Abstract
LINE-1 (L1) retrotransposons represent one of the most successful families of autonomous retroelements, accounting for at least 17% of the human genome. The expression of these elements can be deleterious to a cell. L1 expression has been shown to result in insertional mutagenesis, genomic deletions and rearrangements as well as double-strand DNA breaks. Also, L1 expression has been linked to the induction of apoptosis. These recent discoveries, in addition to correlations of L1 expression with cancer progression, prompted us to further characterize the effect of L1 expression on cellular viability. We show a marked decrease in the overall cellular vitality with expression of the L1 that was primarily dependent on the second open reading frame (ORF2). Both the endonuclease and reverse transcriptase domains of ORF2 can individually contribute to the deleterious effects of L1 expression. L1 decreases cellular viability both by the previously reported apoptotic signaling, but also by inducing a senescence-like state.
Keywords: retrotransposition, DNA Double-strand Breaks (DSBs), senescence, apoptosis, Line-1, retrotransposon
1. Introduction
Long INterspersed Element 1 (LINE 1 or L1), the most abundant and only autonomously active family of non-LTR retrotransposons in the human genome, comprises about 17% of the human genome (Lander et al, 2001). However, due to truncations and mutations, only 80 to 100 of the more than 500,000 copies (Kazazian, Jr. 2004) are still capable of retrotransposition (Sassaman et al, 1997). The few retrotransposition-competent L1s are not likely to facilitate retrotransposition of defective copies as L1 proteins have been shown to display a strong cis-preference, acting primarily on the RNA from which they were translated (Wei et al, 2001).
A fully functional L1 element encodes two proteins, ORF1p and ORF2p. ORF1 encodes a 40 kDa protein with RNA-binding and RNA chaperone activity (Moran et al, 2001; Kolosha and Martin, 1997; Martin and Bushman, 2001; Kolosha and Martin, 2003), while the ORF2 encodes a 150 kDa protein with endonuclease (Feng et al, 1996) and reverse transcriptase (Mathias et al, 1991; Moran et al, 1996) domains. These ORF2 domains play essential roles in Target Primed Reverse Transcription (TPRT), the proposed mechanism for the retrotransposition of L1 and other non-LTR elements (Luan et al, 1993; Luan and Eickbush, 1995; Cost et al, 2002). The expression of L1 proteins, also, has many deleterious effects on cells, initially through insertional mutations (Kazazian Jr., 1998), and later by introducing genome instability through deletions and genomic rearrangements (Gilbert et al, 2002; Ostertag and Kazazian Jr., 2001; Han et al, 2005).
In addition to these deleterious effects, these domains appear to make a large excess of double-strand DNA breaks, DSBs, intermediates expected based on the TPRT model of L1 insertion (Gilbert et al, 2002; Ostertag and Kazazian Jr., 2001; Jurka, 1997). L1 expression results in surprisingly high levels of γ-H2AX foci, an indirect indication of DSBs. Also, Neutral Comet assays more directly show DSB induction upon expression of L1 proteins (Gasior et al, 2006a). Not surprisingly, damage caused by L1 was found to cause an increase in apoptosis in cells in which they are expressed (Belgnaoui et al, 2006).
DSBs have been described as one of the most deleterious types of genomic damage that can occur to eukaryotic genomes. Their repair has a high rate of error leading to the loss of genetic information as well as chromosomal rearrangements (Longhese et al, 2006). DSBs can lead to apoptosis, and it has been hypothesized that this is how L1 expression leads to apoptosis (Haoudi et al, 2004). However, genomic damage can also be associated with other types of cellular response, such as cellular senescence, a cellular endpoint of permanent cell cycle arrest (Gire et al, 2006; Houtgraaf et al, 2006).
An increase in DSBs is also a marker of tumor progression (Bartkova et al, 2005). The DSBs induce the ATM DNA damage response pathway. A study by Bartek et al. (Bartkova et al, 2005) shows an increase in activation of Chk2, an effector kinase in the ATM pathway, in advanced lung and breast cancers (DiTuliio et al, 2002; Lukas et al, 2003; Kastan et al, 2004; Shiloh, 2003). After showing that this induction preceded p53 mutations during the progression of human bladder tumors, Bartek et al. hypothesized that induction of the ATM pathway acted as a selective pressure promoting mutations like those commonly seen in p53 in order to avoid its pro-apoptotic signals (Bartkova et al, 2005). Because mutations of p53 are typically associated with human cancer progression, the induction of this DNA damage response associated with L1 expression may play a role in human cancer progression (Gasior et al, 2006a; Haoudi et al, 2004). While evidence of the deleterious nature of L1 expression is widespread, the exact domains of L1 contributing to the various cellular responses, as well as the full range of cellular responses to L1 have been only poorly characterized to date.
2. Materials and Methods
2.1. Cell Lines, Culture Conditions
MCF7 and MCF7-Bcl2 (Burow et al, 2001) cells were grown in eMEM media supplemented with 5.0% Fetal Bovine Serum (Atlanta Biologicals), 0.5% non-essential amino acids (Invitrogen), 0.5% amino acids (Invitrogen), 0.5% L-glutamine (Invitrogen), and 0.5% sodium pyruvate (Invitrogen) at 37° in a 5% carbon dioxide environment. HeLa cells were grown in eMEM media supplemented with 5.0% Fetal Bovine Serum, 0.5% non-essential amino acids, and 0.5% sodium pyruvate at 37° C in a 5.0% carbon dioxide environment.
