Abstract
Current hypotheses that attempt to rationalize the high degree of base selectivity exhibited by replicative DNA polymerases (pols) concur that ternary complexes formed with incorrect dNTPs are destabilized. Knowing what accounts for this destabilization is likely to be the key to understanding base discrimination. To address this issue, we have determined crystal structures of ternary complexes with all twelve mismatches using an engineered RB69 pol quadruple mutant (qm, L415A/L561A/S565G/Y567A) that enabled us to capture nascent mispaired dNTPs. These structures show that mismatches in the Nascent base-pair Binding Pocket (NBP) of the qm pol differ markedly from mismatches embedded in binary pol-DNA complexes. Surprisingly only 3 of 12 mismatches clash with the NBP when they are modeled into the wt pol. The remaining can fit into a wt pol ternary complex but deviate from normal Watson-Crick base-pairs. Repositioning of the templating nucleotide residue and the enlarged NBP in qm ternary complex play important roles in accommodating incorrect incoming dNTPs. From these structures, we propose additional reasons as to why incorrect dNTP are incorporated so inefficiently by wild type RB69 pol: (i) steric clashes with side chains in the NBP after Fingers closing; (ii) weak interactions or large gaps between the incoming dNTP and the templating base; (iii) burying a protonated base in the hydrophobic environment of the NBP. All of these possibilities would be expected to destabilize the closed ternary complex so that incorporation of incorrect dNTP would be a rare event.
INTRODUCTION
DNA polymerases (pols) are essential for maintaining the genetic integrity of all organisms, making errors only once in every 106 insertion events.1–2 Dysregulation of DNA synthesis, because of mutations in DNA polymerase genes, has profound negative effects, often associated with pathological conditions. 3–6 Clearly, understanding DNA pol base selectivity will likely lead to direct and important medical applications. Current hypotheses that attempt to account for the high degree of base selectivity exhibited by replicative DNA polymerases include geometry of the nascent base-pairs, interbase hydrogen bonds, solvent exclusion and base stacking.7–10 Incorrect dNTPs that do not meet these requirements are rejected before they can be incorporated because the closed ternary complexes with mismatches are destabilized. What accounts for this destabilization is likely to be the key to elucidating the mechanisms used by DNA pols for base discrimination.
A large number of pol structures with correct incoming dNTPs are now available, but structural data with mismatched base-pairs at the insertion site of these pols is extremely limited.11–13 Johnson and Beese captured twelve mismatches at the penultimate base-pair position of a primer/template (P/T) duplex DNA in complex with Bacillus Stearothermophilus DNA pol I fragment (BF).14 Six of the twelve mismatches were either unpaired or too disordered to provide a detailed molecular model. Among the remaining six well-ordered mismatches, three were obtained in the presence of Mn2+, a divalent metal ion that has recently been found to affect tautomerization of the nucleotide residues.12 Along these lines we have shown that the dNTP/dF pair in the Nascent Base-pair binding Pocket (NBP) of RB69pol differed markedly from dN/dF pairs at the penultimate base-pair position in binary complexes with wt RB69pol.15–16 Thus we anticipated that the conformation of the twelve mismatches at the insertion site in ternary complexes of the RB69pol variant would be very different from what has been observed at the penultimate base-pair position of the BF. In addition, the studies of Johnson and Beese addressed how a replicative pol detects mismatches in P/T duplex after chemistry, while the crucial process for incorporating a correct dNTP or rejecting an incorrect dNTP occurrs at the insertion site prior to nucleotide transfer. For this reason, the structures of the twelve mismatches at the insertion site should be highly informative with respect to base discrimination.
We have chosen RB69pol, a high-fidelity model family B pol, for structural studies of pol ternary complexes with nascent base-pair mismatches because of extensive structural and kinetic data available for this pol17–21 and because of sequence similarities with human pols alpha and delta.22 However a comprehensive explanation for base discrimination still remains elusive not only for RB69pol but for other pols as well. One essential requirement for achieving this goal is to have high resolution structures of all twelve mismatches at the pol insertion site as well as pre-steady state kinetic parameters for nucleotide incorporation of the 16 base-pairs combinations. However it has been extremely difficult, perhaps impossible, to capture structures of all twelve mismatches with wt RB69pol because closed ternary complexes with mismatched bases, in general, are unstable.23 Our previous kinetic studies on single, double and triple RB69pol mutants showed a strong correlation between increased NBP volume and loss of base selectivity; i.e. incremental expansion of space in the NBP resulted in a progressive increase in incorporation efficiency of incorrect dNMPs.17, 19 Although we were able to determine structures of ternary complexes of a triple mutant (L561A/S565G/Y567A) with four mismatches at the insertion site;16 we failed to capture purine-purine mismatches. In an attempt to obtain the structures of the remaining mismatches so that we could have a complete set of ternary complexes with mismatched base-pairs, we introduced an additional substitution, L415A, into the triple mutant (Fig. S1). The resulting quadruple mutant (qm) allowed us to obtain closed ternary structures of RB69pol with all sixteen combinations of nascent base-pairs (four correct Watson-Crick base-pairs and twelve mismatches) at the insertion site poised for nucleotide incorporation. We have also determined the pre-steady-state kinetic parameters for nucleotidyl transfer with all sixteen combinations. Together these results have permitted us to not only visualize the conformation of the sixteen base-pairs in the NBP of the ternary complexes but also to correlate the kinetic parameters obtained for each combination of mispaired bases with the corresponding structures. This provides a basis for identifying features of the NBP in the qm that allow insertion of incorrect dNMPs and has prompted us to speculate about the reasons for the inefficient incorporation of incorrect dNMPs by the wt enzyme. Briefly, it appears that steric clashes between mispaired bases and NBP side chains, weak interactions between incoming dNTPs and templating bases, and burying a large gap or protonated base in the NBP are the main reasons that the closed ternary complex of the wt RB69pol, containing a mismatch, is destabilized. In addition, we found that repositioning of the templating nucleotide residue in the qm RB69pol ternary complex plays an important role in accommodating mismatched incoming dNTPs. In contrast, the templating nucleotide residue is rigidly anchored in the wt pol, so the same mismatched base-pairs may require repositioning of the incorrect dNTP instead of the templating base. As a consequence, misalignment of the α phosphorous atom of the incoming dNTP with the 3’-OH at the primer-terminus results in low incorporation efficiency.
RESULTS AND DISCUSSION
Structural Overview of the dNTP/dN-containing RB69pol qm Ternary Complexes
The sixteen structures referred to above were determined with resolutions ranging from 1.87 to 2.20 Å and Rfree values spanning 21.3–26.4% (Table 1, S1–S4). Overall these structures were nearly identical to that of our previously reported 1.8 Å dCTP/dG-containing wt RB69pol ternary complex except for the different nascent base-pairs and for the NBP which had four amino acid substitutions (L415A/L561A/S565G/Y567A). The root-mean-square deviations of Cα atoms varied between 0.20 to 0.26 Å. The electron densities for the incoming dNTP, the P/T duplex DNA and the surrounding network of ordered water molecules in all these structures were well-defined except for the dATP/dG-and dGTP/dG-containing ternary complexes (Fig. 1, S2). With these two structures, the electron densities for the templating dG were less well-defined than for the incoming dATP and dGTP (Fig. 1K, 1L). The triphosphate tails of the incoming dNTPs in all sixteen ternary complexes were coordinated to the B metal ion in the classic α, β, γ-tridendate state. When these sixteen structures were superimposed, the phosphodiester backbones of the P/T duplex DNA were congruent except for the nucleotide residues at the 5’ template overhang where three different conformations were observed. The base of the overhanging nucleotide at position n+1 of the template strand was stabilized by stacking on top of either the indole ring of W574 or the phenyl ring of F359 (Fig. S3A, S3B), except when the templating base was dA, and the incoming dNTP was a purine. In that case, the nucleotide at position n+1 of the template strand flipped back into the NBP (Fig. S3C, S3D). It is worth noting that different conformations of the 5’ template overhang were observed only for complexes with mispaired bases in qm RB69pol. For ternary complexes with Watson-Crick base-pairs, the nucleotide residue at position n+1 of the template strand was always stacked on top of the phenyl ring of F359 (Fig. S3A). Since the twelve mismatches have different hydrogen bonding (HB) patterns and geometric shapes, it is useful to assemble them into three groups: (i) purine-pyrimidine, (ii) pyrimidine-pyrimidine and; and (iii) purine-purine.
