ABSTRACT
The DNA binding activity of the photosystem-specific repressor PpsR is known to be repressed by the antirepressor AppA. AppA contains a blue-light-absorbing BLUF domain and a heme-binding SCHIC domain that controls the interaction of AppA with PpsR in response to light and heme availability. In this study, we have solved the structure of the SCHIC domain and identified the histidine residue that is critical for heme binding. We also demonstrate that dark-adapted AppA binds heme better than light-excited AppA does and that heme bound to the SCHIC domain significantly reduces the length of the BLUF photocycle. We further show that heme binding to the SCHIC domain is affected by the redox state of a disulfide bridge located in the Cys-rich carboxyl-terminal region. These results demonstrate that light, redox, and heme are integrated inputs that control AppA’s ability to disrupt the DNA binding activity of PpsR.
IMPORTANCE
Photosynthetic bacteria must coordinate synthesis of the tetrapyrroles cobalamin, heme, and bacteriochlorophyll, as overproduction of the latter two is toxic to cells. A key regulator controlling tetrapyrrole biosynthesis is PpsR, and the activity of PpsR is controlled by the heme-binding and light-regulated antirepressor AppA. We show that AppA binds heme only under dark conditions and that heme binding significantly affects the length of the AppA photocycle. Since AppA interacts with PpsR only in the dark, bound heme thus stimulates the antirepressor activity of PpsR. This causes the redirection of tetrapyrrole biosynthesis away from heme into the bacteriochlorophyll branch.
Introduction
AppA from Rhodobacter sphaeroides (1) is a blue-light photoreceptor discovered a decade ago that utilizes flavin as a chromophore. It is among the first identified members of a broadly disseminated BLUF (sensors of blue light using FAD) family of photoreceptors (2). Since their identification, there has been extensive research on the BLUF photocycle providing significant insight on how light absorption by a bound flavin leads to a conformational change in the BLUF domain (3). In many cases, it remains unclear how light-driven alterations in the BLUF input domain result in changes in the downstream output module. Among BLUF proteins with a linked output domain, only two examples of downstream signal transduction events have been described at a molecular level. One example is YcgF from Escherichia coli (4, 5) and its homolog BlrP1 in Klebsiella pneumoniae (6) where light excitation of the BLUF domain stimulates phosphodiesterase activity of the EAL output domain that converts c-di-GMP to linear diguanylate (6). The other example is AppA where light excitation of the BLUF domain inhibits an output domain to function as an antirepressor of the photosystem-specific repressor PpsR (1).
PpsR in R. sphaeroides controls heme, bacteriochlorophyll, and photosynthesis gene expression with the activity of the PpsR-AppA system highly regulated by numerous input signals such as light, redox, and heme (1, 7, 8). Several input signals directly affect the activity of PpsR, while others affect the ability of AppA to interact with PpsR. For example, under aerobic growth conditions, two cysteine residues in PpsR are in an oxidized state which stimulates its DNA binding activity (1). This is contrasted by anaerobic conditions where these cysteines are in a reduced state which impedes DNA binding. PpsR also binds heme via a conserved Cys residue present in the DNA-binding domain and a histidine present in a Per-Arnt-Sim (PAS) domain (7). Bound heme affects the interaction between PpsR and its target DNA (7).
The antirepressor AppA consists of an N-terminal BLUF domain that utilizes flavin to sense light, a middle SCHIC (sensor containing heme instead of cobalamin) domain (9) that binds heme and a redox-responding C-terminal Cys-rich domain (Fig. 1A). The Cys-rich carboxyl domain is capable of reducing the redox-active Cys in PpsR under dark anaerobic conditions to lower the DNA binding affinity of PpsR (1, 8). Additionally, dark-adapted AppA is able to form a stable PpsR2-AppA complex that cannot bind to target DNA sequences (1). Interestingly, the interaction of AppA with PpsR is inhibited by blue-light absorption of the BLUF domain, a regulatory feature that imparts light control of photosynthesis gene expression (1). A final aspect of AppA regulation involves binding of heme to the SCHIC domain of AppA that is thought to promote interaction of AppA with PpsR (10). The SCHIC domain has sequence similarity to the well-characterized vitamin B12-binding domain of methyltransferases (9, 10), but unlike methyltransferases, the SCHIC domain differentially binds heme over B12 (9, 10).
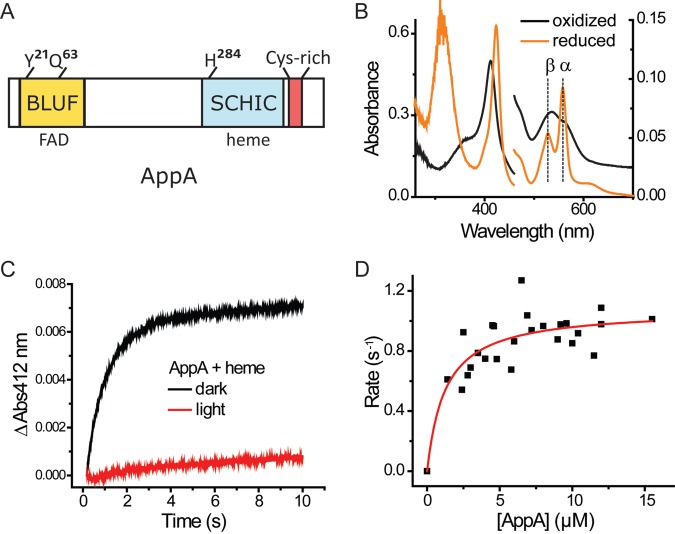
FIG 1 .
AppA binds heme. (A) Domain structure of AppA. (B) UV-visible spectrum of AppA-heme reconstituted under aerobic conditions (the spectrum of AppA with flavin is subtracted). Heme (5 µM) was incubated with 10 µM AppA for at least 20 min before the oxidized spectrum was taken. For the reduced spectrum, 10 mM dithionite was added to the sample of AppA-heme right before the measurement was taken. It should be noted that the oxidized versus reduced spectrum refer to the redox state of heme iron (Fe3+ versus Fe2+), not the presence of oxygen (aerobic versus anaerobic). Absorbance is shown on both the right and left y axes. (C) Dynamics of AppA-heme reconstitution. AppA (12 µM) and 1 µM heme (final concentrations for both) were mixed and monitored in a stopped-flow spectrophotometer in the dark or illuminated with white light. The change in absorbance at 412 nm is shown on the y axis. (D) Kd of AppA-heme interaction. Reaction rates at different AppA concentration were fitted with a hyperbola model.
Several research groups have reported that the SCHIC domain binds heme, but the details of this interaction have not been clarified. One study used an AppA variant that had the C-terminal Cys-rich motif replaced with a maltose-binding protein (MBP). Studies with this construct indicated that heme binds to AppA-MBP only in the absence of oxygen and that heme is gradually discoordinated from AppA as the oxygen level increases (9). A second heme binding study used a His6-tagged AppA variant that lacked the BLUF domain (ΔN-AppA-His6) (10). Since neither study tested full-length untagged AppA, it has remained unclear whether oxygen or light has an effect on heme binding to AppA.