2.2. Transfection Conditions
Approximately 500,000 cells were seeded in each T75 flask. The following day, the cells were transfected with appropriate plasmids, using Lipofectamine and Plus reagent following the manufacturer’s protocol was conducted. The transfection solution was left on the cells for three hours before being replaced with normal growth media, and the cells were allowed to grow for 24 hours. Following this growth period, selection was carried out using the standard growth media with the addition of G418 (400 μg/ml) or Zeocin (200 μg/ml) as appropriate. G418 selection was maintained for 14 days and Zeocin selection was maintained for 7 days to select for G418 resistant colonies or zeocin resistant cells, respectively.
2.3. Plasmid Construction
Expression vectors were created by utilizing a PCR reaction to add a 5′ Hind III site and a 3′ BamH I site to the end of each of the open reading frames to be expressed. These products were then subcloned into TOPO-TA (Invitrogen) before being digested with Hind III and BamH I. The appropriately sized piece was then isolated and ligated into similarly digested pBud vector under control of the CMV promoter.
The sequence used to create the vectors expressing both L1 and L1 ORF2 were generated synthetically and has previously been described (Gasior et al, 2006a; Gasior et al, 2006b).
All primers used in the study are listed in supplemental table 1.
2.4. Site-Directed Mutagenesis
We used the QuikChange Site-Directed Mutagenesis kit (Stratagene) to insert mutations into two previously characterized (Feng et al, 1996; Mathias et al, 1991; Moran et al, 1996), highly conserved domains of the L1 second open reading frame. Endonuclease mutants were made by changing amino acid number 205 in the second open reading frame, from Asp to Ala. Reverse transcriptase mutants were created by changing amino acid number 702 in the second open reading frame, from Asp to Ala.
All primers used in the study are listed in supplemental table 1.
2.5. Cellular Proliferation Assay
Cells were transfected with 3 ug of DNA. Following one week of zeocin selection, cells were collected from the T75 flask by trypsin digestion. 200 μl of this cell solution was added to 800 μl of trypan blue. 10 μl aliquots of the resulting solution were counted in a hemocytometer to determine the relative number of viable cells. All cellular proliferation assays experiments were repeated minimally in triplicate.
2.6. Apoptosis Inhibition
Caspase activity was inhibited using a broad spectrum caspase inhibitor, zVad-Fmk, which binds irreversibly to the caspase active site. Following transfection, cells were grown in appropriate growth or selection media supplemented with 20 μM zVad-Fmk. zVad-Fmk was maintained at this concentration up until the cells were harvested for analysis.
MCF7 cells stably expressing of Bcl2 or co-transfection of 3ug of Bcl2 expression vector in HeLa was also used to inhibit apoptosis.
2.7. Senescence Assay
Senescence assays were conducted using the senescence detection kit from BioVision using recommended protocols. Cells were incubated in Staining Solution Mix for 6 hours instead of overnight. All cellular proliferation assays experiments were repeated minimally in triplicate.
2.8. L1 retrotransposition assay
106 cells were seeded per T75 flask 15–20 hours before transfection. Cells were transfected with 1 μg of DNA (either wild-type L1 tagged or an empty (pCEP) expression vector) 9 μl of Plus Reagent and 4.5 μl of Lipofectamine (Invitrogen). After 3h the transfection cocktail was replaced with the growth media. Selection was added 24h post-transfection and maintained for 3 weeks.
3. Results
3.1. L1 Expression and Toxicity in MCF7 and HeLa Cells
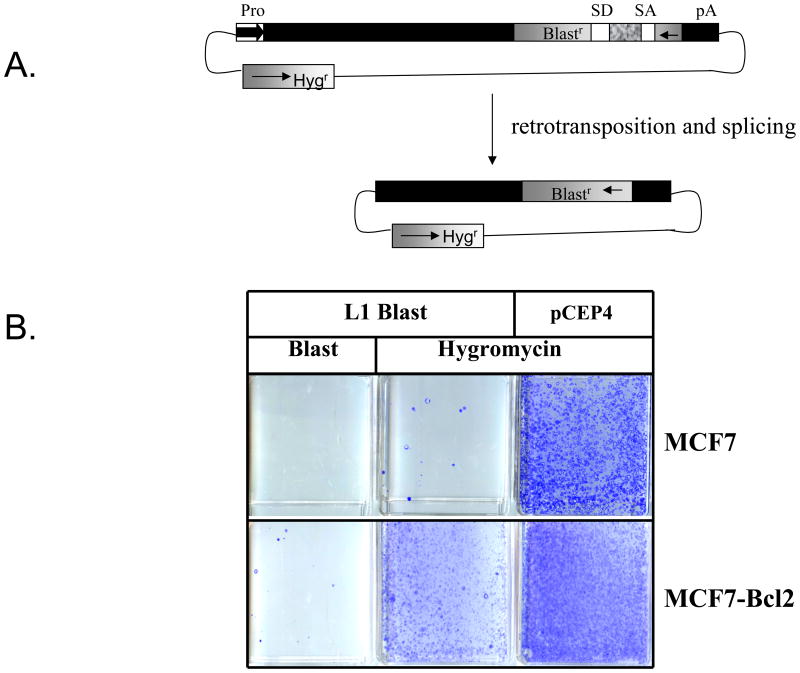
