SUMMARY
Activation of the DNA replication checkpoint by the ATR kinase requires protein interactions mediated by the ATR activating protein, TopBP1. Accumulation of TopBP1 at stalled replication forks requires the interaction of TopBP1 BRCT5 with the phosphorylated SDT repeats of the adaptor protein MDC1. Here we present the X-ray crystal structures of the tandem BRCT4/5 domains of TopBP1 free and in complex with a MDC1 consensus pSDpT phospho-peptide. TopBP1 BRCT4/5 adopts a variant BRCT-BRCT packing interface and recognizes its target peptide in a manner distinct from that observed in previous tandem BRCT-peptide structures. The phosphate-binding pocket and positively charged residues in a variant loop in BRCT5 present an extended binding surface for the negatively charged MDC1 phospho-peptide. Mutations in this surface reduce binding affinity and recruitment of TopBP1 to γH2AX foci in cells. These studies reveal a new mode of phospho-peptide binding by BRCT domains in the DNA damage response.
INTRODUCTION
The DNA replication checkpoint is crucial for the prevention of genomic instability during DNA replication in cells. Activation of the DNA replication checkpoint requires the orchestrated assembly of proteins at the stalled replication fork. Topoisomerase IIβ binding protein 1 (TopBP1) is key to the success of DNA replication checkpoint activation by operating at multiple and distinct steps that contribute to the robust activation of the critical Ser/Thr kinase, ATR (Ataxia telangiectasia and Rad3 related). The abundance of conserved phospho-peptide binding BRCA1 C-terminal (BRCT) domains in TopBP1 provides extraordinary specificity to target different replication fork proteins. The TopBP1 N-terminal BRCT0/1/2 domains recognize the Rad9 C-terminal tail of the Rad9-Rad1-Hus1 (9-1-1) complex to activate ATR via the ATR activation domain (AAD) of TopBP1 (Delacroix et al., 2007; Lee et al., 2007). ATR kinase activity is further potentiated by a secondary interaction between the TopBP1 C-terminal BRCT7/8 domains and auto-phosphorylated ATR (Liu et al., 2011). In an earlier step in checkpoint activation, the TopBP1 BRCT7/8 domains bind BRCA1-associated C-terminal helicase/Fanconi anemia group J protein (BACH1/FANCJ) to regulate the helicase activity of BACH1 and increase single strand DNA and subsequent RPA loading (Gong et al., 2010). Despite these findings, it remained elusive how TopBP1 accumulates at stalled replication forks, since TopBP1 localization is independent of BACH1 and Rad9 interactions (Gong et al., 2010; Yan and Michael, 2009).
We have previously shown that the fifth BRCT domain of TopBP1 is responsible for TopBP1 localization to stalled replication forks (Wang et al., 2011; Yamane et al., 2002). TopBP1 BRCT5 directly interacts with the phosphorylated Ser-Asp-Thr (SDT) repeats in Mediator of DNA damage checkpoint protein 1 (MDC1), and this binding is required for sustaining and amplifying ATR activity for checkpoint activation (Wang et al., 2011). MDC1 is a critical DNA damage response (DDR) adaptor in DNA double strand break (DSB) repair. The rapid phosphorylation of histone H2AX at Ser139 (γ-H2AX) by the Ser/Thr kinase ATM is recognized by the tandem BRCT domains of MDC1, which further functions as a platform to bind various DDR factors such as RNF8 and the MRE11-RAD50-NBS1 (MRN) complex (Huen and Chen, 2010). A region in MDC1 spanning amino acids 210–460 contains six highly conserved SDT motifs that are constitutively phosphorylated by Casein kinase 2 (CK2). These di-phosphorylated motifs are recognized by the FHA-BRCT-BRCT domain repeat in NBS1 (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008; Xu et al., 2008), as well as the FHA domain of Aprataxin (Becherel et al., 2010).
BRCT domains are versatile modules that form various domain assemblies and are implicated in numerous functions, including protein-protein, phospho-peptide, DNA and poly(ADP-ribose) binding (Leung and Glover, 2011). The conserved mode of phospho-peptide recognition by tandem BRCT domains is well established through structural studies in DDR proteins such as BRCA1, MDC1, TopBP1, MCPH1, S. pombe Crb2 and S. pombe Brc1 (Clapperton et al., 2004; Kilkenny et al., 2008; Leung et al., 2011; Shiozaki et al., 2004; Singh et al., 2012; Stucki et al., 2005; Williams et al., 2010; Williams et al., 2004). The tandem BRCT presents an extended phospho-peptide binding surface, with a pSer/pThr binding pocket located at the N-terminal BRCT domain and a secondary pocket at the BRCT-BRCT interface with specificity for +3/+4 residues. Unlike conventional tandem BRCT domains that require both BRCT domains to form a viable phospho-peptide binding surface, only the C-terminal BRCT5 of the tandem BRCT4/5 pair is needed for MDC1 interaction and indeed BRCT4 lacks key amino acids required for phospho-peptide recognition (Rappas et al., 2011). In light of this knowledge, we sought to delineate the molecular basis of TopBP1-MDC1 interaction by characterizing, both structurally and functionally, the interaction between the tandem TopBP1 BRCT4/5 domains and a MDC1 di-phospho-peptide containing a consensus sequence of the SDT repeats. We show that TopBP1 BRCT4/5 adopts an unconventional tandem BRCT repeat structure with a phosphate-binding pocket in the C-terminal BRCT5 domain. The combination of the phosphate-binding pocket and a structured loop in BRCT5 creates an extended positively charged surface that mediates MDC1 SDT di-phospho-peptide binding and TopBP1 accumulation to stalled replication forks.
