ABSTRACT
The majority of emerging zoonoses originate in wildlife, and many are caused by viruses. However, there are no rigorous estimates of total viral diversity (here termed “virodiversity”) for any wildlife species, despite the utility of this to future surveillance and control of emerging zoonoses. In this case study, we repeatedly sampled a mammalian wildlife host known to harbor emerging zoonotic pathogens (the Indian Flying Fox, Pteropus giganteus) and used PCR with degenerate viral family-level primers to discover and analyze the occurrence patterns of 55 viruses from nine viral families. We then adapted statistical techniques used to estimate biodiversity in vertebrates and plants and estimated the total viral richness of these nine families in P. giganteus to be 58 viruses. Our analyses demonstrate proof-of-concept of a strategy for estimating viral richness and provide the first statistically supported estimate of the number of undiscovered viruses in a mammalian host. We used a simple extrapolation to estimate that there are a minimum of 320,000 mammalian viruses awaiting discovery within these nine families, assuming all species harbor a similar number of viruses, with minimal turnover between host species. We estimate the cost of discovering these viruses to be ~$6.3 billion (or ~$1.4 billion for 85% of the total diversity), which if annualized over a 10-year study time frame would represent a small fraction of the cost of many pandemic zoonoses.
IMPORTANCE
Recent years have seen a dramatic increase in viral discovery efforts. However, most lack rigorous systematic design, which limits our ability to understand viral diversity and its ecological drivers and reduces their value to public health intervention. Here, we present a new framework for the discovery of novel viruses in wildlife and use it to make the first-ever estimate of the number of viruses that exist in a mammalian host. As pathogens continue to emerge from wildlife, this estimate allows us to put preliminary bounds around the potential size of the total zoonotic pool and facilitates a better understanding of where best to allocate resources for the subsequent discovery of global viral diversity.
Introduction
The majority of emerging infectious diseases (EIDs) of humans are zoonoses, and the majority of these originate in wildlife (1–3). These diseases are largely viral (e.g., severe acute respiratory syndrome [SARS] and Nipah virus) and represent a significant global health threat. Analyses of trends in EIDs suggest that the rate of infectious disease emergence is increasing (3) and that the emergence of new viruses is not yet constrained by the richness (number of viruses) or diversity (genetic variability) of unknown viruses in wildlife, which is thought to be high. Systematically measuring viral richness, abundance, and diversity (here termed “virodiversity”) in wildlife is hindered by the large number of host species (e.g., around 5,500 mammals), their global distribution and often remote habitats (4), and the expense of collection, sampling, and viral identification or discovery (5), and it has not yet been achieved for even a single host species. In this study, we repeatedly sampled a mammalian host known to harbor emerging zoonotic pathogens (the Indian Flying Fox, Pteropus giganteus) and used PCR with degenerate primers targeting nine viral families to discover a large number and diversity of viruses. We then adapted the techniques normally used to estimate biodiversity in vertebrates and plants to estimate the total viral richness within these nine families in P. giganteus. Our analyses demonstrate proof-of-concept and provide the first statistically supported estimates of the unknown viral richness of a mammalian host and the sampling effort required to achieve it.
RESULTS
Viral discovery.
A total of 12,793 consensus PCR assays were performed for the detection of viruses from nine different families/genera, including coronaviruses (CoVs; n = 1,631), paramyxoviruses (PMVs; n = 1,108), hantaviruses (HTVs; n = 1,108), astroviruses (AstVs; n = 1,348), influenza A viruses (IFAVs; n = 1,108), bocaviruses (BoVs; n = 1,739), adenoviruses (AdVs; n = 1,902), herpesviruses (HVs; n = 1,741), and polyomaviruses (PyVs; n = 1,108) (Table 1). None of the samples were positive for IFAVs or HTVs, despite previous studies documenting their presence in other bat species (6–8); however, a total of 985 viral sequences representing the other seven viral families were detected in these bats. These sequences were segregated into 55 discrete viruses based on distinct monophyletic clustering (see Materials and Methods) (Table 1), and a virus was considered novel if the sequence identity to its closest relative was less than or equal to the identity between the two closest species for a given viral family (9).
TABLE 1 .
Summary of viral discovery performed on P. giganteus
| Virus | No. of samples PCR positive/no. testeda |
||||
|---|---|---|---|---|---|
| Urine | Throat | Feces | Roost urine | Total | |
| Herpesvirus | |||||
| PgHV-1 | 9/926 | 29/711 | 0/78 | 0/26 | 38/1,741 |
| PgHV-2 | 4/926 | 9/711 | 0/78 | 0/26 | 13/1,741 |
| PgHV-3 | 1/926 | 0/711 | 0/78 | 0/26 | 1/1,741 |
| PgHV-4 | 9/926 | 21/711 | 0/78 | 0/26 | 30/1,741 |
| PgHV-5 | 1/926 | 0/711 | 0/78 | 1/26 | 2/1,741 |
| PgHV-6 | 1/926 | 0/711 | 0/78 | 0/26 | 1/1,741 |
| PgHV-7 | 2/926 | 8/711 | 0/78 | 0/26 | 10/1,741 |
| PgHV-8 | 23/926 | 157/711 | 0/78 | 0/26 | 180/1,741 |
| PgHV-9 | 0/926 | 3/711 | 0/78 | 0/26 | 3/1,741 |
| PgHV-10 | 15/926 | 68/711 | 0/78 | 0/26 | 83/1,741 |
| PgHV-11 | 0/926 | 4/711 | 0/78 | 0/26 | 4/1,741 |
| PgHV-12 | 10/926 | 99/711 | 0/78 | 0/26 | 109/1,741 |
| PgHV-13 | 6/926 | 159/711 | 0/78 | 0/26 | 165/1,741 |
| Total | 81/926 | 557/711 | 0/78 | 1/26 | |
| Paramyxovirus | |||||
| PgPMV-1 | 1/598 | 0/510 | NTb | NT | 1/1,108 |
| PgPMV-2 | 2/598 | 0/510 | NT | NT | 2/1,108 |
| PgPMV-3 | 0/598 | 2/510 | NT | NT | 2/1,108 |
| PgPMV-4 | 0/598 | 1/510 | NT | NT | 1/1,108 |
| PgPMV-5 | 0/598 | 3/510 | NT | NT | 3/1,108 |
| PgPMV-6 | 1/598 | 7/510 | NT | NT | 8/1,108 |
| PgPMV-7 | 0/598 | 2/510 | NT | NT | 2/1,108 |
| PgPMV-8 | 1/598 | 0/510 | NT | NT | 1/1,108 |
| PgPMV-9 | 2/598 | 0/510 | NT | NT | 2/1,108 |
| PgPMV-10 | 1/598 | 1/510 | NT | NT | 2/1,108 |
| PgPMV-11 (NiV) | 1/598 | 2/510 | NT | NT | 3/1,108 |
| Total | 9/598 | 18/510 | |||
| Polyomavirus | |||||
| PgPyV-1 | 1/598 | 0/510 | NT | NT | 1/1,108 |
| PgPyV-2 | 0/598 | 3/510 | NT | NT | 3/1,108 |
| PgPyV-3 | 3/598 | 1/510 | NT | NT | 4/1,108 |
| Total | 4/598 | 4/510 | |||
| Coronavirus | |||||
| PgCoV-1 | 8/816 | 1/745 | NT | 5/70 | 14/1,631 |
| PgCoV-2 | 33/816 | 10/745 | NT | 17/70 | 60/1,631 |
| PgCoV-3 (bovine/human-like) | 1/816 | 0/745 | NT | 0/70 | 1/1,631 |
| PgCoV-4 (avian IBV-like) | 0/816 | 1/745 | NT | 0/70 | 1/1,631 |
| Total | 42/816 | 12/745 | 22/70 | ||
| Adenovirus | |||||
| PgAdV-1 | 1/931 | 0/806 | 0/78 | 0/87 | 1/1,902 |
| PgAdV-2 (avian AdV) | 1/931 | 0/806 | 0/78 | 0/87 | 1/1,902 |
| PgAdV-3 | 4/931 | 4/806 | 0/78 | 0/87 | 8/1,902 |
| PgAdV-4 | 0/931 | 1/806 | 0/78 | 0/87 | 1/1,902 |
| PgAdV-5 | 34/931 | 16/806 | 0/78 | 3/87 | 53/1,902 |
| PgAdV-6 | 0/931 | 2/806 | 0/78 | 0/87 | 2/1,902 |
| PgAdV-7 | 11/931 | 1/806 | 0/78 | 1/87 | 13/1,902 |
| PgAdV-8 | 17/931 | 2/806 | 0/78 | 0/87 | 19/1,902 |
| PgAdV-9 | 5/931 | 1/806 | 0/78 | 2/87 | 8/1,902 |
| PgAdV-10 | 1/931 | 0/806 | 0/78 | 0/87 | 1/1,902 |
| PgAdV-11 | 4/931 | 0/806 | 0/78 | 0/87 | 4/1,902 |
| PgAdV-12 | 1/931 | 0/806 | 0/78 | 0/87 | 1/1,902 |
| PgAdV-13 | 22/931 | 3/806 | 0/78 | 6/87 | 31/1,902 |
| PgAdV-14 | 38/931 | 11/806 | 0/78 | 5/87 | 54/1,902 |
| Total | 139/931 | 41/806 | 0/78 | 17/87 | |
| Astrovirus | |||||
| PgAstV-1 | 0/696 | 1/585 | NT | 1/67 | 2/1,348 |
| PgAstV-2 | 1/696 | 0/585 | NT | 0/67 | 1/1,348 |
| PgAstV-3 | 3/696 | 0/585 | NT | 8/67 | 11/1,348 |
| PgAstV-4 | 0/696 | 0/585 | NT | 2/67 | 2/1,348 |
| PgAstV-5 | 0/696 | 0/585 | NT | 3/67 | 3/1,348 |
| PgAstV-6 | 0/696 | 0/585 | NT | 1/67 | 1/1,348 |
| PgAstV-7 | 1/696 | 0/585 | NT | 0/67 | 1/1,348 |
| PgAstV-8 | 0/696 | 0/585 | NT | 15/67 | 15/1,348 |
| Total | 5/696 | 1/585 | 30/67 | ||
| Bocavirus | |||||
| PgBoV-1 (human BoV) | 1/925 | 0/710 | 0/78 | 0/26 | 1/1,739 |
| PgBoV-2 (human BoV) | 0/925 | 1/710 | 0/78 | 0/26 | 1/1,739 |
| Total | 1/925 | 1/710 | 0/78 | 0/26 | |
A total of 55 viruses from seven viral families were discovered. The discovery effort (number of samples tested) and prevalence of each virus is presented.