Previous work has shown that L1 retrotransposition rates correlated to p53 status in various cell lines. This work goes on to suggest that this diminished retrotransposition is the result of Bax-induced apoptosis (Haoudi et al, 2004). Indeed, it has also been shown that L1 expression can induce apoptosis in MCF7 cells, a cell line with wild type p53 (Gasior et al, 2006a; Shiloh 2003; Haoudi et al, 2004). As expected, we observe very little retrotransposition when a tagged L1 retrotransposition cassette is transfected into MCF7 cells, despite relatively high transfection and colony-forming efficiencies as measured by parallel transfection of a hygromycin resistance cassette (Figure 1B). In an effort to establish a direct link between apoptosis and reduced retrotransposition in MCF7 cells, assayed L1 retrotransposition using an isogenic MCF7 cell line carrying an expression cassette for Bcl2, an anti-apoptotic protein that would be expected to suppress Bax-induced apoptosis (Zinkle et al, 2006). Despite seeing similar levels of full length L1 mRNA between the cell lines [data not shown], we saw greatly increased retrotransposition in the cells expressing Bcl2. These data from isogenic cell lines suggest that MCF7 are likely capable of undergoing retrotransposition, but that the process of retrotransposition is toxic inducing high levels of apoptosis, and possibly other forms of toxicity. This highly toxic cellular response results in fewer observed retrotransposition events due to loss of vitality in cells where L1 is expressed. However, Bcl2 expression (Figure 1B) did not show full relief from L1-induced toxicity, never raising the number of hygromycin resistant colonies in the L1 expression vector to the number of hygromycin resistant colonies in the pCEP4A vector. This may be because the Bcl2 expression cannot fully repress the apoptosis, or that other forms of toxicity also contributed. In order to better understand the toxic response of some cell types to L1 activity, we wished to explore the influence of the various components of L1 and whether all of the negative consequences of L1 expression are due to the endonuclease activity.
FIGURE 1. L1 Toxicity Reduces Retrotransposition in MCF7 cells.
A. Rationale of Retrotransposition Assay: A blasticidin resistance cassette at the 3′ end of the L1 is used to score retrotransposition events. During the retrotransposition, the splice donor and acceptor interrupting the expression of the blasticidin resistance cassette will be spliced out, conferring blasticidin resistance to cells where a retrotransposition event has taken place. Blasticidin resistant colonies resulting from the growth of these cells will be used to score retrotransposition.
B. MCF7 and MCF7 Bcl2 cells were transiently transfected with a vector expressing wild-type L1 tagged with the blasticidin resistance cassette (L1 WT Blast) or an empty vector (pCEP4). Both of these vectors confer resistance to Hygromycin. The Blasticidin resistant colonies represent L1 retrotransposition events. Hygromycin resistant colonies represent cells that have been successfully transfected with either L1 Blast or pCEP4. Hygromycin colony formation was used to evaluate colony formation ability and to determine plasmid toxicity in each cell line.
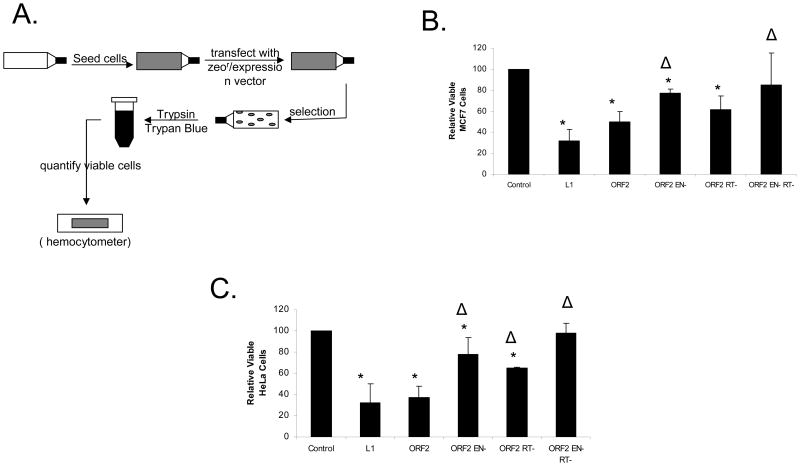
Induction of double stranded DNA breaks (DSBs) has been observed with the expression of both full length L1 and L1 ORF2 (Gasior et al, 2006a). Because studies on the splicing of L1 mRNA show that many cells express a splice product capable of expressing only L1 ORF2 (Belancio et al, 2006), we measured the effect of both L1 and L1 ORF2 in a cellular proliferation assay (Figure 2A). A zeocin resistance-expressing plasmid, along with L1-related or control plasmids are cotransfected. These transfected cells are then selected with zeocin, to ensure that only cells transfected with L1 are assayed, followed by a quantification of viable cells. Thus, anything that leads to cell death, or alters the cellular proliferation rate, will be measured as a reduction of viable cells. Using vectors, which confer Zeocin resistance, designed to optimize expression of L1 (Figure 2B), we saw a reduction in viable MCF7 cells for L1 (68 +/− 11%) that was similar to that seen with the L1 ORF2 (50 +/− 10%).
