ABSTRACT
Many polarly flagellated bacteria require similar two-component regulatory systems (TCSs) and σ54 to activate transcription of genes essential for flagellar motility. Herein, we discovered that in addition to the flagellar type III secretion system (T3SS), the Campylobacter jejuni flagellar MS ring and rotor are required to activate the FlgSR TCS. Mutants lacking the FliF MS ring and FliG C ring rotor proteins were as defective as T3SS mutants in FlgSR- and σ54-dependent flagellar gene expression. Also, FliF and FliG required each other for stability, which is mediated by atypical extensions to the proteins. A FliF mutant that presumably does not interact with the T3SS protein FlhA did not support flagellar gene transcription, suggesting that FliF-T3SS interactions are essential to generate a signal sensed by the cytoplasmic FlgS histidine kinase. Furthermore, the flagellar T3SS was required for FlgS to immunoprecipitate with FliF and FliG. We propose a model whereby the flagellar T3SS facilitates FliF and FliG multimerization into the MS ring and rotor. As a result, these flagellar structures form a cytoplasmic complex that interacts with and is sensed by FlgS. The synthesis of these structures appears to be a regulatory checkpoint in flagellar biogenesis that the FlgS kinase monitors to initiate signal transduction that activates σ54 and expression of genes required for the next stage of flagellation. Given that other polar flagellates have flagellar transcriptional hierarchies that are organized similarly as in C. jejuni, this regulatory checkpoint may exist in a broad range of bacteria to influence similar TCSs and flagellar gene transcription.
IMPORTANCE
Despite the presence of numerous two-component regulatory systems (TCSs) in bacteria, direct signals sensed by TCSs to activate signal transduction are known for only a minority. Polar flagellates, including Pseudomonas, Vibrio, Helicobacter, and Campylobacter species, require a similar TCS and σ54 for flagellar gene transcription, but the activating signals for these TCSs are unknown. We explored signals that activate the Campylobacter jejuni FlgSR TCS to initiate σ54-dependent flagellar gene transcription. Our discoveries suggest that the FlgS histidine kinase monitors the formation of the flagellar type III secretion system and the surrounding MS and C rings. The synthesis of these structures creates a regulatory checkpoint in flagellar biogenesis that is sensed by FlgS to ensure proper transcription of the next set of genes for subsequent steps in flagellation. Given the conservation of flagellar-associated TCSs and transcriptional cascades in polar flagellates, this regulatory checkpoint in flagellar biogenesis likely impacts flagellation in a broad range of bacteria.
Introduction
Signal detection and transduction by two-component regulatory systems (TCSs) are essential in bacteria to link specific external or internal stimuli to correct behavioral responses, such as gene expression (1). In contrast to peritrichous Escherichia coli and Salmonella species, polarly flagellated bacteria, including Pseudomonas, Vibrio, Helicobacter, and Campylobacter species, require a similar TCS to activate the transcription of σ54-dependent flagellar genes (2–8). However, the precise signal sensed by these flagellar-associated TCSs to initiate flagellar gene expression is not known.
One of the best-characterized flagellar-associated TCSs among polar flagellates is the FlgSR TCS of Campylobacter jejuni. In this TCS, the cytoplasmic FlgS histidine kinase autophosphorylates upon sensing a signal and then activates the FlgR response regulator via phosphotransfer to a specific aspartate residue (8–11). FlgR is a member of the NtrC family of transcriptional regulators (12). Once activated, phospho-FlgR positively influences σ54 RNA polymerase (RNAP) holoenzyme to initiate transcription of genes encoding the flagellar rod, ring, and hook proteins and the FlaB minor flagellin. Furthermore, FlgSR and σ54 are required for full expression of σ28, which is required for expression of the FlaA major flagellin to complete flagellar biogenesis and the expression of the Fed proteins (13). The Feds are six proteins that are coexpressed with flagellar proteins but are not required for motility. Five Feds (FedA, FedB, FedC, FedD, and CiaI) are required by C. jejuni for optimal commensal colonization of the natural avian host (13). CiaI is also required for wild-type level of invasion of human intestinal epithelial cells (13, 14).
In addition to FlgSR, we previously identified other C. jejuni proteins that are required for σ54-dependent flagellar gene expression, including flagellar type III secretion system (T3SS) components (i.e., FlhA, FlhB, FliP, and FliR) and FlhF, a GTPase that polarly localizes flagella in C. jejuni and other polar flagellates (15–19). Due to the apparent absence of a master transcriptional regulator atop the flagellar transcriptional regulatory cascade, expression of flgS, flgR, the T3SS genes, and flhF is thought to be constitutive in C. jejuni. Whereas the requirement of FlhF for flagellar gene expression is unknown, we showed that a formed but not fully secretion-competent flagellar T3SS is required to activate the FlgSR TCS and σ54-dependent flagellar gene transcription (2, 10). Based on these findings, we proposed that the signal detected by the FlgS histidine kinase to initiate signal transduction and flagellar gene expression in C. jejuni is a domain of a fully formed flagellar T3SS. However, the specific signal within the T3SS has remained elusive.
In this study, we explored whether other C. jejuni flagellar components are required for transcription of σ54-dependent flagellar genes. We discovered that FliF, which forms the homopolymeric MS ring housing the flagellar T3SS, and the FliG rotor component of the C ring are as essential as the flagellar T3SS for FlgSR- and σ54-dependent flagellar gene transcription. We observed that FliF and FliG are dependent on each other for stability, which is mediated by domains not common to homologues found in well-characterized flagellator systems. We observed that FlgS was present in a complex with FliF and FliG in C. jejuni. Furthermore, immunoprecipitation of FlgS with FliF and FliG was dependent on the flagellar T3SS and contacts likely between FliF and the T3SS. We propose a mechanism whereby the flagellar T3SS is required for multimerization of FliF and FliG into the MS ring and rotor component of the C ring, which results in the formation of a signal within these structures that is sensed by FlgS. The synthesis of the MS ring, rotor and the T3SS creates a regulatory checkpoint that allows FlgS to monitor a step in flagellation to ultimately regulate σ54 activity and expression of the next set of genes in the flagellar transcriptional hierarchy. The presence of this regulatory checkpoint results in linkage of the synthesis of these flagellar structures to activation of σ54 flagellar gene transcription and the production of proteins that are to be secreted by the T3SS. Furthermore, given the presence of similarly ordered flagellar transcriptional regulatory cascades and homologous TCSs in other polar flagellates, MS ring, rotor, and T3SS formation may function as a regulatory checkpoint in flagellar biogenesis for regulating σ54 activity and flagellar gene transcription in a broad range of bacterial species.
RESULTS
The FliF MS ring and FliG rotor proteins are required for σ54-dependent flagellar gene transcription.
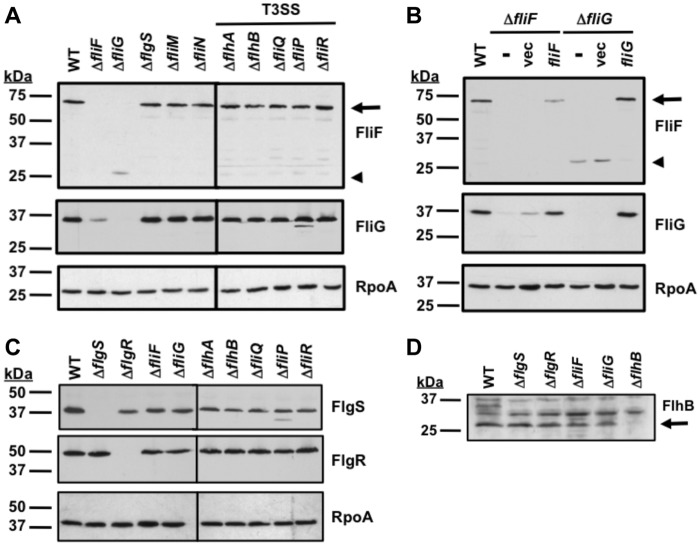
A diagram of the C. jejuni flagellum, which is composed of proteins similar to those forming flagella of other motile bacteria, is presented in Fig. 1A (20). We previously found that components of the flagellar T3SS (i.e., FlhA, FlhB, FliP, and FliR), the FlhF GTPase, and the FlgSR TCS are required for wild-type levels of σ54-dependent flagellar gene expression in C. jejuni (Fig. 1B) (2, 9–11, 16). We analyzed whether other C. jejuni flagellar proteins were also involved in activating FlgSR signal transduction to initiate expression of σ54-dependent flagellar genes. Therefore, we monitored expression of the σ54-dependent flaB::astA transcriptional reporter in C. jejuni mutants lacking specific flagellar genes. Mutants lacking fliF or fliG, which encode the inner-membrane MS ring protein and the cytoplasmic rotor component of the C ring, respectively, expressed flaB::astA at 50-fold-lower levels than wild-type C. jejuni (Fig. 1A and B). This decreased level of flaB::astA expression was similar to the amount of expression in any flagellar T3SS mutant (e.g., ΔflhA or ΔfliQ) (Fig. 1B). Expression of flaB::astA was restored to 60 to 65% of wild-type levels by complementing the ΔfliF and ΔfliG mutants in trans with plasmids to constitutively express fliF or fliG (Fig. 1C).
