule, or if their activity becomes circadian (this happens when they are given a running wheel), their livers now exhibit circadian rhythms of gene expression. Mouse livers do not show this flexibility; if a mouse is given ultradian exposure to food, their livers remain rhythmic, although the amplitude is somewhat dampened. These findings suggest that the rhythmicity of the liver is important for proper metabolic function and that the voles have evolved a system to allow them to normally suppress the circadian aspect of liver function to adapt to their ultradian pattern of food intake.
What are the signals that arise from feeding that entrain the liver clock and other peripheral oscillators? They could be food itself, food-induced metabolites, or hormones whose secretion is controlled by feeding or its absence. The mechanisms involved in the regulation of hepatic circadian oscillators are not yet fully understood but several observations provide some insights into how this system may work. In cultured cell assays, many types of stimuli can induce or reset circadian clocks through regulation of clock gene expression (Balsalobre et al., 2000a; Balsalobre et al., 1998; Balsalobre et al., 2000b). These factors include forskolin, phorbol esters, glucose, and glucocorticoids, indicating that activation of many common signaling pathways can converge to influence the circadian clock. For example, in rat fibroblast cultures, insulin causes an acute induction of Per1 mRNA production (Balsalobre et al., 2000b). Addition of a glucose bolus to the culture medium causes a downregulation of Per1 and Per2 mRNA levels and induces rhythmic gene expression patterns (Hirota et al., 2002). In this culture system, the downregulation of Per expression by glucose appears to be indirect (it is blocked by inhibition of transcription or translation) and seems to depend on glucose metabolism, rather than glucose itself. Interestingly, it was recently reported that leptin causes up-regulation of Per2 and Clock gene expression in mouse osteoblasts, which have endogenous circadian clocks (Fu et al., 2005). Another example of clock regulation can be found in flies where the metabolic transcription factor FOXO has also been shown to modulate expression of clock genes and locomotor activity behavior (Zheng et al., 2007). Thus, multiple signals may be involved in the entrainment of clocks, including nutrients (sterols, lipids and/or carbohydrates), humoral signals (insulin, glucocorticoid, and perhaps incretin), and possibly even signals from vagal efferents that travel from autonomic centers to the liver (Cailotto et al., 2008; Kalsbeek et al., 2004; Pocai et al., 2005).
Nutrient signaling and circadian components
Many transcription factors that are known to respond to food or metabolites, such as nuclear receptors, also regulate components of the circadian clock. As mentioned above, a recent comprehensive survey of nuclear receptor mRNA profiles in several metabolic tissues in mice revealed that more than half of the known nuclear receptors exhibit rhythmic mRNA expression profiles (Yang et al., 2006). These receptors, which sense various lipids, vitamins, and fat-soluble hormones, are known mediators of metabolism. Direct links between some of these rhythmic nuclear receptors and the core clock components have been demonstrated. Therefore, these nuclear receptors may be part of the pathway by which food can entrain the liver clock (Ramsey et al., 2007).
The orphan nuclear receptors RORα and REV-ERBα are themselves considered part of the clock mechanism and contribute to an interlocking feedback loop that controls Bmal1 transcription (Akashi and Takumi, 2005; Preitner et al., 2002; Sato et al., 2004). Interestingly, a critical role has recently been demonstrated for the peroxisome proliferator-activated receptor (PPAR) nuclear receptor co-activator PGC-1α in circadian regulation (Liu et al., 2007). PGC-1α is a not a nuclear receptor itself, but is a transcriptional co-activator that regulates nuclear receptor activity and is a regulator of genes important in oxidative phosphorylation. PGC-1α stimulates Bmal1 expression through coactivation of ROR proteins. Liver-specific knockdown of PGC-1α results in arrhythmicity, suggesting that this protein is required for normal clock function in this tissue. This protein is also of particular interest because it is inducible---sensitive to a variety of environmental signals including nutritional status, activity and temperature ---and is thought to regulate adaptive energy metabolism in multiple tissues (Leone et al., 2005; Lin et al., 2005). PGC-1α therefore may be a key component that couples metabolic signals with clock function.
Other genes have been implicated in clock regulation. For example, Bmal1 is also transcriptionally regulated by the nuclear receptor PPARα, a key regulator of lipid metabolism (Canaple et al., 2006; Inoue et al., 2005). The glucocorticoid receptor has also been linked to the clock mechanism by the demonstration that glucocorticoids acutely induce Per1 in cell culture and can reset peripheral clocks in vivo (Balsalobre et al., 2000a; Reddy et al., 2007). In addition, in vascular tissue, RARα and RXRα have been shown to bind directly to CLOCK and the closely related NPAS2 (MOP4) in a hormone-dependent manner, causing inhibition of CLOCK(NPAS2)/ BMAl1 activity (McNamara et al., 2001). Two recent studies raise interest in the role of the histone deacetylase SIRT1 in the integration of circadian and metabolic transcription networks (Asher et al., 2008; Nakahata et al., 2008). Sirt1 is an ortholog of the yeast Sirt2 gene, a key factor in the longevity response to caloric restriction. SIRT1 has now been shown to interact directly with CLOCK and to deacetylate BMAL1 and PER2. Further studies will be necessary to delineate the physiological intersection between these sirtuin proteins and circadian pathways, and the potential implications for the coordination of feeding, activity, and metabolic homeostasis.
Another possibility that has not been well studied is that CLOCK, NPAS2, BMAL1 or the PERs may themselves act directly as sensors for some feeding related signal. These proteins all contain PAS domains, an ancient sensory domain conserved throughout all kingdoms (reviewed in Gu et al., 2000; Kewley et al., 2004). PAS domains detect redox state, hypoglycemia, oxygen balance, and xenobiotic metabolism. Signals are transduced by PAS domains to the rest of the protein to induce changes in the functional state of the protein. Do these PAS-containing clock proteins have the ability to sense their environment and adjust their transcriptional activity accordingly? There are two interesting observations suggesting that this may be the case.
The first example comes from experiments suggesting that cellular redox status is capable of altering CLOCK/BMAL1 and NPAS2/BMAL1 activity (Rutter et al., 2001). The reduced forms of the nicotinamide adenine dinucleotide cofactors, NADH and NADPH, were shown to stimulate the DNA-binding activity of these clock protein heterodimers, whereas the oxidized cofactors inhibited DNA-binding activity. The conversion from active to inactive forms of CLOCK/BMAL1 (or NPAS2/BMAL1) occurs over a very narrow range of reduced to oxidized NAD ratios, making this a very sensitive switch. Therefore, changes in feeding that alter cellular redox status could use such a mechanism to rapidly affect circadian clock activity. The in vivo importance of this mechanism as part of the food entrainment pathway still remains to be directly demonstrated. , In agreement with this model for regulating circadian clock activity, addition of lactate (which should increase the amount of reduced NAD) to neuroblastoma cells does cause NPAS2/BMAL1 activation (Rutter et al., 2001). However, the addition of pyruvate (which should also increase the reduced form of NAD) to rat fibroblasts does not increase the relative levels of Per1 or Per2 (direct targets of CLOCK/BMAL1 and NPAS2/BMAL1) (Hirota et al., 2002). Clearly, additional experiments are needed to determine whether redox status is an important aspect of circadian regulation by food.
In the second example, NPAS2 was demonstrated to bind to heme and may use this cofactor as a gas sensor (Dioum et al., 2002). In the heme-bound form, the DNA-binding activity of NPAS2 was inhibited by carbon monoxide (CO) in a dose-dependent manner. This effect was distinct from the redox sensing regulation described above, which does not require bound heme. The physiological role for CO sensing as a clock input is unclear, but NPAS2 is known to be important for regulating expression of the rate-limiting enzyme for heme-biosynthesis (aminolevulinic acid synthase; ALAS). This NPAS2 function may form a feedback loop that communicates the status of heme metabolism to the clock mechanism (Dioum et al., 2002; Kaasik and Lee, 2004). Interestingly, recent studies of Rev-erbα have suggested that heme is an endogenous ligand, raising the possibility that heme may play a central role in coordinating circadian function (Raghuram et al., 2007; Yin et al., 2007).
