Abstract
The human body is colonized with a diverse resident microflora that includes viruses. Recent studies of metagenomes have begun to characterize the composition of the human 'virobiota' and its associated genes (the 'virome'), and have fostered the emerging field of host-virobiota interactions. In this Perspective, we explore how resident viruses interact with the immune system. We review recent findings that highlight the role of the immune system in shaping the composition of the virobiota and consider how resident viruses may impact host immunity. Finally, we discuss the implications of virobiota–immune system interactions for human health.
Subject terms: Virology, Microbiota
Main
The human body is colonized by commensal microorganisms that encompass diverse phyla from the three domains of life: Eukarya, Archaea and Bacteria. Most of these microorganisms reside at body surfaces that are in direct contact with the environment, including the intestine, skin and respiratory tract. Research efforts over the past two decades have focused primarily on the bacterial component of the human microbiota and its associated genes (the 'microbiome'). These efforts have yielded a wealth of insight about the composition of human-associated bacterial communities, how these resident bacteria interact with the immune system and how bacteria–immune system interactions are altered in disease1,2.
Recently, it has become apparent that the microbiota of healthy humans also includes viruses, termed the 'virobiota'3. Much of our current knowledge of the virobiota is derived from metagenomics, studies in which the DNA (and sometimes RNA) content of a microbial community is sequenced4,5,6. These studies have revealed that the human microbiome includes many viral genes (the 'virome'). The intestine and the skin, for example, are both associated with viruses that replicate in eukaryotic cells (eukaryotic viruses), and viruses that replicate in bacteria (bacteriophages or phages)5,6,7. The intestinal microbiota also contains viruses that infect plants, which are likely associated with the host diet5,6,8.
Bacteria that inhabit the intestine and skin are generally regarded as stable residents that confer metabolic and/or immune benefits to their hosts9. It is therefore reasonable to ask whether viruses also can be stably associated with healthy human tissues. In the case of bacteriophages, a persistent, nonpathogenic association seems possible as viral replication occurs in bacterial hosts, which can themselves be stable members of the microbiota. In contrast, the relationship of eukaryotic viruses to their human hosts is much less clear. Such viruses require host cells to replicate and in most cases trigger innate and/or adaptive immune responses after entry into host cells. Thus, the eukaryotic viruses sampled in metagenomic studies could include those from acute, acute recurrent or chronic infections, or newly emerged latent viruses. In the absence of detailed longitudinal metagenomic studies of the virobiota, it is difficult to know at this point whether the human eukaryotic virobiota includes truly resident viruses that are stably associated with healthy host tissues.
In this Perspective we will discuss how the virobiota interacts with the immune system and how this could impact host health. We will focus most of our discussion on viruses associated with tissues that interface with the external environment (for example, intestine, skin and respiratory tract). It is important to recognize that the study of the virobiota is an emerging field and that our understanding of how these viruses interact with the immune system is currently minimal. Thus, some of our discussion will be speculative by necessity. However, our aim is to provide a framework for thinking about virobiota–immune system interactions in mammalian hosts and to motivate experimental studies of these interactions.
Bacteriophages that associate with host tissues
Metagenomic studies of microbiota at various tissue sites have revealed that many of the viruses associated with healthy human tissues are bacteriophages5,6,10. Sequencing of the metagenome from human fecal material has revealed that many phages associate with the gastrointestinal microbiota, which can harbor up to 1014 bacterial cells. Phage populations diversify in the intestine as new members of the bacterial community are introduced, suggesting that phage diversity and bacterial diversity are linked11.
Intestinal phages are genetically and morphologically heterogeneous. They include both lysogenic prophages, which are stably integrated into the bacterial chromosome, and lytic phages, which actively infect and lyse bacteria12. Prophages can undergo lytic induction to produce infectious phage particles when bacterial cells encounter environmental stressors or are stimulated by nutrients13,14. Metagenomic sequencing has revealed a few dominant families of intestinal phages that include double- and single-stranded DNA phages5,15. Although there is minimal variation of intestinal phage populations in individuals over time, there is substantial variation between individuals5,6, even when those individuals have similar bacterial community structures9.