Table 1.
Refinement and geometry statistics for all 16 dNTP/dN-containing qm RB69pol ternary complexes.
| Base Pairs | dNTP/dN | PDB code | Resolution (Å) | R(%) | Rfree(%) | d(C1’–C1’) (Å) | λ(dNTP)(°) | λ(dN)(°) |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Correct BP | dATP/dT | 4FJ5 | 2.05 | 18.4 | 23.7 | 10.3 | 59.0 | 57.6 |
| dTTP/dA | 4FJN | 2.00 | 19.9 | 24.1 | 10.4 | 60.3 | 55.2 | |
| dGTP/dC | 4FJH | 2.11 | 17.5 | 22.2 | 10.4 | 56.2 | 55.4 | |
| dCTP/dG | 4FK0 | 2.18 | 20.3 | 25.6 | 10.5 | 57.7 | 54.4 | |
|
| ||||||||
| Py-Py | dCTP/dC | 4FJI | 2.20 | 21.0 | 26.4 | 10.3 | 65.2 | 65.5 |
| dCTP/dT | 4FJ8 | 2.19 | 19.5 | 25.1 | 10.7 | 64.4 | 60.3 | |
| dTTP/dC | 4FJJ | 2.00 | 18.6 | 22.5 | 10.8 | 62.4 | 63.3 | |
| dTTP/dT | 4FJ9 | 1.97 | 19.0 | 23.4 | 9.5 | 69.6 | 40.7 | |
|
| ||||||||
| Pu-Py | dATP/dC | 4FJG | 2.02 | 22.0 | 25.9 | 10.5 | 64.5 | 54.4 |
| dCTP/dA | 4FJM | 2.02 | 19.1 | 23.4 | 10.3 | 68.9 | 48.4 | |
| dGTP/dT | 4FJ7 | 1.90 | 18.1 | 21.3 | 10.5 | 49.5 | 62.4 | |
| dTTP/dG | 4FK2 | 2.00 | 18.6 | 22.5 | 10.3 | 69.1 | 45.0 | |
|
| ||||||||
| Pu-Pu | dATP/dA | 4FJK | 2.00 | 18.2 | 22.5 | 16.3 | 69.7 | 153.2 |
| dATP/dG | 4FJX | 2.12 | 18.3 | 22.5 | 10.9 | 54.2 | 112.3 | |
| dGTP/dA | 4FJL | 1.87 | 17.9 | 22.2 | 16.3 | 67.2 | 153.1 | |
| dGTP/dG | 4FK4 | 1.90 | 18.7 | 22.2 | 10.9 | 54.0 | 113.2 | |
|
| ||||||||
Figure 1.
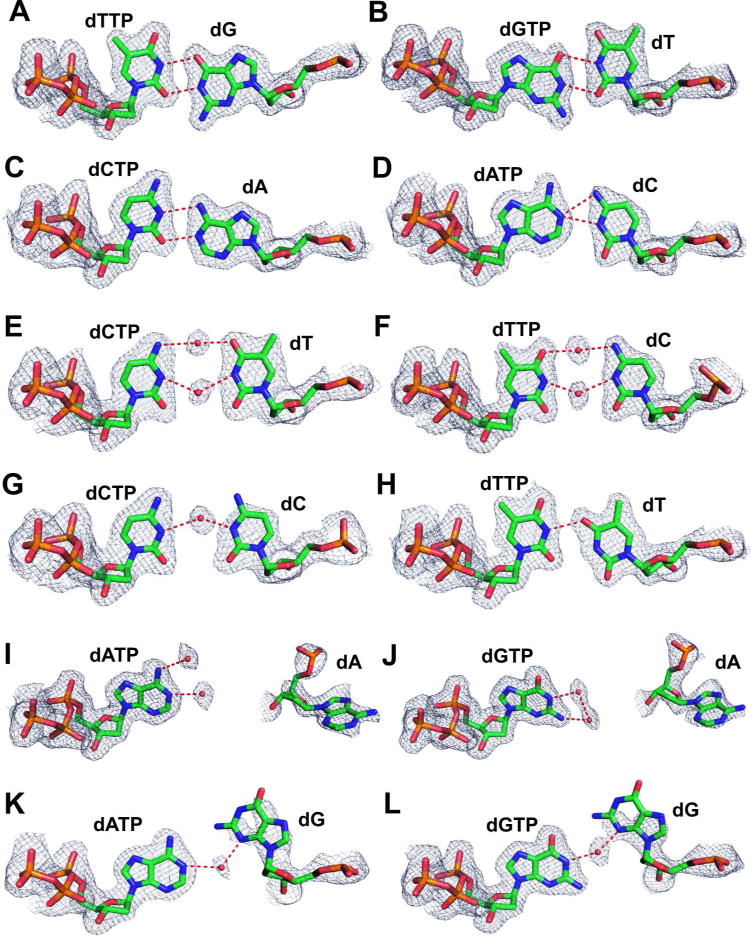
Final 2Fo-Fc electron density map for 12 mismatches at the NBP of qm RB69pol contoured at 1.2 σ: (A) dTTP/dG; (B) dGTP/dT; (C) dCTP/dA; (D) dATP/dC; (E) dCTP/dT; (F) dTTP/dC; (G) dCTP/dC; (H) dTTP/dT; (I) dATP/dA; (J) dGTP/dA; (K) dATP/dG and (L) dGTP/dG.
(i) Purine-Pyrimidine Mismatches in the NBP of qm RB69pol
In the dTTP/dG-containing complex, the pyrimidine base of dTTP is hydrogen bonded to the templating dG with wobble geometry (Fig. 1A). The purine base of dG is displaced toward the minor groove, and occupies the cavity created by the Y567 to Ala substitution. In addition, the Cα hydrogen of G568 appears to be hydrogen bonded to the N3 of the templating dG as the HB distance between Cα of G568 and the N3 of dG is 3.17 Å (Fig. S4A). Similar wobble geometry was observed between dGTP and dT in the dGTP/dT-containing structure (Fig. 1B). The purine base of the incoming dGTP is displaced into the cavity generated by the Y567A mutation (Fig. S4B). The wobble geometry between dGTP and the templating dT causes the pyrimidine base of dT to be tilted slightly toward the major groove. As a result, the distance between the O2 of dT and Cα of G568 is too great (3.6 Å) for a HB (Fig. S4B). In the dCTP/dA-containing complex, the incoming dCTP and templating dA adopt a wobble conformation similar to the dTTP/dG wobble base-pair (Fig. 1C). One direct inter-base HB was observed between the N3 of dCTP and N6-H of dA (Fig. S4C). In addition, the N1 of the templating dA is very likely to be protonated because the separation between the O2 of dCTP and the N1 of dA is only 2.75 Å, a distance too short for favorable polar-polar interactions (Fig. S4C, S5A). In contrast, the incoming dATP forms an inverted-wobble base-pair with the templating dC in the dATP/dC-containing complex (Fig. 1D). The N1 of dATP is within HB distance to both N3 of dC (2.99 Å) and N4 of dC (3.10 Å), suggesting that the N3 of templating dC is protonated forming a bifurcated HB to N1 of the incoming dATP (Fig. S4D, S5B). Thus, for the reasons stated, both the dCTP/dA- and dATP/dC-containing complexes, the templating bases (dC or dA) have a high probability of being protonated, although how this happens is not clear. Also, both templating bases recede into the DNA minor groove so that Cα-H of G568 can form a HB with O2 of dC or N3 of dA (Fig. S4).