RESULTS
Crystal structure of TopBP1 BRCT4/5
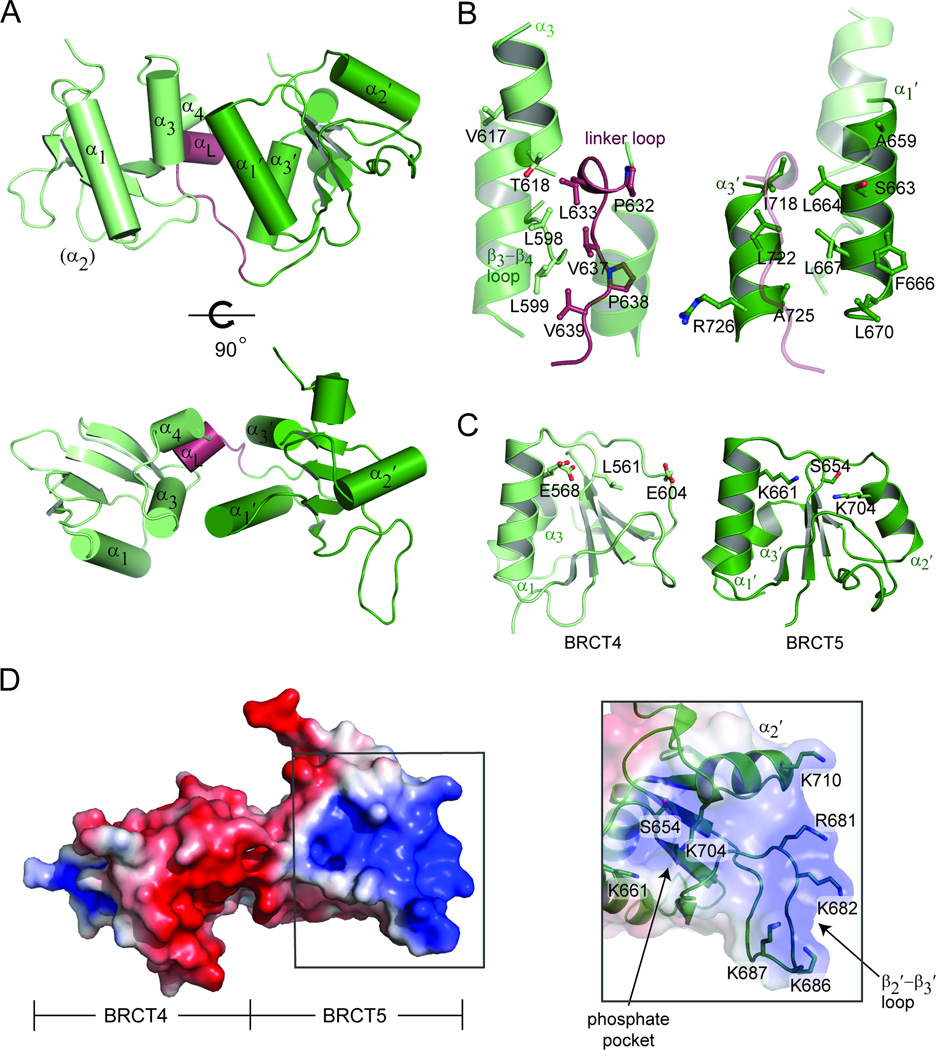
The crystal structure of TopBP1 BRCT4/5 was solved to 1.9 Å resolution (Table 1). The tandem BRCT pair adopts a unique domain packing, where the juxtaposition of the two BRCT domains is head-to-head (where head is defined as the α1-α3 face and tail as the α2 face) rather than the head-to-tail arrangement characteristic of canonical BRCT repeats (Fig. S1A). This is likely driven by a combination of the variant BRCT fold in the N-terminal BRCT4 and a significantly shorter linker region between BRCT4 and BRCT5 (Fig. 1A, Fig. S1B). TopBP1 BRCT4 lacks a α2-helix, which in canonical BRCT repeats houses conserved residues that participate in phosphate-binding and the hydrophobic BRCT-BRCT interface. Instead, the BRCT4 α2 is replaced by a short loop that is solvent exposed rather than being involved at the BRCT-BRCT interface. A short linker helix (αL) composed of three residues (Pro632, Leu633 and Phe634) is also part of a significantly shorter inter-BRCT linker in TopBP1 BRCT4/5 (Fig. 1A). The inter-BRCT linker packs tightly between the adjacent BRCT domains to stabilize the BRCT-BRCT interface.
Table 1.
Data collection, phasing and refinement statistics
| TopBP1 BRCT4/5 | TopBP1 BRCT4/5 peptide complex |
|
|---|---|---|
| Data collection: | ||
| Space group | P2221 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 35.90, 48.80, 126.09 | 58.81, 59.10, 78.31 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 102.05, 98.04, 114.34 |
| Resolution (Å) | 63.04–1.90 | 34.57–2.60 |
| Rsyma | 0.077 (0.497) | 0.059 (0.402) |
| I /σI | 18.0 (2.5) | 11.9 (2.2) |
| Completeness (%) | 99.8 (99.2) | 95.2 (95.2) |
| Redundancy | 3.8 (3.7) | 2.1 (2.1) |
| Refinement: | ||
| Resolution (Å) | 38.60–1.90 | 34.57–2.60 |
| No. reflections | 18137 (927) | 26637 (1330) |
| Rwork / Rfreeb | 0.175/0.224 | 0.190/0.234 |
| No. of atoms | ||
| Protein | 1540 | 5716 |
| Peptide | - | 159 |
| Ligand | 33 | - |
| Water | 238 | 257 |
| Average B factor (Å2) | ||
| Protein | 11.0 | 44.3 |
| Peptide | - | 58.6 |
| Ligand | 31.9 | - |
| Water | 23.7 | 33.6 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.011 | 0.009 |
| Bond angles (°) | 1.347 | 1.132 |
Rsym = Σ|I−〈I〉|/Σ|I|
R = Σ║Fo|−|Fc║/Σ|Fo|Rfree was calculated from 5 % of the data excluded from refinement
Figure 1.