FIGURE 2. L1 Expression has a Deleterious Effect on HeLa and MCF7 cells.
A: Cellular Proliferation Assay: 5.0×105 cells are seeded in a T75 flask. These flasks are transfected with either an empty vector containing zeocin resistance or a plasmid expressing both L1 protein(s) and zeocin resistance. The day following transfection, the cells are maintained under zeocin selection for one week to eliminate untransfected cells. After one week, the cells are harvested and Trypan Blue negative cells were counted to determine the number of viable cells.
B: L1 Expression Lowers Cellular Viability in MCF7 cells: The relative level of viable cells was measured with the expression of L1, ORF2 and ORF2 with mutations in the endonuclease (EN-) and/or reverse transcriptase (RT-) domains. * denotes a significant difference from the control with a p value ≤ 0.05. Δ denotes a significant difference from ORF2 with a p value ≤ 0.05.
C: L1 Expression Lowers Cellular Viability in HeLa cells: The relative level of viable cells was measured with the expression of L1, ORF2 and its mutants. * denotes a significant difference from the control with a p value ≤ 0.05. Δ denotes a significant difference from ORF2 with a p value ≤ 0.05. All statistical differences determined by both student t-test and ANOVA analysis with Tukey post hoc test. Displayed error bars represent standard deviations in this figure.
The highly conserved endonuclease and reverse transcriptase domains of L1 ORF2p have been demonstrated to be necessary for L1 retrotransposition (Feng et al, 1996; Mathias et al, 1991; Moran et al, 1996). We hypothesized that mutation of conserved residues within these domains would diminish the deleterious effect of L1 expression. The effect of mutations in the endonuclease (D205A) and reverse transcriptase (D702A) domains of L1 ORF2 were measured using the cellular proliferation assay (Figure 2A). The mutation in the endonuclease domain (ORF2 EN-) resulted in a significantly increased number of viable cells, while mutations in the reverse transcriptase domain (ORF2 RT-) yielded only a modest increase in viable cells. Neither of these mutations alone reduced the effect of L1 ORF2 to background levels. However, the effect of a double mutant of both the endonuclease and reverse transcriptase (ORF2 ER--) was not significantly different from an empty vector control (Figure 2B).
Having seen evidence of L1 expression-related toxicity in MCF7 cells, we wanted to determine if this effect was specific to this cell line or if it occurred in other cells commonly used to assay L1 activity. The cellular proliferation assay measuring the effect of expression of L1 (68 +/− 17%) and L1 ORF2 (63 +/− 11%) in HeLa cells showed a marked decrease in cellular proliferation (Figure 2C). In addition the decrease in cellular proliferation found in HeLa cells with both L1 and L1 ORF2 were similar to our observations in MCF7 cells. In HeLa cells, mutations to either the endonuclease or reverse transcriptase domains greatly increased the levels of viable cells. However, cellular proliferation was again only returned to background levels with mutations to both domains (Figure 2C).
3.2. LINE-1 Expression Induces Multiple Cellular End Points
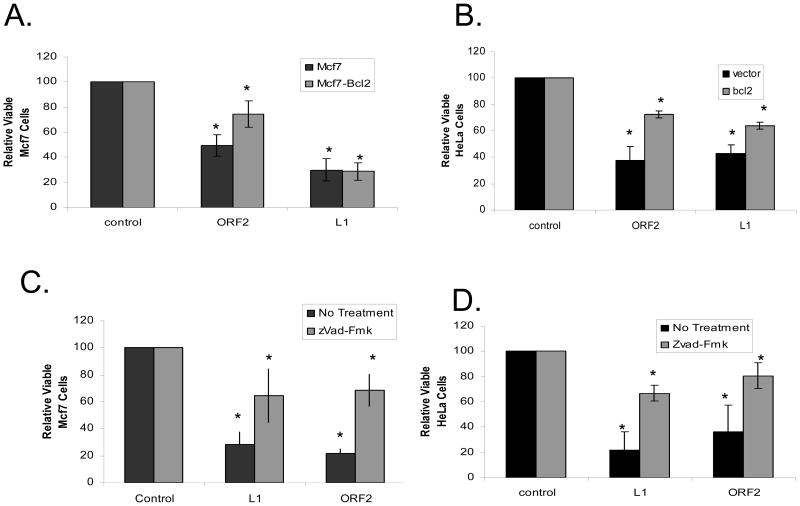
It has previously been shown that L1 expression has the ability to induce apoptosis in MCF7 cells by increased Bax levels as well as Caspase 3 activation (Belgnaoui et al, 2006). We confirmed and extended this result by co-transfecting a Bcl2 expression vector along with either L1 or L1 ORF2 in both HeLa and MCF7 cells to demonstrate a partial increase in cellular proliferation compared to co-expression of an empty vector. Bcl2 expression in both cell lines did not completely relieve the depression of cellular proliferation to background levels with the expression of either L1 (MCF7: 30 +/− 9% to 28 +/− 7% and HeLa: 42 +/− 7% to 64 +/− 2%) or L1 ORF2 (MCF7: 50 +/− 9% to 73 +/−10% and HeLa: 37 +/− 11% to 75 +/− 2%) (Figure 3A, B). Indeed, inhibition of apoptosis by Bcl2 was only able to return cell proliferation rates to background levels when a mutation to functional domains of L1 ORF2 was introduced [Supplemental Figure 1].