(ii) Pyrimidine-Pyrimidine Mismatches in the NBP of the qm RB69pol
In both the dCTP/dT- and dTTP/dC-containing complexes, two highly ordered water molecules were observed at the interface of the base-pairs (Fig. 1E, 1F). One water molecule is hydrogen bonded to the O4 of dT (or dTTP) on one side and to N4-H of dCTP (or dC) on the other side. The second water molecule is hydrogen bonded to both N3-H of dT (or dTTP) and N3 of dCTP (or dC). Superposition of the Cα coordinates of these two complexes shows that: (i) the pyrimidine base of thymine overlays perfectly with the pyrimidine base of cytosine and; (ii) the two water molecules at the base-pair interface are also superimposable. A water molecule mediated inter-base HB was also observed in the dCTP/dC-containing complex. One ordered water molecule is hydrogen bonded to N3 of both the incoming dCTP and the templating dC (Fig. 1G). The HB distance between this water molecule and N3 of dCTP is 3.1 Å, and the HB distance between the same water molecule and N3 of dC is 2.6 Å. In contrast, no water molecules were observed at the interface of the dTTP/dT mismatch. Instead, the N3 hydrogen of the incoming dTTP forms a direct inter-base HB to O4 of the templating dT (Fig. 1H). This wobble geometry causes the base of templating dT to recede into the cavity created by the Y567 to Ala substitution. As a result, the O2 of the templating dT is 2.89 Å from the Cα hydrogen of G568 (Fig. S6D). In contrast, the distance between the O2 of dT (or dC) and Cα of G568 is too great for a HB in the dCTP/dT-, dTTP/dC- or dCTP/dC- containing complexes (Fig. S6). Overall, the space occupied by pyrimidine-pyrimidine mismatches in the NBP, except for the dTTP/dT mismatch, is greater than that required for a W-C base-pair because of the lack of direct HB and deviation from co-planarity between bases.
(iii) Purine-Purine Mismatches in the NBP of the qm RB69pol
Although we were expecting to find an anti-anti or syn-anti conformation for purine-purine mismatches, as reported for heteroduplex DNA,24 neither conformation was observed when these mismatches occupied the NBP of the RB69pol qm. When dA was the templating base and dATP (or dGTP) the incoming dNTP, dA flipped 180° out of its normal position so that it was now opposite the incoming dNTP. It was stabilized there by stacking on top of the indole ring of W574 (Fig. 1I, 1J, S6). As a result, the C1’-C1’ distance of the dATP/dA or the dGTP/dA mismatch was about 16.3 Å which is 5.8 Å greater than the normal 10.5 Å-C1’-C1’ distance for W-C base-pairs (Table 1). When the dATP/dA-containing complex was overlaid on the dGTP/dA-containing complex, the two incoming dNTPs and two templating dAs superimposed perfectly. In the dATP/dG- and dGTP/dG-containing structures, the templating dG did not flip backward, but shifted instead by 30° toward the DNA major groove (Fig. 1K, 1L, S7). In addition, the purine base of dG tilted 20° toward the penultimate base-pair (Fig. S7). One ordered water molecule was observed at the interface of the templating dG and the incoming dNTP. This water molecule was hydrogen bonded to N3 of dG on one side and to N1-H of dATP or to N1-H of dGTP on the other side. Together, these four structures suggest that the qm RB69pol is able to incorporate a purine-purine mismatch by removing the templating base from the NBP and generating an transient abasic templating site.
Pre-steady-state Kinetic Parameters for Incorporation of dNMPs into DNA by the qm RB69pol
To characterize the kinetic behavior of the qm RB69pol, we determined the pre-steady-state kinetic parameters with all sixteen base-pair combinations. As shown in Table 2, the maximum turnover rates (kpol) for incorporation of correct dNMPs ranged from 7 to 11 s−1, which are 15 to 38-fold lower than the values found with wt RB69pol. The apparent dissociation constants (Kd,app) for incorporation of correct dNTPs ranged from 4 to 11 μM, which are 6 to 10-fold lower than values observed with wt RB69pol. Consequently, the corresponding catalytic efficiencies (kpol/Kd,app) with the qm decreased 2 to 4-fold compared to wt RB69pol.
Table 2.
Pre-Steady-State Kinetic Parameters for wt and qm RB69pol with all 16 dNTP/dN combinations.(Included are two dPuTPs vs templating THF)
| kpol (s−1) | Kd,app (μM) | kpol/Kd,app (μM−1 s−1) | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| dNTP/dN | wt* | Qm | wt* | qm | wt* | qm |
|
| ||||||
| dTTP/dA | 270 | 7 ± 0.1 | 42 | 4 ± 0.5 | 6.4 | 1.7 |
| dATP/dA | 0.13 | 360 ± 28 | 810 | 750 ± 100 | 1.6 × 10−4 | 0.48 |
| dGTP/dA | 0.003 | 400 ± 50 | 780 | 2000 ± 430 | 3.8 × 10−6 | 0.2 |
| dCTP/dA | N.A.D | 26 ± 1 | N.A.D | 160 ± 24 | 1.7 × 10−4 | 0.17 |
|
| ||||||
| dGTP/dC | 160 | 11 ± 0.5 | 66 | 11 ± 2 | 2.4 | 1.0 |
| dATP/dC | 0.08 | 27 ± 0.5 | 1800 | 32 ± 4 | 4.4 × 10−5 | 0.85 |
| dTTP/dC | 0.09 | 15 ± 0.1 | 1700 | 31 ± 1 | 5.2 × 10−5 | 0.5 |
| dCTP/dC | N.A.D | 210 ± 20 | N.A.D | 1460 ± 270 | 3.0 × 10−7 | 0.14 |
|
| ||||||
| dCTP/dG | 200 | 7 ± 0.2 | 70 | 9 ± 1 | 2.9 | 0.8 |
| dATP/dG | 0.004 | 370 ± 9 | 910 | 320 ± 24 | 4.4 × 10−4 | 1.1 |
| dGTP/dG | N.A.D | 215 ± 6 | N.A.D | 390 ± 30 | 1.0 × 10−5 | 0.55 |
| dTTP/dG | N.A.D | 17 ± 0.5 | N.A.D | 69 ± 10 | 7.0 × 10−5 | 0.24 |
|
| ||||||
| dATP/dT | 220 | 8 ± 0.2 | 50 | 8 ± 0.9 | 4.4 | 1.0 |
| dCTP/dT | N.A.D | 14 ± 0.3 | N.A.D | 31 ± 3 | 6.0 × 10−6 | 0.45 |
| dTTP/dT | 0.015 | 5 ± 0.05 | 1400 | 16 ± 1 | 1.0 × 10−5 | 0.32 |
| dGTP/dT | 0.04 | 8 ± 0.6 | 730 | 650 ± 90 | 5.4 × 10−5 | 0.01 |
|
| ||||||
| dATP/THF | 0.3 | 14 ± 0.4 | 1400 | 5 ± 0.7 | 2.1 × 10−4 | 2.8 |
| dGTP/THF | 0.17 | 11 ± 1 | 1800 | 8 ± 1 | 9.4 × 10−5 | 1.4 |
|
| ||||||
N.A.D stands for not accurately determined, because the corresponding Kd,app value is greater than 2 mM, therefore only the kpol/Kd,app values is reliable. The kpol and Kd,app values for dNMP incorporation opposite dN by wt RB69pol were from Zhang.H.17 THF stands for tetrahydrofuran templating site (abasic site).
The most surprising feature of this RB69pol variant is that the (kpol) for incorporation of dATP or dGTP opposite dA or dG (purine-purine mismatches) were greater than 300 s−1 which is faster than the kpol for incorporation of a correct dNTP by wt RB69pol. Similar kpol values (210 s−1) values were observed when dCTP was the incoming dNTP and dC was the templating base. This is the only example, to the best of our knowledge, where a replicative DNA pol mutant incorporated an incorrect dNTP faster than a correct dNTP (Table 2). Although the kpol for purine-purine mismatch incorporation by the qm RB69pol was 104–105 fold greater than observed for wt RB69pol, the corresponding values of Kd,app between the two pols (wt and qm RB69pol) are within 3-fold of each other. Thus, the kpol/Kd,app for incorporation of purine-purine mismatches by the qm RB69pol is 103–104-fold greater than that of wt RB69pol, which enabled us to capture ternary complexes of the RB69pol qm containing purine-purine mismatches.