In this study, we demonstrate that dark-adapted AppA binds heme better than light-excited AppA does and that heme bound to the SCHIC domain significantly reduces the length of the BLUF photocycle. We also solved the structure of the SCHIC domain and identified the histidine residue that is critical for heme binding. The interaction between AppA and heme is further shown to be affected by the redox state of a disulfide bridge located in the Cys-rich C-terminal region. These results indicate that light, redox, and heme sensing by AppA are integrated in this single protein and that AppA antirepression of PpsR activity in response to alterations in redox, light, and heme is significantly more nuanced and complex than previously reported.
RESULTS
AppA binds heme under both anaerobic and aerobic conditions.
We examined the interaction of AppA with heme by purifying tagless full-length AppA from an E. coli overexpression system. As previously reported by our group, there were no significant amounts of heme observed when full-length AppA was purified from the E. coli overexpression system (1). However, it does contain flavin adenine dinucleotide (FAD) as the BLUF domain cofactor. To ensure that our experimental conditions covered a wide range of oxygen concentrations, we prepared AppA either in degassed buffer (oxygen level of <1 mg/liter) or in air-saturated buffer (oxygen level of >6 mg/liter) for use in interaction studies with heme. Both samples exhibited almost identical absorption spectra, indicating no oxygen-dependent effect on heme binding by full-length tagless AppA (see Fig. S1 in the supplemental material). This is in contrast to a previous report that AppA binds to heme only when oxygen is absent (9). Calculating the amount of heme binding in replicate reconstitution experiments showed that AppA binds heme at a ratio of 1.17 ± 0.07 heme per 1 AppA, further confirming that full-length AppA can bind heme in the presence of oxygen.
Spectral analysis of the full-length AppA-heme complex showed characteristics similar to those previously reported with His6-tagged AppA that lacked the BLUF domain and AppA that had the Cys-rich carboxyl domain replaced with a maltose-binding domain (9, 10). The spectrum with ferric (Fe3+) heme has a well-defined Soret peak at 412 nm with broad peaks in the 500- to 600-nm region (Fig. 1B). Reduction of heme iron to Fe2+ with 10 mM dithionite yields a Soret peak at 424 nm and α and β peaks at 558 nm and 528 nm, respectively (Fig. 1B). We also performed stopped-flow analysis to measure the kinetics of heme binding by AppA. After mixing AppA with heme under dark aerobic conditions, the time course of heme binding as measured by monitoring ΔA412 could be described by a single exponential equation (Fig. 1C). The heme-binding rate under dark aerobic conditions was fitted with the concentration of AppA, yielding a Kd (dissociation constant) of 1.25 ± 0.44 µM (Fig. 1D). This rather weak binding affinity is similar to values reported for other heme-sensing proteins such as PpsR (~1.9 µM) (7) and R-transferase (~0.15 µM) (11).
Light excitation of the BLUF domain impedes heme binding by the SCHIC domain.
To test whether the light-regulated BLUF domain has a role in regulating SCHIC domain interaction with heme, we monitored the kinetics of heme binding in dark- and light-adapted AppA using stopped-flow analysis. As shown in Fig. 1C, there is no significant binding of heme to lit AppA observed over the course of the experiment. This is in contrast to dark-adapted AppA that exhibits exponential binding of heme as monitored by amplification of ΔA412 (Fig. 1C). This result indicates that AppA binds heme effectively only under dark conditions.
In previous studies, we described several mutations in the BLUF domain (AppAY21F [AppA with a Y-to-F change at position 21] and AppAQ63E) that remain in a lit signaling state irrespective of whether these proteins are under dark or illuminated conditions (12, 13). We assayed heme binding with AppAY21F and AppAQ63E in the dark to confirm whether there is indeed a connection between light sensing and heme binding. As shown in Fig. 2A, incubation of AppAY21F and AppAQ63E with heme for 30 min shows that both proteins bind heme very poorly in the dark as evidenced by large amounts of free unbound heme with a peak at 370 nm and only a slight shoulder of bound heme with a peak at 412 nm. This is in contrast to wild-type AppA, which shows excellent binding under similar dark conditions (Fig. 2A).
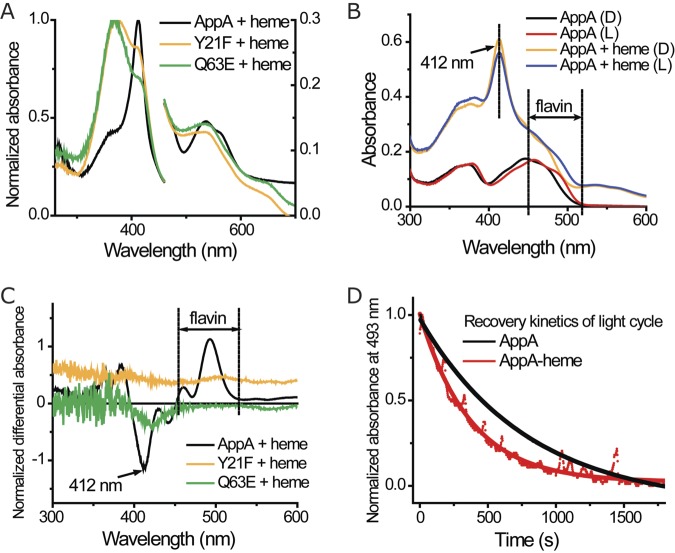
FIG 2 .
Integration of light sensing and heme binding in AppA. (A) Absorption spectrum of heme and different AppA constructs under aerobic conditions. AppA mutants and heme were mixed at a 2:1 ratio and incubated for at least 20 min before the spectrum was taken. The spectrum of the protein with flavin is subtracted. Normalized absorbance is shown on the right and left y axes. (B) Absorption spectrum of AppA and AppA-heme complex in dark (D) and lit (L) states. The spectrum of protein was not subtracted. (C) Differential spectrum of AppA mutants and heme (dark state minus light state). The samples in Fig. 2A were used here. (D) Recovery kinetics of AppA/AppA-heme light cycle. AppA and AppA-heme complex were light excited and put back in dark conditions. Absorbance at 493 nm was measured to monitor the photocycle of the BLUF domain, and the data were fitted to a single-exponential model.
We also observed that light excitation of the BLUF domain affected the spectrum of prebound heme. In this analysis, light excitation of AppA-heme complex resulted in a slight but reproducible reduction of the Soret peak at 412 nm (Fig. 2B), which indicates a weakening of AppA-heme interaction. A dark-minus-light differential spectrum of the AppA-heme sample clearly shows this reduction at 412 nm and an alteration of flavin absorbance at the 500-nm region attributed to the BLUF photocycle (Fig. 2C). Note that similar analysis with BLUF mutants AppAY21F and AppAQ63E did not show alterations in the Soret peak at 412 nm upon exposure to light, as these mutants are in locked in an illuminated state (Fig. 2C).
Bound heme pushes the BLUF domain toward the dark state conformation.