Crystal structure of TopBP1 BRCT4/5. (A) Cartoon representation of TopBP1 BRCT4/5. BRCT4 (light green), BRCT5 (dark green) and the linker region (red) are colored accordingly. Secondary structure elements are labeled. (B) BRCT-BRCT interface of TopBP1 BRCT4/5. Residues involved in the hydrophobic packing for the N-terminal face (left) and C-terminal face (right) are shown as sticks and labeled. (C) Comparison of conserved phosphate-binding residues in BRCT4 (left) and BRCT5 (right). (D) Electrostatic surface representation of TopBP1 BRCT4/5 oriented with BRCT4 on the left and BRCT5 on the right (left). The positively charged surface of BRCT5 is boxed. Residues that make up the BRCT5 phosphate binding pocket and positively charged surface are shown as sticks and labeled (right). See also Figure S1.
As a consequence of the unusual head-to-head domain arrangement, the composition of the N-terminal domain face that contacts the C-terminal domain is significantly different from the one employed in conventional BRCT repeats. The N-terminal domain face consists of residues from α3 (Val617 and Thr618), the β3-β4 loop (Leu598 and Leu599) and linker region (Pro632, Leu633, Val637, Pro638 and Val639) (Fig. 1B, left panel). Contributions from these different regions substitute for the α2-helix typically used in the canonical interface. Conversely, the C-terminal domain face that contacts the N-terminal domain involves the α1′ and α3′ helices, which are the same secondary structure elements utilized in canonical BRCT repeats. Residues that form this hydrophobic face include Ala659, Ser663, Leu664, Phe666, Leu667 and Leu670 of α1 and Ile718, Leu722, Ala725 and Arg726 of α3′ (Fig. 1B, right panel). Together, the α3, β3-β4 loop, linker region, α1′ and α3′-helices form an extensive hydrophobic interface that enables a head-to-head domain packing in TopBP1 BRCT4/5.
To date, the phosphate-binding pockets identified in canonical BRCT repeats are found in the N-terminal BRCT. This enables the characteristic specificity for the +3 residue in a targeted phospho-peptide through a secondary pocket formed at the BRCT-BRCT interface. Besides being positioned on the opposite side of the canonical phospho-peptide binding surface, the putative phosphate-binding pocket in BRCT4 is highly acidic, and conserved phosphate-binding residues are instead substituted with Leu561, Glu568 and Glu604 (Fig. 1C, left panel). Strikingly, an intact phosphate-binding pocket containing the conserved Ser654, Lys661 and Lys704 residues is instead found in the C-terminal BRCT5 (Fig. 1C, right panel). Although the presence of a C-terminal phosphate-binding pocket is perplexing in comparison to other BRCT repeats, the possibility for BRCT5 to recognize a phosphate supports previous findings that BRCT5 interacts with phospho-MDC1 to control the DNA replication checkpoint (Wang et al., 2011).
To probe for potential protein binding surfaces on TopBP1 BRCT4/5, we first examined the electrostatic potential surface of TopBP1 BRCT4/5. Although the BRCT repeat structure carries an overall negative charge, a highly positively charged surface is located in BRCT5 (Fig. 1D, left panel). This region is rich in basic residues that extend from the putative phosphate-binding pocket (K661 and K704) to the extended β2′-β3′ loop (R681, K682, K686 and K687) and C-terminus of α2′ (K710) (Fig. 1D, right panel). Alignments of various tandem BRCT domains indicate that the β2′-β3′ loop is the most variable region in the BRCT family (Glover et al., 2004). In TopBP1 BRCT5, the β2′-β3′ loop adopts an unusually extended, structured protrusion. A series of main chain hydrogen bonds mediated by Asn684, Ala685, Lys687, Gly688, Met689 and Ala691 ensure rigidity of the loop (Fig. S1C). The side chains of Asn684 and Ser683 also participate in hydrogen bonds with the loop main chain. This provides a structural platform for the four basic loop residues (Arg681, Lys682, Lys686 and Lys687) to create a positively charged concave pocket. Furthermore, the loop residues, especially Asn684 and the group of basic residues, are conserved in other species (Fig. S1D).