NT, not tested.
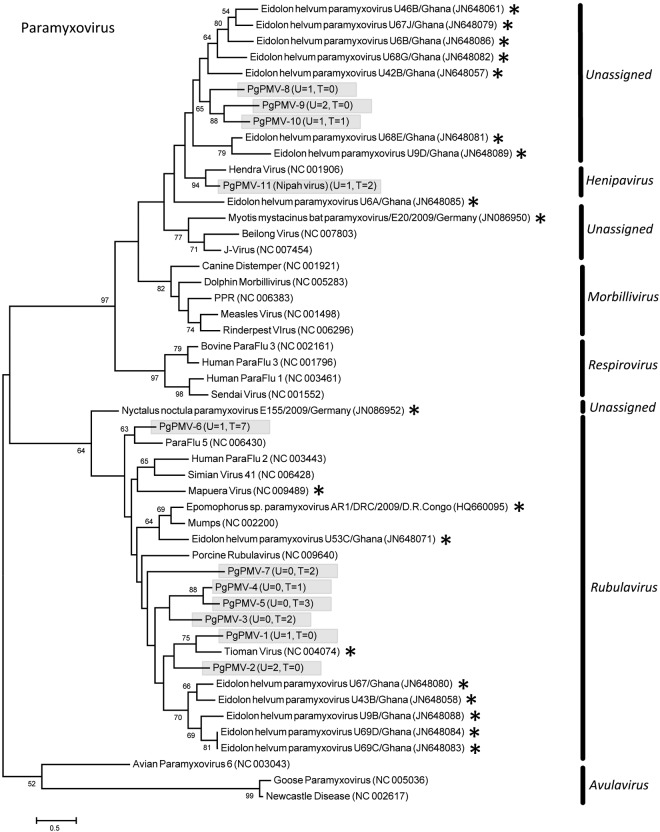
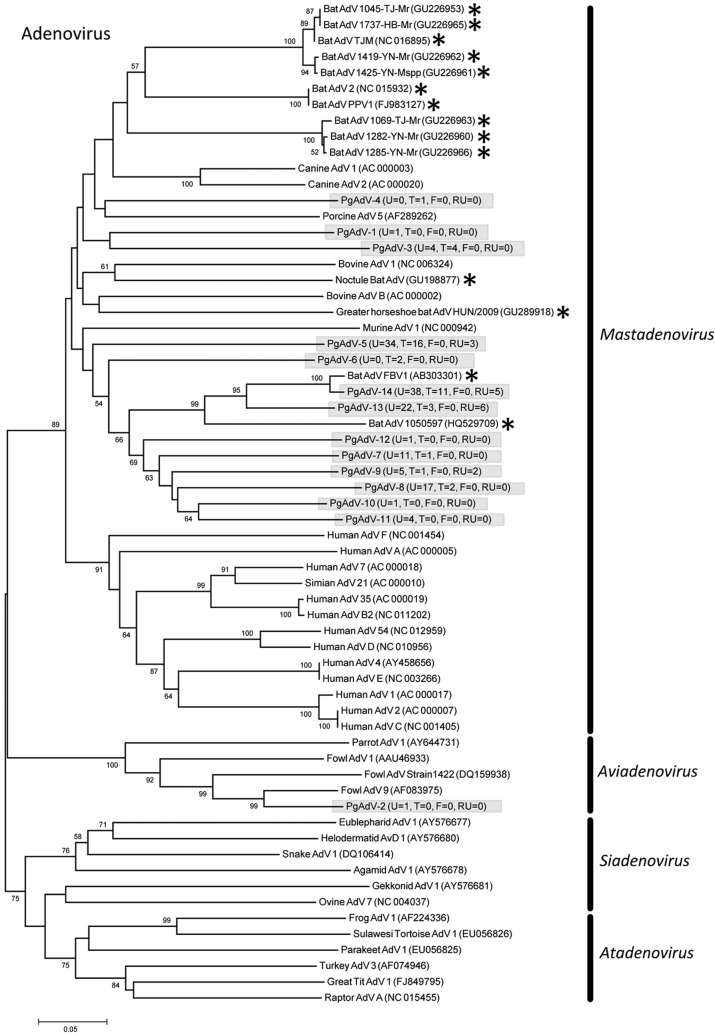
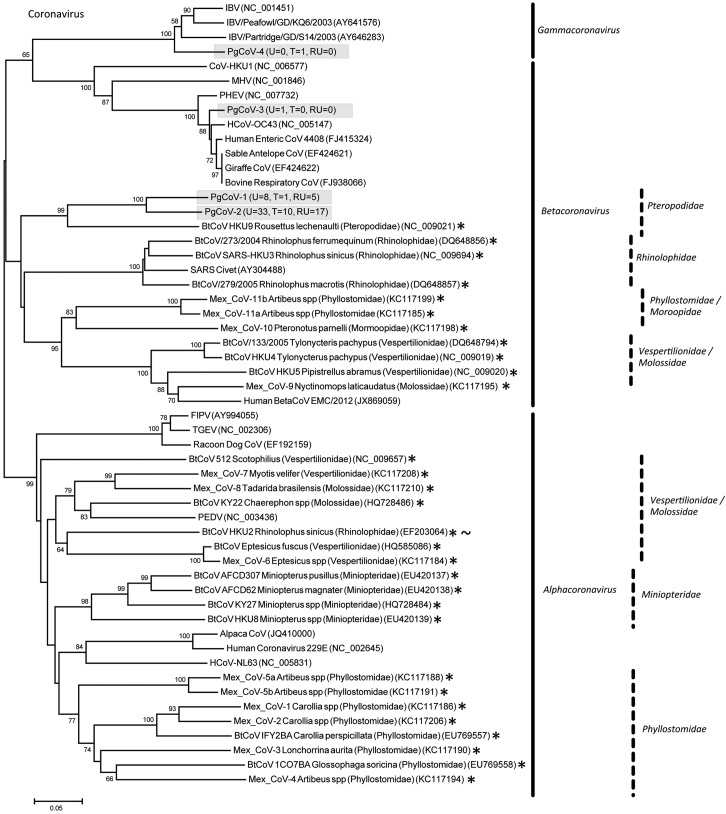
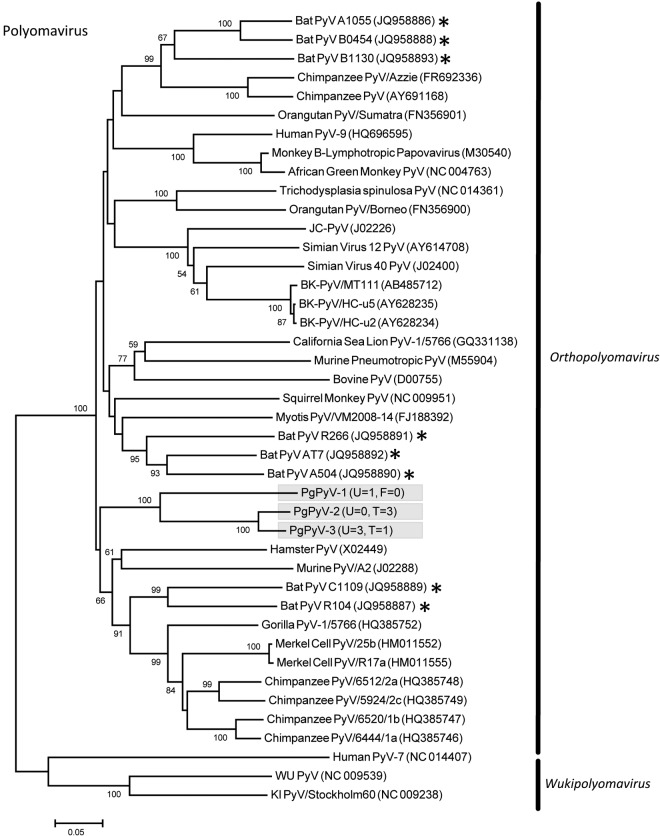
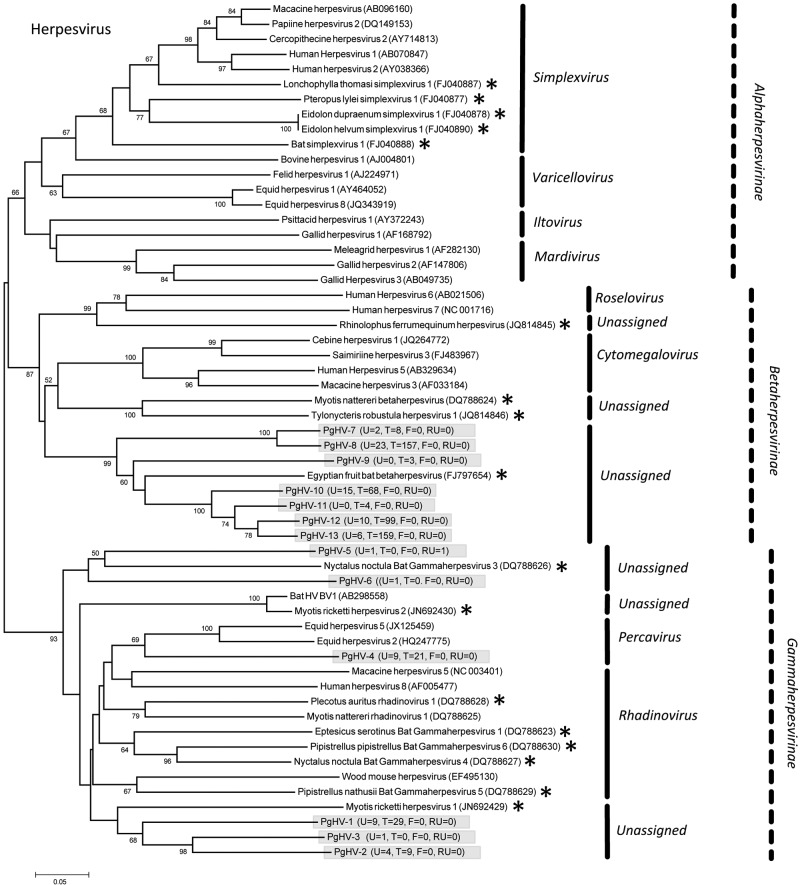
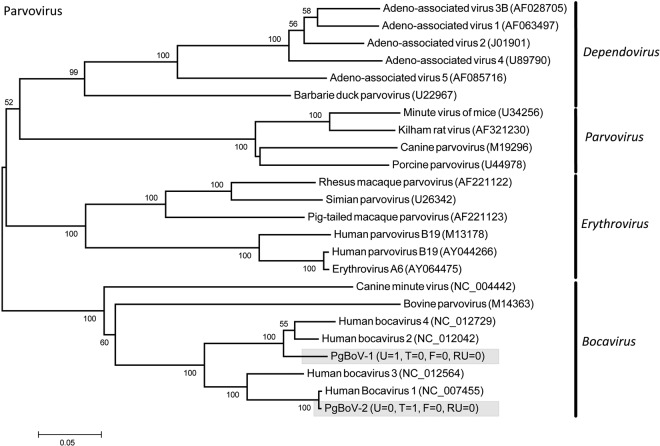
Eleven PMVs were detected, including 10 novel viruses (PMV-1 from P. giganteus [PgPMV-1] to PgPMV-10) and Nipah virus (PgPMV-11). These PMVs exhibited high sequence variation and clustered phylogenetically with either the rubulaviruses or an unassigned group related to the henipaviruses (Fig. 1). Within the AdV family, 14 viruses were discovered (PgAdV-1 to -14). Thirteen were novel mastadenoviruses, while one virus (PgAdV-2) had 98% nucleotide identity to the aviadenovirus Fowl adenovirus E (Fig. 2). Eight different AstVs were found (PgAstV-1 to -8), all of which were novel and clustered within the genus Mamastrovirus (Fig. 3). Within the CoV family, four distinct viruses were discovered. The first two were closely related betacoronaviruses (PgCoV-1 and -2). The third was also a betacoronavirus (PgCoV-3) but was more distantly related and showed 97% nucleotide identity to bovine and human coronaviruses (human strains 4408 and OC43). The fourth CoV was a gammacoronavirus (PgCoV-4) with 91% nucleotide identity (97% at the amino acid level) to the avian Infectious bronchitis virus (Fig. 4). Three novel PyVs were identified (PgPyV-1 to -3), all of which clustered with viruses in the genus Orthopolyomavirus (Fig. 5). A total of 639 HV sequences were detected, which segregated into 13 distinct clades (PgHV 1 to 13) using hierarchical clustering (see Materials and Methods). None could be reliably classified within any existing genus, and they likely represent new groups within the Betaherpesvirinae and Gammaherpesvirinae subfamilies (Fig. 6). One