Nutrient Signals and SCN Oscillators
Feeding signals entrain peripheral clocks with great efficacy, whereas the central SCN clock appears to be largely refractory to these signals (Damiola et al., 2000; Stokkan et al., 2001). However, there is evidence that the SCN can receive and respond to signals from feeding under at least some conditions.
In cases where timed feeding is also coupled with caloric restriction, effects on the rat SCN can be observed by phase shifts in locomotor activity, body temperature, and melatonin rhythms (Challet et al., 1997). Entrainment to light-dark cycles is also altered (as measured both in activity rhythms and molecular rhythms in the SCN) (Mendoza et al., 2005b). In addition, light effects on the SCN are altered during times of low glucose availability (Challet et al., 1999). The rat SCN can also entrain to regular scheduled feeding (without caloric restriction) but this occurs much more slowly, taking nearly 12 weeks to reach stable entrainment (Castillo et al., 2004). The reward aspects of food seem to be important in SCN entrainment, since rat SCNs can entrain to rhythmic palatable diet exposure on a background of ad lib regular diet in constant dark conditions (Mendoza et al., 2005a). Finally, under conditions of constant light that cause the rat circadian rhythms to become arrhythmic, restricted feeding was capable of rescuing both locomotor activity rhythms and Per2 mRNA rhythms in the SCN (Lamont et al., 2005). Similar effects of restricted feeding on locomotor activity were previously also observed in hamsters maintained in constant light (Mistlberger, 1993).
Very little is known about how metabolic signals communicate with the SCN. The nature of the signals are unknown and it is not clear whether this is a direct or indirect regulation. Receptors for leptin and ghrelin are present on SCN cells (Guan et al., 1997; Yi et al., 2006; Zigman et al., 2006), so it is possible that these important metabolic peptides signal directly to these neurons. Indeed, adminstration of ghrelin to SCN slices or SCN explants in vitro caused phase shifts in Per2∷luc reporter gene expression (Yannielli et al., 2007). However, administration of ghrelin to intact mice caused small phase shifts only after 30 hours of food deprivation, with no effect in ad lib fed mice.
Neuropeptide Y (NPY) is a potent appetite transducer and its secretion exhibits both ultradian and circadian rhythms. The relative levels of anorexigenic (leptin) and orexigenic (ghrelin) hormones participate with the circadian clock in the hypothalamus in the coordination of the temporal patterns of NPY secretion. During restricted feeding paradigms, the daily pattern of NPY shifts along with the locomotor activity to anticipate the food availability (reviewed in Kalra and Kalra, 2004; Sindelar et al., 2005). However, in addition to being an “output” of the SCN clock, NPY also acts as an “input” to the SCN, involved in communicating non-photic signals (reviewed in Yannielli and Harrington, 2004). In addition, both histaminergic and serotonergic signaling pathways that influence feeding and energy metabolism in hypothalamus have been shown to modulate both SCN oscillations and sleep (Challet, 2007; Masaki et al., 2004). Therefore, these signaling systems appear to be involved in some sort of feedback loop to link feeding and metabolic state to the SCN.
A Food Entrainable Oscillator
Understanding the effects of food on the circadian system is complicated by the presence of another somewhat mysterious oscillator, the food-entrainable oscillator (FEO). This oscillator controls circadian rhythms of food anticipatory behavior and as its name suggests, is entrainable by food. When rodents are maintained on a restricted feeding regimen (for example, food is available for only a few hours during the middle of the day), within a few days they show increased activity and other behavioral changes shortly before the time that food becomes available. This rhythmic anticipatory activity has the hallmarks of bona fide circadian rhythms. Namely, the rhythms are maintained for several days of total food deprivation (i.e. they “free run” in constant conditions), they show phase-dependent phase-shifts with transients following a shift in mealtime, and they are subject to limits of entrainment (for example, rats can only entrain to food cycles between 22–31 hours in length) (reviewed in (Stephan, 2002)). Despite these similarities to the SCN oscillator, the FEO is anatomically distinct as SCN lesions do not abolish the rhythmic food anticipatory behavior. The anatomical location of the FEO is still not clear. Multiple lesion studies (ablation of the adrenals, hypothalamic structures, hippocampus, amygdala, nucleus accumbens, among others) have failed to abolish this activity.
Although some consideration has been given to the idea that the FEO might reside within the digestive system itself, most studies point to a CNS location (Stephan, 2002). Recently, the dorsal medial hypothalamic (DMH) nucleus has received much attention as a possible oscillator site due to observations from two independent lines of experimentation. Mice lacking a functional Per2 gene (but not those lacking Per1) lose food anticipatory activity rhythms, suggesting that Per2 is a critical component of the FEO system (Feillet et al., 2006). Examination of Per2 expression throughout the brain revealed a robust rhythm in the DMH only when mice were entrained to restricted feeding conditions (Mieda et al., 2006). Furthermore, the DMH rhythm of Per2 persisted through two days of total food deprivation. Recent neurotoxic DMH lesions resulted in significant losses of food anticipatory rhythms (as measured by general cage activity, sleep, and body temperature), also supporting DMH as the location for the FEO (Gooley et al., 2006). However, robust food anticipatory rhythms were maintained in DMH-lesioned animals in an independent study that examined a different behavioral readout (food bin approaches) (Landry et al., 2007). The difficulty in conclusively identifying the location of the FEO has raised the possibility that the FEO may not reside in a single structure, but may rather be distributed among many sites (Davidson, 2006).
Effects of Circadian Control On Metabolism
In view of the fact that metabolic networks are under extensive circadian control at the levels of transcription, translation, and post-translational mediated signaling, does the integration of circadian and metabolic cycles confer adaptive advantage and optimize energy utilization? Answers to this question have begun to emerge from studies in Cyanobacteria and the model plant Arabidopsis thaliana. Observations in these model organisms indicate that synchrony of endogenous circadian period length with the environment may play a direct role in survival and optimize functions ranging from photosynthesis to reproduction (Dodd et al., 2005; Ouyang et al., 1998) . Cyanobacteria strains with circadian periods similar to that of the light-dark cycle had a higher relative fitness than those with periods different from the environmental light cycle. Likewise, plants in which internal period length is aligned with the external light cycle display improved carbon fixation, growth, and reproduction. Those plants in which period length is discordant with the light cycle display reduced fertility and longevity. Interestingly, although circadian clocks have not been identified in yeast, these organisms exhibit robust metabolic cycles when cultured in nutrient-poor conditions (Chen et al., 2007). The periodic cycles of glycolysis and respiration are closely coupled to the replicative phase of the cell cycle. Intriguingly, disruption of the synchrony of these cycles resulted in an increased rate of spontaneous DNA mutation. Together, these studies suggest that rhythmic partitioning of metabolic processes can enhance survival by coupling the cell division cycle with cycles of energy storage and utilization. The availability of mutant mice with different circadian periods now provides the opportunity to formally test the concept that circadian “resonance” (i.e., synchrony between the external light-dark cycle and the internal period), is important in metabolic health and energy balance.
Physiological roles of clock genes
Analysis of mammals with genetic lesions that disrupt circadian rhythms has recently provided insight into the role of several of the major circadian clock genes in physiology. Homozygous Clock mutant mice of the C57BL/6J genetic background have severe alterations in energy balance, with a phenotype with many characteristics of metabolic syndrome, including obesity, hyperlipidemia, hepatic steatosis, high circulating glucose and low circulating insulin (Turek et al., 2005). The feeding rhythm in these mice is dampened, with increased food intake during the day, resulting in significantly increased overall food intake. It is likely that this phenotype results, at least in part, from altered rhythms of neuropeptides in the hypothalamus --- ghrelin, CART, and orexin are all expressed at constitutively low levels in the Clock homozygous mutant mice (Turek et al., 2005). Whether loss of clock function in the peripheral tissues contributes to this metabolic phenotype still remains to be seen. Interestingly, a lean phenotype is observed in Clock mutant mice outbred onto an ICR genetic background. However, this phenotype is caused by impaired lipid absorption in animals with an ICR background (Oishi et al., 2006).