TopBP1 BRCT4/5 binds phosphorylated MDC1 SDT repeats
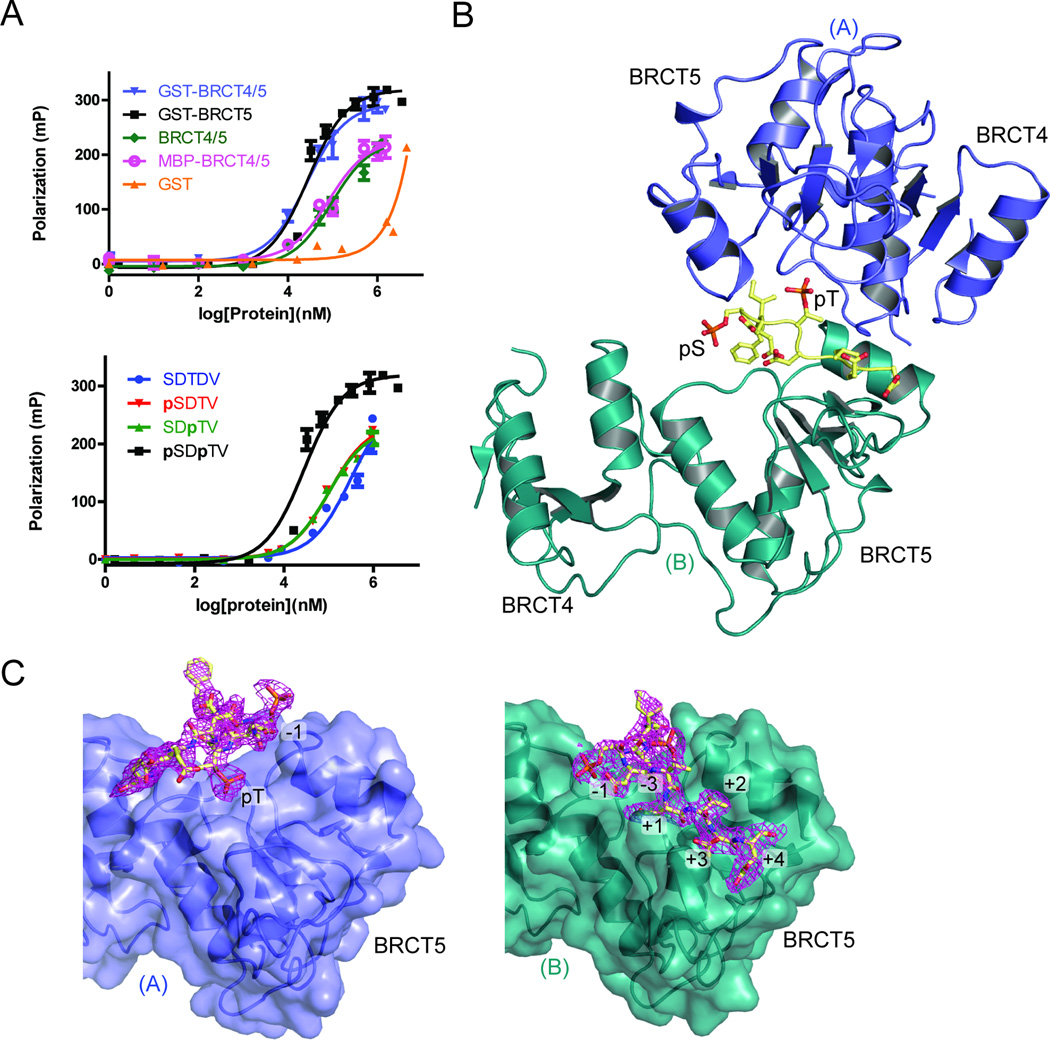
We have previously shown that TopBP1 BRCT5 interacts with a MDC1 di-phospho-peptide encoding a consensus sequence of the six SDT repeats (Fig. S2A) (Wang et al., 2011). To further characterize binding specificities of this interaction in vitro, we used a fluorescence polarization (FP) assay. Using this assay, we tested for the ability of GST fusion proteins of TopBP1 BRCT5 and BRCT4/5 to bind a FITC-labeled MDC1 consensus SDT di-phospho-peptide (GFIDpSDpTDVEEE). GST-BRCT4/5 and GST-BRCT5 bound the phospho-peptide with essentially identical affinities (Kd = 28 ± 4 µM for GST-BRCT5, Kd = 27 ± 4 µM for GST-BRCT4/5), indicating that BRCT4 is not important for the MDC1 interaction (Fig. 2A, upper panel, Table 2). We next tested the importance of Ser/Thr phosphorylation for TopBP1 binding. A non-phosphorylated version of the SDT peptide bound TopBP1 BRCT5, albeit with a significant (~11-fold) reduction in binding affinity (Fig. 2A, lower panel, Table 2). We also compared the affinity of TopBP1 BRCT5 affinity for doubly phosphorylated MDC1 peptides with MDC1 peptides bearing a single phosphate at either the Ser or Thr positions. The results indicate that either singly phosphorylated peptide is bound with a somewhat reduced (~3.5-fold) affinity compared to the doubly phosphorylated version (Fig. 2A, lower panel, Table 2). This result indicates that both residues play a role in TopBP1 binding. The importance of phosphorylation for this interaction appears to be significantly less than for other BRCT – phospho-peptide interactions. For example, TopBP1 BRCT7/8 binds its phosphorylated target peptide from BACH1 ~100-fold more tightly than the dephosphorylated peptide (Gong et al., 2010). This suggests that the mechanism of peptide recognition employed by BRCT4/5 maybe significantly different than other tandem BRCT repeats.
Figure 2.
MDC1 SDT di-phospho-peptide interactions with TopBP1 BRCT4/5. (A) FP binding results for the MDC1 FITC-labeled di-phospho-peptide and various TopBP1 proteins. Triplicate data points are represented in graphs as mean ± SEM. (Above) FITC-peptide binding results for GST, GST-fusion proteins of BRCT4/5 and BRCT5, as well as untagged BRCT4/5 and MBP-BRCT4/5. (Below) FP assay of non-phosphorylated, pSer, pThr or di-phosphorylated MDC1 FITC-labeled peptide with GST-TopBP1 BRCT5. (B) Crystal structure of MDC1 di-phospho-peptide in complex with TopBP1 BRCT4/5. The phosphorylated residues of the peptide (yellow) are labeled. BRCT4/5 protomer A (blue) and B (teal) are designated. (C) Di-phospho-peptide interactions with BRCT5 (represented in surface representation) of protomer A (left panel) and protomer B (right panel). The 2|Fo| - |Fc| electron density map for the di-phospho-peptide is shown in magenta and peptide-interacting residues are labeled. See also Figure S2.
Table 2.