Phages likely have a profound impact on the composition and functional properties of the bacterial microbiota, which in turn could shape development and function of the immune system. First, phages may serve as important reservoirs of genetic diversity in the microbiota by acting as vehicles for the horizontal transfer of virulence, antibiotic resistance and metabolic determinants among bacteria16. Bacterial acquisition of phage genes could modify the functional properties of the microbiota, thereby substantially impacting host metabolism and immunity.
Second, phages may impact the composition of commensal bacterial populations through the predation of susceptible bacterial strains. This was suggested by recent studies of Enterococcus faecalis V583, a commensal of the human intestine. When E. faecalis V583 colonizes the intestines of mice, it produces lytic phages derived from chromosomally encoded prophage elements14. Production of the phage particles confers an advantage to E. faecalis V583 in competition with closely related enterococcus strains. This indicates that phages have the potential to influence the assembly and composition of intestinal bacterial communities14. Phage predation may be widespread, as suggested by the presence of clustered regularly interspaced short palindromic repeat (CRISPR) systems in many human commensal bacteria. CRISPRs are genetic modules in which short repeats of foreign DNA are inserted between spacer sequences in the bacterial chromosome. The spacer sequences are transcribed into short complementary RNAs that target invading DNA for destruction. Bacteria of the gastrointestinal tract and oral mucosa have acquired CRISPR-specific spacer sequences homologous to phage DNA6,17, suggesting that commensal bacteria are preyed upon by lytic phages in the mammalian host.
The ability of phages to shape the genetic diversity and the species diversity of the microbiota is likely to substantially impact the host immune system. It is well established that certain bacterial groups have distinctive effects on maturation of the immune system. For example, individual commensal species in the intestine influence the proportions of lamina propria T lymphocyte subsets that have distinct effector functions18,19. This in turn has profound effects on whether proinflammatory or anti-inflammatory immunity develops. Furthermore, commensal bacteria specifically limit and promote viral infection and transmission in mammalian hosts20,21,22. It is thus interesting to consider whether phage predation of intestinal bacteria could alter community composition in ways that impact function of the immune system and influence the spread of pathogenic viruses.
Resident eukaryotic viruses of body surface tissues
The resident viruses of humans also include viruses that infect eukaryotic cells. Like bacteriophages, populations of eukaryotic viruses in humans are heterogeneous, with distinct groups of viruses predominating in different tissues (Fig. 1). For example, the virobiota associated with adult human skin swabs is composed almost exclusively of polyomaviruses, including Merkel cell polyomavirus, a DNA virus associated with cutaneous cancer7,23. The frequency of human infection by resident polyomaviruses is likely to be high because ∼70% of humans test seropositive for antibodies to Merkel cell polyomavirus23. Human skin also harbors an abundance of human papillomaviruses that are related to the β- and γ-papillomaviruses7.
Figure 1. Anatomical locations of resident viruses in humans.
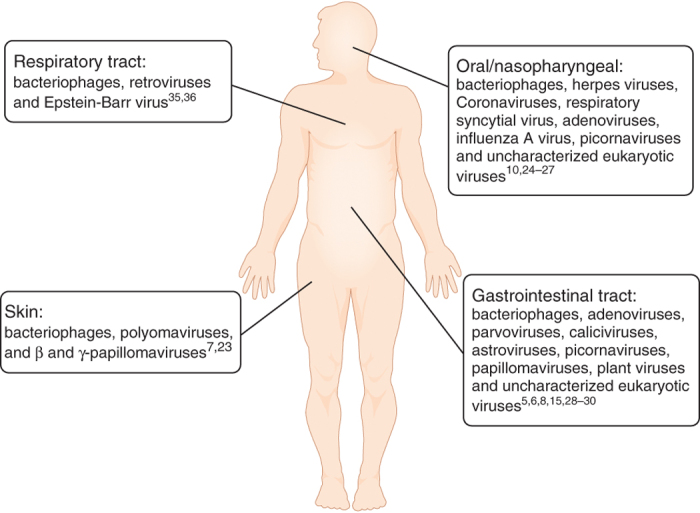
The human body is populated by resident viruses that include both prokaryotic and eukaryotic viruses. These viruses are associated with various tissues, including the oral cavity, nasopharynx, gastrointestinal and respiratory tracts, and the skin surface. Studies that identified viruses in various host tissues are referenced.