Circadian clock function in the periphery
An important metabolic role for circadian clock function in the periphery has also been observed in other genetic models. For example, alterations in lipid and glucose homeostasis occur with mutations in clock-related genes, such as the Nocturnin knock-out mice mentioned above. Although the Nocturnin gene is not itself part of the central clock mechanism, it is involved in post-transcriptional regulation of rhythmic gene expression (Baggs and Green, 2003; Garbarino-Pico et al., 2007). Nocturnin−/− mice have normal feeding behavior as well as normal food intake and activity levels. However, they remain lean on a high fat diet. This is likely due to changes in lipid uptake or utilization, as these mice exhibit a loss of rhythmicity in genes important for these lipid pathways (Green et al., 2007). The Nocturnin−/− mice also have changes in glucose tolerance and improved whole-body insulin sensitivity.
Mice lacking the Bmal1 gene also have altered gluconeogenesis and improved whole-body insulin sensitivity (Ramsey et al., 2007; Rudic et al., 2004); suggesting clock control of these pathways. Furthermore, loss of Bmal1 in pre-adipocytes results in greatly reduced lipid storage and a failure to differentiate. The overexpression of BMAL1 results in induction of several genes involved in lipogenesis and increased lipid synthesis, suggesting that the circadian clock may also be important in proper adipocyte function (Shimba et al., 2005). However, the pleiotropic functions of BMAL1 make assessment of the global knockout phenotype particularly difficult since these animals become cachectic with age and develop both arthritic and myopathic complications (Kondratov et al., 2006).
Mutation in another central clock gene, Per2, results in increased bone density in mice (Fu et al., 2005). Histomorphometric analyses showed that Per2 null mice have an increased ratio of osteoblast to osteoclast activity, in addition to altered expression of Cyclin D1 and c-Myc. These pathways are regulated by leptin and it appears that Per2 may play a role in mediating the leptin-dependent sympathetic regulation of bone formation. Since bone and adipose tissue share a common ontogeny, it is possible that these findings may also have implications for adipogenesis (Gimble et al., 2006). The Per2 gene has also been implicated in cell cycle regulation in thymocytes (Fu et al., 2002). Per2 null mice that have been irradiated with gamma radiation are more prone to tumor development, suggesting that Per2 functions as a tumor suppressor in these cells.
Clock gene effects on metabolic tissues
Several studies have also suggested a link between circadian clock genes and apoptosis that may have an important effect on metabolic tissues. For example, there is increased susceptibility of Clock mutant mice to apoptosis induced by either cyclophosphamide or partial hepatectomy (Gorbacheva et al., 2005; Matsuo et al., 2003). Moreover, both Per1 mutant and Clock mutant animals display reduced expression of c-Myc, cyclin D1, and Wee-1, three cell cycle genes that exhibit pronounced circadian patterns of expression. CLOCK/BMAL1 may also impact cell growth in part through direct effects on chromatin remodeling, or by influencing the generation of reactive oxygen species (Doi et al., 2006; Kondratov et al., 2007).
How might molecular clock gene function influence the production of reactive oxygen species (ROS)? Studies of the clock disruption in myocardium have begun to shed light on the mechanism of ROS generation and its contribution to cardiovascular dysfunction. Under normal growth conditions in myocardial cells, circadian clock expression influences the selection of fuel source and the balance between β-oxidation and glycolysis (Durgan et al., 2006). Increased β-oxidation, which occurs following clock gene disruption in myocardium, contributes to increased production of ROS. Since altered circadian clock function may also limit the availability of glutathione, clock function may be important in ROS inactivation (Kondratov et al., 2006). Interestingly, recent work in Foxo mutant flies has suggested a tie between the generation of ROS and the regulation of the master neural pacemaker. Specifically, flies deficient in the FOXO transcription factor display increased oxidative stress following exposure to paraquat and altered circadian locomotor activity patterns (Zheng et al., 2007). Since the mammalian ortholog of the fly FOXO (Foxo-1) is an essential regulator of insulin signaling in liver and other metabolic tissues (Matsumoto et al., 2006), it will be important to identify upstream factors linking circadian and metabolic cycles. Investigation into the convergence of insulin signaling pathways with circadian systems may ultimately provide insight into the role of clock in β-oxidation and insulin resistance (Boudina et al., 2007).
Emerging evidence indicates that clock gene function also plays a crucial role in many other metabolic tissues (Young, 2006). For example, clocks within skeletal muscle may affect alternation between glycolysis and β-oxidation. Clock expression may also be important in adipogenesis (a process that also effects insulin sensitivity), as well as impact both adrenal and pancreatic physiology. Moreover, clock gene function has been shown to influence catecholamines within the adrenal medulla; circadian function at the level of both brain and peripheral organs may influence the counter-regulatory response to hypoglycemia (Bartness et al., 2001; Oster et al., 2006). These wide-ranging phenotypes of clock gene disruption may reflect physiological problems at the tissue level, or it may be that clock dysfunction alters the phase relationship amongst multiple peripheral metabolic tissue clocks, thereby causing disruption of overall internal circadian resonance. These studies beg the question of what precisely causes the metabolic phenotypes that occur in many of the circadian cycle mutant mice. Are the phenotypes due to a loss of circadian function within specific metabolically-relevant tissues or are they the result of certain metabolic tissues being prone to circadian misalignment-induced dysfunction? In addition, it will be important to compare the impact of multiple clock gene mutations in particular metabolic phenotypes to distinguish between gene-specific effects and the more general effects on specific processes of disrupting the clock network. Soon, studies exploiting both conditional rescue and ablation approaches will provide opportunities to dissect the cell autonomous and multi-tissue role of the clock gene network in metabolic physiology (Hong et al., 2007; McDearmon et al., 2006). Analysis of biological oscillations produced by the clock network provides an ideal model for complex systems biology. A full understanding of the dynamic function of the circadian network will require integration of genetic, cellular, and physiological analyses.
Mutations and the Circadian/Metabolic Systems
Mutations that affect the functions of the circadian system have a broad impact on metabolic function, but the reverse is also true. Mutations or challenges that cause metabolic problems also have been shown to affect the circadian system. Recent studies in high-fat fed animals have shown that introduction of calorically dense chow leads to rapid changes in both the period of locomotor activity in constant darkness, and to increased food intake during the normal rest period under 12 hour light, 12 hour dark conditions (Kohsaka et al., 2007). These changes in behavioral rhythmicity corresponded with altered clock gene expression within peripheral metabolic tissues, and as well as with altered cycling of the nuclear hormone receptors involved in sterol, lipid and carbohydrate metabolism in both liver and visceral white adipose tissue. One implication of these findings is that nutritional status directly affects the phase of the SCN clock, as activity in darkness was lengthened. The findings also show that peripheral clock gene expression is sensitive to exposure to an obesity-inducing environment—a condition that is anticipated to reflect more common causes of clock dysfunction in humans than primary missense mutations in the function of the core clock genes.
In another example, locomotor activity rhythms were affected by disruption of the brain-specific Homeobox Factor Bsx, a gene encoding a recently identified key regulator of hypothalamic Npy/AgRP neuron development (Sakkou et al., 2007). Bsx mutant mice are not only resistant to obesity when crossed with ob mice, they also display attenuated onset and dampened amplitude of nocturnal locomotor activity. Determination of the period length of locomotor activity under constant conditions in metabolic mutant mice will yield greater insight into the role of these signaling pathways in SCN. They will also shed light on extra-SCN clock gene expression and function. Furthermore, understanding the effect of these important mediators of behavior and energy balance on both locomotor activity and sleep will broaden our knowledge of the links between circadian and metabolic systems.