Summary of Fluorescence Polarization Binding Experiments
| Protein | Peptide | Kd (µM) | |
|---|---|---|---|
| GST-BRCT5 | FITC-GFIDSDTDVEEE-NH2 | 310 ± 70 | |
| FITC-GFIDpSDTDVEEE-NH2 | 105 ± 9 | ||
| FITC-GFIDSDpTDVEEE-NH2 | 98 ± 9 | ||
| FITC-GFIDpSDpTDVEEE-NH2 | 28 ± 4 | ||
| FITC-GFIDpSDpTDDEEE-NH2 | 24 ± 3 | ||
| GST-BRCT4/5 | FITC-GFIDpSDpTDVEEE-NH2 | 27 ± 4 | |
| MBP-BRCT4/5 | FITC-GFIDpSDpTDVEEE-NH2 | 82 ± 16 | |
| BRCT4/5 | FITC-GFIDpSDpTDVEEE-NH2 | 94 ± 15 | |
| GST | FITC-GFIDpSDpTDVEEE-NH2 | N.D.a | |
| GST-BRCT5 Mutants |
S654A | FITC-GFIDpSDpTDVEEE-NH2 | 32 ± 3 |
| R681E/K682E | FITC-GFIDpSDpTDVEEE-NH2 | 280 ± 60 | |
| K704A | FITC-GFIDpSDpTDVEEE-NH2 | 210 ± 50 | |
N.D. – not determined
Crystal structure of TopBP1 BRCT4/5 bound to phosphorylated MDC1
To further investigate the TopBP1-MDC1 interaction, we co-crystallized and solved the structure of TopBP1 BRCT4/5 in complex with a MDC1 di-phospho-peptide to 2.6 Å resolution. A single MDC1 di-phospho-peptide is bound in an extended conformation by two BRCT4/5 protomers on opposite sides (represented as protomers A and B, Fig. 2B). The peptide-bound dimer is further related by 2-fold non-crystallographic symmetry with another peptide-bound dimer in the asymmetric unit (Fig. S2B). Comparisons of the apo and bound structures indicate that BRCT4/5 is structurally rigid and does not change significantly upon peptide binding (RMSD for Cα = 0.39 and 0.46 with protomer A and B, respectively). The two protomers are oriented in an orthogonal manner in the dimer and interact indirectly through the MDC1 di-phospho-peptide, except for a single hydrogen bond between the Tyr622 side chain of protomer A and the Gly702 main chain of protomer B. Consistent with previous data for BRCT5-mediated MDC1 binding, the MDC1 di-phospho-peptide exclusively contacts the two BRCT5 domains from each protomer. Although the MDC1 di-phospho-peptide interacts with two BRCT5 domains, their binding interfaces are not symmetrical and differ in size and composition. The majority of interactions with protomer A are contributed by the peptide pThr residue and bury a total solvent accessible surface area of 434 Å2 (Fig. 2C). This relatively small contact interface suggests that peptide interactions with protomer A are unlikely to be stable on its own, and indeed some of the crystal contacts actually bury a larger surface area than the protomer A – peptide interface. In contrast, the interface between the di-phospho-peptide and protomer B is more extensive, spanning residues −3 to +4 relative to the pThr and burying 919 Å2 of solvent accessible surface area (Fig. 2C).
To test the possibility that two protomers of TopBP1 could bind to a MDC1 di-phospho-peptide in solution, we assessed the impact of enforced dimerization on the peptide binding affinity of TopBP1. We compared the MDC1 diphospho-peptide binding affinities of GST-BRCT4/5 or GST-BRCT5, which both exist as dimers in solution, with monomeric forms of TopBP1 BRCT4/5 (Fig. 2A, upper panel). The GST fusion-stabilized dimers bind MDC1 ~3-fold tighter than either the free BRCT4/5 or MBP-BRCT4/5, which are both monomeric as determined by gel filtration chromatography (Kd = 82 ± 16 µM for MBP-BRCT4/5; Kd = 94 ± 15 µM for untagged BRCT4/5). The enhanced affinity of the dimeric forms of TopBP1 over the monomeric forms is consistent with an avidity effect that would be expected in a structure where two protomers bind the MDC1 di-phospho-peptide.
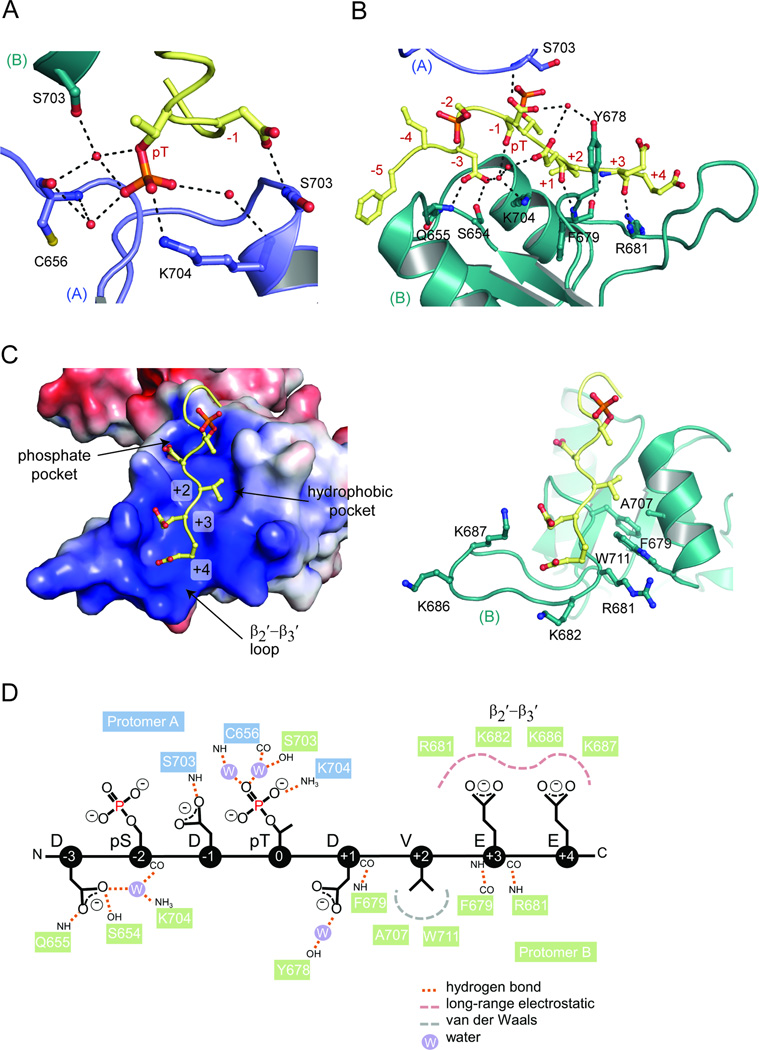
TopBP1 BRCT4/5 - MDC1 SDT repeat binding interactions
Unlike the highly specific phosphate-binding properties of canonical tandem BRCT domain pockets, peptide recognition by the BRCT5 phosphate-binding pocket relies more on general charge-charge interactions and water-mediated contacts. Interactions with BRCT5 of protomer A involve the MDC1 pThr and −1 Asp. The pThr is coordinated in the phosphate-binding pocket, but only participates in a single direct interaction with the conserved Lys704 side chain, although it makes water-mediated interactions with the main chain of Cys656 and Lys704 and side chain of Ser703 of protomer B (Fig. 3A). Besides the pThr, the −1 Asp side chain also hydrogen bonds to the Ser703 main chain of protomer A. Unexpectedly, the −3 Asp, rather than the −2 pSer, points into the phosphate-binding pocket of protomer B (Fig. 3B). The −2 pSer instead lies across the phosphate-binding pocket of protomer B. The −3 Asp side chain hydrogen bonds with the Gln655 main chain and Ser654 side chain, as well as the Lys704 side chain through a water molecule. Other contacts include a main chain-main chain hydrogen bond between the +1 Asp and Phe679 and water-bridged interactions involving the Tyr678 side chain and the −1 and +1 Asp side chains.