FIGURE 3. Inhibition of Apoptosis Partially Relieves L1 Expression Related Toxicity.
A: Bcl2 Does Not Completely Relieve L1 Expression Related Toxicity in MCF7 cells: The relative viable cells were measured with the expression of L1 or ORF2 in the presence and absence of Bcl2 expression. * denotes a significant difference from the control with a p value ≤ 0.05.
B: Bcl2 Does Not Completely Relieve L1 Expression Related Toxicity in HeLa cells: The relative viable cells were measured with the expression of L1 or ORF2 in the presence and absence of Bcl2 expression. * denotes a significant difference from the control with a p value ≤ 0.05. Vector refers to the pBluescript vector used as a transfection control.
C: zVad-Fmk Treatment Does Not Completely Relieve L1 Expression Related Toxicity in MCF7 cells: The relative viable cells were measured with the expression of L1 or ORF2 in the presence and absence of zVad-Fmk treatment. * denotes a significant difference from the control with a p value ≤ 0.05.
D: zVad-Fmk Treatment Does Not Completely Relieve L1 Expression Related Toxicity in HeLa cells: The relative viable cells were measured with the expression of L1 or ORF2 in the presence and absence of zVad-Fmk treatment. * denotes a significant difference from the control with a p value ≤ 0.05. All statistical differences determined by both student t-test and ANOVA analysis with Tukey post hoc test. Displayed error bars represent standard deviations in this figure.
A previous study (Belgnaoui et al, 2006) also showed an induction of caspase 3, a cell-death protease (Zhivotovsky et al, 1996), with L1 expression. Having seen an incomplete reduction of the effect of L1 expression on cellular proliferation with the co-expression of Bcl2, we wished to inhibit the pro-apoptotic activities of the cellular caspases to see if we could restore cellular proliferation to background levels. To test the role of cellular caspases in the L1 related decline in cellular proliferation, we tested the effect of L1 expression in the presence of 20 μM zVad-Fmk, a strong broad spectrum caspase inhibitor (Martinet et al, 2006). Following the pattern seen with the expression of Bcl2, zVad-Fmk’s inhibition of caspase activity significantly reduces the effect of L1 expression. It does not return the cellular proliferation levels to background in either cell line with L1 (MCF7: 29 +/− 9% to 65 +/− 29% and HeLa: 21+/− 15% to 67 +/− 6%) or L1 ORF2 (MCF7: 22+/− 3% to 68 +/− 22% and HeLa: 37+/− 21% to 81 +/− 10%) (Figure 3C, D). In a fashion similar to Bcl2 inhibition of L1-induced apoptosis, the inhibition of caspase activity, by zVad-Fmk, was able to restore cellular proliferation rates to wild type levels with a mutation to the endonuclease domain of L1 ORF2 [Supplemental Figure 1].
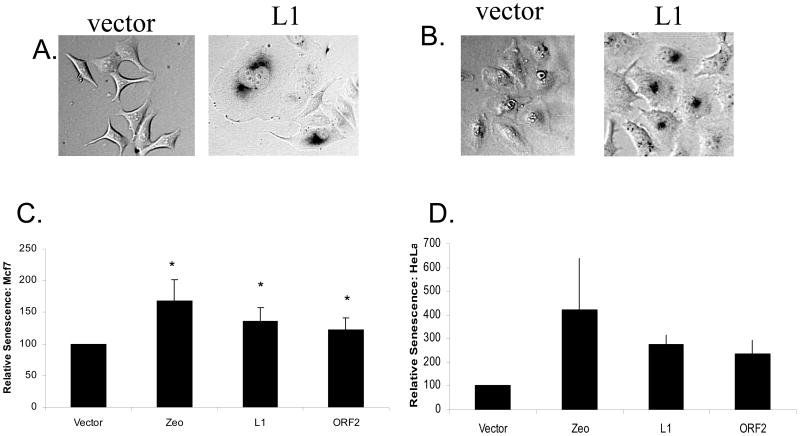
Because two independent approaches to inhibit apoptosis did not return cellular proliferation to background levels and because other sources of DNA damage and DSBs have previously been shown to also induce cellular senescence (Gire et al, 2004;, Houtgraaf et al, 2006), we wished to determine whether L1 expression could be contributing to the lowered proliferation by inducing a senescence-like state. To test for this effect, we assayed for expression of a senescence specific β-galactosidase (Figure 4A, B). Utilizing this assay in both MCF7 and HeLa cell lines, we measured the ability of L1 or L1 ORF2 expression to induce a senescence-like state, as indicated by expression of senescence-specific β-galactosidase. When compared to an empty vector control (100%), senescence-specific β-galactosidase levels seen with both L1 expression (MCF7: 135% +/− 18% and HeLa: 276% +/− 36%) and ORF2 expression (MCF7: 123 +/− 22% and HeLa: 236 +/− 58%) were significantly higher (Figure 4C, D).
Figure 4. L1 Expression Induces Cellular Senescence.
A: L1 Expression Induces Cellular Senescence in MCF7 cells: The senescence detection assay was utilized to stain the nuclei of senescent cells. Images of these cells were then collected at 200x magnification.