Brain-Peripheral Tissue Crosstalk and Energy Balance
Identification of the interactions between transcriptional networks that control circadian, cell, and metabolic cycles may provide a framework to better understand the impact of circadian control not only within individual tissues, but also the cross-talk between brain and peripheral tissues that participate in energy storage and utilization.
Anatomical and molecular studies have co-localized several brain regions in which neural networks involved in circadian and metabolic systems overlap (Figure 5). Importantly, clock genes have been shown to cycle not only within the SCN, but also in several other brain regions. These regions include the forebrain in nuclei surrounding the third ventricle where either orexigenic (NPY/AgRP) or anorexigenic (POMC/CART) neuropeptides are expressed, and the dorsomedial nucleus, a relay site to brainstem regions involved in wakefulness and sleep (Elmquist et al., 1998; Gooley et al., 2006; Mieda et al., 2006). Hypothalamic neuropeptides, particularly AgRP and POMC, are also expressed according to a pronounced diurnal rhythm, although the extent to which these oscillations are entrained by feeding, light, or nutrient signaling remains uncertain (reviewed in Kalra et al., 1999). Infusion of intralipid, which induces elevation of circulating free fatty acids, has been shown to acutely modulate expression of orexin. The activity of orexin neurons has also been tied to glucose metabolism and leptin (Chang et al., 2004; Date et al., 1999; Yamanaka et al., 2003). Interestingly, recent studies in high-fat fed canines suggest that nocturnal elevation (during sleep) of free fatty acids may be an early hallmark of the insulin resistant state (Kim et al., 2007). Taken together, both nutrients and the hormonal signals tied to nutrient balance are likely to impact regions of the hypothalamus important in control of circadian systems, wakefulness, and sleep.
Figure 5. Neural pathways linking circadian and metabolic systems.
Neuroanatomical studies have implicated several nodal points that may connect circadian, sleep, and metabolic centers within the brain. External cues such as light are transmitted from the eyes via the retinohypothalamic tract to the SCN. Projections from the suprachiasmatic nucleus (SCN) extend toward the subparaventricular zone (SPZ) and from the SPZ to the dorsomedial hypothalamus (DMH). Clock gene expression has been identified in DMH neurons, and the DMH has emerged as an important site in the activity response to food (the food entrainable oscillator). The DMH has many outputs to other regions of the brain, including the lateral hypothalamus (LHA), which controls circadian regulation of the sleep/wakefulness and fasting/feeding cycles. (Inset) The LHA also receives neuroendocrine input from the arcuate (ARC) neurons producing anorexigenic and orexigenic neuropeptides .The hormone leptin produced by adipose tissue activates the production of anorexigenic neuropeptides such as αMSH/CART, which in turn blocks production of the orexigenic peptides orexin (ORX) and melanin concentrating hormone (MCH) in the LHA. In the absence of leptin, orexigenic neurons in the ARC produce the neuropeptide NPY/AgRP that stimulates hunger and decreased energy expenditure via signaling to the LHA.
Curiously, projections from the master pacemaker neurons of the SCN also synapse on the subparaventricular zone, a region that in turn projects to the lateral hypothalamic area (LHA) neurons that express hypocretin/orexin, indicating presence of a “hardwired” circuit joining centers involved in rhythmicity, wakefulness, and feeding (Horvath and Gao, 2005). Projections from forebrain regions also reach both the midbrain and the hindbrain to nuclei implicated in dopaminergic-reward aspects of feeding and in cholinergic-and incretin-mediated satiety signaling (Abizaid et al., 2006; Drucker, 2006; Farooqi et al., 2007; Fulton et al., 2006; Hagan et al., 2000; Harris et al., 2005; Williams et al., 2006). In Clock mutant mice, altered expression of hypothalamic neuropeptides within leptin-responsive neurons of the arcuate nucleus correlates with the development of hyperphagia and obesity. Both the temporal pattern and absolute expression levels of orexin and hypothalamic ghrelin are diminished in these animals (Turek et al., 2005). Moreover, Clock mutant animals display alterations in both hedonic drive (cocaine response) and cholinergic signaling (autonomic tone), suggesting additional neural effects of the clock network that may impact body weight and fuel utilization (Curtis et al., 2007; McClung et al., 2005; Reilly et al., 2007). A major goal now will be to unravel the role of clock gene expression within forebrain, midbrain, and hindbrain. It will be crucial to test the effect of clock gene function within neuronal subpopulations that may be important in the spectrum of metabolic and behavioral abnormalities that arise in mice harboring disrupted circadian cycle genes.
Food Ingestion and Circadian/Metabolic Systems
The prevailing model for understanding long-term body weight constancy holds that the tone of hypothalamic orexigenic and anorexigenic centers is dynamically regulated by changes in the levels of peripheral hormones (principally leptin and insulin) that are produced in relationship to nutrient stores and food availability. The discovery of leptin was a watershed in yielding molecular insight into the pathways linking the peripheral and neural systems involved in feeding and energy expenditure. Although the major effect of leptin involves maintenance of body mass constancy during the challenge of famine, leptin itself is expressed in mice according to a diurnal rhythm (with relatively higher levels during the dark period) (Ahima et al., 1998). The absence of leptin during periods of food scarcity results in decreased energy expenditure and quiescent neuroendocrine systems, including the thyroidal and reproductive axes (Ahima and Flier, 2000). More recently, studies on hypothalamic nutrient sensing have uncovered an important role of hypothalamic leptin and nutrient signaling in the regulation of peripheral gluconeogenesis, a process that is particularly important during sleep when the liver serves as the major source of glucose (Parton et al., 2007; Pocai et al., 2005; Sandoval et al., 2007).
Interestingly, although both leptin and melanocortinergic signaling mediate hypothalamic regulation of feeding, energy utilization, and glucose metabolism, the specific hypothalamic neurons involved in the control of gluconeogenesis appear to be distinct from those involved in energy expenditure. This suggests a sort of specialization within the neural energy circuit itself (Claret et al., 2007; Parton et al., 2007). With regard to hypothalamic control of the liver, it has also been demonstrated that mammalian glucose metabolism is subjected to pronounced diurnal variation across the light-dark cycle, with alternating cycles of gluconeogenesis and glycogen synthesis that are coordinated with the rest-activity cycle (reviewed in (Ramsey et al., 2007)). Temporal control of hypothalamic autonomic efferents to liver, fat, and possibly pancreas may play a role in human glucose constancy. Interestingly, in fibroblasts, activation of 5’-AMP-activated kinase (AMPK), an important nutrient sensor in the liver, led to a phase-advance in clock protein expression. Activation of AMPK also correlated with phase advances in mice following treatment with metformin, an anti-hyperglycemic biguanide that is thought to target AMPK (Um et al., 2007). Clearly, it will be important to further examine the effects of the AMPK pathway on both peripheral interactions between circadian and metabolic pathways. AMPK signaling should also be assessed at the level of hypothalamic neurons where AMPK has been implicated in feeding regulation (Martin et al., 2006).
In addition to evidence for interactions between circadian and metabolic systems at the level of adiposity hormones, the discovery of gut-derived polypeptides involved in control of meal size, meal timing, gastric emptying, and endocrine pancreas function, provides an additional window into links between circadian and metabolic systems (Drucker, 2006; Murphy and Bloom, 2006; Nyholm et al., 1999). As mentioned above, studies with ghrelin have indicated effects on both SCN oscillators and locomotor activity patterns in mice, though the effects were dependent upon the nutritional state of the animal (Tang-Christensen et al., 2004; Yannielli et al., 2007). In addition, studies of gastrin releasing peptide, a mediator of both feeding and locomotor activity, indicate an effect on circadian phase that opposes effects of NPY (discussed above) (Gamble et al., 2007; Ladenheim et al., 2002). Peptide tyrosine-tyrosine (PYY3–36), a gut derived member of the pancreatic polypeptide family, has also been shown to not only influence feeding, but to also correlate with alterations to wakefulness and sleep architecture (Akanmu et al., 2006). A link between circadian physiology and responsiveness of endocrine pancreas to the incretin hormone glucagon like peptide-1 has also been suggested in studies showing effects of melatonin directly on pancreatic β-cells (Kemp et al., 2002). Studies of gastrointestinal clock gene expression in rodents have established expression of clock components within both stomach and colon, although clock function within the small intestine (specifically within cells that express anorectic and insulinotropic polypeptides) remains to be examined (Hoogerwerf et al., 2007). Nonetheless, the observation of clock gene expression within the colon is significant with regards to circadian control of incretin production, as there is abundant expression of PYY3–36 and related neuroendocrine polypeptides in the distal colon.