Given that light excitation of the BLUF domain affects heme binding to the SCHIC domain, we asked whether heme bound to the SCHIC domain inversely affects the state of the BLUF domain. To test this possibility, we measured the BLUF photocycle by monitoring spectral changes in the flavin absorption region. Dark-adapted AppA has a broad flavin absorption peak with a maximum at 450 nm that upon light excitation redshifts ~10 nm as a result of a conformational change in the BLUF domain. This excitation spectral shift subsequently falls back to the ground state with a half-life of ~15 min when the protein returns from a lit to a dark conformation (1). The dark and lit state flavin absorption spectra show the same spectral features in both AppA and AppA with bound heme indicating that the BLUF domain has the same general conformation whether or not heme is bound (Fig. 2B). However, the length of the AppA-heme photocycle is considerably shorter, with a decay half-life of ~4 min compared to AppA itself that has a half-life of ~15 min (Fig. 2D). This indicates that the BLUF domain prefers a dark conformation when heme is bound to the SCHIC domain.
The SCHIC domain uses His284 to interact with heme.
We obtained crystals of the SCHIC domain (AppA amino acids 186 to 398) (Fig. 3A; see Table S1 in the supplemental material) without bound heme using hanging drop vapor diffusion. Unfortunately, the SCHIC domain alone does not incorporate heme in stoichiometric amounts, so we were not successful in growing SCHIC crystals containing heme. Due to the inherent problems of heme solubility, we were also unsuccessful in loading heme by soaking protein crystals with heme containing mother liquor. Nevertheless, the solved SCHIC domain apo-structure at 2.04 Å does provide valuable information on a putative heme-binding cleft.
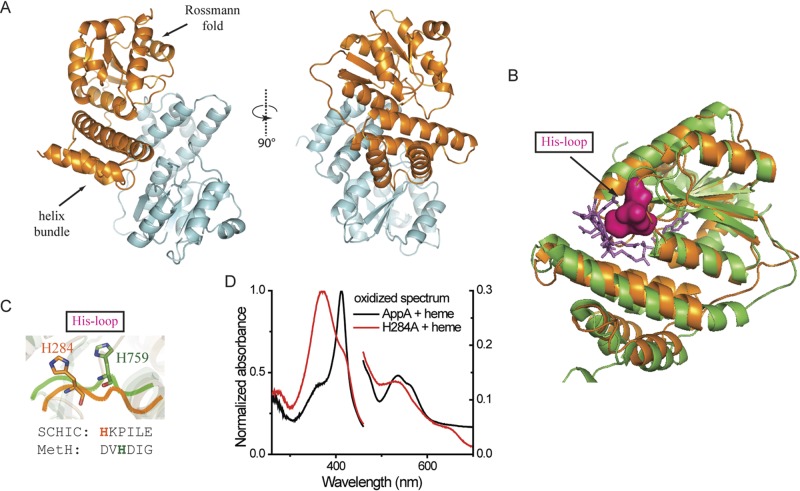
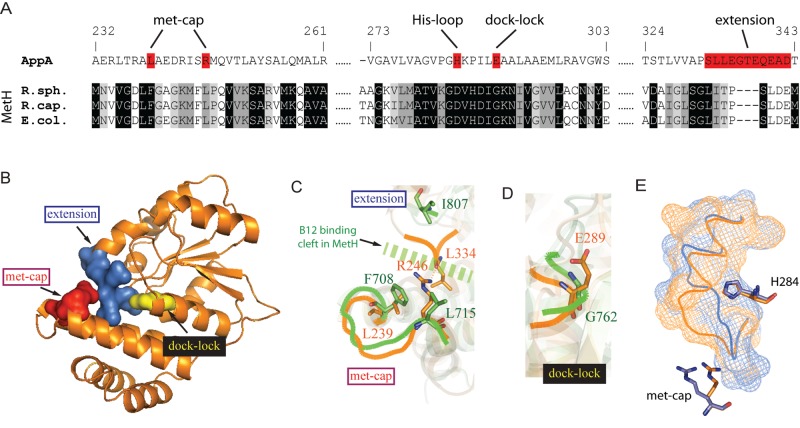
FIG 3 .
Crystal structure of SCHIC domain and His284 as the heme coordinating residue. (A) Crystal structure of SCHIC domain contains two subunits (subunit A shown in orange, and subunit shown B in light blue). (B) SCHIC domain (subunit A [orange]) shares similar overall conformation with E. coli MetH B12-binding domain (green). The His loop in MetH is highlighted in magenta, and the vitamin B12 molecule is shown as purple sticks. Subunit A of SCHIC domain is used for the structure comparison. (C) His284 in SCHIC (orange) and His759 in MetH (green) are located on the His loop region. Note the differences in their positions and orientations. (D) UV-visible spectrum of AppA-heme and AppAH284A-heme under aerobic conditions. The spectrum of AppA/AppAH284A is subtracted. Heme (5 µM) was incubated with 10 µM AppA/AppAH284A for at least 20 min before the spectrum was taken. Normalized absorbance is shown on both the right and left y axes.
The crystal lattice contains two copies of a homodimeric SCHIC domain that have similar conformations with a root mean square deviation (RMSD) of 0.40 Å between the dimers. Individual subunits are comprised of an N-terminal helix bundle followed by a C-terminal α/β Rossmann fold (Fig. 3A). As shown in Fig. 3B, the SCHIC structure superposes very closely with the vitamin B12-binding domain from methionine synthase (MetH, 1BMT.pdb) (Fig. 3B) (14). The MetH-B12 structure contains a well-defined B12-binding cleft, with a loop where a highly conserved histidine residue directly coordinates with the cobalt in B12 (Fig. 3C) (14). The SCHIC domain also contains a similar loop with a potential iron-coordinating histidine (His284) located 2 amino acids further down this loop than for the cobalt-coordinating histidine present in MetH-B12 (Fig. 3C). The imidazole ring in SCHIC His284 is also rotated and moved with an average distance of 6.4 Å from the His759 present in MetH (Fig. 3C). To test whether heme forms a ligand to His284, we mutated His284 to alanine within full-length AppA and then spectrally analyzed the ability of the mutant peptide to bind heme. Incubation of AppAH284A with heme showed a peak at 370 nm, which is the spectral characteristic of free unbound heme compared to the 412-nm Soret peak of heme bound to wild-type AppA (Fig. 3D). This observation indicates that the Ala substitution disrupted heme and that His284 indeed coordinates with the heme iron.
A disulfide bond in the Cys-rich motif affects the SCHIC-heme interaction.
Even though full-length AppA is capable of binding heme in the presence of molecular oxygen, this result does not rule out that changes in the redox state of the Cys-rich C-terminal domain can affect heme binding. Previous studies from our laboratory have shown that AppA contains a single redox active pair of cysteines that function as a dithiol/disulfide couple with a midpoint potential (Em) of −325 mV (8). This potential is very similar to the Em −320 mV potential of a disulfide pair in PpsR that AppA is capable of reducing (1, 8). To determine which cysteines are redox active, we performed matrix-assisted laser desorption ionization−time of flight (MALDI-TOF) analysis of trypsin-digested iodoacetamide (IAM)-labeled AppA. The tryptic peptide AppA397-409 was further analyzed using liquid chromatography coupled to electrospray ionization tandem mass spectrometric analysis (LC-ESI-MS-MS) which shows the presence of a Cys399–Cys406 disulfide bridge (Fig. 4; see Fig. S2 in the supplemental material).