B: L1 Expression Induces Cellular Senescence in HeLa cells: The senescence detection assay was utilized to stain the nuclei of senescent cells. Images of these cells were then collected at 200x magnification.
C: Quantification of L1-Induced Cellular Senescence in MCF7 cells: The senescent cells seen in the images collected from MCF7 were quantified and set relative to an empty vector control. Zeo represents treatment with Zeocin, a DNA damaging agent used as a positive control for DNA damage-induced senescence. Cells were placed in appropriate media containing 200 μg/ml zeocin, a DNA damaging agent. Cells expressing a cellular senescence specific β-galactasidase were stained, and scored visually. No Treat signifies cells which were not transfected or treated with Zeocin. * denotes a significant difference in stained cells from the control with a p value ≤ 0.05.
D: Quantification of L1-Induced Cellular Senescence in Hela cells: The senescent cells seen in the images collected from HeLa were quantified and set relative to an empty vector control. Zeo represents treatment with Zeocin, a DNA damaging agent used as a positive control for DNA damage-induced senescence. Cells were placed in appropriate media containing 200 μg/ml zeocin, a DNA damaging agent. Cells expressing a cellular senescence specific β-galactasidase were stained, and scored visually. * denotes a significant difference from the control with a p value ≤ 0.05. All statistical differences determined by both student t-test and ANOVA analysis with Tukey post hoc test. Displayed error bars represent standard deviations in this figure.
4. Discussion
Mobile elements have long been known to be responsible for genomic mutations and rearrangements through their insertion in dispersed genomic loci (Kazazian Jr., 1998). Several recent studies have suggested that expression of the human L1 mobile element can have even more immediate, negative consequences for the cell (Gasior et al, 2006a; Belgnaoui et al, 2006; Haoudi et al, 2004). These cellular impacts include cell cycle arrest and apoptosis (Gasior et al, 2006a; Belgnaoui et al, 2006) and seem to be at least partially dependent on the endonuclease activity of the L1 ORF2 protein. This endonuclease activity has been demonstrated to cause a large excess of DNA DSBs in the cell that might be expected to contribute to both cell cycle arrest and apoptosis (Gasior et al, 2006a). However, there were also some indications that the negative consequences to cell growth and viability were not solely associated with the endonuclease activity (Gasior et al, 2006a).
Most previous studies have measured the influence of expression of both L1 proteins simultaneously from an intact L1 element. This left the question as to whether consequences, such as apoptosis, were induced by genomic disruptions associated with the retrotransposition process, or whether L1 might supply ORF2 separately from the active L1 complex to cause damage. We have confirmed that over expression of ORF2 alone inhibited the level of viable cells by 60% and 50% in cellular proliferation in HeLa and MCF7 cell, respectively (Figure 2). Because an ORF2 expression construct by itself is a very poor substrate for retrotransposition (Wei et al, 2001), this demonstrates that retrotransposition itself is not needed to cause toxicity. The level of ORF2 expression is almost certainly much higher when expressed by itself instead of as the second ORF in a bicistronic L1 RNA. Thus, our data suggest that, while toxic by itself, ORF2 is probably more toxic when expressed as part of the full-length L1. One likely explanation is that the full-length L1 incorporates the ORF2p into a particle that is more efficiently transported to the nucleus, or is more active than when it is expressed by itself. This is likely due to an association with the ORF1 product in the complex.
Mutation of the ORF2 endonuclease domain only partially removed the negative influence of this protein on cell growth. The RT domain had a slightly smaller, but significant impact in the viability (Figure 2). Mutation of both the endonuclease and the RT domains removed essentially all the negative consequences of ORF2 expression. These data are consistent with the anecdotal evidence that high levels of ORF2 could only be expressed in cells when both domains were mutated (Goodier et al, 2004). The negative consequences of the endonuclease domain are almost certainly due to the excess DNA DSBs caused by this activity. It is more difficult to assess why the RT domain would be detrimental to cells, but evidence has shown that L1 elements can interact with random genomic breaks (Morrish et al, 2002); Sen et al, 2007). Perhaps, the interaction of L1 RT with random genomic breaks inhibits their repair.
One of the important implications of the damage caused by ORF2 expression alone is that studies of the splicing of L1 RNA have demonstrated that alternative RNA forms are produced that splice out the ORF1 region and would be capable of expressing only ORF2 (Belancio et al, 2006). Proteins translated from these spliced RNAs are also capable of driving Alu retroposition [VP Belancio Unpublished Data]. These splice products would be expected to have negative consequences for the cell while still having little if any capability of driving L1 retrotransposition. Thus, even L1 elements with defective ORF1 coding regions might make an RNA that splices to express a functional ORF2 with a number of negative consequences for the cell.
Our studies on the loss of cellular viability caused by L1 expression confirm and extend previous studies (Belgnaoui et al, 2006; Haoudi et al, 2004) that L1 can trigger apoptosis. In addition, our finding that Bcl2 expression greatly decreased the loss of viability is consistent with studies suggesting apoptosis through a Bax-induced apoptosis. However, we also found that Bcl2 expression was not able to completely inhibit the loss of viability from L1 expression. Furthermore, zVAD-FMK, a broad spectrum caspase inhibitor (An et al, 1996), was also unable to completely return cells to control levels of growth. The evidence gathered utilizing the cellular proliferation assay in the presence of these anti-apoptotic proteins suggests that L1 might be reducing cellular proliferation by a means other than apoptosis.