The kinetic parameters for incorporation of pyrimidine-pyrimidine mismatches by the qm RB69pol are similar to one another except for the dCTP/dC pair. In that case, the kpol is 210 s−1 but the Kd,app is 1.4 mM resulting in an incorporation efficiency of 0.14 μM−1s−1, which is at least 105-fold greater than that found for wt RB69pol (Table 2). For the other three pyrimidine-pyrimidine mismatches (dTTP/dC, dTTP/dT and dCTP/dT), the kpol values ranged from 5 to 15 s−1, and the Kd,app values were between 16 and 31 μM giving kpol/Kd,app values from 0.32 to 0.5 μM−1s−1 (104 to 105-fold greater than wt RB69pol). There was an asymmetric pattern for the incorporation kinetics of purine-pyrimidine mismatches by the qm RB69pol (dTTP opposite dG was inserted 24-fold more efficiently than dGTP opposite dT). Similarly, the efficiency for incorporation of dATP opposite dC was 5-fold greater for dCTP opposite dA. These differences are mainly due to Kd,aps which differ by 10-fold between dTTP/dG and dGTP/dT, and 5-fold between dATP/dC and dCTP/dA. Overall, the qm variant exhibits a decrease in base selectivity in the range of 103 to 106-fold compared to wt RB69pol.
The structures of qm RB69pol ternary complexes suggested that this pol may generate a transient abasic site and use this mechanism to incorporate purine-purine mismatches, as originally suggested by Wilson’s group with pol β.25 To see if this explanation would apply to the RB69pol qm, we determined the kpol and Kdapp for incorporation of a purine dNTP opposite an abasic site and found that the kpol values ranged from 11 to 14 s−1, which were 20 fold lower than the kpol for incorporation of purine-purine mismatches (Table 2). In fact, both the kpol and Kd,app values are very close to those found for the incorporation of correct dNTPs by the qm RB69pol. Thus, the transient abasic site mechanism alone cannot account for our results with the RB69pol qm variant.
Structural Basis for the Kinetic Behavior of the RB69pol qm
Our previous structural studies with wt RB69pol have shown that there is a rigid HB network at the minor groove of the primer-template junction that involves several residues of the pol.16, 18–20, 26 This network includes four ordered water molecules (labeled w1 to w4 in Fig. 2A), and the side chain γ-hydroxyl groups of Y416, Y567 and T622 (Fig. 2A). The Y567 to Ala substitution in the qm RB69pol not only generates a cavity below the templating base but also disrupts this HB network. As shown in Fig 2B, the water molecule (w1) was absent in the NBP of the qm RB69pol. In addition, the carboxyl group of D411 is coordinated to both metal ion A and B in the wt structure, but only to metal ion B in the qm structure (Fig. 2D, 2E). Superposition of the dCTP/dG-containing wt and qm structures show that: (i) the templating base recedes into the DNA minor groove by 0.6 Å (Fig. 2C); (ii) the main chain of A567 shifts 0.6 Å vertically toward the minor groove and 0.8 Å laterally toward the Y416 side chain relative to the situation that obtains with the wt pol (Fig. 2C); (iii) the side chain of L412 tilts toward the cavity generated by the L415 to Ala substitution (Fig. 2F); (iv) the side chain of D411 adopts a rotamer conformation that differs from its orientation in wt RB69pol (Fig. 2F); (v) metal ion A shifts vertically by 0.4 Å away from the primer-terminus (Fig. 2F); (vi) the carboxyl group of D623 also shifts vertically by 0.3 Å away from the incoming dNTP (Fig. 2F).
Figure 2.
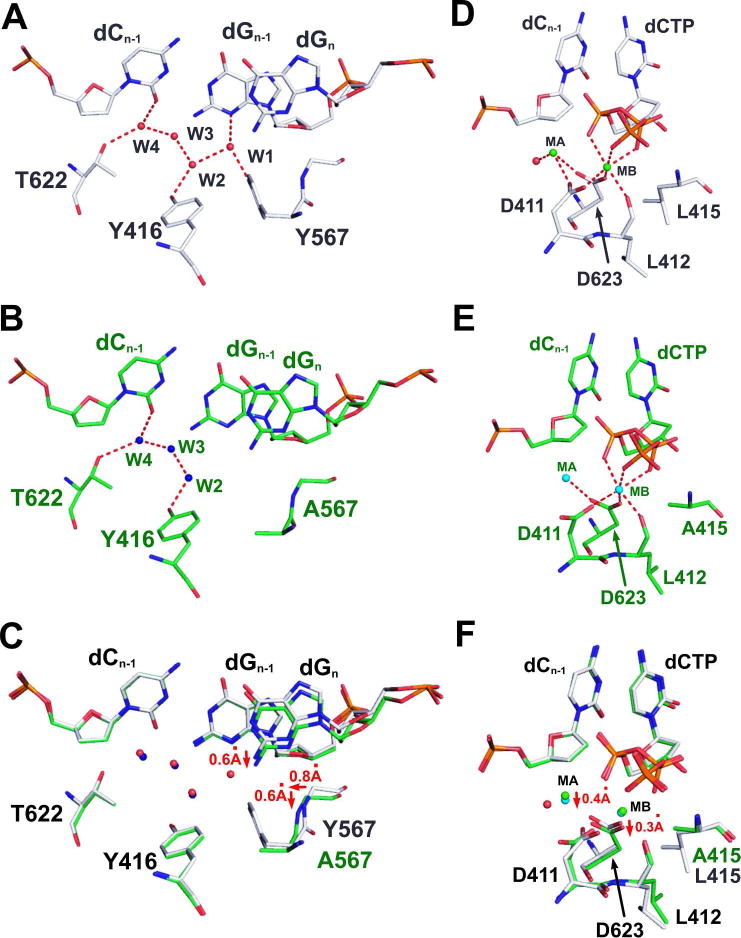
HB networks at the minor groove of the P/T junction and at the two metal ion binding sites. (A) Overview of the rigid HB network with ordered water molecules labeled as W1 through W4 in the NBP of wt RB69pol. (B) Overview of HB network in the NBP of qm RB69pol. (C) Superposition of A (shown in grey sticks) and B (shown in green sticks). (D) HB network at the metal ion binding site of wt RB69pol. (E) HB network at the metal ion binding site of qm RB69pol. (F) Superposition of D (shown in grey sticks) and E (shown in green sticks).
The differences observed between the dCTP/dG-containing qm and dCTP/dG-containing wt ternary complexes likely extend to the differences between qm and wt structures containing the other three Watson-Crick base-pairs. Metal ion A is responsible for bringing the 3’ hydroxyl group at the primer-terminus closer to the α-phosphorus atom (Pα) of the incoming dNTP enabling phosphodiester bond formation. The coordination bond length of metal ion A determines how close the 3’-OH at the primer terminus can approach Pα. Thus, the downward shift of metal ion A from its optimal position in the wt pol would be expected to reduce the rate of phosphodiester bond formation. This could explain why the maximum turnover rates for incorporation of a correct dNTP by the qm RB69pol are 20 to 30-fold lower than observed with wt RB69pol. Because the turnover rate decreases so dramatically for correct dNTPs, the open and closed forms of the ternary complex very likely approach equilibrium before chemistry. As a consequence the Kdapp values for correct dNTP incorporation by the qm were 6 to 10-fold lower than the Kdapp values found with the wt enzyme.27–28
One puzzling aspect of the kinetic data for the qm is that the kpol for incorporation of purine-purine mismatches and incorporation of dCTP opposite dC is 210 s−1, even though metal ion A is shifted so that it is no longer coordinated to D411 in the qm structures. Previous studies by Berdis, et al have shown that base stacking plays an important role in stabilizing the binding of an incoming dNTP,29 but here the incorrect incoming dNTP stacks equally well with the penultimate base-pair as the correct incoming dNTP (Fig. S8). In fact, it is the templating bases of the mismatches that failed to stack well against the penultimate base-pair, especially for Pu-Pu mismatches. Therefore, base stacking alone cannot explain the high kpol values for Pu-Pu misincorporation. Among the sixteen base-pairs, HB interactions between the templating base and the incoming dNTPs were as follows: (i) nine of these nascent base-pairs have at least one direct inter-base HB; (ii) two of the nascent base-pairs have two inter-base HBs mediated by two water molecules; (iii) the dATP/dG, dGTP/dG and dCTP/dC base-pairs have one inter-base HB mediated by a single water molecule; (iv) the dATP/dA and dGTP/dA base-pairs are the only ones without inter-base HBs. Thus, the four purine-purine mismatches, together with the dCTP/dC base-pair, have the weakest inter-base interactions, which is consistent with the high Kd,app values. Another common feature among these five structures is that the distance between N3 or O2 of the templating base and Cα of G568 is too great for a HB. Also, the B factors of the nucleotide residues at the n and n+1 positions of the template strand in these five structures are twice as high as the average B factors for the ternary complex itself (Table S5). In contrast, the B factors for the templating base in the other nine structures are lower than the average B factors for the remaining ternary complexes (Table S5). Thus, it is possible that dynamic interactions between the base-pairs of purine-purine and dCTP/dC mismatches may cause a slight vertical shift of incoming dNTPs after Fingers closing, which could restore the optimal octahedral geometry of metal ion A allowing accelerated rates of nucleophilic attack by the 3’-OH on the α phosphorus atom of the incoming dNTP. This speculation is consistent with our kinetic data for incorporation of dATP or dGTP opposite an abasic site, where the kpol is only 11 s−1, rather than the >300 s−1 observed for incorporation of purine-purine mismatches. With respect to the correct W-C base-pairs and the other seven mismatches, the interactions between the incoming dNTPs and the templating base is relatively strong due either to direct inter-base HB or to the involvement of at least two HBs mediated by two ordered water molecules. As a result, the correct W-C base-pairs and the other seven mismatches can easily fit into the enlarged NBP of the qm RB69pol without requiring additional adjustments of either the incoming dNTP or the templating base after Fingers closing.