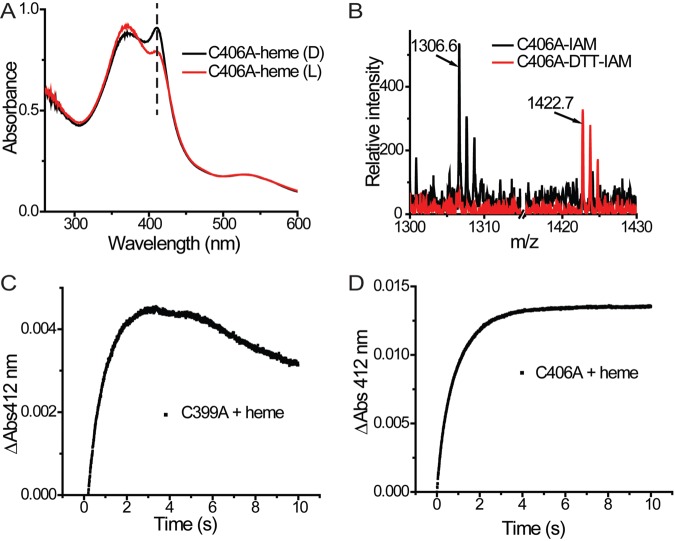
FIG 4 .
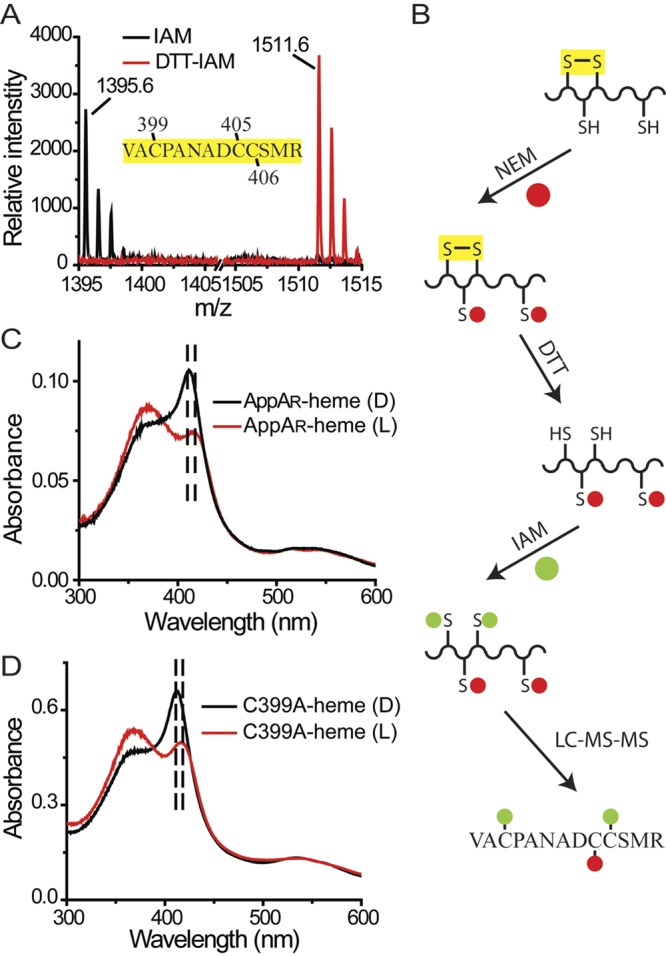
The disulfide bridge in the Cys-rich motif affects the SCHIC-heme interaction. (A) A disulfide bridge was identified within peptide AppA397-409. Under oxidized conditions, neither Cys399 nor Cys406 can be modified by iodoacetamide (IAM). Under reduced conditions, the disulfide bridge formed between Cys399 and Cys406 is reduced, and both residues can be modified by iodoacetamide (DTT-IAM). (B) LC-MS-MS with double-labeled AppA identified the Cys399-Cys406 disulfide bond (see Fig. S2 in the supplemental material). (C) The cysteine residues in AppA are reduced by DTT (AppAR). When light excited, the Soret peak of AppAR-heme is redshifted from 412 nm to 418 nm. (D) When light excited, the Soret peak of AppAC399A-heme is redshifted from 412 nm to 418 nm.
To address whether the Cys399–Cys406 disulfide bond affects the AppA-heme interaction, we reduced the disulfide bond with dithiothreitol (DTT) and then analyzed the heme spectrum. The heme spectrum of reduced AppA exhibits a normal Soret peak at 412 nm in the dark state. However, when the BLUF domain is light excited, the heme Soret peak in reduced AppA exhibits a 6-nm redshift along with a decrease in intensity (Fig. 4C). This is distinctly different than that of oxidized wild-type AppA where the heme Soret peak decreases without a redshift upon light excitation (Fig. 2B). To confirm that this effect is a result of the reduced disulfide bridge, we constructed a Cys399-to-Ala point mutation. Purified AppAC399A also exhibited the same 6-nm redshift along with a decrease in intensity as described above for reduced AppA (Fig. 4D). Interestingly, AppA with a Cys406 Ala mutation (the other residue of the disulfide pair) exhibited a dark/light spectrum that is similar to the spectrum of oxidized wild-type AppA (Fig. 5A). Further analysis showed that a disulfide bond can form between Cys399 and a neighboring Cys (Cys405) in this construct (Fig. 5B), which appears to function like the original Cys399–Cys406 disulfide bond present in wild-type AppA. Taken together, these results indicate that the presence of a disulfide bridge in the Cys-rich motif has an effect on the heme binding properties of the SCHIC domain.
FIG 5 .
(A) When light excited, the Soret peak of AppAC406A-heme remains at 412 nm (indicated by the vertical broken black line). (B) A disulfide bond exists in peptide AppA397-409 in the AppAC406A construct which can only be Cys399-Cys405. (C) Dynamics of AppAC399A-heme reconstitution. (D) Dynamics of AppAC406A-heme reconstitution.
The effect of this reduced disulfide on heme binding can also be demonstrated by assaying absorbance changes using stopped-flow experiments. As discussed above, heme binding with wild-type AppA can be analyzed by stopped-flow analysis by measuring the rise in absorbance at 412 nm that can be described in a single exponential equation (Fig. 1C). Loss of the Cys399–Cys406 disulfide bridge in AppAC399A results in a similar rapid rise at 412 nm, followed first by a short plateau and then by a slow decrease of the Soret peak (Fig. 5C). As is the case of wild-type AppA, AppAC406A, which forms the alternative Cys399–Cys405 disulfide bond, does not exhibit a slow decrease in the Soret peak after binding heme (Fig. 5D). We interpret this result as evidence that the binding of heme in reduced AppA first involves the formation of the 412-nm Soret spectral species followed by slow decay to an alternate state that is capable of undergoing the 6-nm redshift upon light excitation of the BLUF domain.