Circadian Disruption and Metabolic Disease
At the clinical and epidemiologic level, several lines of evidence suggest that circadian disruption is associated with cardiovascular and metabolic complications across large segments of the human population (reviewed in Laposky et al., 2007) (Figure 6). Cross-sectional studies have uncovered an increased prevalence of metabolic syndrome, high body mass index, and cardiovascular events in shift-workers (Ellingsen et al., 2007; Karlsson et al., 2001). These observations raise the possibility that chronic misalignment between the cycles of rest and activity, and those of fasting and feeding, may contribute to the initiation and progression of obesity and metabolic syndrome. With regard to feeding, intriguing behavioral studies in humans suggest that nocturnal feeding patterns (“night eating syndrome”) may represent an independent risk for metabolic disease (Allison et al., 2007). These findings are reminiscent of recent experimental rodent studies showing that excess energy intake during diet-induced obesity leads to increased energy intake only during the rest period and not during the active period when the animal normally eats (Kohsaka et al., 2007). Thus, it appears that the capacity to defend long-term energy constancy is related both to the time-of-day when food is consumed and to the relationship between meal (or snack) time and the sleep-wake cycle.
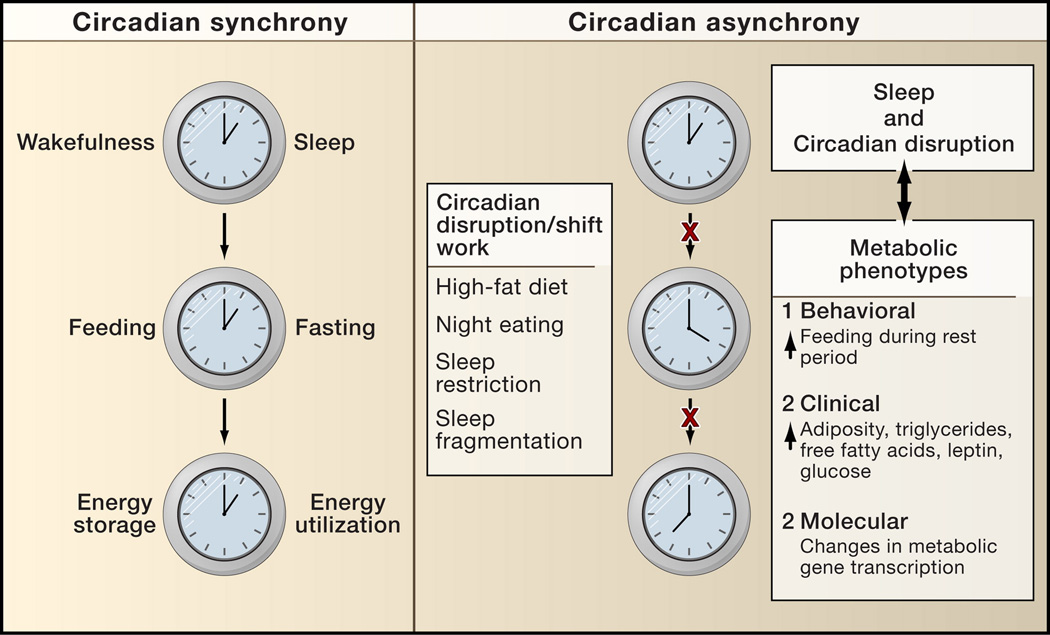
Figure 6. Circadian synchrony and metabolic disease.
Many aspects of metabolic physiology are known to occur at specific times each day. Gene expression patterns corresponding to periods of energy storage and energy utilization have been tied to the function of the peripheral clock in the liver and are shown as outputs of the clock (the cycle of energy storage and utilization). Many disorders such as myocardial infarction peak at certain times during a 24 hr day, suggesting a potential link between disruption of circadian rhythms and disease pathology. Emerging evidence suggests that disruption of synchrony between periods of rest/activity with feeding/fasting and energy storage/utilization may be tied to dysregulation of not only body weight but also diverse metabolic processes such as glucose metabolism, vascular reactivity, thrombosis, and lipid homeostasis.
How might circadian misalignment impact the metabolic co-morbidities of obesity: diabetes and cardiovascular disease? Several lines of evidence suggest that circadian dysregulation may exert a broad impact not only on glucose control, but also on inflammation, fibrinolysis, fluid balance and vascular reactivity. Indeed, circadian control of glucose metabolism in humans is a well-recognized aspect of clinical diabetes management, and an alteration in the normal cyclic pattern of glucose tolerance is a hallmark of type 2 diabetes (Holterhus et al., 2007). This phenomenon of circadian control of glucose metabolism is perhaps most familiar to individuals with type 1 diabetes mellitus. These patients must adjust their daily insulin requirements around the light-dark cycle according to fluctuations in the basal requirement for insulin, as well as in response to changes in glycemic excursion after meals in the morning and at night (Van Cauter et al., 1997). Consideration of the physiological and molecular basis for changes in the circadian pattern of glucose tolerance, and for variation in the counterregulatory response to hypoglycemia, may ultimately lead to better strategies for diabetes management.
In addition to control of glucose metabolism, circadian systems may also participate in additional components of the metabolic syndrome. A central node linking metabolic and circadian pathways involves the nuclear receptor superfamily, as reviewed above. The pathways regulated include those downstream of Rev-erbα and the RORs that modulate the core clock and diverse metabolic processes ranging from adipogenesis to inflammation and thrombosis (reviewed in (Duez and Staels, 2008). Indeed, PAI-1, an important component of the fibrinolytic cascade that signals the breakdown of coagulation products, is subjected to circadian control and is a downstream target of Rev-erbα and RORα. Further studies are needed to address whether the strong clustering of acute cardiovascular catastrophes within restricted times of the day may be related to disruptions in normal circadian regulation of these pathways within liver, adipose tissue, or perhaps the vasculature.
In experimental animal models, repeated misalignment of internal phase with the light cycle, a so-called “jet lag” paradigm, has also been associated with accelerated cardiomyopathy and premature mortality (Davidson et al., 2006; Penev et al., 1998). Both epidemiologic and clinical studies have begun to probe whether restricted sleep, or altered cycling of sleep-wakefulness states, may contribute to hyperphagia and metabolic dysregulation. Reduced sleep time is associated with increased body mass index (BMI), and forced sleep restriction in humans is tied to alterations of neuroendocrine control of appetite and glucose tolerance (Spiegel et al., 2004; Taheri et al., 2004; Tasali et al., 2008).
Although previous work has focused on the impact of sleep loss on cognitive and mood disorders, further investigation is needed to examine the link between altered circadian synchrony and human health. To achieve this goal, it will be necessary to establish standard methodologies for the analysis of circadian parameters in studies of feeding and glucose metabolism in human subjects. Furthermore, experimental genetic strategies need to be applied in animal models to pinpoint the underlying pathways important in coordinating these systems. The availability of temporal and metabolic phenotype data in humans will ultimately enable association studies to evaluate the contribution of circadian gene variation to human diabetes and obesity. Biological oscillations represent an area of metabolic physiology that, although familiar to most of us, is often overlooked in both clinical trial design and in experimental genetic studies. It seems opportune to consider timing and the clock system as we search for new mechanism-based treatments for metabolic disease.