FIG 1 .
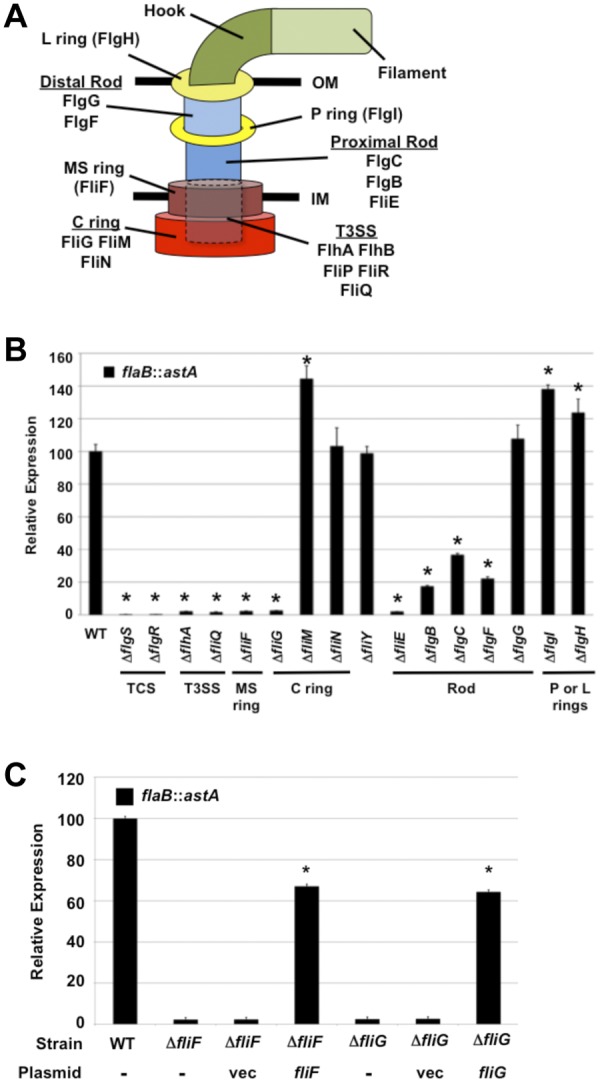
Analysis of expression of the σ54-dependent flaB::astA transcriptional reporter in C. jejuni flagellar mutants. (A) Organization of the C. jejuni bacterial flagellum. The T3SS (dotted-line cylinder) is located in the inner membrane (IM) and surrounded by the MS and C rings. Proteins that compose each substructure are indicated. OM, outer membrane; IM, inner membrane. (B) Results of arylsulfatase assays examining expression of the flaB::astA transcriptional fusion in wild-type C. jejuni and isogenic mutants lacking different flagellar genes. The level of flaB::astA expression in each mutant is relative to the level in wild-type C. jejuni, which was set to 100 units. Error bars indicate standard deviations of the average arylsulfatase activities analyzed from three samples. *, indicates the mutant had significantly different (increased or decreased) reporter activity than the wild-type strain (P < 0.05). (C) Results of arylsulfatase assays examining flaB::astA expression in wild-type C. jejuni and isogenic ΔfliF or ΔfliG mutants. The ΔfliF or ΔfliG mutants were not complemented (-) or were complemented in trans with plasmid alone (vec) or a plasmid to express fliF or fliG. The genotype and the plasmid used for complementation are listed for each strain. The level of flaB::astA expression in each mutant is relative to the level in wild-type C. jejuni, which was set to 100 units. Error bars indicate standard deviations of the average arylsulfatase activities analyzed from three samples. *, indicates the complemented mutant had a significantly different level of reporter activity than the respective mutant lacking any plasmid (P < 0.05).
In contrast to FliF or FliG, the FliM and FliN switch proteins of the C ring and FliY, a putative phosphatase that likely influences chemotaxis, were not required for flaB::astA expression (Fig. 1B). In fact, a C. jejuni ΔfliM mutant expressed approximately 40% more flaB::astA than wild-type C. jejuni. Analysis of rod proteins revealed that the lack of FliE, the most proximal rod component, caused a 50-fold reduction in flaB::astA expression, but mutation of other rod genes (flgB, flgC, and flgF) only reduced flaB::astA expression 3- to 5-fold (Fig. 1B). In addition, a FlgG distal rod mutant showed no change in flaB::astA expression relative to that of wild-type C. jejuni. Mutants lacking the FlgH L ring protein or the FlgI P ring component demonstrated a 20 to 40% increase in flaB::astA expression compared to the level in the wild-type strain (Fig. 1B). In summary, these data indicated that all of the flagellar T3SS components, the FliF MS ring protein, the FliG rotor protein of the C ring, and select rod proteins were required for wild-type levels of σ54-dependent flagellar gene expression in C. jejuni.
FliF, FliG, and the flagellar T3SS require each other for FlgSR signal transduction.
To understand the relationship between the FlgSR TCS, FliF, FliG, and the flagellar T3SS that facilitates signal transduction, we examined whether production of these proteins was affected in different C. jejuni flagellar mutants. Immunoblot analysis of C. jejuni lysates revealed wild-type levels of FliF and FliG in FlgSR TCS or flagellar T3SS mutants (Fig. 2A). We also detected wild-type levels of the FlgS histidine kinase and FlgR response regulator in mutants lacking FliF, FliG, and T3SS proteins (Fig. 2C). In both assays, the levels of RpoA (the α subunit of RNAP) were similar in all strains, verifying that similar amounts of proteins from lysates were analyzed (Fig. 2A and C). Due to small amounts of flagellar T3SS proteins in C. jejuni, it was difficult to assess their abundance. However, we were able to monitor FlhB production (Fig. 2D). FlhB undergoes autoproteolytic cleavage during flagellar biogenesis (10, 21). We detected similar levels of processed FlhB in the wild-type strain and mutants lacking FlgSR, FliF, or FliG (Fig. 2D). These data indicated that the flagellar proteins required for signal transduction via the FlgSR TCS were produced independently of each other, yet alone they were not sufficient to initiate signaling. Instead, FliF, FliG, and the flagellar T3SS are all required to activate σ54-dependent flagellar gene expression via the FlgSR TCS in C. jejuni.
FIG 2 .
Examination of FliF, FliG, FlgS, FlgR, and FlhB production in C. jejuni flagellar mutants. (A and B) Immunoblot analysis of FliF, FliG, and RpoA production in whole-cell lysates of wild-type C. jejuni (WT) or isogenic mutants lacking different flagellar genes (A) or wild-type C. jejuni and isogenic ΔfliF or ΔfliG mutants either not complemented (-) or complemented in trans with vector alone (vec) or a plasmid to express fliF or fliG (B). Arrows indicate full-length FliF, and arrowheads indicate a truncated FliF protein. (C) Immunoblot analysis of FlgS, FlgR, and RpoA production in whole-cell lysates of wild-type C. jejuni or isogenic mutants lacking different flagellar genes. (D) Immunoblot analysis of FlhB production in total-membrane preparations of wild-type C. jejuni and isogenic mutants lacking different flagellar genes. Arrow indicates the processed form of FlhB. For all immunoblots, each protein was detected by specific antiserum.
FliF and FliG depend on each other for stability.
In contrast to the FlgSR TCS or T3SS mutants, only a minor amount of the full-length 63-kDa FliF protein was detected in C. jejuni ΔfliG, which was only visible upon overexposure of the immunoblot shown in Fig. 2A (data not shown). Instead, a smaller protein of 25 kDa was detected, suggesting that FliF was mostly unstable without FliG (Fig. 2A). Similarly, the level of the full-length 38-kDa FliG protein was greatly reduced in the C. jejuni ΔfliF mutant (Fig. 2A). In contrast, FliF and FliG levels were not affected in ΔfliM or ΔfliN mutants, which each lacked a switch component of the C ring. Production of FliG and FliF was restored to wild-type level in the ΔfliG C. jejuni mutant complemented in trans with fliG (Fig. 2B). In the ΔfliF mutant, production of FliG was restored to the wild-type level upon in trans complementation with fliF, but the level of FliF was reduced relative to wild-type C. jejuni, suggesting partial complementation. Unlike in other flagellar systems where FliF and FliG are stable in the absence of the other protein (22), our data indicated that C. jejuni FliF and FliG were dependent on each other for stability.