Figure 3.
TopBP1 BRCT5 di-phospho-peptide binding interactions. (A) MDC1 peptide interactions with protomer A. Hydrogen bonding and electrostatic interactions are indicated by dotted lines and waters are shown as red spheres. Different TopBP1 protomers are designated as (A) and (B). (B) MDC1 peptide interactions with protomer B. (C) Specificity of MDC1 peptide C-terminal residues by BRCT5 of protomer B in electrostatic surface representation (left) and cartoon (right). (D) Schematic diagram of MDC1 peptide interactions with both TopBP1 BRCT5 protomers. See also Figure S3.
Perhaps the most extensive interaction surface involves the recognition of the C-terminal residues of the conserved MDC1 SDT motif. As part of the larger binding interface established by protomer B, the +2 to +4 residues are recognized by the BRCT5 basic surface that extends from the phosphate-binding pocket to the β2′-β3′ loop. The +2 Val sits in a small hydrophobic pocket situated between the basic phosphate-binding pocket and β2′-β3′ loop (Fig. 3C). Residues that contribute to this pocket include Ala707 and Trp711 from α2′ and Phe679 from the β2′-β3′ loop. The conserved +3 and +4 Glu residues are cradled in the positively charged β2′-β3′ loop. The +3 main chain hydrogen bonds with the main chain of Phe679 and Arg681 (Fig. 3B), while the +3 and +4 acidic side chains make electrostatic interactions with Lys687 and Lys682, respectively (Fig. 3C). Overall, the makeup of the BRCT5 binding surface matches the conservation of a small hydrophobic residue at +2 and acidic residues at +3 and +4 positions of the MDC1 SDT repeats (Fig. S2A). The interactions between both protomers and the MDC1 di-phospho-peptide are summarized in Fig. 3D.
To test the importance of the conserved hydrophobic Val residue at the peptide +2 position, we compared the binding affinity of the wild type MDC1 phospho-peptide with that of a peptide in which the +2 Val is substituted with an Asp. While a Val is the most common residue at this position, one of the MDC1 SDT repeats harbours an Asp at this position. We reasoned that if interactions between the di-phospho-peptide and BRCT5 are purely electrostatic, then an increase in the overall negative charge of the peptide should enhance binding affinity. If instead the hydrophobic interactions involving the +2 residue are critical, then the substitution should reduce binding affinity. We observe, however, that the substitution results in no significant change in binding affinity (Fig. S3C, Table 2). This may indicate that the loss in hydrophobic interactions in the mutant is balanced by long-range electrostatic interactions between the substituted Asp and the surrounding positively charged surface of BRCT5 and indicates that each of the SDT repeats are equally capable of binding TopBP1. The results suggest that the pocket may function to restrict the +2 residue to either a small hydrophobic residue or an Asp.