Future Perspectives
The past decade has witnessed major strides in our understanding of the neurobehavioral basis of both feeding and circadian timing. Interestingly, both circadian and metabolic systems involve extensive crosstalk between CNS and peripheral tissues through humoral, nutrient, and direct neural wiring. At the organismal level, physiologic observations suggest that the circadian clock may be important in a wide range of pathologies that cluster at specific times of day, including myocardial infarction, arrhythmogenicity, congestive heart failure, thrombosis, and carbohydrate and lipid turnover. Yet, we still have little knowledge of the identities of the internal environmental sensors that couple circadian systems with processes ranging from nutrient homeostasis to coagulation, cytokine production, respiration, and myocardial contractility. In addition, circadian clock proteins, such as CLOCK and BMAL1, may also have functions independent of their roles as components of the circadian oscillator. Thus, both inter-and intra-organ asynchrony may also contribute to chronic pathologies such as diabetes, obesity, and cardiovascular disease. Current gaps in our understanding of the circadian system include the contributions of CNS oscillators versus peripheral oscillators for regulating circadian homeostasis and the detailed mechanisms by which peripheral organ clocks modulate tissue physiology. Studies of the circadian clock have opened a window on the molecular control of behavior, and now open a unique opportunity to probe the molecular links between behavior and metabolism.
Acknowledgments
We thank K. M. Ramsey and B. Marcheva for figure assistance. J.S. Takahashi is an Investigator of the Howard Hughes Medical Institute.
References
- Abizaid A, Liu ZW, Andrews ZB, Shanabrough M, Borok E, Elsworth JD, Roth RH, Sleeman MW, Picciotto MR, Tschop MH, et al. Ghrelin modulates the activity and synaptic input organization of midbrain dopamine neurons while promoting appetite. J Clin Invest. 2006;116:3229–3239. doi: 10.1172/JCI29867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahima RS, Flier JS. Leptin. Annu Rev Physiol. 2000;62:413–437. doi: 10.1146/annurev.physiol.62.1.413. [DOI] [PubMed] [Google Scholar]
- Ahima RS, Prabakaran D, Flier JS. Postnatal leptin surge and regulation of circadian rhythm of leptin by feeding. Implications for energy homeostasis and neuroendocrine function. J Clin Invest. 1998;101:1020–1027. doi: 10.1172/JCI1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akanmu MA, Ukponmwan OE, Katayama Y, Honda K. Neuropeptide-Y Y2-receptor agonist, PYY3-36 promotes non-rapid eye movement sleep in rat. Neurosci Res. 2006;54:165–170. doi: 10.1016/j.neures.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Allison KC, Crow SJ, Reeves RR, West DS, Foreyt JP, Dilillo VG, Wadden TA, Jeffery RW, Van Dorsten B, Stunkard AJ. Binge eating disorder and night eating syndrome in adults with type 2 diabetes. Obesity (Silver Spring) 2007;15:1287–1293. doi: 10.1038/oby.2007.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Baggs JE, Green CB. Nocturnin, a Deadenylase in Xenopus laevis Retina. A Mechanism for Posttranscriptional Control of Circadian-Related mRNA. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000a;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000b;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Bartness TJ, Song CK, Demas GE. SCN efferents to peripheral tissues: implications for biological rhythms. J Biol Rhythms. 2001;16:196–204. doi: 10.1177/074873040101600302. [DOI] [PubMed] [Google Scholar]
- Boudina S, Sena S, Theobald H, Sheng X, Wright JJ, Hu XX, Aziz S, Johnson JI, Bugger H, Zaha VG, et al. Mitochondrial energetics in the heart in obesity-related diabetes: direct evidence for increased uncoupled respiration and activation of uncoupling proteins. Diabetes. 2007;56:2457–2466. doi: 10.2337/db07-0481. [DOI] [PubMed] [Google Scholar]
- Cailotto C, van Heijningen C, van der, Vliet J, van der Vliet G, Habold C, Kalsbeek A, Pevet P, Buijs RM. Daily rhythms in metabolic liver enzymes and plasma glucose require a balance in the autonomic output to the liver. Endocrinology. 2008;149:1914–1925. doi: 10.1210/en.2007-0816. [DOI] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- Castillo MR, Hochstetler KJ, Tavernier RJ, Jr, Greene DM, Bult-Ito A. Entrainment of the master circadian clock by scheduled feeding. Am J Physiol Regul Integr Comp Physiol. 2004;287:R551–R555. doi: 10.1152/ajpregu.00247.2004. [DOI] [PubMed] [Google Scholar]
- Challet E. Minireview: Entrainment of the suprachiasmatic clockwork in diurnal and nocturnal mammals. Endocrinology. 2007;148:5648–5655. doi: 10.1210/en.2007-0804. [DOI] [PubMed] [Google Scholar]
- Challet E, Losee-Olson S, Turek FW. Reduced glucose availability attenuates circadian responses to light in mice. Am J Physiol. 1999;276:R1063–1070. doi: 10.1152/ajpregu.1999.276.4.R1063. [DOI] [PubMed] [Google Scholar]
- Challet E, Pevet P, Vivien-Roels B, Malan A. Phase-advanced daily rhythms of melatonin, body temperature, and locomotor activity in food-restricted rats fed during daytime. J Biol Rhythms. 1997;12:65–79. doi: 10.1177/074873049701200108. [DOI] [PubMed] [Google Scholar]
- Chang GQ, Karatayev O, Davydova Z, Leibowitz SF. Circulating triglycerides impact on orexigenic peptides and neuronal activity in hypothalamus. Endocrinology. 2004;145:3904–3912. doi: 10.1210/en.2003-1582. [DOI] [PubMed] [Google Scholar]
- Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- Claret M, Smith MA, Batterham RL, Selman C, Choudhury AI, Fryer LG, Clements M, Al-Qassab H, Heffron H, Xu AW, et al. AMPK is essential for energy homeostasis regulation and glucose sensing by POMC and AgRP neurons. J Clin Invest. 2007;117:2325–2336. doi: 10.1172/JCI31516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Cheng Y, Kapoor S, Reilly D, Price TS, Fitzgerald GA. Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc Natl Acad Sci U S A. 2007;104:3450–3455. doi: 10.1073/pnas.0611680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date Y, Ueta Y, Yamashita H, Yamaguchi H, Matsukura S, Kangawa K, Sakurai T, Yanagisawa M, Nakazato M. Orexins, orexigenic hypothalamic peptides, interact with autonomic, neuroendocrine and neuroregulatory systems. Proc Natl Acad Sci U S A. 1999;96:748–753. doi: 10.1073/pnas.96.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ. Search for the feeding-entrainable circadian oscillator: a complex proposition. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1524–1526. doi: 10.1152/ajpregu.00073.2006. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16:R914–R916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Drucker DJ. The biology of incretin hormones. Cell Metab. 2006;3:153–165. doi: 10.1016/j.cmet.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Duez H, Staels B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diabetes & vascular disease research : official journal of the International Society of Diabetes and Vascular Disease. 2008;5:82–88. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- Durgan DJ, Trexler NA, Egbejimi O, McElfresh TA, Suk HY, Petterson LE, Shaw CA, Hardin PE, Bray MS, Chandler MP, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem. 2006;281:24254–24269. doi: 10.1074/jbc.M601704200. [DOI] [PubMed] [Google Scholar]
- Ellingsen T, Bener A, Gehani AA. Study of shift work and risk of coronary events. Journal of the Royal Society of Health. 2007;127:265–267. doi: 10.1177/1466424007083702. [DOI] [PubMed] [Google Scholar]