Identification of FliF and FliG domains required for stability and signal transduction.
Previous investigations from our laboratory suggested that formation of the flagellar T3SS may be a direct signal sensed by the cytoplasmic FlgS histidine kinase to initiate signal transduction. However, considering recent findings from analyses of the biogenesis of flagellar systems and bacterial injectisome systems with similar T3SSs, we considered altering our original hypothesis. Although more details remain to be elucidated, formation of bacterial T3SSs begins with T3SS proteins forming an inner membrane complex (23–25). In flagellar T3SSs, interactions between FlhA and FliF appear to recruit FliF and, subsequently, C ring components (FliG, FliM, and FliN) to the T3SS (24, 26). FliF and FliG (and other C ring proteins) then assist each other to multimerize into the MS and C rings (24). Interactions between the FliF C-terminal cytoplasmic domain and the FliG N-terminal domain tether the C ring to the cytoplasmic base of the MS ring, thereby encasing most of the T3SS structure (Fig. 1A) (22, 27).
Considering these studies and our data presented above that indicates the requirement of FliF and FliG for FlgSR- and σ54-dependent flagellar gene expression, we hypothesized that FliF and FliG may form a signal sensed by FlgS to stimulate signal transduction upon multimerization into the MS ring and rotor component of the C ring. Because FliF and FliG have cytoplasmic domains that are more accessible to FlgS than the flagellar T3SS, we considered that FliF and FliG may polymerize into the MS ring and rotor in a T3SS-dependent manner to form cytoplasmic structures that are sensed by FlgS. This hypothesis implies that the T3SS is only indirectly involved in the activation of FlgS and signal transduction. An alternative hypothesis includes the possibility that the flagellar T3SS has a direct role in signaling to FlgS that is dependent on MS ring and rotor formation.
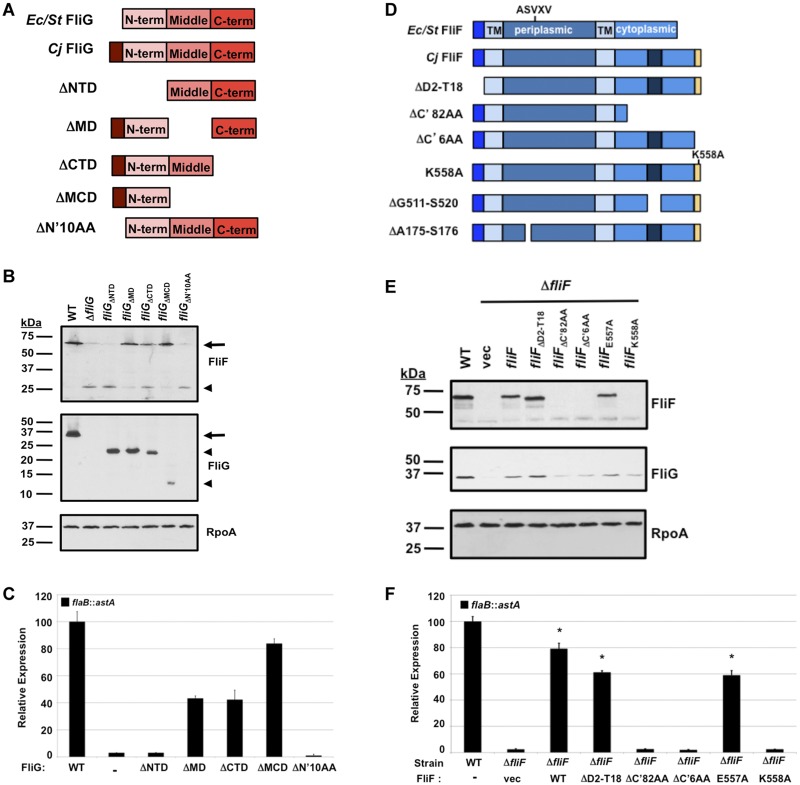
We performed a domain analysis of C. jejuni FliG and FliF to determine which regions of these proteins are required to activate the FlgSR TCS. This approach also allowed us to assess the domains required for the stability of both proteins. Using well-studied FliG proteins as models (28–30), C. jejuni FliG is likely organized into 3 functional domains: an N-terminal domain of ~110 amino acids that interacts with FliF, a middle domain to interact with the FliM switch protein, and a C-terminal domain to interact with both FliM and stator proteins of the motor (Fig. 3A). C. jejuni FliG also contains an N-terminal extension of 11 amino acids that is absent from FliG of E. coli and Salmonella species (Fig. 3A and see Fig. S1A in the supplemental material). This extension is also present in FliG of H. pylori, a closely related epsilonproteobacterium (Fig. S1A). To examine domains of FliG required for flaB::astA expression and stability of FliF and FliG, we replaced wild-type fliG on the chromosome with fliG mutants encoding proteins lacking one or more domains. C. jejuni bacteria producing FliG lacking the middle domain (FliGΔMD) or the C-terminal domain (FliGΔCTD) stably produced both FliF and truncated FliG proteins and expressed flaB::astA at approximately 40% of the wild-type level (Fig. 3A to C). Moreover, C. jejuni producing FliGΔMCD, which lacked the middle and C-terminal domains and only produced the N-terminal domain of FliG, produced full-length FliF with only a 20% reduction in flaB::astA expression (Fig. 3A to C). These data suggested that the middle and C-terminal domains of FliG were not required for the stability of FliF or FliG or activation of the FlgSR TCS for σ54-dependent flagellar gene expression.
FIG 3 .
Analysis of FliF and FliG domains required for stability and flaB::astA expression. (A) Organization of the E. coli (Ec), S. Typhimurium (St), and C. jejuni (Cj) FliG proteins. The FliG proteins possess N-terminal, middle, and C-terminal domains, shown in various shades of pink. The atypical N-terminal extension of C. jejuni FliG is shown in dark red. Shown below wild-type C. jejuni FliG are mutant FliG proteins with in-frame deletions of specific regions analyzed in this study. (B) Immunoblot analysis of FliF, FliG, and RpoA production in whole-cell lysates of wild-type C. jejuni or fliG mutants. Arrows indicate full-length FliF or FliG, and arrowheads indicated truncated FliF and FliG proteins. Each protein was detected by specific antiserum. (C) Results of arylsulfatase assays examining expression of the flaB::astA transcriptional fusion in wild-type C. jejuni and isogenic fliG mutants. The level of flaB::astA expression in each fliG mutant is relative to the level in wild-type C. jejuni, which was set to 100 units. Error bars indicate standard deviations of the average arylsulfatase activities analyzed from three samples. All mutants showed significantly different levels of reporter activity than the wild-type strain (P < 0.05). (D) Organization of the E. coli, S. Typhimurium, and C. jejuni FliF proteins. The FliF proteins are predicted to possess an N-terminal cytoplasmic domain (bright blue), two transmembrane (TM) domains, a periplasmic domain, and a C-terminal cytoplasmic domain. In the C-terminal cytoplasmic domain of C. jejuni FliF, an additional domain of 10 amino acids is shown in dark blue and the C-terminal extension is shown in yellow. The ASVXV motif that is conserved in FliF proteins is shown above the periplasmic domain. Shown below the wild-type C. jejuni protein are mutant FliF proteins with in-frame deletions of specific regions analyzed in this study. (E) Immunoblot analysis of FliF, FliG, and RpoA production in whole-cell lysates of wild-type C. jejuni or ΔfliF mutant complemented with vector alone (vec) or plasmid containing wild-type fliF or mutant alleles. Each protein was detected by specific antiserum. (F) Results of arylsulfatase assays examining expression of the flaB::astA transcriptional fusion in wild-type C. jejuni or ΔfliF mutant complemented with vector alone (vec) or plasmid containing wild-type fliF or mutant alleles. The level of flaB::astA expression in each complemented fliF mutant is relative to the level in wild-type C. jejuni, which was set to 100 units. Error bars indicate standard deviations of the average arylsulfatase activities analyzed from three samples. *, the complemented mutant had a significantly different level of reporter activity than the ΔfliF mutant harboring vector alone (P < 0.05).