A complex viral population is also associated with the mucosal surfaces of the oral cavity and the respiratory tract. Both PCR and metagenome sequencing have been used to characterize the composition of the viral communities associated with the oral mucosa. These studies show a consistent presence of DNA from human herpes viruses and related viruses24,25. Metagenomic studies of the nasopharynx microbiota also suggest that the respiratory tract is rich in viruses. Analysis of nasopharyngeal aspirates from patients experiencing respiratory-tract infections revealed the presence of many respiratory viruses, including human respiratory syncytial virus, influenza A virus and rhinoviruses. Viral nucleic acid is also present in respiratory aspirates of healthy children and indicates the presence of adenoviruses, picornaviruses and coronaviruses26,27.
Several studies of the human intestinal virome have suggested that eukaryotic viruses are rare relative to bacterial viruses5,6,8. However, genetic signatures of eukaryotic single-stranded RNA viruses, single-stranded DNA viruses, double-stranded DNA viruses and retroviruses have been detected in the fecal viromes of healthy individuals5. Human papillomavirus also has been detected but only in a single subject28. Genetic signatures of many plant viruses have been detected in metagenomic studies of the human intestinal virome5,8. In particular, metagenomic studies of RNA viruses of the human intestine suggest that plant viruses are prevalent in this group and are likely derived from the diet8. Eukaryotic viral sequences are more prevalent in the intestines of diseased individuals. For example, feces from individuals with diarrheal disease and acute flaccid paralysis harbor a dominant population of DNA and RNA eukaryotic viral sequences derived from picornaviruses and parvoviruses29,30.
Much remains to be done to fully define the composition of the viral communities associated with human mucosal tissues and skin. For example, most metagenomic studies of human-associated viral populations have focused on analyzing viral DNA content. A more complete characterization of RNA viruses associated with humans will undoubtedly provide a more complete picture of the virobiota. Furthermore, as noted above, it remains unclear whether the eukaryotic viruses associated with healthy human mucosal tissues are the result of chronic or latent infections or whether they arise from acute but transient viral infections. Careful longitudinal metagenomic studies of viral populations associated with mucosal tissue will be required to resolve these issues and to identify those viruses that stably inhabit human tissues.
Immune system control of resident viruses
The study of resident viruses is an emerging field, and thus our current understanding of how these viruses interact with the immune system is limited. However, our knowledge of how the bacterial microbiota interacts with the immune system provides a useful framework for our initial consideration of virobiota–immune system interactions. Here we consider key concepts that have emerged from studies of the bacterial microbiota and how they might promote our understanding of host-associated viruses and their interactions with the mammalian immune system.
Studies of commensal bacteria–host interactions have suggested that a key driving force in the evolution of the mammalian immune system has been the need to maintain homeostatic relationships with resident bacteria31,32. In the intestine, for example, the immune system has a central role in controlling the density and the composition of resident bacterial communities33,34. Recent studies in primates suggest that there is a similar relationship between the immune system and the intestinal virobiota. Simian immunodeficiency virus (SIV), a primate virus related to the human immunodeficiency virus (HIV), causes AIDS in rhesus monkeys. SIV infection of rhesus monkeys is associated with damage to the intestinal barrier (enteropathy), which promotes AIDS progression. Until recently, the causes of this enteropathy have not been clear. A metagenomic study of the virome of SIV-infected rhesus monkeys showed that SIV infection is associated with a dramatic expansion of the intestinal virome4. This viral expansion was associated with enteropathy, particularly in monkeys that harbored adenoviruses (Fig. 2). These findings suggest that virome expansion is linked to the pathology observed in AIDS and highlights the role of the immune system in controlling virus populations in the intestine.