Mismatches in the NBP of the qm Polymerase Differ from Mismatches in the Penultimate Position of P/Ts Complexed with the Bst pol fragment (BF)
In the twelve structures of covalently incorporated DNA mismatches complexed with the Bacillus DNA pol I fragment (BF), it was found that mismatches at the penultimate base-pair position of the P/T duplex DNA disrupted the insertion site and stalled the pol.14 Our structures of mismatches at the NBP of the RB69pol qm now provide an excellent opportunity to examine how the environment of the pol active site affects mismatched base-pair interactions. Among the twelve mismatches at penultimate base-pair position (position n-1) of the P/T complexed with the BF, only six are well ordered, the others are either not base-paired or are disordered. For this reason we have only compared the six well ordered mismatches at the n-1 position of the P/T-BF binary complex with the corresponding mismatches in the NBP of the qm RB69pol. To simplify this comparison, we denote the penultimate base-pairs in BF as dN/dN whereas the dNTP/dN notation refers solely to the nascent base-pairs in the RB69pol qm.
Among the purine-purine mismatches, dA/dG retains an anti-anti conformation with two direct inter-base HBs (Fig. 3B). This anti-anti dA/dG conformation has also been reported for an isolated heteroduplex DNA.24 Unlike dA/dG, the dG/dG mismatch adopts a syn-anti conformation, as the primer base rotates 180°, relative to the sugar moiety, into a syn conformation (Fig. 3D). Interestingly, neither the anti-anti nor the syn-anti conformation was observed for the nascent dATP/dG or dGTP/dG mismatches in the RB69 pol qm complex. Instead, they both have only one inter-base HB mediated by a single ordered water molecule (Fig. 3A, 3B).
Figure 3.
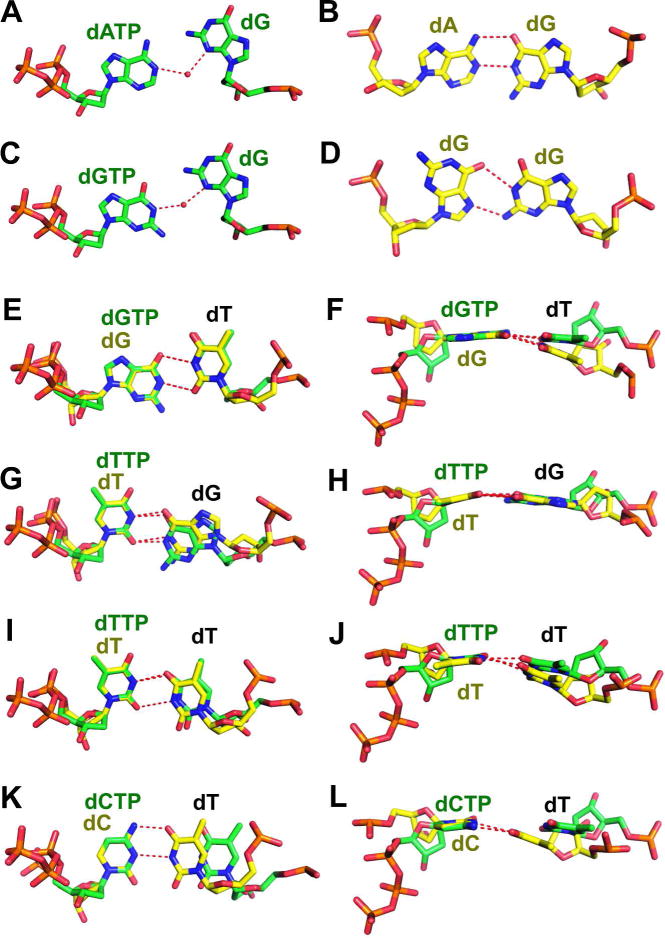
Mismatches in the NBP of qm RB69 pol (shown in green sticks) and in the penultimate position of P/T complexed with BF (shown in yellow sticks): (A) dATP/dG (RB69pol); (B) dA/dG (BF);14 (C) dGTP/dG (RB69pol); (D) dG/dG (BF);14 (E) Superposition of dGTP/dT (RB69pol) with dG/dT (BF); (F) orthogonal view of E; (G) Superposition of dTTP/dG (RB69pol) with dT/dG (BF); (H) orthogonal view of E; (I) Superposition of dTTP/dT (RB69pol) with dT/dT (BF); (J) orthogonal view of I; (K) Superposition of dCTP/dT (RB69pol) with dC/dT (BF); (L) orthogonal view of K.
For purine-pyrimidine mismatches, both dGTP/dT in the qm and dG/dT in BF are in a wobble conformation with two inter-base HBs (Fig. 3E). Overlaying these two mismatches showed that the purine base of dG was not coplanar with the pyrimidine base of dT in the dG/dT mismatch (Fig. 3F). In contrast, the base of dGTP was perfectly aligned and coplanar with the dT in the qm (Fig. 3F). Like the dGTP/dT and dG/dT mismatches, both dTTP/dG and dT/dG mismatches adopt wobble conformations (Fig. 3G, 3H). However the major difference is that the templating dG of the dTTP/dG mismatch has receded more deeply into the DNA minor groove than dT/dG in BF. The inter-atomic distance between C1’ of dTTP and N2 of dG is 5.5 Å, and the distance between C1’ of dT and N2 of dG is 6.1 Å.
For pyrimidine-pyrimidine mismatches, a wobble conformation was observed for both dTTP/dT and dT/dT. Two inter-base HBs were formed between the dT/dT pair in duplex DNA, but only one inter-base HB was observed for dTTP/dT (Fig. 3I). The distance between O2 of dTTP and N3 of dT, 3.6 Å, is too great for a HB. Similar to dG/dT in the DNA duplex, the two dT bases are not coplanar (Fig. 3J). By comparison, the pyrimidine base of dTTP is in the same plane as the base of the templating dT (Fig. 3J). Although the dT/dT pair in BF resembles dTTP/dT in the qm, the conformation of dC/dT in BF differs significantly from the nascent dCTP/dT pair in the qm. Two direct inter-base HBs were observed with dC/dT in BF (Fig. 3K). In contrast, the two inter-base HBs formed between dCTP/dT were mediated by two water molecules in the qm. The distance between N4 of dCTP and O4 of dT is 5.4 Å, whereas the distance between N4 of dC and O4 of dT is 2.7 Å. In addition, the bases of dC and dT deviate somewhat more from coplanarity compared to dCTP/dT (Fig. 3L). Overall, mismatches at the n-1 position in the BF P/T complex are highly constrained by the phosphodiester-linkages present in the duplex but absent in the nascent base-pairs. Thus, the interactions of mismatched bases in the NBP of the qm pol are very different from mismatches in the duplex portion of the P/T complexed with BF.