Further support for the N-terminal domain of FliG being essential for FliF stability and FlgS signal transduction was gained by analyzing C. jejuni fliG mutants that lacked either residues 2 to 11 in the atypical N-terminal extension (FliGΔN’10AA) or the entire N-terminal domain (FliGΔNTD; Fig. 3A and see Fig. S1A in the supplemental material). Whereas FliGΔNTD was stable in C. jejuni, FliGΔN’10AA was not, and neither protein supported the production of full-length FliF (Fig. 3B). Importantly, flaB::astA expression in either mutant was as defective as in the C. jejuni ΔfliG mutant (Fig. 3C).
The topology of FliF in the inner membrane of a bacterium has not entirely been experimentally verified. Based on predictions for other FliF proteins, C. jejuni FliF is likely organized into a short N-terminal cytoplasmic domain, followed by two transmembrane domains with an intervening periplasmic domain and a C-terminal cytoplasmic domain (Fig. 3D) (31). We also noted that C. jejuni FliF contains an additional 10 amino acids within its C-terminal cytoplasmic domain and is extended by 7 residues at its extreme C terminus, neither of which are present in E. coli or Salmonella FliF (Fig. 3D and see Fig. S1B in the supplemental material). These features were also present in FliF of H. pylori (Fig. S1B).
Because cytoplasmic domains of FliF are known to interact with FliG and could conceivably generate a signal for the cytoplasmic FlgS histidine kinase, we initially examined FliF proteins lacking most of the N-terminal cytoplasmic domain (FliFΔD2-T18) or a large region of the C-terminal cytoplasmic domain (FliFΔC’82AA) (Fig. 3D and see Fig. S1B in the supplemental material). For these analyses, wild-type FliF or mutant proteins were examined for their ability to complement in trans a C. jejuni ΔfliF mutant to restore FliG production and flaB::astA expression. Expression of wild-type fliF and fliFΔD2-T18 restored similar levels of FliF and FliG proteins and flaB::astA expression to the ΔfliF mutant (Fig. 3E and F), indicating that the N-terminal cytoplasmic domain of FliF is dispensable for either process. In contrast, FliFΔC’82AA was unstable in C. jejuni, which caused a reduction in FliG levels and a lack of expression of flaB::astA (Fig. 3E and F). These data indicated that the FliF C-terminal cytoplasmic domain was vital for stability of FliF and FliG and σ54-dependent flagellar gene expression. Furthermore, production of FliFΔC’6AA that lacked six of the seven residues of the atypical C-terminal extension failed to restore FliF and FliG production and flaB::astA expression (Fig. 3D to F and Fig. S1B). Alanine-scanning mutagenesis of the C-terminal residues revealed that only K558 was essential for the stability of FliF and FliG and flaB::astA expression (Fig. 3D to F and Fig. S1B). The remaining residues of the C-terminal extension could be altered and not affect the production of FliF or FliG or σ54-dependent flagellar gene expression (as an example, the results from expression of FliFE557A, which has an E-to-A change at position 557, are shown in Fig. 3E and F).
We next investigated whether a region within the C-terminal cytoplasmic domain of FliF was essential for activating FlgSR and σ54-dependent flagellar gene expression. To this end, we constructed a series of fliF mutants that encoded proteins lacking discrete portions of the C terminus of FliF but retaining the C-terminal extension that is necessary for the stability of FliF and FliG. Unfortunately, the only stable FliF mutant protein obtained was FliFΔG511-S520, which lacks 10 residues within the C-terminal cytoplasmic domain that are absent from FliF of E. coli and Salmonella species (Fig. 3D and see Fig. S2A in the supplemental material). This protein restored FliG production and flaB::astA expression to a ΔfliF mutant (Fig. S2A and B), indicating that this domain is dispensable for FlgS activation. However, FliFΔG511-S520 did not restore motility to the C. jejuni ΔfliF mutant (Fig. S2C), suggesting that this domain assists in flagellar motor function.
Although we were unable to delineate further C-terminal cytoplasmic regions of FliF specifically required for FlgSR activation, our data identified the atypical extensions of FliF and FliG as requirements for stability of both proteins, which allowed them to promote flagellar gene expression. Furthermore, we localized the minimal requirements of FliG to stabilize FliF and FliG to the N-terminal domain, which encompasses the initial 110 residues of the protein. This domain is also either directly or indirectly required for the signal transduction leading to wild-type levels of flagellar gene expression.
Evidence for the requirement of FliF-FlhA interactions for FlgSR signal transduction.
As described above, analysis of the biogenesis of T3SSs in E. coli and Salmonella suggested that the FlhA-like proteins of T3SSs interact with FliF homologues, which presumably recruits FliF to a nascent T3SS for subsequent multimerization into the MS ring (24, 26, 28). Currently, it is unknown how FlhA and FliF may interact or how FliF monomers contact each other to multimerize. However, the highly conserved ASVXV motif that is predicted to reside within the FliF periplasmic domain has been implicated in contributing to FliF-FlhA interactions (Fig. 3D) (31). Deletion of the alanine and serine residues in this motif in Salmonella enterica serovar Typhimurium FliF resulted in a nonmotile phenotype that could be suppressed by the mutation of different residues in FlhA (31). Together, these studies suggested that direct FliF-FlhA interactions likely occur and are important for the biogenesis of a mature T3SS, MS ring, and C ring complex.
We sought evidence for a presumed interaction between the C. jejuni flagellar T3SS and FliF as a requirement for signal transduction via the FlgSR TCS and expression of σ54-dependent flagellar genes. We hypothesized that the ASVXV motif of C. jejuni FliF may be essential for FliF-FlhA interactions and, subsequently, the multimerization of FliF and FliG into the MS ring and rotor to compose a domain to activate FlgSR. Therefore, we tested whether a chromosomal mutation in fliF that removed A175 and S176 from the ASVXV motif within C. jejuni FliF supported σ54-dependent flagellar gene expression (Fig. 3D). We observed that FliFΔA175-S176 was stable in C. jejuni and supported wild-type levels of the FliG protein (Fig. 4A). In contrast to the wild-type strain, C. jejuni fliFΔA175-S176 did not express flaB::astA (Fig. 4B). Considering previous suppositions regarding the FliF ASVXV motif, these results suggested that FliF interactions with the flagellar T3SS (likely through FlhA) are essential for generating a specific signal to activate the FlgSR TCS and σ54-dependent flagellar gene expression.
FIG 4 .
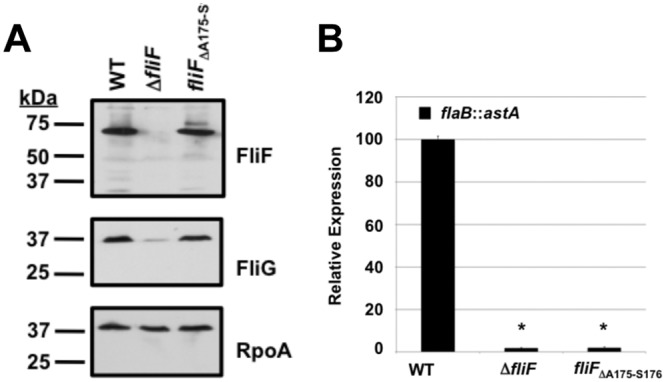
Analysis of the ability of FliFΔA175-S176 to support σ54-dependent expression of flaB::astA. (A) Immunoblot analysis of FliF, FliG, and RpoA production in whole-cell lysates of wild-type C. jejuni or the ΔfliF or fliFΔA175-S176 mutant. Each protein was detected by specific antiserum. (B) Results of arylsulfatase assays examining expression of the flaB::astA transcriptional fusion in wild-type C. jejuni or the ΔfliF or fliFΔA175-S176 mutant. The level of flaB::astA expression in each fliF mutant is relative to the level in wild-type C. jejuni, which was set to 100 units. Error bars indicate standard deviations of the average arylsulfatase activities analyzed from three samples. Both mutants showed a significantly different level of reporter activity than the wild-type strain (P < 0.05), which is indicated by *.
The flagellar T3SS is required for FliF, FliG, and FlgS to form a complex in C. jejuni.
The data presented above suggested that the flagellar T3SS, FliF, and FliG function together in C. jejuni to activate the FlgSR TCS. Our leading hypothesis is that FliF and/or FliG form an activating signal for the cytoplasmic FlgS histidine kinase after the proteins have multimerized into the MS ring and rotor with the assistance of the flagellar T3SS. To test the validity of this hypothesis, we performed coimmunoprecipitation analyses with C. jejuni to determine whether FlgS is present in vivo in the same complex as FliF and/or FliG. These experiments also allowed us to monitor interactions between FliF and FliG, as these proteins are known to interact in flagella.