Mutational analysis of BRCT5 binding interface
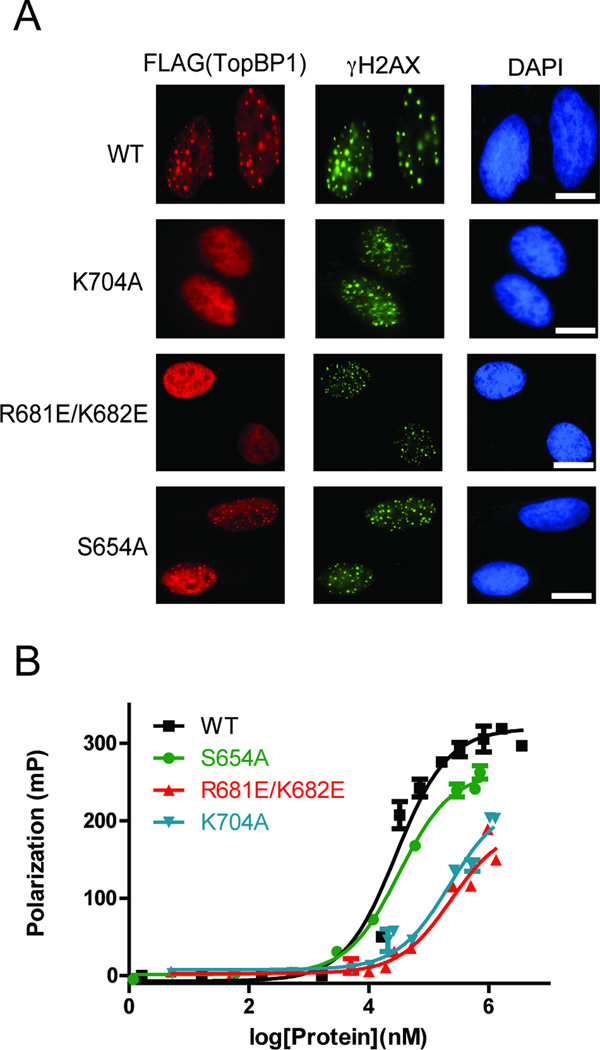
Since TopBP1 BRCT5 facilitates TopBP1 localization at stalled replication forks, we first generated several mutants within the BRCT5 domain and tested their abilities to form hydroxyurea (HU)-induced foci in cells. Mutations in the putative phosphate-binding pocket (K704A) or in the β2′-β3′ loop (R681E/K682E) abolished foci formation, supporting the requirement for the positively charged BRCT5 surface (Fig. 4A). In contrast, the S654A mutation did not disrupt TopBP1 foci formation but did appear to give less intense foci compared to background nuclear fluorescence in these cells. This is in agreement with the observation that the conserved Ser654 in the BRCT5 phosphate-binding pocket does not appear to have a major role in binding the MDC1 di-phospho-peptide in the crystal structure. In contrast, the analogous Ser/Thr is required for phospho-peptide binding in several conventional tandem BRCT domains such as BRCA1, MDC1, and TopBP1 BRCT7/8, highlighting the distinct mechanism of phospho-peptide recognition employed by TopBP1 BRCT4/5 (Leung and Glover, 2011). We next performed FP studies on various BRCT5 mutants to test whether the conserved positively charged surface is responsible for interactions with MDC1 in vitro. Consistent with TopBP1 localization, mutations in either the putative phosphate-binding pocket (K704A) or β2′-β3′ loop (R681E/K682E) in GST-fusion proteins of BRCT5 significantly reduced binding to the MDC1 di-phospho-peptide compared with wild type (Kd ~ 210 ± 50 µM for K704A, Kd ~ 280 ± 60 µM for R681E/K682E) (Fig. 4B). In contrast, the S654A mutant bound the MDC1 di-phospho-peptide with an affinity very similar to wild-type (Kd = 32 ± 3 µM). Overall, the specificity for the MDC1 SDT motif correlates with our mutational analysis and provides a rationale for the unusually structured and positively charged β2′-β3′ loop exclusive to TopBP1 BRCT5.
Figure 4.
Mutational analysis of TopBP1 BRCT5 binding interface. (A) Replication stress-induced focus formation of wild-type and TopBP1 mutants. U2OS cells transfected with plasmids encoding SFB-tagged WT or mutants of TopBP1 were exposed to 2 mM HU for 3 hours. Cells were fixed and immunostained with anti-FLAG and anti-γ-H2AX antibodies. Bar: 10 µm. (B) FP binding studies of MDC1 FITC-labeled di-phospho-peptide with GST-fusion proteins of wild type BRCT5 and various missense variants. Triplicate data points are represented in graphs as mean ± SEM.
DISCUSSION
The recognition of MDC1 by TopBP1 is critical for DNA replication checkpoint control in response to replication stress. TopBP1 BRCT5 directly binds to the conserved SDT repeats of MDC1, and this interaction is necessary for sustaining and amplifying ATR activation. In the context of phospho-peptide recognition by BRCT domains, the interaction between TopBP1 BRCT5 and the MDC1 SDT motifs was intriguing for several reasons. As part of a tandem BRCT pair, the functional requirement for only the C-terminal BRCT5 domain suggested that TopBP1 BRCT4/5 does not follow the canonical BRCT repeat mode of recognition (Wang et al., 2011; Yamane et al., 2002). The MDC1 SDT repeats are established di-phospho-peptide motifs that are also targets for the FHA domains of NBS1 and Aprataxin in DNA repair (Becherel et al., 2010; Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008;Xu et al., 2008). Since BRCT domain phosphate-binding pockets can bind to both pSer and pThr peptides (Leung et al., 2011), TopBP1 BRCT5 could potentially recognize only the pSer, pThr or even both phosphorylated residues. Here we present the molecular basis for TopBP1 BRCT5 recognition of a MDC1 di-phospho-peptide containing a consensus SDT repeat sequence. Our structural and functional analyses not only provides insight into the questions raised above, but also reveal other surprising aspects of BRCT phospho-peptide binding that were previously unknown.
TopBP1 BRCT4/5 contains a number of structural features that diverges from a conventional BRCT repeat. An unexpected BRCT-BRCT packing interface results in a head-to-head arrangement of the BRCT domains. This is a consequence of an absent α2-helix and constraints imposed by the relatively short inter-BRCT linker region. Rather than the α2-α1′-α3′ triple helix bundle associated with typical BRCT repeat interfaces, TopBP1 BRCT4/5 incorporates the same α1′-α3′ helices from BRCT5 and a different surface composed of α3 and the β3-β4 loop from BRCT4. Unconventional BRCT-BRCT interfaces have also been observed in the triple BRCT repeat, TopBP1 BRCT0/1/2, which coincidently also contains relatively shorter inter-BRCT linkers. However, the interfaces between BRCT0/1 and BRCT1/2 are distinct from BRCT4/5 and their respective C-terminal domain faces do not involve the α1′ and α3′ helices (Fig. S1A) (Rappas et al., 2011). Another difference is the presence of a phosphate-binding pocket in BRCT5 rather than the N-terminal BRCT4. Although this is rare in tandem BRCT domains, it is also found in the BRCT1/2 repeats in PAX-interacting protein 1 (PTIP) for example (Sheng et al., 2011). It is not clear, however, whether PTIP BRCT1/2 can bind phospho-peptides, and further structural and functional work will be needed to provide evidence for a common group of BRCT repeats that recognize phospho-peptides via a C-terminal BRCT pocket. Another possibility is that BRCT5 phospho-peptide recognition may in fact resemble phospho-peptide binding of single BRCT domains, a function that still remains unclear.