Figure 2. Pathogenic viral infection results in the expansion of the enteric virobiota.
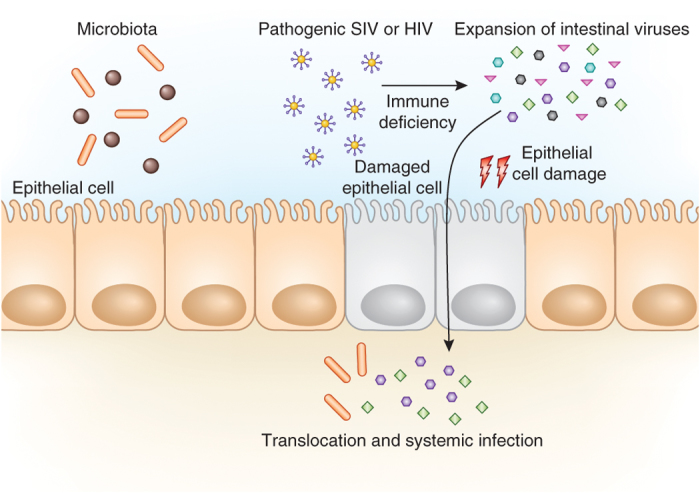
Pathogenic immunodeficiency viruses including SIV and possibly HIV can influence the expansion of resident viral populations in the intestinal tract. Expansion is likely promoted by immune suppression imposed upon the host by the pathogenic virus. Immune suppression results in a global host immune deficiency, allowing select enteric viruses to overpopulate the intestinal tract. Expansion of resident viruses is associated with damage to intestinal epithelial cells4. This subsequently allows translocation of enteric viruses, commensal bacteria and bacterial antigens across the epithelial surface, resulting in inflammation and systemic infection4. These findings highlight the role of the immune system in controlling intestinal virus populations.
Viral populations associated with mucosal tissue are also altered in other disease states. In the human lung, cystic fibrosis or acute respiratory infection can result in the expansion of certain viral species35,36. The lungs of patients with cystic fibrosis have many retrotranscribing RNA viruses and human herpes viruses, including Epstein-Barr virus, which has been linked to poor clinical outcomes in children with cystic fibrosis. The lung virome of patients with cystic fibrosis is highly variable as compared to healthy human lungs, where virome composition is relatively constant36. It has been proposed that viruses in healthy lungs may cause short-term infections that are rapidly cleared by the host immune system, whereas in the lung of a patient with cystic fibrosis these viruses may be more persistent36, perhaps as a result of compromised host immunity. The metabolic profile of the lung of a patient with cystic fibrosis suggests that it is enriched in aromatic amino acids and phosphate36. This may influence the outgrowth of bacterial species that can shape the virobiota and possibly alter the severity of disease. It is intriguing to consider that competition among bacteria in the lungs and the success of certain species based on the metabolic environment may modulate the composition of virobiota in the lungs, ultimately leading to progression of disease if host immunity is compromised.
Does the virobiota shape host immunity?
Early comparisons of germ-free and conventionally colonized mice revealed a profound effect of the microbiota on the formation of lymphoid tissue and subsequent development of the immune system1. Since then, many studies have established the importance of resident bacteria in promoting normal development and function of the immune system18,19. Commensal bacteria also can prime immune responses that provide cross-protection for pathogenic infections37,38. Given the presence of diverse families of eukaryotic and prokaryotic viruses at multiple human body sites, it is likely that viruses associated with mucosal tissue also have a pronounced influence on development and function of the immune system and could impact the host's ability to fight pathogenic viral infections.