To analyze potential in vivo interactions between FlgS, FliF, and FliG, FLAG-FliF was expressed in the C. jejuni ΔfliF strain and FliG-FLAG was expressed in the C. jejuni ΔfliG strain. The C. jejuni strains were cross-linked before lysis to trap transient protein interactions. As expected, native FliG coimmunoprecipitated FLAG-FliF and native FliF coimmunoprecipitated FliG-FLAG (Fig. 5A and B). As described above, the FliG levels were low in the C. jejuni ΔfliF mutant (Fig. 2A and B; see Fig. S3 in the supplemental material). As expected, these low levels of FliG did not immunoprecipitate with the anti-FLAG resin when the FLAG tag alone was expressed in the ΔfliF strain (Fig. 5A). We also observed a lack of coimmunoprecipitation of low levels of FliF with the resin when only the FLAG tag was expressed in the C. jejuni ΔfliG strain (Fig. 5B and Fig. S3).
FIG 5 .
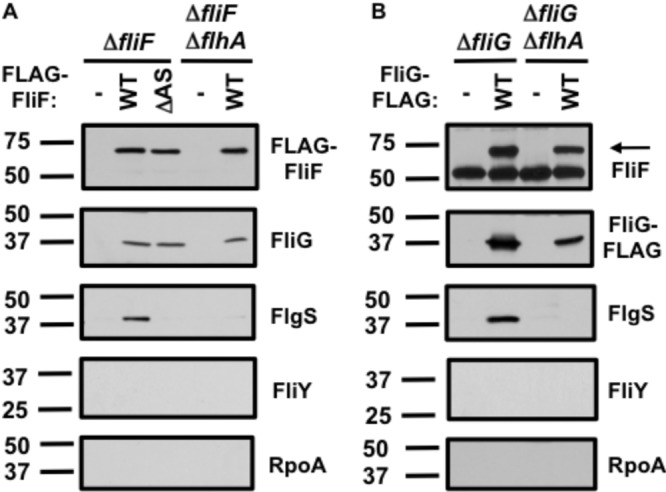
Detection of in vivo interactions between FlgS and FliF or FliG in C. jejuni. (A) Analysis of FliF-coimmunoprecipitated C. jejuni proteins. The C. jejuni ΔfliF or ΔfliF ΔflhA mutant contained a plasmid to express the FLAG tag alone (-), FLAG-FliF (wild type [WT]), or FLAG-FliFΔA175-S176 (ΔAS). (B) Analysis of FliG-coimmunoprecipitated C. jejuni proteins. The C. jejuni ΔfliG or ΔfliG ΔflhA mutant contained a plasmid to express the FLAG tag alone (-) or wild-type FliG-FLAG (WT). For both panels, FliF, FliG, FlgS, FliY, and RpoA were detected by specific antisera. The band in the FliF immunoblot that was present in all samples in panel B is a fragment of the murine antibody of the anti-FLAG resin that was detected by the secondary antibody. The arrow in the FliF immunoblot in panel B indicates the native C. jejuni FliF protein.
Importantly, we found that native FlgS immunoprecipitated with FLAG-FliF and FliG-FLAG, indicating that FlgS was present in the same complex with FliF and FliG in vivo (Fig. 5A and B). The specificity of these interactions was verified when we observed that the putative FliY phosphatase of the flagellar chemotaxis regulatory system or RpoA did not immunoprecipitate with the FLAG-tagged FliF or FliG (Fig. 5A and B). Furthermore, FlgS did not immunoprecipitate with the anti-FLAG resin when only the FLAG tag was expressed in the ΔfliF or the ΔfliG mutant (Fig. 5A and B). These results suggested that FlgS was specifically present as part of a complex with FliF and FliG in C. jejuni.
Our genetic analysis described above suggested that the formation of a FliF- and FliG-dependent activating signal for FlgS only occurs in the presence of the flagellar T3SS and the FliF ASVXV motif, which is presumably required for FliF-FlhA interactions (Fig. 1B and 4B). Therefore, we determined whether FliF and FliG could be found in a complex with FlgS in C. jejuni in a flagellar T3SS mutant by expressing FLAG-tagged FliF or FliG in C. jejuni ΔflhA strains that also lacked native fliF or fliG. In these mutants, FliF and FliG coimmunoprecipitated each other, indicating that FliF and FliG interacted independently of the flagellar T3SS in C. jejuni (Fig. 5A and B). However, FlgS did not coimmunoprecipitate with either FLAG-FliF or FliG-FLAG in the ΔflhA mutants (Fig. 5A and B). Furthermore, when we expressed FLAG-tagged FliFΔA175-S176, which contains a deletion in the ASVXV motif and does not support FlgSR- or σ54-dependent flaB::astA expression (Fig. 4B), FliG immunoprecipitated with the mutant FliF protein but FlgS did not (Fig. 5A, ΔAS). These results combined indicated that FliF and FliG required not only the T3SS but also interactions between FliF and the flagellar T3SS to form a complex with FlgS. Our studies suggest that that the flagellar T3SS likely facilitates multimerization of FliF and FliG into the MS ring and rotor component of the C ring, resulting in formation of a flagellar complex that interacts with FlgS and promotes the FlgSR signal transduction that is required for σ54-dependent flagellar gene expression.
DISCUSSION
TCSs are nearly ubiquitous in bacterial species, but the specific signals directly sensed by the histidine kinases of these systems are only known for a relatively small number. In the polarly flagellated Vibrio, Pseudomonas, Helicobacter, and Campylobacter species, similar TCSs are required to activate σ54-dependent flagellar gene expression (2–8). However, the direct signals sensed by the TCSs in these motile bacteria are unknown. Based on previous analysis of the C. jejuni FlgSR TCS, we proposed that a signal sensed by FlgS to activate σ54-dependent flagellar gene expression may emanate from the flagellar T3SS, as all components of the T3SS were required for transcription of these genes (2, 10).
In this study, we discovered that FliF, which forms the MS ring housing the flagellar T3SS, and FliG, the rotor component of the C ring, are also required to activate FlgSR- and σ54-dependent flagellar gene expression. Furthermore, we detected that FlgS immunoprecipitated with both FliF and FliG and found that the T3SS was essential for coimmunoprecipitation of these proteins. In a flagellar T3SS mutant, FliF and FliG were produced and interacted with each other, and yet a FliF-FliG complex failed to coimmunoprecipitate FlgS or stimulate σ54-dependent flagellar gene expression. Therefore, based on model systems for the biogenesis of bacterial T3SSs, the C. jejuni flagellar T3SS is likely essential for FliF-FliG complexes to polymerize into the MS ring and rotor structures of the C ring (Fig. 6). We attempted to monitor the formation of these structures in C. jejuni but were unable to do so even in the wild-type strain. FliF-FliG complexes likely contact the T3SS through FliF-FlhA interactions, with the conserved FliF ASVXV motif either directly or indirectly involved in FliF-T3SS associations. Mutation of this motif in C. jejuni FliF abolished σ54-dependent flagellar gene expression and disrupted in vivo interactions between FliF and FlgS but not FliG.
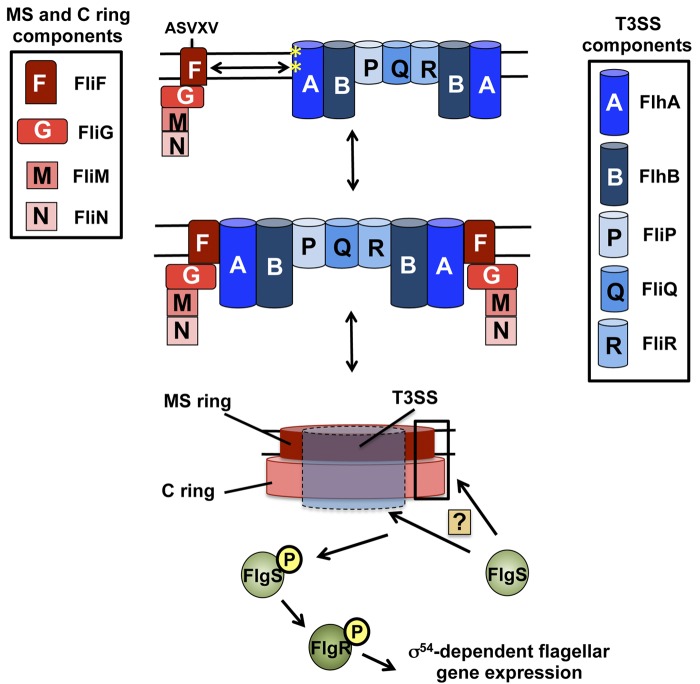
FIG 6 .