virus, PgHV-11, appears to be a recombinant between PgHV-10 and PgHV-13, with a breakpoint evident at approximately nucleotide 90. Upstream from this breakpoint, the sequences for PgHV-11 are related to PgHV-10, while downstream from the breakpoint, they are related to PgHV-13. Finally, two different BoVs were discovered (PgBoV-1 and -2), both of which showed >98% nucleotide identity to known human BoVs (Fig. 7).
FIG 1 .
Phylogenetic tree (ML) of PMV large gene (RdRp). Alignment length, 534 bp of nucleotide sequence. PgPMV-1 to -10 were discovered in this study. PgPMV-11 is Nipah virus. The number of samples that tested positive for each respective virus in urine (U) and throat (T) is indicated in parentheses. *, published bat PMV sequences. Novel viruses detected in this study are identified with the prefix Pg (Pteropus giganteus) and were assigned accession numbers KC692403 to KC692412
FIG 2 .
Phylogenetic tree (ML) of AdV polymerase. Alignment length, 301 bp of nucleotide sequence. PgAdV-1 to -14 were discovered in this study. The number of samples that tested positive for each respective virus in urine (U), throat (T), feces (F), and roost urine (RU) is indicated in parentheses. *, published bat AdV sequences. Viruses detected in this study are identified with the prefix Pg (Pteropus giganteus) and were assigned accession numbers KC692417 to KC692430
FIG 3 .
Phylogenetic tree (ML) of AstV RdRp. Alignment length, 320 bp of nucleotide sequence. PgAstV-1 to -8 were discovered in this study. The number of samples that tested positive for each respective virus in urine (U), throat (T), and roost urine (RU) is indicated in parentheses. *, published bat AstV sequences. Viruses detected in this study are identified with the prefix Pg (Pteropus giganteus) and were assigned accession numbers KC692431 to KC692437
FIG 4 .