Repositioning of the Templating Nucleotide Residue Plays an Important Role in Accommodating Incorrect dNTPs in a qm Ternary Complex
With these sixteen qm structures, we were able to compare base-pairs with the same incoming dNTP and four different templating bases and vice-versa to investigate variations of base-pair interactions. After superimposing the four different dATP-containing qm RB69pol ternary complexes, we were surprised to find that the four incoming dATPs overlaid perfectly with one another (Fig. 4A) but the conformation of the four corresponding templating nucleotide residues varied dramatically. Whereas the templating dT is the W-C partner of an incoming dATP and had the expected, normal conformation; the templating dC almost overlaps with the templating dT (Fig. 4C); the templating dG shifted by 20° toward the DNA major groove (Fig. S7); and the templating dA flipped completely out of the NBP (Fig. S3C, S3D). A similar scenario was also observed for each of the other four dNTP-containing ternary complexes (Fig. 4A, 4B, 4D, 4E, 4F). Overall, the conformation of incoming dNTP was more rigid than the conformation of the corresponding templating residue. The same conclusion was reached when we superimposed four structures with the same templating residue and different incoming dNTPs. As shown in Figure 4E–4H, the four different incoming dNTPs opposite the same templating base can be almost perfectly overlaid. In particular, the sugar moieties and triphosphate tails of four different incoming dNTPs were completely superimposable. In contrast, the conformations of the same templating bases varied dramatically.
Figure 4.
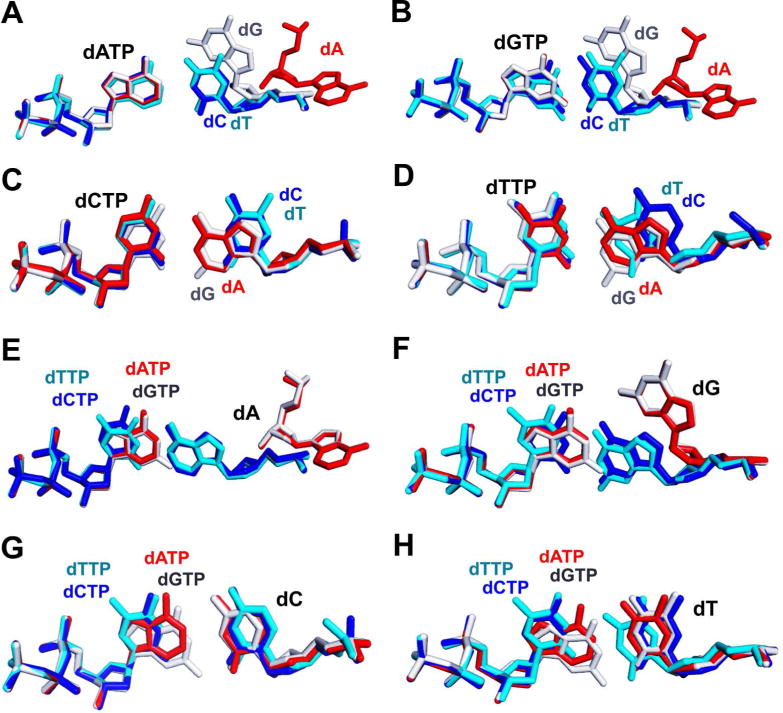
Superposition of four qm structures with the same incoming dNTP (templating dA shown in red stick, templating dG shown in white stick, templating dC shown in blue stick and templating dT shown in cyan): (A) four dATP/dN-containing structures; (B) four dGTP/dN-containing structures; (C) four dCTP/dN-containing structures and (D) four dTTP/dN-containing structures. Superposition of four qm structures with the same templating base dN (dATP shown in red stick, dGTP shown in white stick, dCTP shown in blue stick and dTTP shown in cyan): (E) four dNTP/dA-containing structures; (F) four dNTP/dG-containing structures; (G) four dNTP/dC-containing structures and (H) four dNTP/dT-containing structures.
The rigidity of incoming dNTPs in the NBP of the qm RB69pol is supported by our structural data. As shown in Figure 5A–5B, the ribosyl moiety of the incoming dNTP stacks on top of the phenyl ring of Y416. Also, the backbone amides of S414, L415, and Y416 are hydrogen bonded to the γ, β-phosphates and the 3’-OH of the incoming dNTP. Substitution of L415 with Ala did not affect the rigidity of the incoming dNTPs, as the backbone amide of A415 still forms HBs with the β-phosphate of the incoming dNTPs. Therefore, the flexibility of the phosphodiester backbone or glycosidic bond of the templating base plays a critical role in allowing it to adopt different conformations opposite a rigidly anchored incoming dNTP. In the NBP of wt RB69pol, the side chain of Y567 lies adjacent to the minor groove of the templating base, thus preventing it from receding into the DNA minor groove (Fig. 5C). In contrast, the replacement of Y567 with Ala in the qm RB69pol generates a large cavity right below the minor groove of the templating base (Fig. 5D). In addition, although the side chain of L561 is at the back of major groove of the templating base, it does not block the templating base from shifting toward major groove. Replacing L561 with Ala, together with the Y567 to Ala and S568 to Gly substitutions, provides the phosphodiester backbone or glycosidic bond of the templating base with a greater degree of freedom, allowing it to shift its position in response to different mismatched incoming dNTPs. Thus, repositioning of the templating nucleotide residues plays an important role in accommodating incorrect dNTPs in a qm complex. Conversely, the wt RB69pol rigidly anchors the templating base by the hydroxyl side chain of S565 via a nonpolar-polar interaction.26 Therefore, the same mismatch in wt Rb69pol now requires repositioning of the incoming incorrect dNTP. This results in misalignment of incorrect dNTP with the 3’-OH at the primer terminus, and furthermore leads to their low incorporation efficiency.
Figure 5.
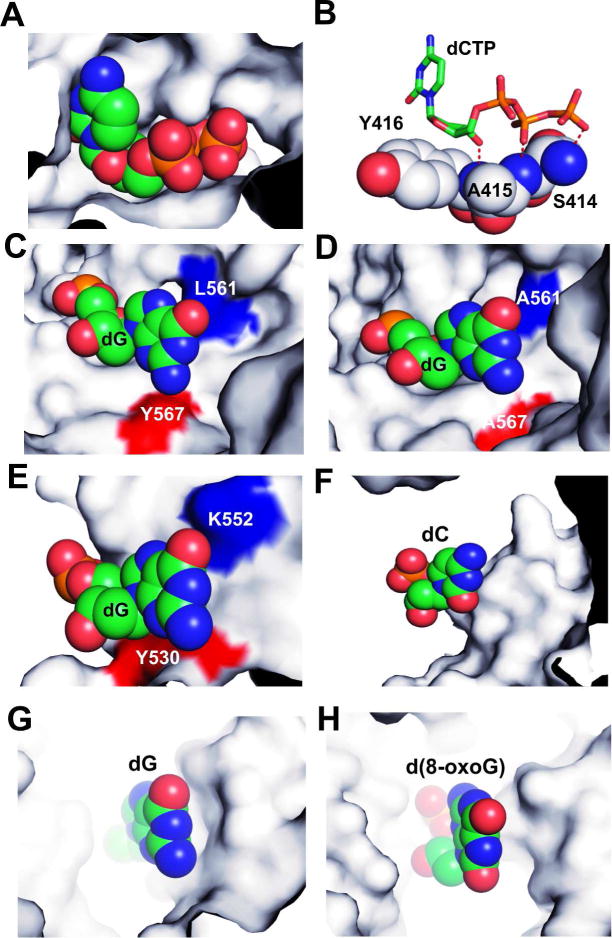
The NBP of various pols in a space-filling mode. (A) the NBP surrounding the incoming dCTP of qmRB69pol. (B) HB interactions between dCTP and protein side chains at the NBP of qm RB69pol. (C) the NBP around the templating dG of wt RB69pol. (D) the NBP around the templating dG of qm RB69pol. (E) the NBP around the templating dG of T7 DNA pol. (F) the NBP around the templating dC of pol Dbh. (G) the NBP around the templating dG of Dpo4. (F) the NBP around the templating 8-oxoG of pol κ.