Finally, as discussed earlier, the presence of heme in AppA that has Cys399–Cys405 in its oxidized disulfide state shortens the BLUF photocycle half-life to ~4 min, which is significantly faster than the 15-min half-life observed with oxidized AppA that does not contain heme (Fig. 2D). Interestingly, heme-induced reduction of the BLUF photocycle is absent when the disulfide in AppA is reduced with DTT. Under this condition, the photocycle half-life is ~11 min with or without bound heme. The latter result suggests that the redox state of this disulfide affects signal communication between the BLUF domain and the heme-binding SCHIC domain.
DISCUSSION
Numerous studies show that Rhodobacter sphaeroides uses the PpsR-AppA regulatory system to coordinate biosynthesis of heme and bacteriochlorophyll with synthesis of photosystem apoproteins (1, 15, 16). In this study, we demonstrate that tagless full-length AppA can bind heme under both anaerobic and aerobic conditions (Fig. 1B). This finding is in contrast to a report by Moskvin et al. (9), which indicated that oxygen can discoordinate heme from AppA in a concentration-dependent manner. This apparent difference likely stems from the fact that our study utilized full-length tagless AppA, while the study by Moskvin et al. utilized an MBP-tagged AppA that also contained a truncation of the Cys-rich motif.
If heme is not an oxygen sensor, then what is its function when it is bound to the SCHIC domain of AppA? The absorption characteristics of the flavin region in both dark and light states are identical with or without bound heme, indicating that the BLUF domain undergoes a similar hydrogen bond rearrangement in the presence or absence of heme (Fig. 2B). However, the length of the AppA photocycle is dramatically reduced when oxidized AppA contains bound heme relative to that observed with AppA alone. The consequence of a quicker photocycle is that it would increase the population of AppA that is present in the dark state. Given that only AppA in the dark state is capable of interacting with PpsR, the binding of heme would therefore increase the antirepressor activity of AppA on PpsR (Fig. 6A). This would result in increased bacteriochlorophyll and light-harvesting gene expression to promote maximal synthesis of the photosystem (Fig. 6B). The involvement of heme in controlling the AppA photocycle appears to be a means to ensure that increased synthesis of the photosystem does not occur unless there is sufficient heme available to handle the essential function of electron transfer. In purple bacteria, cytochrome biosynthesis is essential, while synthesis of the photosystem is not. Furthermore, electrons derived from photosynthesis need to be shuttled back to the photosystem via heme-containing cytochromes. Consequently, it is not surprising that heme has a role in controlling the amount of photosystem synthesis via the regulation of AppA activity.
FIG 6 .
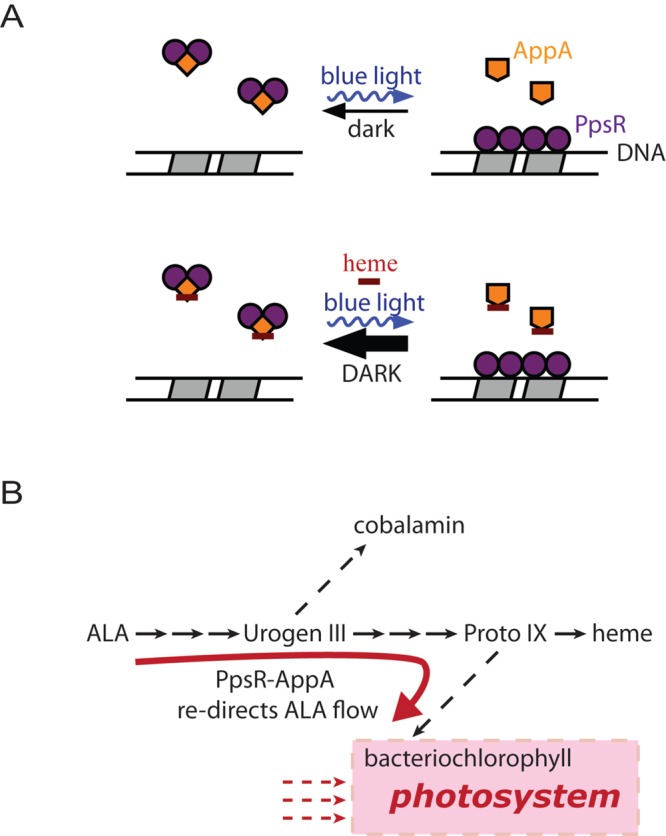
Proposed model of heme function. (A) Dark-adapted AppA can form the PpsR2-AppA complex and inactivate PpsR, thus enhancing the target gene transcription. When light excited, AppA is incapable of interacting with PpsR, leaving PpsR bound to its target DNA. The presence of heme pushes AppA toward its dark state and thus promotes the formation of the PpsR2-AppA complex. (B) Diagram of tetrapyrrole biosynthesis and photosynthesis. The presence of heme could potentially redirect the flow of 5-aminolevulinic acid (ALA) away from the heme branch and into the bacteriochlorophyll branch. Urogen III, uroporphyrinogen III; Proto IX, protoporphyrin IX.
The solved crystal structure of the SCHIC domain shows remarkable sequence similarity with the vitamin B12-binding domain in MetH (9, 10). Heme and vitamin B12 have significant different structures and central metals. For example, heme b has methyl and formyl side groups with the bound iron free to form axial ligands. This is in contrast to vitamin B12 where the corrin ring has 7 propionamide side groups and 2 alcohol side groups as well as a bulky attached dimethylbenzimidazol ribonucleotide group that forms a lower axial ligand to the bound cobalt. B12 also contains a second upper axial cobalt (Co) ligand typically comprised of a methyl or 5′-deoxyadenosine group. Thus, it is surprising that the crystal structure of the SCHIC domain is so similar to the structure of the B12-binding domain from MetH (Fig. 2B).
Despite these similarities, there are several subtle changes that likely contribute to tetrapyrrole selectivity (Fig. 7A). For example, the loop in MetH that forms a lower axial ligand of His to Co is altered in the SCHIC structure such that the His is displaced by two residues (Fig. 3C). In addition, MetH-B12 has Phe708 and Leu715 located at one end of the second helix in the helix bundle that serves as a cap to cover the reactive methyl ligand of cobalamin (met cap) (14). Located in a similar position of this helix in the SCHIC structure is Leu239 and a positive charged Arg246 that are in contact with a loop extended from the Rossmann fold (extension) (Fig. 7B). These changes would block the binding of Met- or Adeno-B12 to the SCHIC domain (Fig. 7C). This extension also forms a hydrogen bond network with the His loop in the SCHIC domain that locks the orientation of Fe-binding His284 in a position (see Fig. S4 in the supplemental material) that is distinct from the Co-coordinating His759 in MetH-B12 (14). Finally, there is additional space in the core of the MetH-B12 Rossmann fold that anchors the bulky attached nucleotide tail of vitamin B12 (14). In the SCHIC domain, the access to this docking space is blocked by Glu289 which resides on the His loop (dock lock [Fig. 7B and D]). These changes likely constitute selectivity of these similar tetrapyrrole binding domains for heme versus B12.
FIG 7 .