Phylogenetic tree (ML) of CoV RdRp. Alignment length, 310 bp of nucleotide sequence. PgCoV-1 to -4 were discovered in this study. The number of samples that tested positive for each respective virus in urine (U), throat (T), and roost urine (RU) is indicated in parentheses. *, published bat CoV sequences. Bat coronaviruses cluster based on the host family (indicated). ~, HKU2 seems anomalously positioned as it was detected in Rhinolophus sinicus, which is unrelated to bats from the families Vespertilionidae or Molossidae. The reason for this is unknown. Viruses detected in this study are identified with the prefix Pg (Pteropus giganteus), and were assigned accession numbers KC692413 to KC692416. IBV, infectious bronchitis virus; MHV, mouse hepatitis virus; PHEV, porcine hemagglutinating encephalomyelitis virus; HCoV, human CoV; BtCoV, bat CoV; FIPV, feline infectious peritonitis virus; TGEV, transmissible gastroenteritis coronavirus; PEDV, porcine epidemic diarrhea virus.
FIG 5 .
Phylogenetic tree (ML) of PyV VP1 (major capsid protein). Alignment length, 320 bp of nucleotide sequence. PgPyV-1 to -3 were discovered in this study. The number of samples that tested positive for each respective virus in urine (U) and throat (T) is indicated in parentheses. *, published bat PyV sequences. Viruses detected in this study are identified with the prefix Pg (Pteropus giganteus) and were assigned accession numbers KC692400 to KC692402.
FIG 6 .
Phylogenetic tree (ML) of HV polymerase. Alignment length, 211 bp of nucleotide sequence. PgHV-1 to -13 were discovered in this study. The number of samples that tested positive for each respective virus in urine (U), throat (T), feces (F), and roost urine (RU) is indicated in parentheses. *, published bat HV sequences. Viruses detected in this study are identified with the prefix Pg (Pteropus giganteus) and were assigned accession numbers KC692438 to KC692450.
FIG 7 .
Phylogenetic tree (ML) of BoV NS1. Alignment length, 287 bp of nucleotide sequence. PgBoV-1 and -2 were detected in this study. The number of samples that tested positive for each respective virus in urine (U), throat (T), feces (F), and roost urine (RU) is indicated in parentheses. Viruses detected in this study are identified with the prefix Pg (Pteropus giganteus) and were assigned accession numbers KC692451 to KC692452.
Viral discovery curves and estimates of viral richness.
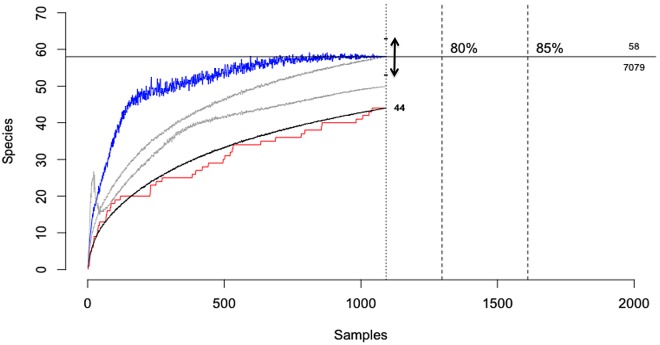
Asymptotic viral richness was estimated from observed detections using three statistical models, Chao2, ICE, and Jackknife (10). To ensure internal consistency, only those samples screened for the full complement of nine viral families/genera were included (n = 1,092), which accounted for 44/55 viruses identified in this study. The relative frequency of these viruses is presented in Fig. S1 in the supplemental material. Of the 1,092 samples included, 766 were negative for all viruses. There were 595 viral detections from 326 positive samples, with 167 samples containing >1 virus. When all 44 viruses were considered, the accumulative discovery curve began to show signs of saturation (Fig. 8). The Chao2 estimator demonstrated asymptotic behavior as early as 500 samples (<50% of tested samples) and was highly stable (low variance) by 1,000 samples (Fig. 8). The total viral richness within our sample population was estimated to be 58 viruses (limited to viral families tested for), and the required sampling effort to discover all 58 was estimated to be 7,079 samples. Asymptotic estimates of viral richness were also calculated individually for PMVs, AstVs, HVs, and AdVs (see Fig. S2 in the supplemental material). For HVs, AstVs, and PMVs, estimates were stable or stabilizing and showed that most of the predicted viral diversity had been identified. For AdVs, none of the estimators stabilized, probably due to a high singleton-to-doubleton ratio (i.e., the rate of discovery of new viruses was still high). Individual assessments were not performed for CoVs, PyVs, or BoVs because a lack of single or double detections prevented meaningful estimates (10).
FIG 8 .
Viral discovery curve. A subset of 1,092 samples were tested for all nine viral families, and 44 viruses (out of a total of 55) are represented. The 11 viruses not considered were PgHV-2, -5, -6, and -9; PgAdV-1 and -10, PgAstV-4, -5, -6, and -8, and PgBoV-1. Black line, the rarefaction curve; red line, collector curve showing accumulation of novel viruses over samples tested; blue line, Chao2 estimator at every sample point, with arrow indicating 95% confidence intervals; gray lines, ICE and Jackknife estimators at every sample point; dashed vertical lines, required sampling effort to discover an arbitrary proportion of the total diversity (including 100%); horizontal line, total estimated diversity, 58 viruses, and effort required to discover 100% of the estimated diversity, 7,079 samples.
Coinfection.
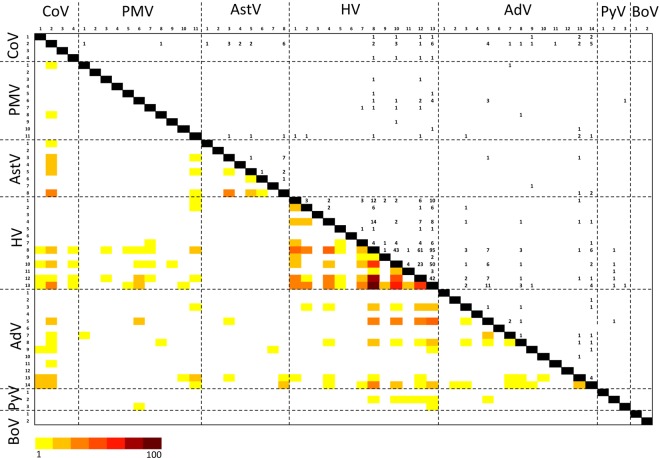
Positive PCR products were cloned and sequenced to look for the presence of cooccurring viruses. A total of 276 samples contained >1 virus, including urine (n = 56), throat swabs (n = 199), and roost urine (n = 56). Between 2 and 5 viruses were found to coexist, and both intrafamilial (n = 223/276) and interfamilial (n = 93/276) viral family cooccurrences were observed (Fig. 9). Intraspecific codetections were limited to the families Herpesviridae and Adenoviridae (Fig. 9 and see Table S1 in the supplemental material). The patterns of HV cooccurrence were significantly nonrandom (P = <0.001 with C score [11, 12]), and positive pairwise associations were observed between PgHV-13 and PgHV-10 and between PgHV-11 and PgHV-10 (P = <0.001). A negative association was also observed between PgHV-13 and -12, where the observed frequency of cooccurrence was below what would be expected by chance, given the prevalence of both viruses (P = <0.001). The patterns of cooccurrence for different adenoviruses were less structured, and no robustly supported positive or negative associations were observed. The same is true for analyses of interfamilial codetections.
FIG 9 .
Coinfection matrix of the 55 viruses identified or discovered in P. giganteus. Heat map of coinfections is presented in lower left section. Data used to generate the heat map (numbers of pairwise incidences) are presented in upper right section.
DISCUSSION
In this study, we combined virological and ecological techniques to describe the virodiversity of a known zoonotic reservoir, P. giganteus, including the first-ever estimate of viral richness and the sampling effort required to detect any proportion of it. The article also includes a phylogenetic description of 55 viruses identified in this species and an ecological description of the positive and negative occurrences between them. This unprecedented description of the potential zoonotic pool not only establishes a framework for comparison of virodiversity among host populations in different geographic regions and ecological settings but also provides important a priori knowledge should a novel pathogen emerge or have emerged. For example, sequence data from this study are already being used to develop serological assays to test people with a range of symptoms and known high risk of exposure to bats. This will allow us to identify cases of bat-to-human viral spillover and, potentially, their health consequences.
Estimate of total viral diversity (richness) in P. giganteus in Bangladesh.