Cellular senescence represents a cellular endpoint where the cell enters a permanent cell cycle arrest, which can be triggered by DSBs (Gire et al, 2004; Houtgraaf et al, 2006). We tested for expression of senescence-specific β-galactosidase to estimate the number of senescent cells after L1 expression. Both HeLa and MCF7 cells showed a significant increase in senescence-specific β-galactosidase levels with both L1 and ORF2 expression. In HeLa cells, these levels were approximately three-fold greater than the control. Thus, the induction of a senescence-like state is likely to explain most of the loss of viability that was not due to apoptosis.
An indirect sign of the potential for cellular damage caused by L1 elements is the myriad of strategies cells use to protect themselves from its activity. This includes extensive transcription repression by methylation [(Roman-Gomez et al, 2005); (Bourc’his et al, 2004); (Webster et al, 2005), as well as post-transcriptional regulation by premature poly-adenylation (Perepelitsa et al, 2005), aberrant splicing (Belancio et al, 2006) and RNAi (Yang et al, 2006). In addition, there are other cellular proteins, such as the APOBEC3 family of proteins that appear to inhibit L1 retrotransposition (Stenglein and Harris, 2006; Bogerd et al, 2006; Muckenfuss et al, 2006; Kubo et al, 2006).
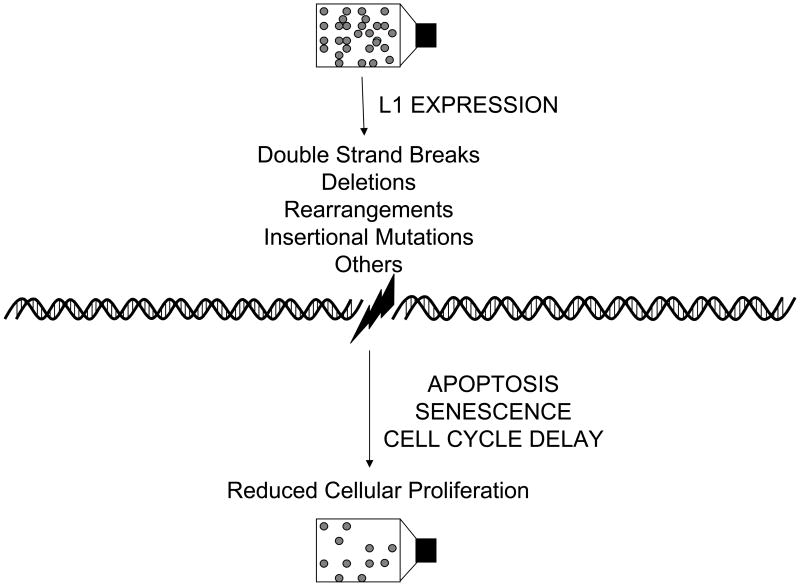
Figure 5 summarizes the pathways induced by the deleterious nature of L1 expression and some of the means by which L1 can induce these pathways. L1 expression results in genomic deletions, rearrangements, and double stranded breaks in DNA (Kazazian Jr., 1998; Gilbert et al, 2002; Ostertag and Kazazian Jr., 2001; Belgnaoui et al, 2006; Longhese et al, 2006). It is very likely that different cells will have widely different responses to L1 activity, with different propensities to either tolerate L1 activity, or respond with apoptosis or cellular senescence. In each of the latter cases, the cellular response would result in a minimization of the negative (i.e. mutagenic) consequences of the mobile element activity on the viability of the organism. Thus, these responses may be among the natural defenses used by the organism to minimize damage from either germ line or somatic (Kubo et al, 2006; van den Hurk et al, 2007) expression of L1 elements. It is interesting that even in cancer cell lines, that are both resistant to apoptosis and growth immortalized, L1 is still capable of inducing both apoptosis and a senescence-like state. Because L1 expression is often increased in transformed cells, these mechanisms may remain important in minimizing genetic instability due to L1 activity even in tumors.
Figure 5. Summary of L1 Induced Pathways to Decreased Cellular Proliferation.
Flow chart of L1 expression and subsequent cellular endpoints. In this diagram, the circles represent cells growing in a flask both before and after L1 expression with a listing of the factors that may influence the cell viability and proliferation.
Supplementary Material
Supplemental Figure 1: Inhibition of Apoptosis in Combination with Mutations to L1 ORF2 Relieves L1 Expression Related Toxicity
Supplemental Table 1: Contains the Primers used in the Generation of Mutations to L1 ORF2.
Acknowledgments
We would like to thank Matthew Burow (Tulane University) and Barbara Beckman (Tulane University) for their generous gift of the MCF7 and isogenic MCF7 line carrying a Bcl2 expression cassette.
This work was supported by grants to PD from the USPHS grant R02GM45668, NIH P20 RR020152, National Science Foundation EPS-0346411. NW was supported in part by a student grant from the Cancer Association of Greater New Orleans (CAGNO) 2005 and LEQSF (2003-08)-GF-25.