Model for the formation of an activating signal within the C. jejuni flagellar MS ring, C ring, and T3SS that is sensed by FlgS for initiation of signal transduction and σ54-dependent flagellar gene expression. The flagellar T3SS components first nucleate into a nascent complex in the inner membrane of C. jejuni. The FliF MS ring protein and the FliG C ring rotor protein, possibly also in complex with the FliM and FliN C ring switch proteins, are then recruited to the flagellar T3SS complex. We expect that at least the conserved ASVXV motif within FliF is required for FliF to interact with residues of FlhA (*) and, possibly, other flagellar T3SS components. Once recruited to the flagellar T3SS, FliF, FliG, FliM, and FliN multimerize into mature MS and C rings to surround the flagellar T3SS. The formation of the MS and C rings may create a cytoplasmic domain on the surface of these structures that is sensed by FlgS through a direct interaction to result in autophosphorylation of FlgS. In this model, a domain to activate FlgS may be composed by FliF monomers, FliG monomers, or FliF-FliG complexes within the mature MS and C rings. Alternatively, the formation of the MS and C rings may promote full assembly or maturation of the flagellar T3SS, which creates a domain that is sensed by FlgS to initiate signal transduction. After autophosphorylation of FlgS, signal transduction proceeds, resulting in phosphorylation of the FlgR response regulator and the expression of σ54-dependent flagellar genes.
Considering our data, we envision two possible models for how the flagellar T3SS, FliF, and FliG form an activating signal that is detected by FlgS to initiate signal transduction and σ54-dependent flagellar gene expression (Fig. 6). In the first model, formation of the MS ring and rotor in a T3SS-dependent manner may allow neighboring FliF and/or FliG proteins in the MS ring and rotor to form a cytoplasmic domain that is sensed by and interacts with the cytoplasmic FlgS histidine kinase to initiate the signal transduction necessary for flagellar gene expression. Whereas our data indicated that FlgS is present in the same complex as FliF and FliG, we do not know if FlgS interacted directly with FliF and/or FliG. Additionally, we do not know if other proteins, such as those composing the flagellar T3SS, were also present in this complex and contributed directly to interactions with FlgS. An alternative model is that FliF and FliG multimerization into the MS ring and rotor may be required for full assembly or maturation of the flagellar T3SS, which may directly interact with FlgS to initiate signal transduction. Due to a paucity of reagents to examine T3SS proteins, we were not able to determine whether flagellar T3SS proteins could coimmunoprecipitate FlgS and were present in a complex with FliF, FliG, and FlgS. Although we cannot exclude the latter model, we believe there is more support for the former model. Based on the architecture of the MS ring, C ring, and T3SS revealed by electron cryotomography of the C. jejuni flagellum (32), the MS ring and rotor structures are more accessible to FlgS in the cytoplasm than the flagellar T3SS, which is mostly encased by the MS and C rings. If this model is correct, our study implicates the MS ring and rotor in the stimulation of a bacterial TCS, in addition to their traditional roles in motility.
This mechanism for activating FlgS via formation of the MS ring, rotor, and flagellar T3SS appears to be an important regulatory checkpoint that allows FlgS to monitor the status of flagellar biosynthesis before initiating FlgSR signal transduction that ultimately controls σ54 activity. Flagellar biogenesis in bacteria requires the ordered expression of many genes so that flagellar proteins are produced in the correct temporal manner for proper steps in flagellation (33). As originally shown in Salmonella, many bacteria, including C. jejuni, use a regulatory checkpoint in flagellar biogenesis to monitor hook synthesis before the alternative σ factor, σ28, initiates transcription (13, 34–36). This checkpoint involves assessing hook formation through the presence of FlgM, the σ28 repressor, in the cytoplasm. Once the hook is completed, FlgM is secreted from the cytoplasm via the nascent flagellum, which relieves σ28 from repression to promote the expression of genes necessary for filament synthesis. Our data suggest the existence of a regulatory checkpoint in C. jejuni to ensure the activation of another alternative σ factor, σ54, via the FlgSR TCS only upon the formation of the MS ring, rotor, and the flagellar T3SS (Fig. 6). This mechanism would ensure that production of certain rod, ring, and hook proteins, which are all encoded in the σ54 regulon, is prevented before a mature flagellar T3SS, MS ring, and C ring required to secrete these proteins are made. Therefore, this mechanism allows the FlgS histidine kinase to monitor an important stage in flagellar biogenesis and link proper σ54-dependent gene expression to a structural step in flagellation.
We also observed that the FliE rod protein is required for FlgSR- and σ54-dependent flagellar gene expression. Little is known about FliE in any flagellar system, but this protein is thought to be the most proximal rod protein that connects the T3SS and MS ring to the rod (37, 38). As such, it is not accessible to the cytoplasmic FlgS histidine kinase. We speculate that FliE may assist in contacts between the flagellar T3SS and the MS ring or aid FliF on the periplasmic side in polymerizing into the MS ring. Future experiments will further elucidate the role of FliE in generating a stimulus to activate the FlgSR TCS.
Another unusual finding in our study was that C. jejuni FliF and FliG required each other for stability, which is not observed in other motile bacteria (15). The requirements for stability of these proteins were localized to K558 at the FliF C terminus and the N-terminal 10 residues of FliG, both of which are absent in E. coli and Salmonella FliF and FliG but present in the respective proteins of Helicobacter pylori. These findings may suggest that FliF and FliG interact through these domains in C. jejuni (and perhaps in H. pylori as well), which would be a different mode of interaction between these proteins than is seen in other flagellar systems. As such, it is possible that these altered FliF-FliG interactions in the C. jejuni flagellar motor may cause certain aspects of flagellar rotation to be different from the flagellar rotation of other motile bacteria.
The results of our study in C. jejuni have potentially broad implications for understanding signals that stimulate σ54-dependent flagellar gene expression in a variety of polar flagellates, including Vibrio, Pseudomonas, and Helicobacter species. In these bacteria, fliF, fliG, and genes for the flagellar T3SS are expressed before the regulon that is dependent on the respective FlgSR-like TCSs is transcribed (3–5). Thus, it is possible that multimerization of FliF and FliG into the MS ring and rotor in a T3SS-dependent manner in these bacteria also results in the formation of a similar direct signal in these flagellar complexes that is detected by a FlgS homologue. Thus, our study may have revealed a common regulatory checkpoint in flagellar biogenesis in a wide range of polar flagellates that is mediated by a histidine kinase and a domain within the flagellar MS and C ring structures of flagella to accurately control σ54 activity.
MATERIALS AND METHODS
Bacterial strains and growth.
C. jejuni 81-176 strains and procedures for generating mutants are described in the supplemental material, including in Tables S1 and S2. For all experiments, C. jejuni strains were initially grown from freezer stocks on Mueller-Hinton (MH) agar containing 10 µg/ml trimethoprim for 48 h under microaerobic conditions at 37°C. After the initial growth, strains were restreaked onto appropriate media and grown for another 16 h for use in experiments. Chloramphenicol, kanamycin, or streptomycin was added to the medium at 10 µg/ml, 50 µg/ml, or 0.1 to 2 mg/ml, respectively, when necessary.
astA transcriptional reporter assays.
Arylsulfatase assays were used to measure the level of expression of the flaB::astA transcriptional fusion on the chromosome of C. jejuni ΔastA strains as previously described (see Table S1 in the supplemental material) (2, 39, 40). Each strain was analyzed in triplicate, and each assay was performed three times. The level of expression of the transcriptional fusion in each strain was calculated relative to the expression in the wild-type C. jejuni ΔastA strain, which was set to 100 units.
Generation of recombinant proteins and polyclonal murine antisera.
The C. jejuni 81-176 fliF coding sequence from codon 2 to the stop codon was amplified by PCR with primers containing in-frame 5′ BamHI sites. The DNA fragment was inserted into BamHI-digested pGEX-4T-2 to create pDRH2266. The C. jejuni 81-176 fliG and fliY coding sequences from codon two to the stop codon were amplified by PCR with primers containing in-frame 5′ BamHI sites. In addition, one primer contained the sequence for the addition of a C-terminal 6×His tag between the last codon and the stop codon. The DNA fragments were inserted into BamHI-digested pT7-7 to create pJMB1506 (pT7-7::fliG-6×His) and pJMB1977 (pT7-7::fliY-6×His). The correct construction of all plasmids was verified by DNA sequencing. Plasmids were transformed into E. coli BL21(DE3).