The eukaryotic virobiota and the immune system
One way that the virobiota could impact host immunity is by triggering immune responses that protect against pathogenic viral and/or bacterial infections. It has been estimated that the average human has ∼8–12 chronic viral infections at any given time39. These infections are caused by a variety of different viruses, including papillomaviruses, herpes viruses and polyomaviruses, that often reside in tissues of healthy individuals (Fig. 1). Certain resident eukaryotic viruses can cause acute or latent infections that transition to chronic infections, where viral particles are shed throughout life. For example, herpes viruses infect most people during childhood, and latent herpes infection can lead to shedding of viral particles throughout the lifetime of an individual. Although latency and reactivation of herpes virus infection is considered pathogenic, chronic herpes virus infection has been shown to protect the host from viral and bacterial infections40. Protection is conferred by chronic infection with select types of herpes viruses, including γ-herpes viruses, which increase basal interferon γ (IFN-γ) expression and facilitate activation of macrophages. This in turn limits the spread of other infecting bacteria and viruses.
Some chronic viral infections lead to a decrease in host immunity that promotes immune suppression and greater susceptibility to infection. As discussed above, SIV can trigger AIDS in some primate species, which results in an expansion of the intestinal virome4. In another example, chronic infection by lymphocytic choriomeningitis virus hinders the expression of type I interferons and promotes the opportunistic invasion of murine cytomegalovirus41.
These studies highlight a fine balance between viral symbiosis and pathogenesis during chronic infections. As discussed above, the community of resident human associated viruses is not yet clearly defined, but viruses that maintain chronic infections may represent bona fide stable residents of the human virobiota. The nature of the association of particular viruses with the host (chronic or transient) will likely have a profound impact on the immune response to viral and bacterial pathogen challenge. This suggests that when analyzing host immune status, composition of the resident viral community and the potential existence of chronically associated viruses must be considered.
Bacteriophages and the immune system
Bacteriophages are a long-overlooked component of the virobiota that have the potential to shape mammalian immunity. Several studies have shown that phages can trigger a host immune response or can modulate host immunity. It is not surprising that humoral antibody responses can be mounted to phages that have been used to immunize animals at high titers42,43. Phages can also inhibit activation and proliferation of human T cells in vitro through an unknown mechanism44. Finally, phage and their nucleic acids alter the expression of innate immune genes in mouse tissues45,46. Despite these intriguing findings, virtually nothing is known about whether phages can influence innate and adaptive immunity during natural associations with mammals.
There are several possible routes by which phages could become exposed to mammalian tissues and trigger immune responses. For example, it is known that oral administration of phages to animals results in the translocation of phages to systemic tissues47,48,49. This suggests that mammals have mechanisms for uptake and delivery of phage that may allow intestinal phages to elicit innate and adaptive immune responses. One possible uptake route involves dendritic cells, which are known to sample intestinal luminal contents50 and can actively phagocytize phage particles in culture51. Phage engulfment renders dendritic cells incapable of further phagocytosis, suggesting that phage sampling by dendritic cells could negatively impact their proinflammatory functions52. Alternatively, phage antigens could be presented by dendritic cells to T cells, resulting in the development of cell-mediated immunity and cytokine release. Intestinal phages associate with the intestinal mucosal surface in larger numbers in patients afflicted with Crohn's disease53, raising the question of whether phages may influence the development of intestinal inflammatory diseases.
It is also interesting to consider whether bacteriophages might elicit antiviral innate immune responses. Mammalian cells are endowed with the ability to detect viral nucleic acids through several pattern-recognition receptors that are positioned to detect viral entry into cells (Fig. 3). These pattern-recognition receptors include viral RNA sensors, such as the endosomal Toll-like receptors TLR7 and TLR8, and RIG-I, a cytoplasmic double-stranded RNA helicase54. There are also DNA sensors, including TLR9, which detects endosomal DNA, and the cytoplasmic DNA sensor cyclic-GMP-AMP (cGAMP) synthase55,56. RIG-I can also indirectly sense viral DNA with the aid of RNA polymerase III, which converts cytoplasmic DNA into 5′-phosphorylated RNA that can be recognized by RIG-I (ref. 54). Each of these sensors initiates signaling cascades that activate expression of type I interferon, inflammatory cytokines such as interleukin 6 (IL-6) and IL-1β and chemokines including IL-8 and CXCL-10. If resident bacteriophages can enter host cells, it is possible that they would activate one or more of these pathways, as their nucleic acids would likely be exposed to either the endosomal or cytoplasmic viral nucleic acid sensors (Fig. 3). This could induce a tonic stimulation of antiviral immunity that might confer an advantage to the host by protecting against pathogenic viral infections.