- Elmquist JK, Ahima RS, Elias CF, Flier JS, Saper CB. Leptin activates distinct projections from the dorsomedial and ventromedial hypothalamic nuclei. Proc Natl Acad Sci U S A. 1998;95:741–746. doi: 10.1073/pnas.95.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqi IS, Bullmore E, Keogh J, Gillard J, O’Rahilly S, Fletcher PC. Leptin regulates striatal regions and human eating behavior. Science. 2007;317:1355. doi: 10.1126/science.1144599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feillet CA, Ripperger JA, Magnone MC, Dulloo A, Albrecht U, Challet E. Lack of food anticipation in Per2 mutant mice. Curr Biol. 2006;16:2016–2022. doi: 10.1016/j.cub.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Fulton S, Pissios P, Manchon RP, Stiles L, Frank L, Pothos EN, Maratos-Flier E, Flier JS. Leptin regulation of the mesoaccumbens dopamine pathway. Neuron. 2006;51:811–822. doi: 10.1016/j.neuron.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Gamble KL, Allen GC, Zhou T, McMahon DG. Gastrin-releasing peptide mediates light-like resetting of the suprachiasmatic nucleus circadian pacemaker through cAMP response element-binding protein and Per1 activation. J Neurosci. 2007;27:12078–12087. doi: 10.1523/JNEUROSCI.1109-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, Green CB. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME. Playing with bone and fat. J Cell Biochem. 2006;98:251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- Gooley JJ, Schomer A, Saper CB. The dorsomedial hypothalamic nucleus is critical for the expression of food-entrainable circadian rhythms. Nat Neurosci. 2006;9:398–407. doi: 10.1038/nn1651. [DOI] [PubMed] [Google Scholar]
- Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- Guan XM, Hess JF, Yu H, Hey PJ, van der Ploeg LH. Differential expression of mRNA for leptin receptor isoforms in the rat brain. Mol Cell Endocrinol. 1997;133:1–7. doi: 10.1016/s0303-7207(97)00138-x. [DOI] [PubMed] [Google Scholar]
- Hagan MM, Rushing PA, Pritchard LM, Schwartz MW, Strack AM, Van Der Ploeg LH, Woods SC, Seeley RJ. Long-term orexigenic effects of AgRP-(83---132) involve mechanisms other than melanocortin receptor blockade. Am J Physiol Regul Integr Comp Physiol. 2000;279:R47–R52. doi: 10.1152/ajpregu.2000.279.1.R47. [DOI] [PubMed] [Google Scholar]
- Harris GC, Wimmer M, Aston-Jones G. A role for lateral hypothalamic orexin neurons in reward seeking. Nature. 2005;437:556–559. doi: 10.1038/nature04071. [DOI] [PubMed] [Google Scholar]
- Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem. 2002;277:44244–44251. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- Holterhus PM, Odendahl R, Oesingmann S, Lepler R, Wagner V, Hiort O, Holl R, Initiative GAD, Group GPCW. Classification of distinct baseline insulin infusion patterns in children and adolescents with type 1 diabetes on continuous subcutaneous insulin infusion therapy. Diabetes Care. 2007;30:568–573. doi: 10.2337/dc06-2105. [DOI] [PubMed] [Google Scholar]
- Hong HK, Chong JL, Song W, Song EJ, Jyawook AA, Schook AC, Ko CH, Takahashi JS. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS Genet. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogerwerf WA, Hellmich HL, Cornelissen G, Halberg F, Shahinian VB, Bostwick J, Savidge TC, Cassone VM. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133:1250–1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Horvath TL, Gao XB. Input organization and plasticity of hypocretin neurons: possible clues to obesity’s association with insomnia. Cell Metab. 2005;1:279–286. doi: 10.1016/j.cmet.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Inoue I, Shinoda Y, Ikeda M, Hayashi K, Kanazawa K, Nomura M, Matsunaga T, Xu H, Kawai S, Awata T, et al. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb. 2005;12:169–174. doi: 10.5551/jat.12.169. [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Kalra SP, Dube MG, Pu S, Xu B, Horvath TL, Kalra PS. Interacting appetite-regulating pathways in the hypothalamic regulation of body weight. Endocr Rev. 1999;20:68–100. doi: 10.1210/edrv.20.1.0357. [DOI] [PubMed] [Google Scholar]
- Kalra SP, Kalra PS. NPY and cohorts in regulating appetite, obesity and metabolic syndrome: beneficial effects of gene therapy. Neuropeptides. 2004;38:201–211. doi: 10.1016/j.npep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, La Fleur S, Van Heijningen C, Buijs RM. Suprachiasmatic GABAergic inputs to the paraventricular nucleus control plasma glucose concentrations in the rat via sympathetic innervation of the liver. J Neurosci. 2004;24:7604–7613. doi: 10.1523/JNEUROSCI.5328-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occupational and environmental medicine. 2001;58:747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp DM, Ubeda M, Habener JF. Identification and functional characterization of melatonin Mel 1a receptors in pancreatic beta cells: potential role in incretin-mediated cell function by sensitization of cAMP signaling. Mol Cell Endocrinol. 2002;191:157–166. doi: 10.1016/s0303-7207(02)00064-3. [DOI] [PubMed] [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Kim SP, Catalano KJ, Hsu IR, Chiu JD, Richey JM, Bergman RN. Nocturnal free fatty acids are uniquely elevated in the longitudinal development of diet-induced insulin resistance and hyperinsulinemia. Am J Physiol Endocrinol Metab. 2007;292:E1590–E1598. doi: 10.1152/ajpendo.00669.2006. [DOI] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita Y, Shiozawa M, Jin W, Majewski RR, Besharse JC, Greene AS, Jacob HJ. Implications of circadian gene expression in kidney, liver and the effects of fasting on pharmacogenomic studies. Pharmacogenetics. 2002;12:55–65. doi: 10.1097/00008571-200201000-00008. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Gorbacheva VY, Antoch MP. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol. 2007;78:173–216. doi: 10.1016/S0070-2153(06)78005-X. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladenheim EE, Hampton LL, Whitney AC, White WO, Battey JF, Moran TH. Disruptions in feeding and body weight control in gastrin-releasing peptide receptor deficient mice. J Endocrinol. 2002;174:273–281. doi: 10.1677/joe.0.1740273. [DOI] [PubMed] [Google Scholar]
- Lamont EW, Diaz LR, Barry-Shaw J, Stewart J, Amir S. Daily restricted feeding rescues a rhythm of period2 expression in the arrhythmic suprachiasmatic nucleus. Neuroscience. 2005;132:245–248. doi: 10.1016/j.neuroscience.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Landry GJ, Yamakawa GR, Mistlberger RE. Robust food anticipatory circadian rhythms in rats with complete ablation of the thalamic paraventricular nucleus. Brain Res. 2007;1141:108–118. doi: 10.1016/j.brainres.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Laposky AD, Bass J, Kohsaka A, Turek FW. Sleep and circadian rhythms: Key components in the regulation of energy metabolism. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.06.079. [DOI] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian Circadian Biology: Elucidating Genome-Wide Levels of Temporal Organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TL, Alquier T, Asakura K, Furukawa N, Preitner F, Kahn BB. Diet-induced obesity alters AMP kinase activity in hypothalamus and skeletal muscle. J Biol Chem. 2006;281:18933–18941. doi: 10.1074/jbc.M512831200. [DOI] [PubMed] [Google Scholar]
- Masaki T, Chiba S, Yasuda T, Noguchi H, Kakuma T, Watanabe T, Sakata T, Yoshimatsu H. Involvement of hypothalamic histamine H1 receptor in the regulation of feeding rhythm and obesity. Diabetes. 2004;53:2250–2260. doi: 10.2337/diabetes.53.9.2250. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Han S, Kitamura T, Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest. 2006;116:2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Identification of the Circadian Transcriptome in Adult Mouse Skeletal Muscle. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, Nestler EJ. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci U S A. 2005;102:9377–9381. doi: 10.1073/pnas.0503584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P, Seo SB, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Mendoza J, Angeles-Castellanos M, Escobar C. A daily palatable meal without food deprivation entrains the suprachiasmatic nucleus of rats. Eur J Neurosci. 2005a;22:2855–2862. doi: 10.1111/j.1460-9568.2005.04461.x. [DOI] [PubMed] [Google Scholar]