Abbreviations
- Bax
Bcl2 Associated X protein
- Bcl2
B-cell Cll Lymphoma 2
- kDa
Kilodalton
- LTR
Long Terminal Repeat
- ORF
Open Reading Frame
- zVad-Fmk
Z-Val-Ala-Asp-fluoromethylketone
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at xxx.
Footnotes
Supplemental Data are available at Gene Online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An S, Yap D, Knox KA. Ligation of CD40 rescues Ramos-Burkitt lymphoma B cells from calcium ionophore- and antigen receptor-triggered apoptosis by inhibiting activation of cysteine protease CPP32/Yama and cleavage of its substrate PARP. FEBS Lett. 1996;386:115–22. doi: 10.1016/0014-5793(96)00427-9. [DOI] [PubMed] [Google Scholar]
- Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger PL. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–21. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui SM, Gosden RF, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci, USA. 2006;103:8780–5. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TG. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Burow ME, Weldon CB, Tang Y, McLachlan JA, Beckman BS. Oestrogen-mediated suppression of tumour necrosis factor alpha-induced apoptosis in MCF-7 cells: subversion of Bcl-2 by anti-oestrogens. J Steroid Biochem Mol Biol. 2001;78:409–18. doi: 10.1016/s0960-0760(01)00117-0. [DOI] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO. 2002;21:5899–910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTullio RA, Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bertek J, Halazonetis TD. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nature Cell Biol. 2002;4:998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–16. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Wakeman TP, Xu B, Deininger PL. The Human LINE-1 Retrotransposon Creates DNA Double-strand Breaks. J Mol Biol. 2006a;257:1383–93. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Palmisano M, Deininger PL. Alu-linked hairpins efficiently mediate RNA interference with less toxicity than do H1-expressed short hairpin RNAs. Anal Biochem. 2006b;349:41–48. doi: 10.1016/j.ab.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Gire V, Roux P, Wynford-Thomas D, Brondello JM, Dulic V. DNA damage checkpoint kinase Chk2 triggers replicative senescence. EMBO. 2004;23:2554–63. doi: 10.1038/sj.emboj.7600259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, et al. A potential role for the nucleolus in L1 retrotransposition. Hum Mol Genet. 2004;13:1041–8. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- Han K, Sen SK, Wang J, Callinan PA, Lee J, Cordaux R, Liang P, Batzer MA. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33:4040–52. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haoudi A, Semmes OJ, Mason JM, Cannon RE. Retrotransposition-Competent Human LINE-1 Induces Apoptosis in Cancer Cells With Intact p53. J Biomed Biotechnol. 2004;4:85–94. doi: 10.1155/S1110724304403131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtgraaf JH, Versmissen J, van der Giessen WJ. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc Revasc Med. 2006;7:165–72. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hohjoh H, et al. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements and disease. Curr Opin Genet Dev. 1998;8:343–50. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile Elements: Drivers of Genome Evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci USA. 1997;94:10155–60. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosha VO, Martin SL. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1) J Biol Chem. 2003;278:8112–7. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- Kubo S, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci USA. 2006;103:8036–41. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Longhese MP, Mantiero D, Clerici M. The cellular response to chromosome breakage. Mol Microbiol. 2006;60:1099–108. doi: 10.1111/j.1365-2958.2006.05186.x. [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995;15:3882–91. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nature Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet W, De Meyer GR, Timmermans JP, Herman AG, Kockx MM. Macrophages but not smooth muscle cells undergo enzyloxycarbonyl-Val-Ala-DL-Asp(O-Methyl) –fluoromethylketone-induced non-apoptotic cell death depending on receptor-interacting protein 1 expression: Implications for the stabilization of macrophage-rich atherosclerotic plaques. The Journal of Pharmacology and Experimental Therapeutics. 2006;317:1356–64. doi: 10.1124/jpet.106.102970. [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–10. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;187:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–65. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversion in L1 retrotransposition. Genome Res. 2001;11:2059–65. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V, Deininger PL. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2005;35:363–6. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–23. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Sen SK, Huang CT, Han K, Batzer MA. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007;35:6741–51. doi: 10.1093/nar/gkm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 etrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–41. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- van den Hurk JA, et al. L1 retrotroposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–92. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- Webster KE, et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci USA. 2005;102:4068–73. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Jr, Boeke JD, Moran JV. Human L1 Retrotransposition: cis Preference versus trans Complementation. Mol Cell Biol. 2001;21:1429–39. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Burgess DH, Orrenius S. Proteases in apoptosis. Experientia. 1996;52:968–78. doi: 10.1007/BF01920106. [DOI] [PubMed] [Google Scholar]
- Zingler N, Willhoeft U, Brose HP, Schoder V, Jahns T, Hanschmann KM, Morrish TA, Lower J, Schumann GG. Analysis of 50 junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 50-end attachment requiring microhomology-mediated end joining. Genome Res. 1997;15:780–9. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkle S, Gross A, Yang E. BCL2 family in DNA damage and cell cycle control. Cell Death Differ. 2006;13:1351–9. doi: 10.1038/sj.cdd.4401987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Inhibition of Apoptosis in Combination with Mutations to L1 ORF2 Relieves L1 Expression Related Toxicity
Supplemental Table 1: Contains the Primers used in the Generation of Mutations to L1 ORF2.