Figure 3. Model for bacteriophage recognition by antiviral innate immune sensors.
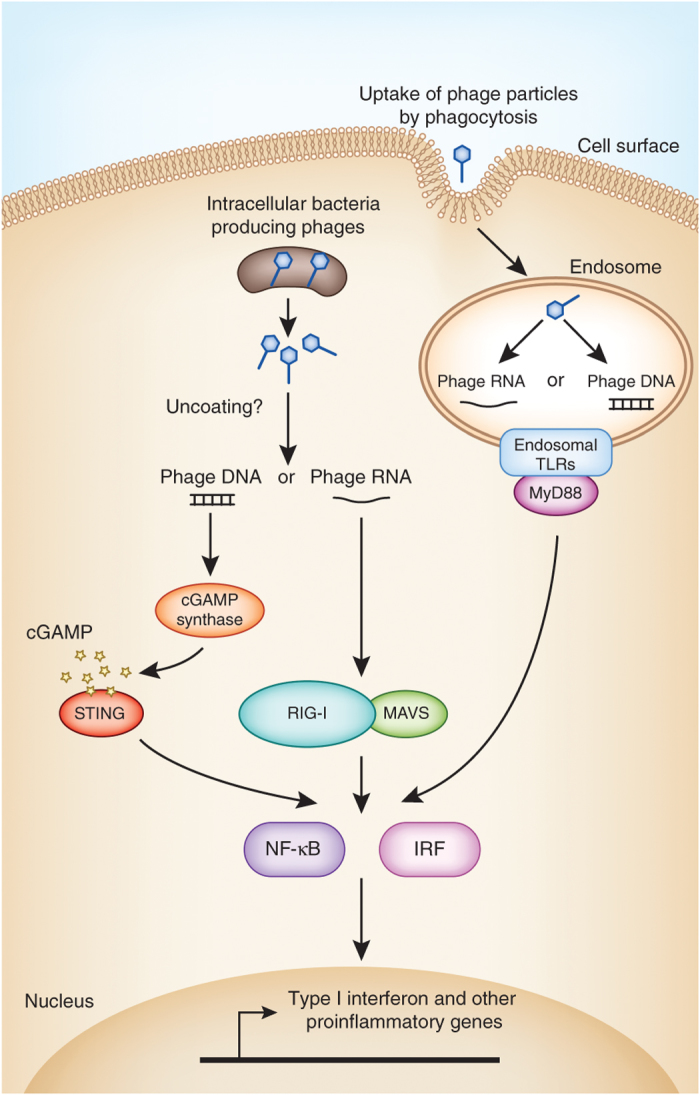
DNA and RNA phages could be sensed by components of the mammalian innate immune system if cells directly phagocytize phage particles or if phages are delivered to the intracellular environment by phage-producing bacteria. After degradation of the phage particle, phagocytized phage nucleic acids could be sensed by endosomal Toll-like receptors such as TLR7 or TLR9. If intracellular phages are uncoated in the cytoplasm, the released nucleic acids could be sensed by the sensors cGAMP synthase (DNA) or RIG-I (RNA), which signal through stimulator of interferon genes (STING) and mitochondrial antiviral-signaling protein (MAVS), respectively. These viral nucleic acid sensors activate the transcription factors NF-κB, IRF3 and IRF7 to promote the transcription of antiviral effectors such as IFN-β and proinflammatory cytokines.
Another way in which phages could become exposed to the mammalian immune system is through their association with commensal bacteria. Many commensal bacteria harbor prophages in their genomes and can induce these elements to produce phage particles in vivo14. Under certain conditions, such as host immunodeficiency, commensal bacteria from the intestine can enter epithelial cells or become engulfed by phagocytic cells. Phage particles associated with these bacteria could be exposed to the cytoplasm or to endocytic compartments of cells and could thus prime innate or adaptive immune responses (Fig. 3). In this way, bacteria could be delivery vehicles for phages that are sensed by the immune system.