Purification of glutathione S-transferase (GST)-FliF was achieved by inoculating 3 liters of LB broth containing 100 µg/ml ampicillin with a 1:40 dilution of overnight culture and growing the culture at 37°C with shaking to an optical density at 600 nm (OD600) of 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 100 µM, and the culture was incubated for another 4 h. Bacteria were recovered by centrifugation at 6,000 rpm for 10 min, and the bacteria were resuspended in cold phosphate-buffered saline (PBS) with Complete protease inhibitor (Roche). Bacteria were passaged through an EmulsiFlex-C5 disrupter (Avesin) 3 times at 15,000 to 20,000 lb/in2. The soluble fraction was recovered by centrifugation at 13,000 rpm for 2 h. GST-FliF was purified with glutathione Sepharose 4B beads according to the manufacturer’s instructions (GE Healthcare).
Purification of FliG-6×His and FliY-6×His was achieved by inoculating 1 liter of LB broth containing 100 µg/ml ampicillin with a 1:40 dilution of overnight culture and growing the culture at 37°C to an OD600 of 0.6. IPTG was added to a final concentration of 1 mM, and the culture was incubated for another 4 h at 30°C. Bacteria were recovered by centrifugation at 6,000 rpm for 10 min, and the bacteria were resuspended in lysis buffer according to the manufacturer’s instructions (Qiagen). Bacteria were passaged through an EmulsiFlex-C5 disrupter (Avesin) 3 times at 15,000 to 20,000 lb/in2. The soluble fraction was recovered by centrifugation at 13,000 rpm for 2 h. FliG-6×His and FliY-6×His were purified with Ni-nitrilotriacetic acid (NTA) beads according to the manufacturer’s instructions (Qiagen).
The purified GST-FliF, FliG-6×His, and FliY-6×His proteins were used to immunize mice by standard procedures for antiserum generation by a commercial vendor (Cocalico Biologicals).
Motility assays.
C. jejuni strains were grown on MH agar with 10 µg/ml trimethoprim for 16 h at 37°C in microaerobic conditions. Strains were suspended from agar plates in MH broth and diluted to an OD600 of 0.8. Strains were then inoculated into semisolid MH motility medium (containing 0.4% agar) and incubated for 24 h at 37°C in microaerobic conditions.
Immunoblotting analysis.
Preparation of C. jejuni strains for whole-cell lysates or total membrane isolation was performed as previously described (10). All immunoblotting analysis was performed with equal amounts of samples from C. jejuni strains after SDS-PAGE by standard procedures. Proteins were detected with specific murine or rabbit antisera generated previously or as described above. The primary antisera were used at the following concentrations: FliF M1 (1:1,000) or M201 (1:4,000), FliG M161 (1:5,000), RpoA M59 (1:3,000) (10), FlgS Rab11 (1:10,000) (41), FlgR Rab13 (1:10,000) (41), FlhB Rab476 (1:1,000) (10), and FliY M197 (1:4,000).
In vivo immunoprecipitation of C. jejuni proteins.
C. jejuni strains expressing FLAG-tagged FliF or FliG proteins were grown on MH agar with appropriate antibiotics for 16 h at 37°C in microaerobic conditions. Bacteria were suspended to an OD600 of 1.0, washed once with PBS, and resuspended in 18 ml of PBS. Bacteria were cross-linked by the addition of 2 ml 1% formaldehyde as previously described (42). After incubation for 30 min at 37°C, cross-linking was quenched with 4 ml of 1 M glycine for 10 min at 25°C. Bacteria were washed once with PBS and then disrupted by osmotic lysis (43). After incubation on ice for 10 min, 5 ml of solubilization solution (50 mM Tris, pH 8.0, 10 mM MgCl2, 2% Triton X-100) was added to the lysate. Samples were incubated on ice for 30 min and then centrifuged at 16,000 × g for 10 min. The supernatant was retained after a second centrifugation step at 160,000 × g for 1 h.
Anti-FLAG M2 affinity gel resin was used to immunoprecipitate FLAG-tagged proteins according to the manufacturer’s instructions (Sigma-Aldrich), with slight modifications. Briefly, 1 ml of cross-linked C. jejuni lysate was mixed with 5 µl anti-FLAG M2 affinity gel resin for 3 h at 4°C with agitation. The resin was pelleted by centrifugation at 4°C for 10 min at 13,000 rpm, washed 3 times with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100), and then washed once with 50 mM Tris (pH 8.0). The resin was resuspended in 20 µl Laemmli buffer, boiled for 5 min, and analyzed by 10% SDS-PAGE and immunoblotting with specific antisera.
Statistical analysis.
Tests for significance of the differences in expression from transcriptional reporter assays were conducted using Student’s t test (two-tailed distribution with two-sample, equal-variance calculations). As indicated in figures or figure legends, statistically significant differences between relevant strains possessed P values of <0.05.
SUPPLEMENTAL MATERIAL
Methods for constructing bacterial strains and plasmids used in this study. Download
Alignment of the N-terminal FliG and C-terminal FliF domains from E. coli, Salmonella, C. jejuni, and H. pylori. (A) ClustalW alignment of the N-terminal domains of FliG from E. coli strain K-12 (BAA15764), S. Typhimurium strain LT2 (AAL20882), C. jejuni 81-176 (EAQ73253), and H. pylori strain G27 (ACI27095). The N-terminal extension present in C. jejuni and H. pylori FliG is indicated in a box. (B) ClustalW alignment of the C-terminal transmembrane (TM) and cytoplasmic domains of FliF proteins from E. coli strain K-12 (YP_490192), S. Typhimurium LT2 (AAL20881), C. jejuni 81-176 (EAQ73453), and H. pylori G27 (ACI27094). The transmembrane (TM) domain predicted in all proteins is boxed. The domain of C. jejuni FliF encompassed by G511 to S520 that is required for motility but not signal transduction and the C-terminal extension present in C. jejuni FliF are indicated with boxes. Download
Analysis of the ability of the C. jejuni FliFΔG511-S520 mutant to support flaB::astA expression and motility. Wild-type C. jejuni or C. jejuni ΔfliF complemented with vector alone (vec or -) or plasmid encoding wild-type fliF or fliFΔG511-S520 was analyzed for production of FliF and FliG (A), expression of flaB::astA (B), or motility (C). (A) Immunoblot analysis of FliF, FliG, and RpoA production in whole-cell lysates of C. jejuni strains. Proteins were detected by specific antisera. (B) Arylsulfatase assays examining expression of the flaB::astA transcriptional fusion in wild-type C. jejuni or ΔfliF complemented with vector alone (vec) or plasmid encoding wild-type fliF or fliFΔG511-S520. The level of flaB::astA expression in each complemented fliF mutant is relative to the level in wild-type C. jejuni, which was set to 100 units. Error bars indicate standard deviations of the average arylsulfatase activities analyzed from three samples. *, indicates the complemented mutant had a significantly different level of reporter activity than ΔfliF harboring vector alone (P < 0.05). (C) Motility of strains after 24 h following inoculation in semisolid MH agar and incubation at 37°C in microaerobic conditions. Download
Analysis of production of proteins in C. jejuni strains used in coimmunoprecipitation assays. Immunoblot analysis of production of wild-type FliF, FLAG-FliF (wild type), FLAG-FliFΔA175-S176, wild-type FliG, wild-type FliG-FLAG, FlgS, FliY, and RpoA in whole-cell lysates of C. jejuni strains. The C. jejuni strains included ΔfliF, ΔfliF ΔflhA, ΔfliG, or ΔfliG ΔflhA mutants containing plasmid that expressed the FLAG-tag alone (-), FLAG-FliF (wild type), FLAG-FliFΔA175-S176 (ΔAS), or wild-type FliG-FLAG. FLAG-tagged FliF proteins were only expressed in the ΔflhF and ΔfliF ΔflhA mutants, and wild-type FliG-FLAG was only expressed in the ΔfliG and ΔfliG ΔflhA mutants. All proteins were detected with specific antisera. Arrow indicates wild-type FliF or FLAG-wild-type FliF, and arrowhead indicates a truncated FliF protein. Download
Bacterial strains used in this study.
Plasmids used in this study.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01AI065539 and R21AI103643. J.M.B. was supported by NIH training grant T32 AI007520.