Previous attempts have been made to predict the total number of unknown viruses in humans by analyzing temporal trends in viral discovery (13, 14). However, these assessments principally consider the emergence of disease-causing agents rather than an ecological assessment of virodiversity. Here, we measured viral richness using nonparametric species discovery curves, which are commonly used in biodiversity studies (10, 13, 15–17) and rely on the frequency of rarely occurring species to measure the completeness of discovery (15) and make statistical estimations of the undiscovered fraction (10). Our estimate of the viral carrying capacity of P. giganteus as 58 viruses (for the nine viral families assayed) was robustly supported, with the cumulative number of new viruses slowing toward an asymptotic trajectory and with statistical estimators showing reliably asymptotic behavior. Given that our total discovery effort revealed 55 of the estimated 58 viruses, we suggest that most have now been identified and that the remaining viruses in P. giganteus in Bangladesh are extremely rare. However, we make the qualification that this estimation of richness is likely a minimum and that additional viruses will almost certainly be found through the expansion of viral family testing and the use of high-throughput sequencing.
While we cannot infer a great deal about the biology or taxonomic relatedness of the undiscovered diversity, our estimate of viral richness does allow useful considerations of the efficacy of viral discovery efforts in this species. In general, surveys for undiscovered diversity present diminishing returns, since the commonly occurring species are quickly identified, while the rare species require an increasingly large sampling effort. Here, the Chao 2 estimator predicted that 85% of the total richness could be achieved if another ~500 samples were screened but that a further ~5,500 would then be required to find the remaining 15%. Considering that only very rare viruses would exist within this final fraction and assuming that this rarity reduces both exposure and the chance of transmission, the public health advantage gained by knowledge of their biological properties or taxonomy may not be sufficiently high to justify the cost of their discovery. Similar estimations were made for each of the viral families individually. For example, 13 HVs were discovered in a total of 1,741 samples, with an estimated 8,503 additional samples needed to identify just 2 more predicted viruses (100% of the estimated). Equally, 11 PMVs were identified in 1,108 samples, with one additional virus projected from a further 773 samples. In both cases, an extremely limited return is expected from a costly discovery effort.
Estimates of unknown viral diversity (richness) in all mammals and the cost of discovery.
Mammals are the reservoir hosts of the majority of emerging zoonoses (2, 3, 18). If we assume that all 5,486 described mammalian species (19) harbor an average of 58 viruses in the nine families of interest (as estimated here in P. giganteus) and that these viruses exhibit 100% host specificity, the total richness of mammalian viruses awaiting discovery exceeds ~320,000. We used the data on expenditures for surveillance and pathogen discovery in this study to calculate the direct cost of discovering all 58 viruses in P. giganteus (see the supplemental material for details of this cost analysis). We estimate this cost to be $1.2 million, including collection and laboratory testing of 7,079 samples. Assuming expenditures to be equal for all host species, the cost of sampling and viral discovery for all mammalian viruses would be approximately $6.3 billion. Accounting for diminishing returns means that discovering 85% of the estimated diversity would be disproportionately cheaper at approximately $1.4 billion. Our estimates of virodiversity and cost of discovery are preliminary; however, we include them to demonstrate (i) how a systematic estimation of total viral diversity could be used to inform better surveillance through strategic resource allocation and (ii) that, given our cost estimates, the discovery of the majority of potential zoonotic viruses is not an unattainable goal over the next few decades. The generation of sequence data will not, of course, in itself prevent pandemics. However, it does provide data that refine our knowledge of the functional relationship between host and viral diversity, including traits associated with increased risk of spillover and subsequent emergence (e.g., viruses closely related to and sharing receptor binding domains with known lethal agents [20]) and, also, facilitates the development of rapid diagnostic tests for intervention and control.
Several important limitations must be considered in our extrapolations, including (i) the assumption that a mean of 58 viruses per species is a reasonable estimate and that host populations are panmictic with respect to viral transmission (such that expanded geographic sampling would not influence viral detections), (ii) the assumption that viruses are not shared by more than one host species, (iii) that only those viruses within the nine families are considered in this estimation, (iv) that the results are limited by the sensitivity and specificity of our tests, and (v) that a similar mean cost of sample collection is incurred across all species. Clearly, many of these limitations and assumptions require additional exploration. For example, while including more viral families in our survey would increase the viral richness estimate, accounting for species turnover (viral sharing between species) would reduce it. Also, while the cost of sample collection in Bangladesh is relatively low because of logistical simplicities, in some regions (e.g., tropical montane forests of Africa and Southeast Asia), the cost of transportation is much higher. Better estimates of the total number of viruses in mammals (and the cost of their discovery) will be achieved iteratively as other hosts are more extensively sampled and tested, additional viral families are included, and the limits of viral detection increase.
Novel viruses.
The current study significantly enhances our knowledge of the viruses harbored by P. giganteus, for which only two viruses had been previously described, Nipah virus and a GB virus-like flavivirus (21, 22). A total of 50/55 of the viruses discovered in this study were considered novel, while 5/55 have been reported previously (PgBoV-1 and -2, PgCoV-3 and -4, and PgPMV-11). Additional discussion of the 50 novel viruses is provided in the supplemental material. Here, we discuss a number of important limitations that must be considered in the interpretation of these results. First, the use of consensus PCR limits surveillance and discovery to viruses related to those targeted in these assays. Second, variations in virus concentration can also influence the probability of detection. Third, we evaluated the diversity of viruses in a limited set of compartments and tissues, and unbiased sequencing was not used as a secondary method of capturing diversity. The classification of viruses is also significant, as redefining the genetic limits between one virus and another would change the total number and prevalence of viruses discovered and would impact our estimations of viral richness. We have tried to address this by using monophyletic clades as a taxonomic surrogate, which obviates the variable and polythetic criteria set by the ICTV for species demarcations.
Coinfection.
The identification of coexisting microbes is important to a description of virodiversity because of the positive and negative associations that can occur between them (23–28). Here, we report a large number of intra- and interfamilial cooccurrences in P. giganteus and show that as many as five different viruses can exist in a single sample. Not only does this reveal information about the carrying capacity and composition of discrete viral niches within an individual bat, it also demonstrates the number of different viruses that could potentially spill over to a new host from a single exposure event.
The most common intrafamilial codetections were observed within the subfamily Herpesviridae, supporting previous studies demonstrating coinfection of HVs in bats (29). Statistically supported associations were observed between PgHV-10, -11, and -13, which phylogenetically cluster within a presumptive new genus of the betaherpesvirus subfamily. It is not known whether these detections represent coinfection of the same cell or a group of viral variants with segregated cell tropism. It is also unknown why these viruses should so readily coexist, though ecological mechanisms such as simultaneous transmission (codispersal), the availability of requisite resources, and/or shared benefits associated with host immunomodulation by one or more of these viruses may explain the observed cooccurrence. Recombination is also a possible consequence of coinfection and is a common feature in the ecology and evolution of herpesviruses (30–36). PgHV-11 was identified as a recombinant lineage derived from the strongly associated PgHV-10 and PgHV-13, and all three viruses were detected in the same sample or compartment (throat) multiple times, suggesting that true coinfection does occur, albeit with unknown frequency.
A negative association was also observed between PgHV-12 and -13, suggesting that mechanisms might also exist to reduce cooccurrence. These two viruses are very closely related, and we speculate that cooccurrence may offer little benefit to the viral population because of increased competition for resources coupled with minimal potential for fitness gains via recombination. Even though previous studies showed a lack of immune recognition in betaherpesviruses (37), we suggest this might act as an effective mechanism for reducing the coexistence of closely related viruses by preventing sequential infections. Such a mechanism would not completely preclude cooccurrence due to codispersal and would therefore serve to explain why some codetections were still observed between these two viruses.
Viral spillover.
Our discovery efforts revealed five viruses that appear to represent spillover events. These included two human bocaviruses (PgBoV-1 and -2), an avian adenovirus (PgAdV-2), a human/bovine betacoronavirus (PgCoV-3), and an avian gammacoronavirus (PgCoV-4). In each case, these viruses were only observed once and showed strong phylogenetic association to viruses found in humans, birds, or ruminants. The interface by which these viruses were able to move from these disparate hosts into bats is unclear. However, on several occasions, we have observed P. giganteus in Bangladesh drinking from bodies of water (rivers and ponds) that are used by people, livestock, domestic animals, and wildlife for drinking, bathing, and in some cases, sewage, and we hypothesize that shared water sources may be a source of exposure. Viral spillover (and/or host switching) is an example for which the concept of virodiversity in defined animal host populations might be particularly important. Such processes precede many emergence events (3, 38); however, there is almost certainly additional asymptomatic movement of viruses between hosts, the frequency and impact of which remain poorly understood.