The rigidity of the NBP around the templating residue has also been observed in other high fidelity pols, such as T7 DNA pol, a member of the A family.30–31 As shown in Figure 5E, the side chains of Y530 and K522 of T7 DNA pol are located right at the minor and major grooves of the templating residue. Since Y567 and Y530 are highly conserved residues in both the B and A families,22 the rigidity of the NBP around the templating residue is likely to be a common feature of high fidelity pols. In contrast, the templating residue has greater freedom in the NBP of repair pols, such as Dbh pol,32 Dpo4 33 and pol κ.34 As shown in Figure 5, there are no protein side chains directed at either the minor or major groove of the templating base in the NBPs of pol Dbh (Fig. 5F), Dpo4 (Fig. 5G) and pol κ (Fig. 5H). Therefore, repositioning of the templating nucleotide residue in the NBP to accommodate an incorrect dNTP maybe a general feature of repair pols.
The Majority of Mismatches Can be Modeled into the NBP of wt RB69pol without Steric Clashes
It has been suspected that a high fidelity pol rejects an incorrect dNTP because mismatched base-pairs would sterically clash with protein side chains in the NBP. With all twelve mismatched structures now available, we have been able to superimpose each mismatch-containing structure with a high resolution structure of a dGTP/dC ternary complex of wt RB69pol to see if the mismatched base-pair fit into the NBP of wt RB69pol. Unexpectedly, only three of the twelve mismatches (dTTP/dG, dGTP/dT and dTTP/dT) actually clash with NBP side chains (Fig. 6A, 6B, 6C), whereas the rest can be modeled perfectly well into the NBP of wt RB69pol. As shown in Figure 6A and 6B, the base of the templating dG or the incoming dGTP shifts toward the minor groove to form HBs with dTTP or dT. This wobble geometry leads to a steric clash between the guanine base and the side chain of Y567. Similarly, the wobble geometry of the dTTP/dT base-pair causes a steric clash between the thymine base and the backbone carbonyl of G568 (Fig. 6C). In contrast, the O2 or the N3 of the templating base is within HB distance of the Cα of G568 for the dCTP/dT, dTTP/dC, dCTP/dA and dATP/dC mismatches (Fig. 6D, 6E, 6G, 6H). Also the distance between the O2 or the N3 of the templating base and Cα of G568 is too great to form a HB with the remaining five mismatched base-pairs (dCTP/dC, dATP/dG, dGTP/dG, dATP/dA and dGTP/dA). Based on this, it would seem that wt RB69pol should be able to adopt a closed conformation with mismatched base-pairs, except for the dTTP/dG, dGTP/dT and dTTP/dT pairs. The intriguing question then is why are incorrect dNTPs incorporated so inefficiently by the wt RB69pol?
Figure 6.
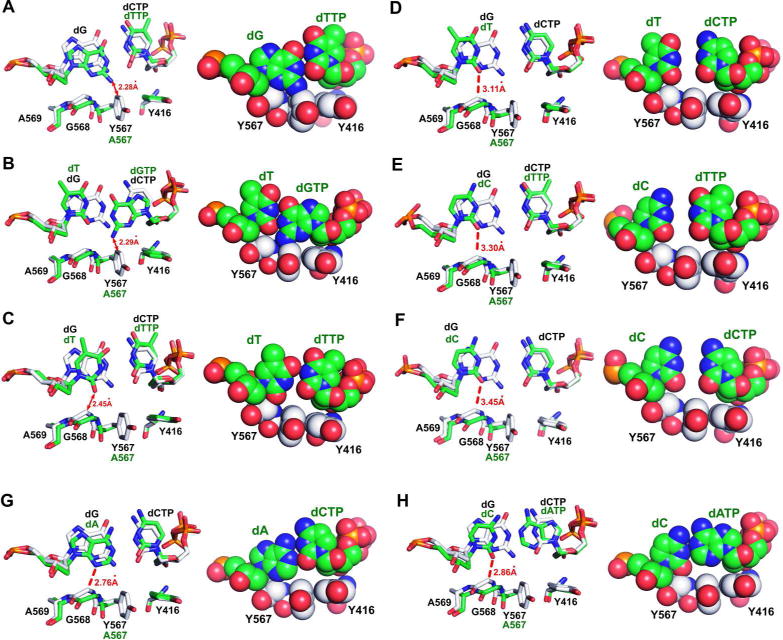
Superposition of mismatch-containing structures of qm RB69pol onto the dCTP/dG-containing wt RB69pol (stick mode in the left and space-filling mode in the right). The structure with a mismatch is shown in green and the wt structure is shown in white. (A) dG/dTTP; (B) dT/dGTP; (C) dT/dTTP; (D) dT/dCTP; (E) dC/dTTP; (F) dC/dCTP; (G) dA/dCTP and (H) dC/dATP.
We propose several possible reasons to account for this, the most probable being that wt RB69pol has a very tight, geometrically constrained NBP with a rigid HB network at the minor groove of the P/T duplex. Although the majority of the mismatches observed at the insertion site of the qm RB69pol can be modeled into the NBP of wt RB69pol without steric clashes, the closed ternary complex of wt pol with mismatched base-pairs is not stable, perhaps because the energy cost in forming the closed complex is too high. It seems that only a W-C base-pair can significantly decrease this energy barrier. Based on the set of structures described above we suggest some other reasons for the inefficient incorporation of incorrect dNTPs by wt RB69pol as follows: (i) steric clashes with protein side chains in the closed ternary complex; (ii) weak interactions between an incorrect dNTP and the templating base; (iii) presence of a large gap between bases; (iv) burying a protonated base in the highly hydrophobic insertion site. Obviously, the dTTP/dG, dGTP/dT and dTTP/dT mismatches belong in the first category because either the purine or pyrimidine bases will cause a steric clash with protein side chains (Fig. 6A–6C). The four purine-purine mismatches (dATP/dG, dGTP/dG, dATP/dA an dGTP/dA) belong in the second category because the templating base either shifts toward the major groove of DNA or is flipped completely out of the NBP generating a cavity opposite the incoming dNTP. As a result, there are no strong interactions between the incoming dNTP and the templating base. The “tightness of fit” idea, originally proposed by Kool et al, stated that a tight complementary fit between the nascent base-pair and NBP in pols is the most important factor governing selection of a dNTP for incorporation.35 Although that idea cannot account for all examples of base discrimintation,15–16 it is clear that a snug fit between the base-pair and the NBP is as important as Watson-Crick HB for base discrimination. As shown in Figure 6D–6F, for dCTP/dT, dTTP/dC and dCTP/dC mismatches, there is a big gap between the incoming dNTP and the templating base, which could destabilize the ternary complex.31, 36 This is probably the main reason that incorporation efficiency is so low for pyrimidine-pyrimidine mismatches.
The dCTP/dA and dATP/dC mismatches belong in the fourth category. The templating bases appeare to be protonated in both structures based on distance and geometry. Unlike the quadruple mutant, the NBP is completely desolvated at the minor groove of nascent base-pairs in the wt RB69pol ternary complex. So, burying a positive charge in the highly hydrophobic NBP of the wt pol exacts a considerable energetic penalty which would be sufficient to destabilize the corresponding ternary complex. Thus, dCTP/dA and dATP/dC mismatches are not incorporated efficiently by wt RB69pol. Previously, Beese et al have reported a dCTP/dA mismatch that included a minor tautomer with Watson-Crick geometry at the NBP of BF.12 If tautomerization within the pol complex involves a transient protonation/deprotonation step,37 then rejection of protonated bases in the NBP of wt pol could also serve as a mechanism to reject tautomerization. Although, these possibilities appear to adequately account for the degree of base discrimination exhibited by wt RB69pol, they do not exclude other explanations that may prove equally valid.
EXPERIMENTAL PROCEDURES
Enzyme and DNA substrates
The qm RB69pol, in an exonuclease-deficient background (D222A and D327A), was constructed, expressed, purified, and stored as previously described. 19, 26 All oligonucleotides were synthesized at the Yale Keck facilities and purified via polyacrylamide gel electrophoresis. The sequence of the primertemplates (P/Ts) used in this study are shown in Fig S9 and S10. For kinetic studies, the primer was 5’ labeled with 32P as previously described.19 For crystallization, dideoxy-terminated primers were used to prevent phosphoryl transfer.