Architectural basis of heme/B12 differentiation. (A) Sequence alignment of AppA SCHIC and selected MetH vitamin B12-binding domains. Three sequences, the R. sphaeroides, Rhodobacter capsulatus, and E. coli sequences, are shown. The MetH B12-binding domains were aligned first. Residues that are identical, highly similar, or less similar were shown with black, medium gray, or light gray background, respectively. The alignment was then compared to the SCHIC sequence based on sequence and structure homology. The regions in AppA SCHIC domain that are critical for heme/B12 differentiation are shown on a red background. (B) Crystal structure of SCHIC domain (subunit A). Regions critical for heme/B12 differentiation other than the His loop are highlighted as followed: extension (blue), met cap (red), and dock lock (yellow). (C) Structure of the extension and met cap regions in SCHIC (orange) and MetH (green). Note that these two regions are in contact in SCHIC (orange) (hydrogen bond between R246 and L334) but far away from each other in MetH (green). The contact in SCHIC would block the B12 binding cleft in MetH (broken green line). (D) Structure of the dock lock in SCHIC (orange) and MetH (green). Note that E289 occupies more space in SCHIC (orange) than G762 does in MetH (green). (E) The extension loop in SCHIC is flexible and interacts differently with the met cap and His284 in molecule A (orange) and molecule B (blue) in the crystallographic dimer. The detailed hydrogen networks are shown in Fig. S3 in the supplemental material.
Interestingly, the described regions critical for heme/B12 differentiation map to an extension in the SCHIC domain Rossmann fold that exhibit different conformations in the two subunits of the SCHIC crystal structure (Fig. 7B and E). In one subunit, part of this extension is moved ~7 Å, forming different interactions with the methyl cap and the His loop (see Fig. S4 in the supplemental material). This could represent the “empty” state versus an “en route” position toward heme-bound conformation. As a heme-sensing module, SCHIC is probably not at its most stable state with and without bound heme, since it does not bind heme tightly. This might explain our failure to grow crystals of SCHIC-heme complex or to reconstitute SCHIC crystals with heme.
While this article was under review, several truncated structures of AppA and PpsR were also published by Winkler et al. (17). A co-complex of the SCHIC domain to a truncated region of PpsR that contained the amino terminus through the first PAS domain was obtained. This suggests that the SCHIC domain interacts with this region of PpsR (17). What remains unknown is the structure of the Cys-rich C terminus of AppA and the structures of full-length AppA and PpsR proteins. The Cys-rich domain of AppA contains six cysteine residues within a 21-amino-acid stretch that has a sequence pattern similar to Cys-rich motif found in granulin, a protein critical for neurodegeneration (18, 19), as well as several plant proteases (20–23). The solution structure of carp granulin-1 shows a hairpin stack fold that is stabilized by disulfide bridges (19). The redox-active cysteines in the region are thought to reduce the redox-active Cys in PpsR, which affects its DNA binding activity. Our results provide evidence that the redox state of AppA Cys-rich motif can affect signal transfer between the light-sensing BLUF domain and the heme-sensing SCHIC domain. The structures of full-length AppA and/or the intact PpsR-AppA complex will be necessary to determine how AppA is able to control the redox state of PpsR and to form a complex that inhibits its activity.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strain BL21(DE3) (Novagen) was used for protein overexpression in E. coli. Luria broth (LB) was used for agar-solidified plates and liquid cultures for E. coli. The antibiotics used and their final concentrations were as follows: ampicillin, 100 µg ml−1; kanamycin, 50 µg ml−1 for agar-solidified plates and 25 µg ml−1 for liquid cultures.
Plasmid construction.
Wild-type AppA, AppA Cys mutants (AppAC399A and AppAC406A) and AppA His mutants (AppAH284A, AppAH308A, and AppAH386A) were expressed in E. coli using modified small ubiquitin-like modifier SUMO-I (LifeSensors Inc.) overexpression system. The coding region was cloned from pTY-AppA (24) and inserted into the NdeI-NotI restriction sites of pSUMO. The pTY-AppA plasmid was subjected to site-directed mutagenesis (Herculase II; Stratagene) to create point mutations in the BLUF domain (AppAY21F and AppAQ63E). All point mutations were created by the standard QuikChange (Agilent Technologies) protocol.
Protein purification for biochemical experiments.
SUMO-tagged full-length AppA was overexpressed and purified as described previously (7). Proteins cloned into the pTY plasmid (AppAY21F and AppAQ63E) were purified using chitin beads and the manufacturer’s protocol (New England Biolabs) followed by gel filtration chromatography using Sephacryl S-200 HP resin (GE Healthcare). All purified proteins were purified in a buffer of 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 5% glycerol, and all experiments were performed in the same buffer unless otherwise noted.
Protein-heme interactions.
Heme reconstitution with AppA and AppA mutants was performed as described previously (7). The sample was either degassed or air bubbled when needed with dissolved oxygen concentration measured using an oxygen CHEMets kit (CHEMetrics). UV-visible absorption spectrum was recorded using a Beckman DU 640 spectrophotometer as described previously (7). To convert AppA into a light-excited state, dark-adapted samples were illuminated with strong white light for 30 s immediately before spectral analysis unless noted otherwise. Note that the heme spectrum itself stays identical in dark and illuminated conditions (see Fig. S4 in the supplemental material).
Purification of SCHIC for crystallization.
To construct an SCHIC domain expression vector, AppA with amino acids 186 to 398 (AppA186-398) was cloned into pTYB12 plasmid (New England BioLabs) with NotI-NdeI sites. E. coli BL21(DE3) cells were freshly transformed, grown on autoinducing medium LBE-5052 (25) for 3 h at 37°C, and then transferred to 16°C for protein expression overnight. Cells were collected and resuspended in chitin binding buffer (50 mM HEPES [pH 7.0], 50 mM NaCl, 0.2 mM EDTA) and lysed using the cell cracker. After centrifugation, the cell extract was passed through chitin beads at room temperature, and the cleavage of chitin tag was initiated with fresh 50 mM DTT in the chitin binding buffer. The next day, tagless protein was eluted and loaded onto Superose 12 for the final purification step. The final buffer was 50 mM HEPES and 400 mM NaCl. Pure (>95%) and soluble protein was obtained and concentrated to ~15 mg/ml for crystallization.
Crystallization of SCHIC.
The purified SCHIC was first screened for potential crystallization conditions using the Index screen by Hampton Research. The Index screen provided initial hits (e.g., N 70, 71, 78, and 79), and we refined the original condition 71 to obtain single crystals. The crystals grew within a few days to ~0.1 to 0.2 mm but were very unstable when cryoprotectants were tested. Therefore, an attempt to add a cryoprotectant in the crystallization buffer was made, and we obtained crystals in 23% polyethylene glycol PEG 3350, 10% glycerol, Bis-Tris (pH 6.0), and 0.2 M NaCl that were suitable for immediate freezing and data collection.
Growing SCHIC protein on minimal medium and selenomethionine.
For phasing purposes, we grew SCHIC on the M9 minimal medium containing 100 mg/liter of selenomethionine and amino acid mixture that inhibits methionine synthesis (F, K, T, V, L, and I). The cells were slower to grow and once induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the synthesis of protein with selenomethionine was allowed to proceed for 24 h. Purification proceeded as described above, and the best crystals grew in the same condition as native crystals; however, to prevent oxidation of selenium, we added 5 mM DTT to the crystallization buffer.
Data collection and processing.