An additional consideration is that any of the 55 viruses found in P. giganteus may have already spilled over into the human population. Annual outbreaks of Nipah virus in Bangladesh demonstrate that human exposure to viruses from these bats persists (39–44), and there are a significant number of undiagnosed morbidities and mortalities in this region that may well have resulted from the spillover of one of these other viruses. Subclinical movement is equally possible, as demonstrated with Tioman virus in Malaysia (45, 46), and investigating these spillover events may help to refine our understanding of disease emergence in novel hosts.
Conclusions.
Our work illustrates the power of using ecological approaches to characterize virodiversity and estimate viral richness and can be considered part of a strategy to better target surveillance to identify agents that pose zoonotic risks before they emerge in people (3). The projected $1.4 billion cost of discovering 85% of the estimated diversity is far less than the economic impact of even a single pandemic like SARS, which has been estimated at $16 Billion (47). If annualized over a 10-year period, the discovery of 85% of mammalian viral diversity would be just $140 million/year, which is both a one-off cost and a fraction of the cost of globally coordinated pandemic control programs such as the “One World, One Health” program, estimated at $1.9 to 3.4 billion per year, recurring (64). While these programs will not themselves prevent the emergence of new zoonotic viruses, they will further contribute to pandemic preparedness by enhancing our understanding of viral ecology and the mechanisms of disease emergence and by providing sequences and other insights that reduce the morbidity, mortality, and economic impact of emerging infectious diseases by expediting recognition and intervention.
MATERIALS AND METHODS
Samples and PCR screening.
Samples (n = 1,897) were collected from apparently healthy P. giganteus bats throughout Bangladesh between 2006 and 2010, as described previously (48). This included urine (n = 926), throat swabs (n = 806), feces (n = 78), and roost urine (n = 97). All samples were collected by trained veterinarians, and all animals were released unharmed. Samples were collected directly into lysis buffer (bioMérieux, Inc.) and stored at −80°C until transfer to the Center for Infection and Immunity at Columbia University. Roost urine samples were also obtained by suspending 3- by 2-m polyethylene sheets underneath roosting colonies, which collected urine (with possible fecal contamination) from the bats roosting above. Total nucleic acid was extracted from all samples using the EasyMag (bioMérieux, Inc.) platform, and cDNA synthesis performed using SuperScript III first-strand synthesis supermix (Invitrogen), all according to the manufacturer’s instructions. Viral discovery was performed using broadly reactive consensus PCR assays targeting coronaviruses (49), paramyxoviruses (50), astroviruses (51), influenza A viruses (38), adenoviruses (52), polyomaviruses (53), bocaviruses (54), and herpesviruses (55). Consensus primers for hantaviruses were modified from an existing protocol (56) in order to increase the degeneracy of the assay, and the assay validated for its ability to detect diverse hantaviruses, including Andes, Puumala, Sin Nombre, Prospect Hill, Seoul, and Thottapalayam hantaviruses. The modified primer sequences were UHantaF1 (GWGGVCARACWGCHGAYT) and UHantaR1 (CCWGGTGTDADYTCHTCWGC) (expected amplicon, 250 bp), and the annealing temperature was 52°C. All PCR products of the expected size were cloned into Strataclone PCR cloning vector, and 12 white colonies sequenced using standard M13R primers.
Trace sequences were analyzed and edited using Geneious (version 6.0.3). Sequences were aligned with ClustalW and MUSCLE, and phylogenetic trees constructed with neighbor-joining (p-distance, pairwise deletion, 1,000 bootstraps), maximum-likelihood (1,000 bootstraps), and Bayesian (Mr Bayes) algorithms. Models of evolution were selected using ModelTest, and a tree representing a consensus of the different methods is presented. Sequence identity (p-distance, pairwise deletion) was calculated in Mega 5.
Virus classification.
For the purposes of this study, we avoided the use of taxonomic concepts such as species or genotype because of the variable criteria used for such distinctions (9) and because the degree of sequence conservation used to establish such distinctions can vary across the genome and may be affected by the relatively short sequence fragments generated in this study. We focused instead on collections of viral sequences that form distinct monophyletic clades within a particular family, and we considered a virus novel if the sequence identity to its closest relative is less than or equal to the identity between the two closest species for a given viral family. Due to the very large number of herpesvirus sequences identified in this study (n = 650), we used hierarchical clustering to segregate sequences for this particular family. To do this, we first extracted 598 polymerase sequences from published complete genomes (downloaded from NCBI on 14 September 2012) and combined them with the sequences generated in this study (total of 1,248 sequences). Coding sequences were translated and aligned using MUSCLE (version 3.8.31) (57) with the default settings. The nucleotide alignment was constructed by replacing each amino acid with the codon that gave rise to it. Columns containing gaps in more than 1,000 of the 1,248 sequences were removed. The genetic distance between HV species was subsequently established using the published sequences in the alignment only, as described previously (58), and a >7% nucleotide difference (Hamming distance) was used to define HV clusters. PgHV sequences were then segregated using hierarchical clustering, as implemented in the SciPy package (59) using average linkage clustering.
Virus richness and sample estimation.
We implemented models from the biodiversity literature that utilize incidence distributions to estimate virus richness (number of unique viruses) and, hence, to estimate the number of undetected viruses in the assemblage (60, 61). Incidence data result where each virus detected in the assemblage is noted in each sample as either present (verified detection) or absent (not detected, which could result due to the virus being absent or being present but not detected by the test, i.e., false absence).
From our samples, we first constructed virus accumulation and rarefaction curves for visualization. The asymptote of the rarefaction curve provides the estimate of the number of viruses that characterizes the assemblage. However, sampling to reach this asymptote is impractical, as the number of samples required may be prohibitively large (61). We thus used statistical methods to estimate the asymptote from the data at hand.
We used the nonparametric asymptotic estimator, Chao2 (15, 10), and also calculated ICE and Jackknife statistics for comparison. Unlike conventional curve-fitting procedures, the nonparametric estimators make no assumptions of an underlying abundance distribution, do not require ad hoc or a priori model fitting, are relatively robust to spatial autocorrelation and scale, and frequently outperform other methods of richness estimation (61). They rely on the principle that the frequencies of the rarest species in a set of samples can be used to estimate the frequencies of undetected species and provide a minimum richness estimate.
All analyses were conducted with the fossil package (62) implemented in R (63). We followed Chao et al. (10) to calculate how many additional samples would be required to detect any proportion (including 100%) of the asymptotic virus richness. All statistics were incorporated into a single plot.
Cooccurrence.
Patterns of association/disassociation were explored with the Fortran software program PAIRS (11), utilizing the C score statistic as our measure of species cooccurrence. PAIRS implements a Bayesian approach (Bayes M criterion) to detect nonrandom associations between pairs of species (12).
Assumptions and caveats
We considered the detection and discovery of viruses akin to the problem of detection and discovery of biodiversity, as is frequently the goal of ecological studies. The basic mechanism of species detection occurs from drawing samples by collection from some larger assemblage (61). In this context, our samples are as described above, urine, throat, fecal, or roost urine taken from an individual bat or bat roost, which represent the biomes for our assemblage of interest. These methods require the assemblage of viruses under sampling to be closed for valid inference, that is, that the assemblage size and composition remained stable throughout the course of the study, an assumption we felt was justified. Although each of these sample types targets a unique biome of potential viral habitat from the host species, each with potentially differing efficacy for detecting any given virus, for the purposes of our analyses, we considered each sample a random and equivalent draw from the assemblage of viruses associated with this host species. We also assumed sample independence, even though multiple samples (e.g., urine and throat) were often drawn from the same individual host and sampled bat populations are likely to be geographically nonrandom. The consequence of this sampling strategy is that our analysis is blind to this additional source of geographical variation and occasional pseudoreplication, which means our virus accumulation results are specific to our sampling methodology and our extrapolations assume ongoing sampling with a similar average composition of samples. The results of additional analyses in which we isolated sample types and individuals and considered geographic variation are not presented herein.
Nucleotide sequence accession numbers.
The GenBank accession numbers for viruses discovered in this study are KC692400 to KC692452.