Crystallization of dNTP/dN-containing Ternary Complexes of qm RB69pol
The qm RB9pol was mixed in an equimolar ratio with a freshly annealed dideoxy-terminated P/T to give a final protein concentration of 120 μM. The incoming dATP, dCTP or dTTP was then added to give a final concentration of 2 mM. The complex with an incoming dGTP was obtained using the soaking-replacement method from the dATP-containing crystals. A solution of 150 mM CaCl2, 10% (w/v) PEG 350 monomethyl ether (MME), and 100 mM sodium cacodylate pH 6.5 was mixed with an equal volume of the protein complex. Square rod-shaped crystals grew within a week at 20 °C and had typical dimensions of approximately 200 × 50 × 50 μm. Crystals were transferred to a cryoprotectant solution where the concentration of PEG 350 MME was increased to 30% w/v prior to freezing in liquid nitrogen.
Data Collection and Structure Determination
Data were collected at 110K using the synchrotron radiation sources at beam line 24ID-E of the Northeast Collaborative Access Team (NECAT), Advanced Photon Source (APS), Argonne National Laboratory (ANL, Chicago, IL). Data were processed using the HKL2000 program suite.38 The structures were determined by molecular replacement with a previously determined structure of dCTP/dG-containing wt RB69pol (3NCI), and refined using REFMAC5.39–40 The P/T duplexes and dNTPs were built using the program COOT.41 Structure refinement statistics are summarized in Table 1–4. All figures were prepared in Pymol (Schrodinger, LLC).42
Chemical Quench Experiments
All experiments were performed at 23 °C using a KinTek RFQ-3 instrument with a buffer solution containing 66 mM Tris-HCl (pH 7.4) and 10 mM MgSO4. A 10-fold excess of RB69pol over the P/T was used to ensure single-turnover conditions. The final concentration of pol after mixing was 1 μM and the [P/T] was 83 nM. Products were separated using 15% denaturing 8 M urea polymacrylamide sequencing gels, imaged on an MD Strom 860 imager (Molecular Imaging), and quantified using ImageQuaNT software. For each Kd,app and kpol determination, seven different [dNTP] were used. Kd,app was defined as the [dNTP] that provides an incorporation rate that is one half of the maximum rate attained at saturating concentrations of substrate. Kpol was defined as maximum rate of dNMP incorporation. Data from the single turnover experiments were fit to a single exponential equation as previously described.19 The corresponding standard deviations were calculated from data fitting using Grafit5.0 (Erithacus Inc.).
ACCESSION NUMBERS
Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4FJ5, 4FJ7, 4FJ8, 4FJ9, 4FJG, 4FJH, 4FJI, 4FJJ, 4FJK, 4FJL, 4FJM, 4FJN, 4FJX, 4FK0, 4FK2 and 4FK4.
Supplementary Material
Acknowledgments
We thank the staff of the NE-CAT beamline 24-ID-E at the Advanced Photon Source of Argonne National Laboratory. This work was supported by grants to W.H.K from NIH RO1-GM063276-09 and to J.W from SCSB-GIST.
Footnotes
This work was supported by NIH RO1-GM063276-09 (to W.H.K.) and SCSB-GIST (to J.W.).
SUPPLEMENTAL DATA
Supplemental Data include 10 figures and 5 tables, and can be found with this article online.
References
- 1.Kunkel TA, Bebenek K. Biochim Biophys Acta. 1988;951:1. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 2.Drake JW. Proc Natl Acad Sci U S A. 1991;88:7160. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flohr T, Dai JC, Buttner J, Popanda O, Hagmuller E, Thielmann HW. Int J Cancer. 1999;80:919. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 4.Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, Preston BD. Nat Med. 2001;7:638. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 5.Venkatesan RN, Treuting PM, Fuller ED, Goldsby RE, Norwood TH, Gooley TA, Ladiges WC, Preston BD, Loeb LA. Mol Cell Biol. 2007;27:7669. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Nat Struct Mol Biol. 2009;16:979. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echols H, Goodman MF. Annu Rev Biochem. 1991;60:477. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel TA, Bebenek K. Annu Rev Biochem. 2000;69:497. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 9.Joyce CM, Benkovic SJ. Biochemistry. 2004;43:14317. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 10.Kunkel TA. J Biol Chem. 2004;279:16895. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 11.Bebenek K, Pedersen LC, Kunkel TA. Proc Natl Acad Sci U S A. 2011;108:1862. doi: 10.1073/pnas.1012825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Hellinga HW, Beese LS. Proc Natl Acad Sci U S A. 2011;108:17644. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirmizialtin S, Nguyen V, Johnson KA, Elber R. Structure. 2012;20:618. doi: 10.1016/j.str.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson SJ, Beese LS. Cell. 2004;116:803. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 15.Xia S, Konigsberg WH, Wang J. J Am Chem Soc. 2011;133:10003. doi: 10.1021/ja2021735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia S, Eom SH, Konigsberg WH, Wang J. Biochemistry. 2012;51:1476. doi: 10.1021/bi2016487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Beckman J, Wang J, Konigsberg W. Biochemistry. 2009;48:6940. doi: 10.1021/bi900422b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia S, Wang M, Blaha G, Konigsberg WH, Wang J. Biochemistry. 2011;50:9114. doi: 10.1021/bi201260h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia S, Wang M, Lee HR, Sinha A, Blaha G, Christian T, Wang J, Konigsberg W. J Mol Biol. 2011;406:558. doi: 10.1016/j.jmb.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia S, Christian TD, Wang J, Konigsberg WH. Biochemistry. 2012;51:4343. doi: 10.1021/bi300416z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia S, Vashishtha A, Bulkley D, Eom SH, Wang J, Konigsberg WH. Biochemistry. 2012;51:4922. doi: 10.1021/bi300296q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braithwaite DK, Ito J. Nucleic Acids Res. 1993;21:787. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai YC, Johnson KA. Biochemistry. 2006;45:9675. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennard O, Hunter WN. Q Rev Biophys. 1989;22:327. doi: 10.1017/s0033583500002997. [DOI] [PubMed] [Google Scholar]
- 25.Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Mol Cell. 2008;30:315. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Xia S, Blaha G, Steitz TA, Konigsberg WH, Wang J. Biochemistry. 2011;50:581. doi: 10.1021/bi101192f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellinger MW, Johnson KA. Proc Natl Acad Sci U S A. 2010;107:7734. doi: 10.1073/pnas.0913946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellinger MW, Johnson KA. Biochemistry. 2011;50:5008. doi: 10.1021/bi200204m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reineks EZ, Berdis AJ. Biochemistry. 2004;43:393. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Dutta S, Doublie S, Bdour HM, Taylor JS, Ellenberger T. Nat Struct Mol Biol. 2004;11:784. doi: 10.1038/nsmb792. [DOI] [PubMed] [Google Scholar]
- 31.Kim TW, Brieba LG, Ellenberger T, Kool ET. J Biol Chem. 2006;281:2289. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 32.Wilson RC, Pata JD. Mol Cell. 2008;29:767. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 33.Trincao J, Johnson RE, Wolfle WT, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Nat Struct Mol Biol. 2004;11:457. doi: 10.1038/nsmb755. [DOI] [PubMed] [Google Scholar]
- 34.Suh HY, Kim JH, Woo JS, Ku B, Shin EJ, Yun Y, Oh BH. Proteins. 2012;80:2099. doi: 10.1002/prot.24093. [DOI] [PubMed] [Google Scholar]
- 35.Kool ET, Sintim HO. Chem Commun (Camb) 2006:3665. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 36.Kim TW, Delaney JC, Essigmann JM, Kool ET. Proc Natl Acad Sci U S A. 2005;102:15803. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia S, Beckman J, Wang J, Konigsberg WH. Biochemistry. 2012;51:4609. doi: 10.1021/bi300241m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Methods Enzymology. 1997:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J Appl Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.The PyMOL Molecualr Graphics System, V1.2rpre. Vol. 2010. Schrodinger, LLC; New York: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