Human intestinal phages have diversity-generating mechanisms that may facilitate evasion of host adaptive immunity. Sequencing of the metagenome of the human intestinal virome revealed that phage genomes are rich in regions of hypervariability28. Most of these hypervariable regions map to phage tail-fiber genes that are diversified to allow the encoded proteins to bind variable phage receptors28. Other hypervariable regions have been observed in genes encoding predicted immunoglobulin superfamily proteins, suggesting that such proteins could act as scaffolds for the presentation of diversified phage peptide sequences. The phage diversity-generating mechanism relies on error-prone reverse transcription28,57. Although the physiological relevance of these hypervariable regions is not yet clear, such a diversity-generating mechanism could allow phages to evade antibodies that target the phage particles.
Future perspectives
Recent advances in high-throughput sequencing have led to the discovery that viruses are an important component of the microbial communities that inhabit healthy human tissues. Emerging information about the interactions between resident viruses and the immune system suggests that these interactions are as intricate and as coevolved as those involving the bacterial microbiota.
Going forward, it is imperative that resident host-associated viruses be incorporated into models of host-microbiota interactions. It will be important to perform careful longitudinal studies of the viral populations associated with healthy human tissues to determine the stability of these populations and how they may evolve over time. There is a need for further studies aimed at understanding how resident prokaryotic phages and eukaryotic viruses shape commensal bacterial communities and how this impacts host immunity. Future work should also focus on exploring the extent to which resident phages and eukaryotic viruses activate host innate and adaptive immunity. Examination of how resident viruses interact with host cells may reveal new host immune factors that lie outside of currently known canonical pathways of antiviral immunity.
Future experimental explorations of virobiota–immune system interactions will undoubtedly be challenging and will require new tools and approaches. However, such studies hold the promise of exciting insight into the complex relationships between humans and their associated microbial communities and may yield new strategies for enhancing human health.
Acknowledgements
Supported by a Ruth L. Kirschstein National Research Service Award F32 DK089718 (to B.A.D.), The Howard Hughes Medical Institute (to L.V.H.), US National Institutes of Health grant R01 DK070855 (to L.V.H.) and a Burroughs Wellcome Foundation Investigators in the Pathogenesis of Infectious Diseases award (to L.V.H.).
Competing interests
The authors declare no competing financial interests.
References
- 1.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White DW, Suzanne Beard R, Barton ES. Immune modulation during latent herpesvirus infection. Immunol. Rev. 2012;245:189–208. doi: 10.1111/j.1600-065X.2011.01074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handley SA, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minot S, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foulongne V, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pride DT, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6:915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitbart M, et al. Viral diversity and dynamics in an infant gut. Res. Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 13.DeMarini DM, Lawrence BK. Prophage induction by DNA topoisomerase II poisons and reactive-oxygen species: role of DNA breaks. Mutat. Res. 1992;267:1–17. doi: 10.1016/0027-5107(92)90106-c. [DOI] [PubMed] [Google Scholar]
- 14.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc. Natl. Acad. Sci. USA. 2012;109:17621–17626. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MS, Park EJ, Roh SW, Bae JW. Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 2011;77:8062–8070. doi: 10.1128/AEM.06331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 2003;6:417–424. doi: 10.1016/s1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 17.Pride DT, Salzman J, Relman DA. Comparisons of clustered regularly interspaced short palindromic repeats and viromes in human saliva reveal bacterial adaptations to salivary viruses. Environ. Microbiol. 