SUPPLEMENTAL MATERIAL
Supplemental discussion. Download
Relative distribution of viruses included in discovery curve analyses (see also Fig. 8). A subset of samples (n = 1,092) was used for discovery curve analysis. Only those samples that were screened for all nine viral families were included, ensuring internal consistency. Eleven of the 55 identified viruses had zero abundance in this subset and were therefore not considered in the analysis. The 11 omitted viruses were PgHV-2, -5, -6, and -9, PgAdV-1 and -10, PgAstV-4, -5, -6, and -8, and PgBoV-1. Forty-four viruses were therefore retained in our estimates, and the relative frequency of each is presented here. Download
Viral discovery curves are presented for (i) all viruses (see also Fig. 8), (ii) PMV, (iii) AstV, (iv) HV, and (v) AdV. Discovery effort (number of samples tested) is indicated by the horizontal dotted line. Red line, collector curve showing accumulation of novel viruses over samples tested; blue line, Chao2 estimator at every sample point, with arrow indicating 95% confidence intervals; gray lines, ICE and Jackknife estimators at every sample point; Φ, estimated total diversity; dashed horizontal lines, required sampling effort to discover an arbitrary proportion of the total diversity; Ω, effort required to discover 100% of the estimated diversity. Download
Summary of intraspecific coinfections. The total number of samples testing positive for each family is presented, together with the number of the samples containing >1 virus of the same family.
ACKNOWLEDGMENTS
We acknowledge funding from the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT project, cooperative agreement number GHN-A-OO-09-00010-00, an NIAID nonbiodefense Emerging Infectious Disease Research Opportunities award (R01-AI079231 [P.D.]), an NIH/NSF Ecology and Evolution of Infectious Diseases award from the Fogarty International Center (R01-TW005869 [P.D.]), K08AI067549 (J.H.E.), and NIH-AI57158 (W.I.L.).
We thank the Bangladesh Forest Department and Ministry of Environment and Forest for permission to catch bats and conduct this study. We thank Pitu Biswas, Gofur Sheikh, and Jim Desmond (EcoHealth Alliance), Jahangir Hossain, Emily Gurley, Salah Uddin Khan, Ausraful Islam, and Najmul Haider (ICDDR,B), as well as Kawthar Muhammad (CII), for their help with sample collection and project management.
The contents of this paper are the responsibility of the authors and do not necessarily reflect the views of USAID or the United States Government.
Author contributions were as follows. Study design was by S.J.A., K.A.M., I.N.-M., N.C.A., T.L.B., W.B.K., T.G., S.P.L., S.S.M., J.A.K.M., P.D., and W.I.L. Laboratory experiments were performed by S.J.A., I.N.-M, M.D.S.-L., and R.O.F. Phylogenetic analyses were performed by S.J.A., T.G., and W.I.L. Ecological analyses were performed by S.J.A., K.A.M., C.M.Z.-T., A.S., P.H., T.L.B., P.D., and W.I.L. Samples were collected by J.H.E., A.I., S.A.K., K.J.O., and P.D. The paper was written by S.J.A., K.A.M., I.N.-M., R.O.-F., N.C.A., A.S., T.L.B., J.H.E., K.J.O., T.G., S.P.L., S.S.M., J.A.K.M., P.D., and W.I.L.
Footnotes
Citation Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JAK, Daszak P, Lipkin WI. 2013. A strategy to estimate unknown viral diversity in mammals. mBio 4(5):e00598-13. doi:10.1128/mBio.00598-13.
REFERENCES
- 1. Morse SS. 1995. Factors in the emergence of infectious diseases. Emerg. Infect. Dis. 1:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woolhouse ME, Gowtage-Sequeria S. 2005. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11:1842–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rondinini C, Di Marco M, Chiozza F, Santulli G, Baisero D, Visconti P, Hoffmann M, Schipper J, Stuart SN, Tognelli MF, Amori G, Falcucci A, Maiorano L, Boitani L. 2011. Global habitat suitability models of terrestrial mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:2633–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lipkin WI. 2013. The changing faces of pathogen discovery and surveillance. Nat. Rev. Microbiol. 11:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sumibcay L, Kadjo B, Gu SH, Kang HJ, Lim BK, Cook JA, Song JW, Yanagihara R. 2012. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Côte d’Ivoire. Virol. J. 9:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weiss S, Witkowski PT, Auste B, Nowak K, Weber N, Fahr J, Mombouli JV, Wolfe ND, Drexler JF, Drosten C, Klempa B, Leendertz FH, Kruger DH. 2012. Hantavirus in bat, Sierra Leone. Emerg. Infect. Dis. 18:159–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. 2012. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA. [Google Scholar]
- 10. Chao A, Colwell RK, Lin CW, Gotelli NJ. 2009. Sufficient sampling for asymptotic minimum species richness estimators. Ecology 90:1125–1133 [DOI] [PubMed] [Google Scholar]
- 11. Ulrich W. 2008. Pairs—a FORTRAN program for studying pair-wise species associations in ecological matrices. http://www.uni.torun.pl/*ulrichw
- 12. Gotelli NJ, Ulrich W. 2010. The empirical Bayes approach as a tool to identify non-random species associations. Oecologica 162:463–477 [DOI] [PubMed] [Google Scholar]
- 13. Woolhouse ME, Howey R, Gaunt E, Reilly L, Chase-Topping M, Savill N. 2008. Temporal trends in the discovery of human viruses. Proc. Biol. Sci. 275:2111–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woolhouse M, Scott F, Hudson Z, Howey R, Chase-Topping M. 2012. Human viruses: discovery and emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:2864–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chao A. 2005. Species estimation and applications. In Balakrishnan N, Read CB, Vidakovic B, Encyclopedia of statistical sciences. Wiley, New York, NY. [Google Scholar]
- 16. Mora C, Tittensor DP, Myers RA. 2008. The completeness of taxonomic inventories for describing the global diversity and distribution of marine fishes. Proc. Biol. Sci. 275:149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Magurran AE, McGill BJ. 2011. Biological diversity: frontiers in measurement and assessment. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 18. Morse SS, Mazet JA, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. 2012. Prediction and prevention of the next pandemic zoonosis. Lancet 380:1956–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson DE, Reeder DM. 2005. Mammal species of the world. Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- 20. Ge X-Y, Li J-L, Yang X-L, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang Y-J, Luo C-M, Tan B, Wang N, Zhu Y, Crameri G, Zhang S-Y, Wang L-F, Daszak P, Shi Z-L. First isolation and characterization of bat SARS-like coronaviruses that use the ACE2 receptor. Nature, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Epstein JH, Prakash V, Smith CS, Daszak P, McLaughlin AB, Meehan G, Field HE, Cunningham AA. 2008. Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg. Infect. Dis. 14:1309–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Epstein JH, Quan PL, Briese T, Street C, Jabado O, Conlan S, Ali Khan S, Verdugo D, Hossain MJ, Hutchison SK, Egholm M, Luby SP, Daszak P, Lipkin WI. 2010. Identification of GBV-D, a novel GB-like flavivirus from old world frugivorous bats (Pteropus giganteus) in Bangladesh. PLoS Pathog. 6:e1000972. 10.1371/journal.ppat.1000972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stapleton JT, Williams CF, Xiang J. 2004. GB virus type C: a beneficial infection? J. Clin. Microbiol. 42:3915–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lawn SD, Bekker LG, Middelkoop K, Myer L, Wood R. 2006. Impact of HIV infection on the epidemiology of tuberculosis in a peri-urban community in South Africa: the need for age-specific interventions. Clin. Infect. Dis. 42:1040–1047 [DOI] [PubMed] [Google Scholar]
- 25. Joseph SB, Hanley KA, Chao L, Burch CL. 2009. Coinfection rates in φ6 bacteriophage are enhanced by virus-induced changes in host cells. Evol. Appl. 2:24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Techasaensiri B, Techasaensiri C, Mejías A, McCracken GH, Jr, Ramilo O. 2010. Viral coinfections in children with invasive pneumococcal disease. Pediatr. Infect. Dis. J. 29:519–523 [DOI] [PubMed] [Google Scholar]
- 27. Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson S, Begon M. 2010. Species interactions in a parasite community drive infection risk in a wildlife population. Science 330:243–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Newman CM, Cerutti F, Anderson TK, Hamer GL, Walker ED, Kitron UD, Ruiz MO, Brawn JD, Goldberg TL. 2011. Culex flavivirus and West Nile virus mosquito coinfection and positive ecological association in Chicago, United States. Vector Borne Zoonotic Dis. 11:1099–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mühldorfer K, Speck S, Kurth A, Lesnik R, Freuling C, Müller T, Kramer-Schadt S, Wibbelt G. 2011. Diseases and causes of death in European bats: dynamics in disease susceptibility and infection Rates. PLoS One 6:e29773. 10.1371/journal.pone.0029773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer-König U, Ebert K, Schrage B, Pollak S, Hufert FT. 1998. Simultaneous infection of healthy people with multiple human cytomegalovirus strains. Lancet 352:1280–1281 [DOI] [PubMed] [Google Scholar]