Footnotes
Citation Boll JM, Hendrixson DR. 2013. A regulatory checkpoint during flagellar biogenesis in Campylobacter jejuni initiates signal transduction to activate transcription of flagellar genes. mBio 4(5):e00432-13. doi:10.1128/mBio.00432-13.
REFERENCES
- 1. Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hendrixson DR, DiRita VJ. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687–702 [DOI] [PubMed] [Google Scholar]
- 3. Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, Dehio C, Stack A, Meyer TF, Suerbaum S, Josenhans C. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52:947–961 [DOI] [PubMed] [Google Scholar]
- 4. Prouty MG, Correa NE, Klose KE. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595–1609 [DOI] [PubMed] [Google Scholar]
- 5. Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809–824 [DOI] [PubMed] [Google Scholar]
- 6. Correa NE, Lauriano CM, McGee R, Klose KE. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743–755 [DOI] [PubMed] [Google Scholar]
- 7. Spohn G, Scarlato V. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wösten MM, Wagenaar JA, van Putten JP. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214–16222 [DOI] [PubMed] [Google Scholar]
- 9. Joslin SN, Hendrixson DR. 2008. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J. Bacteriol. 190:2422–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joslin SN, Hendrixson DR. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J. Bacteriol. 191:2656–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boll JM, Hendrixson DR. 2011. A specificity determinant for phosphorylation in a response regulator prevents in vivo cross-talk and modification by acetyl phosphate. Proc. Natl. Acad. Sci. U. S. A. 108:20160–20165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bush M, Dixon R. 2012. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol. Mol. Biol. Rev. 76:497–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barrero-Tobon AM, Hendrixson DR. 2012. Identification and analysis of flagellar coexpressed determinants (Feds) of Campylobacter jejuni involved in colonization. Mol. Microbiol. 84:352–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buelow DR, Christensen JE, Neal-McKinney JM, Konkel ME. 2011. Campylobacter jejuni survival within human epithelial cells is enhanced by the secreted protein CiaI. Mol. Microbiol. 80:1296–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balaban M, Hendrixson DR. 2011. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog. 7:e1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Balaban M, Joslin SN, Hendrixson DR. 2009. FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J. Bacteriol. 191:6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kazmierczak BI, Hendrixson DR. 2013. Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol. Microbiol. 88:655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Green JC, Kahramanoglou C, Rahman A, Pender AM, Charbonnel N, Fraser GM. 2009. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J. Mol. Biol. 391:679–690 [DOI] [PubMed] [Google Scholar]
- 19. Murray TS, Kazmierczak BI. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188:6995–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65:389–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minamino T, Macnab RM. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182:4906–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kubori T, Yamaguchi S, Aizawa S. 1997. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J. Bacteriol. 179:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wagner S, Königsmaier L, Lara-Tejero M, Lefebre M, Marlovits TC, Galán JE. 2010. Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U. S. A. 107:17745–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Sourjik V. 2011. Assembly and stability of flagellar motor in Escherichia coli. Mol. Microbiol. 80:886–899 [DOI] [PubMed] [Google Scholar]
- 25. Diepold A, Wiesand U, Cornelis GR. 2011. The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol. Microbiol. 82:502–514 [DOI] [PubMed] [Google Scholar]
- 26. McMurry JL, Van Arnam JS, Kihara M, Macnab RM. 2004. Analysis of the cytoplasmic domains of Salmonella FlhA and interactions with components of the flagellar export machinery. J. Bacteriol. 186:7586–7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. U. S. A. 89:6304–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kihara M, Miller GU, Macnab RM. 2000. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 182:3022–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paul K, Gonzalez-Bonet G, Bilwes AM, Crane BR, Blair D. 2011. Architecture of the flagellar rotor. EMBO J. 30:2962–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D. 2010. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466:996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kihara M, Minamino T, Yamaguchi S, Macnab RM. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 183:1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Müller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J. 30:2972–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karlinsey JE, Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa SI, Hughes KT. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220–1231 [DOI] [PubMed] [Google Scholar]
- 35. Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280 [DOI] [PubMed] [Google Scholar]
- 36. Wösten MM, van Dijk L, Veenendaal AK, de Zoete MR, Bleumink-Pluijm NM, van Putten JP. 2010. Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol. Microbiol. 75:1577–1591 [DOI] [PubMed] [Google Scholar]
- 37. Minamino T, Yamaguchi S, Macnab RM. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J. Bacteriol. 182:3029–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hirano T, Minamino T, Namba K, Macnab RM. 2003. Substrate specificity classes and the recognition signal for Salmonella type III flagellar export. J. Bacteriol. 185:2485–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Henderson MJ, Milazzo FH. 1979. Arylsulfatase in Salmonella typhimurium: detection and influence of carbon source and tyramine on its synthesis. J. Bacteriol. 139:80–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yao R, Guerry P. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 178:3335–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hendrixson DR. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646–1659 [DOI] [PubMed] [Google Scholar]
- 42. Sham LT, Barendt SM, Kopecky KE, Winkler ME. 2011. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc. Natl. Acad. Sci. U. S. A. 108:E1061–E1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson TL, Scott ME, Sandkvist M. 2007. Mapping critical interactive sites within the periplasmic domain of the Vibrio cholerae type II secretion protein EpsM. J. Bacteriol. 189:9082–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods for constructing bacterial strains and plasmids used in this study. Download
Alignment of the N-terminal FliG and C-terminal FliF domains from E. coli, Salmonella, C. jejuni, and H. pylori. (A) ClustalW alignment of the N-terminal domains of FliG from E. coli strain K-12 (BAA15764), S. Typhimurium strain LT2 (AAL20882), C. jejuni 81-176 (EAQ73253), and H. pylori strain G27 (ACI27095). The N-terminal extension present in C. jejuni and H. pylori FliG is indicated in a box. (B) ClustalW alignment of the C-terminal transmembrane (TM) and cytoplasmic domains of FliF proteins from E. coli strain K-12 (YP_490192), S. Typhimurium LT2 (AAL20881), C. jejuni 81-176 (EAQ73453), and H. pylori G27 (ACI27094). The transmembrane (TM) domain predicted in all proteins is boxed. The domain of C. jejuni FliF encompassed by G511 to S520 that is required for motility but not signal transduction and the C-terminal extension present in C. jejuni FliF are indicated with boxes. Download
Analysis of the ability of the C. jejuni FliFΔG511-S520 mutant to support flaB::astA expression and motility. Wild-type C. jejuni or C. jejuni ΔfliF complemented with vector alone (vec or -) or plasmid encoding wild-type fliF or fliFΔG511-S520 was analyzed for production of FliF and FliG (A), expression of flaB::astA (B), or motility (C). (A) Immunoblot analysis of FliF, FliG, and RpoA production in whole-cell lysates of C. jejuni strains. Proteins were detected by specific antisera. (B) Arylsulfatase assays examining expression of the flaB::astA transcriptional fusion in wild-type C. jejuni or ΔfliF complemented with vector alone (vec) or plasmid encoding wild-type fliF or fliFΔG511-S520. The level of flaB::astA expression in each complemented fliF mutant is relative to the level in wild-type C. jejuni, which was set to 100 units. Error bars indicate standard deviations of the average arylsulfatase activities analyzed from three samples. *, indicates the complemented mutant had a significantly different level of reporter activity than ΔfliF harboring vector alone (P < 0.05). (C) Motility of strains after 24 h following inoculation in semisolid MH agar and incubation at 37°C in microaerobic conditions. Download
Analysis of production of proteins in C. jejuni strains used in coimmunoprecipitation assays. Immunoblot analysis of production of wild-type FliF, FLAG-FliF (wild type), FLAG-FliFΔA175-S176, wild-type FliG, wild-type FliG-FLAG, FlgS, FliY, and RpoA in whole-cell lysates of C. jejuni strains. The C. jejuni strains included ΔfliF, ΔfliF ΔflhA, ΔfliG, or ΔfliG ΔflhA mutants containing plasmid that expressed the FLAG-tag alone (-), FLAG-FliF (wild type), FLAG-FliFΔA175-S176 (ΔAS), or wild-type FliG-FLAG. FLAG-tagged FliF proteins were only expressed in the ΔflhF and ΔfliF ΔflhA mutants, and wild-type FliG-FLAG was only expressed in the ΔfliG and ΔfliG ΔflhA mutants. All proteins were detected with specific antisera. Arrow indicates wild-type FliF or FLAG-wild-type FliF, and arrowhead indicates a truncated FliF protein. Download
Bacterial strains used in this study.
Plasmids used in this study.