Diffraction data were collected at the Berkeley beamline Advanced Light Source (ALS) 4.2.2. using remote data collection. The best anomalous data set was collected using selenium peak (4 selenium atoms in the SCHIC molecule) at 2.2-Å resolution. Data collection proceeded for 210° with a step of 1°. The crystal mosaicity was ~0.8. The best diffraction native data set was collected at 2.05-Å resolution. Initial indexing by HKL2000 provided primitive monoclinic space group P21. The initial phase estimates using SeMet data set were computed using Phaser (26). Autobuild software of Phenix (27) then provided the beginning model. Coot (28) and Phenix were used for model building and final refinement. Final refinement statistics are shown in Table S1 in the supplemental material.
Stopped-flow spectroscopy.
Dynamics of the AppA-heme reaction was measured using a KinTek stopped-flow SF-300 instrument. Various concentrations of AppA and heme were mixed in the reaction chamber with absorbance at 412 nm monitored. For analysis of the reaction of heme with dark state AppA, AppA was kept in the dark for at least 20 min before the reaction was initiated. For the reaction of heme with lit state AppA, dark state AppA was loaded into a clear glass injection syringe and then illuminated for at least 20 s before and during the time course of the monitored reaction. The Kd of AppA-heme interaction was calculated using the standard model as reported before (29).
Mapping cysteine residues in AppA.
The presence of reduced Cys sulfhydryl residues in AppA was determined by incubating ~10 µM AppA (either oxidized or 10 mM DTT-reduced) with 10 or 20 mM iodoacetamide at 22° C for 10 min. Excess iodoacetamide was then removed by passing through a MidiTrap G-25 (GE Healthcare) desalting column or a Zeba spin desalting column with a molecular weight cutoff of 7,000 (7K MWCO) (Thermo Scientific), depending on the sample volume. The samples were then treated with trypsin (protein/trypsin ratio is 10:1) at 37°C overnight. The digestion was stopped by the addition of trifluoroacetic acid (TFA) to a final concentration of 0.1% and desalted using ZipTip pipettes with C18 resin (Millipore). The samples eluted from ZipTip pipettes by 60% acetonitrile and 0.1% TFA were used for MALDI experiments with a Bruker Autoflex III MALDI-TOF mass spectrometer. To determine which cysteine residues are forming a disulfide bridge, 5 µM AppA was treated with 5 mM N-ethylmaleimide (NEM) for 45 min at 22°C, then with 10 mM DTT for 10 min at 22°C, and finally with 20 mM iodoacetamide for 30 min at 22°C. The double-labeled tryptic digest was analyzed by capillary-scale liquid chromatography coupled to tandem mass spectrometry (LC-MS-MS) (30). An Eksigent two-dimensional (2D) Ultra liquid chromatograph (AB Sciex, Foster City, CA) injected 5 µl of extract onto a trapping column (100 by 0.1 mm) packed with 5-µm Michrom Magic C18 particles and desalted for 8 min at a flow rate of 4 µl/min using LC buffer A (0.1% [vol/vol] formic acid [Sigma-Aldrich], 3% [vol/vol] acetonitrile [EM Science] in high-performance liquid chromatography [HPLC]-grade water [EM Science]). The trapping column was then placed in line with a Picofrit column (150 by 0.075 mm) with an integrated ESI tip (New Objective, Woburn, MA) packed with the same material as in the trap. A 35-min gradient from 10 to 85% LC buffer B (0.1% [vol/vol] formic acid, 3% [vol/vol] water in acetonitrile) separated the peptide mixture prior to tandem mass spectrometry in an LTQ-Orbitrap XL mass spectrometer (Thermo, Waltham, MA). The mass/charge ratios of the intact peptide ions were measured at a resolving power of 15,000 in the orbitrap; the fragment ions were generated, and their masses were measured in the LTQ ion trap. The resulting mass spectra were manually interpreted; the MS-Product module of ProteinProspector (http://prospector.ucsf.edu) (31) was used to predict the fragment ion masses.
Protein structure accession number.
The final structure has been deposited in the Protein Data Bank (PDB) under accession number 4HEH.
SUPPLEMENTAL MATERIAL
UV-visible spectrum of AppA-heme under aerobic (A) and anaerobic (B) conditions. Heme (5 µM) was incubated with 10 µM AppA for at least 20 min before the spectrum was taken. Download
The ESI−collision-induced dissociation (CID) (30) tandem mass spectrum of the tryptic peptide VAC399PANADC405C406SMR from the AppA protein after the double labeling experiment. The observed b-type and y-type fragment ions (32) are labeled (b ions from the N terminus and y ions from the C terminus). The inset shows the mass spectrum of the intact peptide ion prior to fragmentation. The charge state is +2 based on the isotope spacing, and the observed monoisotopic mass/charge ratio is within 2.1 ppm of the predicted value (790.3097) for the indicated peptide structure. The y4 and y5 ions have mass/charge ratios of 553.2 Thomson (Th) and 781.3 Th, respectively, and these are within 0.1 Th of those predicted given the peptide sequence shown in the figure. The mass of fragment y4 specifically indicates that Cys406 has a 57.02-Da adduct (from iodoacetamide added after disulfide reduction); the mass of y5 infers that Cys405 has a 125.05-Da adduct (from N-ethylmaleimide added prior to disulfide reduction). The mass of ion b3 (331.1 Th) is consistent with Cys399 modified by iodoacetamide. These results combined with the MALDI data in Fig. 4B demonstrate that a disulfide bond existed between Cys399 and Cys406 while Cys405 was a free thiol when the protein was isolated. Download
Hydrogen bond network in SCHIC extension region. (A) Hydrogen bond network around the extension loop in SCHIC crystal structure chain A. (B) Hydrogen bond network around extension loop in SCHIC crystal structure chain B. Download
Absorption spectrum of heme in dark and lit states. Download
Refinement statistics of SCHIC domain structure. The RCSB PDB accession number of the native SCHIC domain is 4HEH.
ACKNOWLEDGMENTS
We thank Jay Nix at the ALS, Berkeley, CA, for help during crystallographic data collection. We also thank Spencer Anderson and Hua Yuan for advice and help with data processing of various SCHIC crystal forms and Jared Cochran for help with stopped-flow kinetic studies.
This work was supported by NIH grant R37 GM040941 to C.E.B.
Footnotes
Citation Yin L, Dragnea V, Feldman G, Hammad LA, Karty JA, Dann CE, Bauer CE. 2013. Redox and light control the heme-sensing activity of AppA. mBio 4(5):e00563-13. doi:10.1128/mBio.00563-13.