- 31. Haberland M, Meyer-König U, Hufert FT. 1999. Variation within the glycoprotein B gene of human cytomegalovirus is due to homologous recombination. J. Gen. Virol. 80:1495–1500 [DOI] [PubMed] [Google Scholar]
- 32. Thiry E, Meurens F, Muylkens B, McVoy M, Gogev S, Thiry J, Vanderplasschen A, Epstein A, Keil G, Schynts F. 2005. Recombination in alphaherpesviruses. Rev. Med. Virol. 15:89–103 [DOI] [PubMed] [Google Scholar]
- 33. Norberg P, Kasubi MJ, Haarr L, Bergström T, Liljeqvist JA. 2007. Divergence and recombination of clinical herpes simplex virus type 2 isolates. J. Virol. 81:13158–13167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Greenwood AD, Tsangaras K, Ho SY, Szentiks CA, Nikolin VM, Ma G, Damiani A, East ML, Lawrenz A, Hofer H, Osterrieder N. 2012. A potentially fatal mix of herpes in zoos. Curr. Biol. 22:1727–1731 [DOI] [PubMed] [Google Scholar]
- 35. Roycroft E, Rose L, Scallan MF, Crowley B. 2012. Molecular characterization of varicella-zoster virus clinical isolates from 2006 to 2008 in a tertiary care hospital, Dublin, Ireland, using different genotyping methods. J. Med. Virol. 84:1672–1679 [DOI] [PubMed] [Google Scholar]
- 36. Smith LM, McWhorter AR, Shellam GR, Redwood AJ. 2012. The genome of murine cytomegalovirus is shaped by purifying selection and extensive recombination. Virology 435:258–268 [DOI] [PubMed] [Google Scholar]
- 37. Boppana SB, Rivera LB, Fowler KB, Mach M, Britt WJ. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366–1371 [DOI] [PubMed] [Google Scholar]
- 38. Anthony SJ, St Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, Navarrete-Macias I, Sanchez-Leon M, Saliki JT, Pedersen J, Karesh W, Daszak P, Rabadan R, Rowles T, Lipkin WI. 2012. Emergence of fatal avian influenza in New England harbor seals. mBio 3(4):e00166-12. 10.1128/mBio.00166-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ, Ksiazek TG, Mishra A. 2006. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 12:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. ICDDRB 2008. Outbreaks of Nipah virus in Rajbari and Manikgonj. Health Sci. Bull. 6:12–13 [Google Scholar]
- 41. Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA, Kenah E, Ksiazek TG, Rahman M. 2009. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg. Infect. Dis. 15:1229–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. ICDDRB 2010. Nipah outbreak in Faridpur District, Bangladesh, 2010. Health Sci. Bull. 8:6–11 [Google Scholar]
- 43. Arankalle VA, Bandyopadhyay BT, Ramdasi AY, Jadi R, Patil DR, Rahman M, Majumdar M, Banerjee PS, Hati AK, Goswami RP, Neogi DK, Mishra AC. 2011. Genomic characterization of Nipah virus, West Bengal, India. Emerg. Infect. Dis. 17:907–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rahman MA, Hossain MJ, Sultana S, Homaira N, Khan SU, Rahman M, Gurley ES, Rollin PE, Lo MK, Comer JA, Lowe L, Rota PA, Ksiazek TG, Kenah E, Sharker Y, Luby SP. 2012. Date palm sap linked to Nipah virus outbreak in Bangladesh, 2008. Vector Borne Zoonotic Dis. 12:65–72 [DOI] [PubMed] [Google Scholar]
- 45. Chua KB, Wang LF, Lam SK, Crameri G, Yu M, Wise T, Boyle D, Hyatt AD, Eaton BT. 2001. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology 283:215–229 [DOI] [PubMed] [Google Scholar]
- 46. Yaiw KC, Crameri G, Wang L, Chong HT, Chua KB, Tan CT, Goh KJ, Shamala D, Wong KT. 2007. Serological evidence of possible human infection with Tioman virus, a newly described paramyxovirus of bat origin. J. Infect. Dis. 196:884–886 [DOI] [PubMed] [Google Scholar]
- 47. Brahmbhatt M, Dutta A. 2008. On SARS type economic effects during infectious disease outbreaks. Policy research working paper WPS 4466. World Bank, East Asia and Pacific Region, Chief Economist's Office [Google Scholar]
- 48. Newman SH, Field H, de Jong CE, Epstein JH. 2011. Investigating the role of bats in emerging zoonoses: balancing ecology, conservation and public health interests. FAO Animal production and health manual no. 12. FAO, Rome, Italy. [Google Scholar]
- 49. Quan PL, Firth C, Street C, Henriquez JA, Petrosov A, Tashmukhamedova A, Hutchison SK, Egholm M, Osinubi MOV, Niezgoda M, Ogunkoya AB, Briese T, Rupprecht CE, Lipkin WI. 2010. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. mBio 1(4):e00208-10. 10.1128/mBio.00208-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. 2008. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J. Clin. Microbiol. 46:2652–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Atkins A, Wellehan JF, Jr, Childress AL, Archer LL, Fraser WA, Citino SB. 2009. Characterization of an outbreak of astroviral diarrhea in a group of cheetahs (Acinonyx jubatus). Vet. Microbiol. 136:160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wellehan JF, Johnson AJ, Harrach B, Benkö M, Pessier AP, Johnson CM, Garner MM, Childress A, Jacobson ER. 2004. Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J. Virol. 78:13366–13369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Johne R, Enderlein D, Nieper H, Müller H. 2005. Novel polyomavirus detected in the feces of a chimpanzee by nested broad-spectrum PCR. J. Virol. 79:3883–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kapoor A, Mehta N, Esper F, Poljsak-Prijatelj M, Quan PL, Qaisar N, Delwart E, Lipkin WI. 2010. Identification and characterization of a new bocavirus species in gorillas. PLoS One 5:e11948. 10.1371/journal.pone.0011948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aitichou M, Saleh SS, McElroy AK, Schmaljohn C, Ibrahim MS. 2005. Identification of Dobrava, Hantaan, Seoul, and Puumala viruses by one-step real-time RT-PCR. J. Virol. Methods 124:21–26 [DOI] [PubMed] [Google Scholar]
- 57. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maes P, Klempa B, Clement J, Matthijnssens J, Gajdusek DC, Krüger DH, Van Ranst M. 2009. A proposal for new criteria for the classification of hantaviruses, based on S and M segment protein sequences. Infect. Genet. Evol. 9:813–820 [DOI] [PubMed] [Google Scholar]
- 59. Jones E, Oliphant T, Peterson P. 2001. SciPy: open source scientific tools for Python. http://www.scipy.org
- 60. Gotelli NJ, Colwell RK. 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4:379–391 [Google Scholar]
- 61. Gotelli NJ, Colwell RK. 2011. Estimating species richness, p. 39–54 In Magurran AE, McGill BJ, Biological diversity: Frontiers in measuring biodiversity. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 62. Vavrek MJ. 2012. Package “fossil.” In Palaeoecological and palaeogeographical analysis tools. [Google Scholar]
- 63. R Development Core Team 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 64. The World Bank 2012. People, pathogens and our planet. Volume 2: The economics of One Health. The World Bank, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental discussion. Download
Relative distribution of viruses included in discovery curve analyses (see also Fig. 8). A subset of samples (n = 1,092) was used for discovery curve analysis. Only those samples that were screened for all nine viral families were included, ensuring internal consistency. Eleven of the 55 identified viruses had zero abundance in this subset and were therefore not considered in the analysis. The 11 omitted viruses were PgHV-2, -5, -6, and -9, PgAdV-1 and -10, PgAstV-4, -5, -6, and -8, and PgBoV-1. Forty-four viruses were therefore retained in our estimates, and the relative frequency of each is presented here. Download
Viral discovery curves are presented for (i) all viruses (see also Fig. 8), (ii) PMV, (iii) AstV, (iv) HV, and (v) AdV. Discovery effort (number of samples tested) is indicated by the horizontal dotted line. Red line, collector curve showing accumulation of novel viruses over samples tested; blue line, Chao2 estimator at every sample point, with arrow indicating 95% confidence intervals; gray lines, ICE and Jackknife estimators at every sample point; Φ, estimated total diversity; dashed horizontal lines, required sampling effort to discover an arbitrary proportion of the total diversity; Ω, effort required to discover 100% of the estimated diversity. Download
Summary of intraspecific coinfections. The total number of samples testing positive for each family is presented, together with the number of the samples containing >1 virus of the same family.