2012;14:2564–2576. doi: 10.1111/j.1462-2920.2012.02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Kuss SK, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane M, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller CS, Avdiushko SA, Kryscio RJ, Danaher RJ, Jacob RJ. Effect of prophylactic valacyclovir on the presence of human herpesvirus DNA in saliva of healthy individuals after dental treatment. J. Clin. Microbiol. 2005;43:2173–2180. doi: 10.1128/JCM.43.5.2173-2180.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazarevic V, et al. Analysis of the salivary microbiome using culture-independent techniques. J. Clin. Bioinforma. 2012;2:4. doi: 10.1186/2043-9113-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogaert D, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nokso-Koivisto J, Kinnari TJ, Lindahl P, Hovi T, Pitkaranta A. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J. Med. Virol. 2002;66:417–420. doi: 10.1002/jmv.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. Hypervariable loci in the human gut virome. Proc. Natl. Acad. Sci. USA. 2012;109:3962–3966. doi: 10.1073/pnas.1119061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Victoria JG, et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkbeiner SR, et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 33.Vaishnava S, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lysholm F, et al. Characterization of the viral microbiome in patients with severe lower respiratory tract infections, using metagenomic sequencing. PLoS ONE. 2012;7:e30875. doi: 10.1371/journal.pone.0030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willner D, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 39.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 40.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 41.Zuniga EI, Liou LY, Mack L, Mendoza M, Oldstone MB. Persistent virus infection inhibits type I interferon production by plasmacytoid dendritic cells to facilitate opportunistic infections. Cell Host Microbe. 2008;4:374–386. doi: 10.1016/j.chom.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inchley CJ, Howard JG. The immunogenicity of phagocytosed T4 bacteriophage: cell replacement studies with splenectomized and irradiated mice. Clin. Exp. Immunol. 1969;5:189–198. [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson J, Ormrod DJ, Wilson D, Miller TE. Host immune status in uraemia III. Humoral response to selected antigens in the rat. Clin. Exp. Immunol. 1980;42:234–240. [PMC free article] [PubMed] [Google Scholar]
- 44.Gorski A, et al. Bacteriophages and transplantation tolerance. Transplant. Proc. 2006;38:331–333. doi: 10.1016/j.transproceed.2005.12.073. [DOI] [PubMed] [Google Scholar]
- 45.Eriksson F, et al. Tumor-specific bacteriophages induce tumor destruction through activation of tumor-associated macrophages. J. Immunol. 2009;182:3105–3111. doi: 10.4049/jimmunol.0800224. [DOI] [PubMed] [Google Scholar]
- 46.Mori K, Kubo T, Kibayashi Y, Ohkuma T, Kaji A. Anti-vaccinia virus effect of M13 bacteriophage DNA. Antiviral Res. 1996;31:79–86. doi: 10.1016/0166-3542(96)00951-5. [DOI] [PubMed] [Google Scholar]
- 47.Duerr DM, White SJ, Schluesener HJ. Identification of peptide sequences that induce the transport of phage across the gastrointestinal mucosal barrier. J. Virol. Methods. 2004;116:177–180. doi: 10.1016/j.jviromet.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 48.Hamzeh-Mivehroud M, Mahmoudpour A, Rezazadeh H, Dastmalchi S. Non-specific translocation of peptide-displaying bacteriophage particles across the gastrointestinal barrier. Eur. J. Pharm. Biopharm. 2008;70:577–581. doi: 10.1016/j.ejpb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Keller R, Engley FB., Jr. Fate of bacteriophage particles introduced into mice by various routes. Proc. Soc. Exp. Biol. Med. 1958;98:577–580. doi: 10.3181/00379727-98-24112. [DOI] [PubMed] [Google Scholar]
- 50.Rescigno M, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 51.Barfoot R, et al. Some properties of dendritic macrophages from peripheral lymph. Immunology. 1989;68:233–239. [PMC free article] [PubMed] [Google Scholar]
- 52.Gorski A, et al. Bacteriophage translocation. FEMS Immunol. Med. Microbiol. 2006;46:313–319. doi: 10.1111/j.1574-695X.2006.00044.x. [DOI] [PubMed] [Google Scholar]
- 53.Lepage P, et al. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut. 2008;57:424–425. doi: 10.1136/gut.2007.134668. [DOI] [PubMed] [Google Scholar]
- 54.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat. Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doulatov S, et al. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]