- Mendoza J, Graff C, Dardente H, Pevet P, Challet E. Feeding cues alter clock gene oscillations and photic responses in the suprachiasmatic nuclei of mice exposed to a light/dark cycle. J Neurosci. 2005b;25:1514–1522. doi: 10.1523/JNEUROSCI.4397-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Williams SC, Richardson JA, Tanaka K, Yanagisawa M. The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc Natl Acad Sci U S A. 2006;103:12150–12155. doi: 10.1073/pnas.0604189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE. Effects of scheduled food and water access on circadian rhythms of hamsters in constant light, dark, and light:dark. PhysiolBehav. 1993;53:509–516. doi: 10.1016/0031-9384(93)90145-6. [DOI] [PubMed] [Google Scholar]
- Murphy KG, Bloom SR. Gut hormones and the regulation of energy homeostasis. Nature. 2006;444:854–859. doi: 10.1038/nature05484. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm B, Walker M, Gravholt CH, Shearing PA, Sturis J, Alberti KG, Holst JJ, Schmitz O. Twenty-four-hour insulin secretion rates, circulating concentrations of fuel substrates and gut incretin hormones in healthy offspring of Type II (non-insulin-dependent) diabetic parents: evidence of several aberrations. Diabetologia. 1999;42:1314–1323. doi: 10.1007/s001250051444. [DOI] [PubMed] [Google Scholar]
- Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, Ishida N. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci U S A. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Parton LE, Ye CP, Coppari R, Enriori PJ, Choi B, Zhang CY, Xu C, Vianna CR, Balthasar N, Lee CE, et al. Glucose sensing by POMC neurons regulates glucose homeostasis and is impaired in obesity. Nature. 2007;449:228–232. doi: 10.1038/nature06098. [DOI] [PubMed] [Google Scholar]
- Penev PD, Kolker DE, Zee PC, Turek FW. Chronic circadian desynchronization decreases the survival of animals with cardiomyopathic heart disease. Am J Physiol. 1998;275:H2334–2337. doi: 10.1152/ajpheart.1998.275.6.H2334. [DOI] [PubMed] [Google Scholar]
- Pocai A, Obici S, Schwartz GJ, Rossetti L. A brain-liver circuit regulates glucose homeostasis. Cell Metab. 2005;1:53–61. doi: 10.1016/j.cmet.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Marcheva B, Kohsaka A, Bass J. The clockwork of metabolism. Annu Rev Nutr. 2007;27:219–240. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Maywood ES, Karp NA, King VM, Inoue Y, Gonzalez FJ, Lilley KS, Kyriacou CP, Hastings MH. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology. 2007;45:1478–1488. doi: 10.1002/hep.21571. [DOI] [PubMed] [Google Scholar]
- Reilly DF, Westgate EJ, FitzGerald GA. Peripheral circadian clocks in the vasculature. Arterioscler Thromb Vasc Biol. 2007;27:1694–1705. doi: 10.1161/ATVBAHA.107.144923. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. Regulation of CLOCK and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Sakkou M, Wiedmer P, Anlag K, Hamm A, Seuntjens E, Ettwiller L, Tschop MH, Treier M. A role for brain-specific homeobox factor bsx in the control of hyperphagia and locomotory behavior. Cell Metab. 2007;5:450–463. doi: 10.1016/j.cmet.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Sandoval D, Cota D, Seeley RJ. The Integrative Role of CNS Fuel-Sensing Mechanisms in Energy Balance and Glucose Regulation. Annu Rev Physiol. 2007 doi: 10.1146/annurev.physiol.70.120806.095256. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals RORalpha as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Genetics and Neurobiology of Circadian Clocks in Mammals. Cold Spring HarbSympQuantBiol. 2007;72:251–259. doi: 10.1101/sqb.2007.72.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar DK, Palmiter RD, Woods SC, Schwartz MW. Attenuated feeding responses to circadian and palatability cues in mice lacking neuropeptide Y. Peptides. 2005;26:2597–2602. doi: 10.1016/j.peptides.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: Sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann Intern Med. 2004;141:846–850. doi: 10.7326/0003-4819-141-11-200412070-00008. [DOI] [PubMed] [Google Scholar]
- Stephan FK. The “other” circadian system: food as a Zeitgeber. J Biol Rhythms. 2002;17:284–292. doi: 10.1177/074873040201700402. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Storch K, Lipan O, Leykin I, Viswanathan N, Davis F, Wong W, Weitz C. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Sun S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- Taheri S, Lin L, Austin D, Young T, Mignot E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 2004;1:e62. doi: 10.1371/journal.pmed.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang-Christensen M, Vrang N, Ortmann S, Bidlingmaier M, Horvath TL, Tschop M. Central administration of ghrelin and agouti-related protein (83–132) increases food intake and decreases spontaneous locomotor activity in rats. Endocrinology. 2004;145:4645–4652. doi: 10.1210/en.2004-0529. [DOI] [PubMed] [Google Scholar]
- Tasali E, Leproult R, Ehrmann DA, Van Cauter E. Slow-wave sleep and the risk of type 2 diabetes in humans. Proc Natl Acad Sci U S A. 2008;105:1044–1049. doi: 10.1073/pnas.0706446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5’-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- Van Cauter E, Polonsky KS, Scheen AJ. Roles of circadian rhythmicity and sleep in human glucose regulation. Endocrine Reviews. 1997;18:716–738. doi: 10.1210/edrv.18.5.0317. [DOI] [PubMed] [Google Scholar]
- van der Veen DR, Minh NL, Gos P, Arneric M, Gerkema MP, Schibler U. Impact of behavior on central and peripheral circadian clocks in the common vole Microtus arvalis, a mammal with ultradian rhythms. Proc Natl Acad Sci U S A. 2006;103:3393–3398. doi: 10.1073/pnas.0507825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DL, Baskin DG, Schwartz MW. Leptin regulation of the anorexic response to glucagon-like peptide-1 receptor stimulation. Diabetes. 2006;55:3387–3393. doi: 10.2337/db06-0558. [DOI] [PubMed] [Google Scholar]
- Yamanaka A, Beuckmann CT, Willie JT, Hara J, Tsujino N, Mieda M, Tominaga M, Yagami K, Sugiyama F, Goto K, et al. Hypothalamic orexin neurons regulate arousal according to energy balance in mice. Neuron. 2003;38:701–713. doi: 10.1016/s0896-6273(03)00331-3. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yannielli P, Harrington ME. Let there be “more” light: enhancement of light actions on the circadian system through non-photic pathways. Prog Neurobiol. 2004;74:59–76. doi: 10.1016/j.pneurobio.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Yannielli PC, Molyneux PC, Harrington ME, Golombek DA. Ghrelin effects on the circadian system of mice. J Neurosci. 2007;27:2890–2895. doi: 10.1523/JNEUROSCI.3913-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CX, van der Vliet J, Dai J, Yin G, Ru L, Buijs RM. Ventromedial arcuate nucleus communicates peripheral metabolic information to the suprachiasmatic nucleus. Endocrinology. 2006;147:283–294. doi: 10.1210/en.2005-1051. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ME. The circadian clock within the heart: potential influence on myocardial gene expression, metabolism, and function. Am J Physiol Heart Circ Physiol. 2006;290:H1–H16. doi: 10.1152/ajpheart.00582.2005. [DOI] [PubMed] [Google Scholar]
- Zheng X, Yang Z, Yue Z, Alvarez JD, Sehgal A. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci U S A. 2007;104:15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigman JM, Jones JE, Lee CE, Saper CB, Elmquist JK. Expression of ghrelin receptor mRNA in the rat and the mouse brain. J Comp Neurol. 2006;494:528–548. doi: 10.1002/cne.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]