REFERENCES
- 1. Masuda S, Bauer CE. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110:613–623 [DOI] [PubMed] [Google Scholar]
- 2. Gomelsky M, Klug G. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497–500 [DOI] [PubMed] [Google Scholar]
- 3. Losi A, Gärtner W. 2012. The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. Annu. Rev. Plant Biol. 63:49–72 [DOI] [PubMed] [Google Scholar]
- 4. Hasegawa K, Masuda S, Ono TA. 2006. Light induced structural changes of a full-length protein and its BLUF domain in YcgF(Blrp), a blue-light sensing protein that uses FAD (BLUF). Biochemistry 45:3785–3793 [DOI] [PubMed] [Google Scholar]
- 5. Rajagopal S, Key JM, Purcell EB, Boerema DJ, Moffat K. 2004. Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochem. Photobiol. 80:542–547 [DOI] [PubMed] [Google Scholar]
- 6. Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018 [DOI] [PubMed] [Google Scholar]
- 7. Yin L, Dragnea V, Bauer CE. 2012. PpsR, a regulator of heme and bacteriochlorophyll biosynthesis, is a heme-sensing protein. J. Biol. Chem. 287:13850–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim SK, Mason JT, Knaff DB, Bauer CE, Setterdahl AT. 2006. Redox properties of the Rhodobacter sphaeroides transcriptional regulatory proteins PpsR and AppA. Photosynth. Res. 89:89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moskvin OV, Kaplan S, Gilles-Gonzalez MA, Gomelsky M. 2007. Novel heme-based oxygen sensor with a revealing evolutionary history. J. Biol. Chem. 282:28740–28748 [DOI] [PubMed] [Google Scholar]
- 10. Han Y, Meyer MH, Keusgen M, Klug G. 2007. A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol. Microbiol. 64:1090–1104 [DOI] [PubMed] [Google Scholar]
- 11. Hu RG, Wang H, Xia Z, Varshavsky A. 2008. The N-end rule pathway is a sensor of heme. Proc. Natl. Acad. Sci. U. S. A. 105:76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dragnea V, Arunkumar AI, Lee CW, Giedroc DP, Bauer CE. 2010. A Q63E Rhodobacter sphaeroides AppA BLUF domain mutant is locked in a pseudo-light-excited signaling state. Biochemistry 49:10682–10690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dragnea V, Arunkumar AI, Yuan H, Giedroc DP, Bauer CE. 2009. Spectroscopic studies of the AppA BLUF domain from Rhodobacter sphaeroides: addressing movement of tryptophan 104 in the signaling state. Biochemistry 48:9969–9979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drennan CL, Huang S, Drummond JT, Matthews RG, Lidwig ML. 1994. How a protein binds B12: a 3.0 A X-ray structure of B12-binding domains of methionine synthase. Science 266:1669–1674 [DOI] [PubMed] [Google Scholar]
- 15. Elsen S, Ponnampalam SN, Bauer CE. 1998. CrtJ bound to distant binding sites interacts cooperatively to aerobically repress photopigment biosynthesis and light harvesting II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 273:30762–30769 [DOI] [PubMed] [Google Scholar]
- 16. Ponnampalam SN, Bauer CE. 1997. DNA binding characteristics of CrtJ. A redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 272:18391–18396 [DOI] [PubMed] [Google Scholar]
- 17. Winkler A, Heintz U, Lindner R, Reinstein J, Shoeman RL, Schlichting I. 2013. A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression. Nat. Struct. Mol. Biol. 20:859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eriksen JL, Mackenzie IR. 2008. Progranulin: normal function and role in neurodegeneration. J. Neurochem. 104:287–297 [DOI] [PubMed] [Google Scholar]
- 19. Hrabal R, Chen Z, James S, Bennett HP, Ni F. 1996. The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat. Struct. Biol. 3:747–752 [DOI] [PubMed] [Google Scholar]
- 20. Schaffer MA, Fischer RL. 1988. Analysis of mRNAs that accumulate in response to low temperature identifies a thiol protease gene in tomato. Plant Physiol. 87:431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Watanabe H, Abe K, Emori Y, Hosoyama H, Arai S. 1991. Molecular cloning and gibberellin-induced expression of multiple cysteine proteinases of rice seeds (oryzains). J. Biol. Chem. 266:16897–16902 [PubMed] [Google Scholar]
- 22. Granell A, Harris N, Pisabarro AG, Carbonell J. 1992. Temporal and spatial expression of a thiolprotease gene during pea ovary senescence, and its regulation by gibberellin. Plant J. 2:907–915 [DOI] [PubMed] [Google Scholar]
- 23. Berti PJ, Storer AC. 1995. Alignment/phylogeny of the papain superfamily of cysteine proteases. J. Mol. Biol. 246:273–283 [DOI] [PubMed] [Google Scholar]
- 24. Kraft BJ, Masuda S, Kikuchi J, Dragnea V, Tollin G, Zaleski JM, Bauer CE. 2003. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: a novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry 42:6726–6734 [DOI] [PubMed] [Google Scholar]
- 25. Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234 [DOI] [PubMed] [Google Scholar]
- 26. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu M, Tanaka WN, Zhu H, Xie G, Dooley DM, Lei B. 2008. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem. 283:6668–6676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang XJ, Thibault P, Boyd RK. 1993. Fragmentation reactions of multiply-protonated peptides and implications for sequencing by tandem mass spectrometry with low-energy collision-induced dissociation. Anal. Chem. 65:2824–2834 [DOI] [PubMed] [Google Scholar]
- 31. Clauser KR, Baker P, Burlingame AL. 1999. Role of accurate mass measurement (± 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871–2882 [DOI] [PubMed] [Google Scholar]
- 32. Martin SA, Biemann K. 1987. A comparison of keV atom bombardment mass spectra of peptides obtained with a two-sector mass spectrometer with those obtained from a four sector mass spectrometer. Int. J. Mass Spectrom. Ion Processes 78:213–228 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
UV-visible spectrum of AppA-heme under aerobic (A) and anaerobic (B) conditions. Heme (5 µM) was incubated with 10 µM AppA for at least 20 min before the spectrum was taken. Download
The ESI−collision-induced dissociation (CID) (30) tandem mass spectrum of the tryptic peptide VAC399PANADC405C406SMR from the AppA protein after the double labeling experiment. The observed b-type and y-type fragment ions (32) are labeled (b ions from the N terminus and y ions from the C terminus). The inset shows the mass spectrum of the intact peptide ion prior to fragmentation. The charge state is +2 based on the isotope spacing, and the observed monoisotopic mass/charge ratio is within 2.1 ppm of the predicted value (790.3097) for the indicated peptide structure. The y4 and y5 ions have mass/charge ratios of 553.2 Thomson (Th) and 781.3 Th, respectively, and these are within 0.1 Th of those predicted given the peptide sequence shown in the figure. The mass of fragment y4 specifically indicates that Cys406 has a 57.02-Da adduct (from iodoacetamide added after disulfide reduction); the mass of y5 infers that Cys405 has a 125.05-Da adduct (from N-ethylmaleimide added prior to disulfide reduction). The mass of ion b3 (331.1 Th) is consistent with Cys399 modified by iodoacetamide. These results combined with the MALDI data in Fig. 4B demonstrate that a disulfide bond existed between Cys399 and Cys406 while Cys405 was a free thiol when the protein was isolated. Download
Hydrogen bond network in SCHIC extension region. (A) Hydrogen bond network around the extension loop in SCHIC crystal structure chain A. (B) Hydrogen bond network around extension loop in SCHIC crystal structure chain B. Download
Absorption spectrum of heme in dark and lit states. Download
Refinement statistics of SCHIC domain structure. The RCSB PDB accession number of the native SCHIC domain is 4HEH.