A striking feature of the peptide-bound structure is the lack of significant, tight contacts between either the pSer or pThr and the consensus phosphate binding pocket of BRCT5. This is in contrast to all other structures of tandem BRCT domains bound to phospho-peptides where the phosphate is coordinated in a highly conserved manner (Leung and Glover, 2011). Indeed, FP results indicate that removal of both phosphate groups only results in an ~11-fold reduction in binding affinity, suggesting phosphorylation is less critical for TopBP1 BRCT4/5 – phospho-peptide recognition than for other tandem BRCT – target phospho-peptide interactions. The lack of importance of the canonical phosphate binding pocket is further underlined by the fact that mutation of the conserved Ser654 does not significantly impact phospho-peptide binding affinity or TopBP1 foci formation (Fig. 4). This is in contrast with the analogous Ser1655 in the BRCA1 BRCT phosphate binding pocket, which is critical for phospho-peptide binding and for BRCA1 BRCT domain function (Lee et al., 2010; Williams et al., 2004). Instead, phospho-peptide binding appears to largely rely on electrostatic interactions involving not only the phosphate binding pocket, but also the extended β2’-β3’ hairpin that contacts the conserved C-terminal acidic tail of the MDC1 phospho-peptide motif.
Perhaps the most surprising aspect of phospho-peptide recognition by TopBP1 BRCT5 is the apparent dimerization of BRCT5 induced by MDC1 binding. While the interface between protomer B and the phospho-peptide is quite large and specific mutations in its binding surface abrogate in vitro binding and foci formation, the interface with protomer A is much smaller and could be an artefact of crystal packing. We were unable to isolate or trap the peptide-induced dimer in solution using gel-filtration chromatography, chemical cross-linking, or EMSA. This would suggest that either the binding of two BRCT4/5 protomers to a single MDC1 di-phospho peptide does not occur in solution, or that the interactions driving dimer formation with the phospho-peptide may be too transient to form a tight complex. On the other hand, we were able to demonstrate significantly higher MDC1 di-phospho-peptide binding affinities for dimeric GST-fusion proteins of BRCT4/5 than for monomeric MBP-BRCT4/5 or BRCT4/5 alone (Fig. 2A). An explanation for this result could be that GST-induced dimerization indirectly stabilizes two BRCT4/5 domains in a state that favours the formation of the peptide-induced dimer observed in the crystal structure. If pairs of TopBP1 BRCT5 domains do bind phospho-peptide targets in the context of the intact full length protein, then this would imply that other regions in TopBP1 might be important to stabilize TopBP1 oligomerization. Indeed, TopBP1 has been shown to oligomerize through a tandem BRCT7/8-mediated recognition of an Akt-dependent internal TopBP1 pSer (pS1159) motif to stabilize the interaction between phospho-E2F1 and the single BRCT6 domain of TopBP1 (Liu et al., 2006). TopBP1 BRCT4/5 has also been shown to be important for the colocalization of TopBP1 with 53BP1 at DNA double strand breaks, where it may participate in the G1 DNA damage checkpoint (Cescutti et al., 2010). While the details of this interaction have not been elucidated, it is intriguing that colocalization and binding of the isolated BRCT4/5 with 53BP1 in cells appears to be dependent on ATM as well as fusion of the BRCT4/5 to a tetramerization domain. It is tempting to speculate that BRCT4/5 may bind one or more of the highly acidic ATM phosphorylation sites of 53BP1 (Jowsey et al., 2007) via a dimeric mechanism similar to that observed in the TopBP1 BRCT4/5 – MDC1 complex.
The highly conserved MDC1 SDT repeats are not only recognized by TopBP1 but are also bound by the pThr-specific FHA binding domains of Aprataxin and the FHA-tandem BRCT module of NBS1. Structural and biochemical studies of these interactions reveal a primary recognition of pThr within the FHA phosphate-binding pocket, with more limited electrostatic interactions involving the pSer as well as neighbouring acidic residues in the pSDpT motif (Becherel et al., 2010; Lloyd et al., 2009; Williams et al., 2009). TopBP1 BRCT4/5, however, is unique in its recognition of the conserved +2 to +4 residues. This suggests that the conservation of these residues in each of the SDT repeats in MDC1 is not solely for CK2 phosphorylation, but is also critical for TopBP1 binding. Clearly, further investigation is needed to understand how these unique di-phospho-peptide motifs in MDC1 are recognized by a host of proteins in DDR.
Like the TopBP1-BACH1 interaction, the interaction between TopBP1 and MDC1 is also crucial for DNA replication checkpoint control. This study provides the structural basis that underlies another key TopBP1-mediated interaction that contributes to ATR activation and checkpoint signalling. Moreover, MDC1 binding by TopBP1 BRCT5 uncovers novel aspects of BRCT domain phospho-peptide recognition that further illustrates the diversity of BRCT domain function in the DNA damage response.
EXPERIMENTAL PROCEDURES
Cloning, expression and purification
TopBP1 BRCT5 (641–746) was cloned into pGEX-4T-1 and BRCT4/5 (549–746) was cloned into pGEX-6P-1 (GE Healthcare) to create GST fusion proteins. BRCT4/5 (549–746) was cloned into pKM596 (New England Biolabs) to create an MBP fusion protein. BRCT5 mutants were engineered from BRCT5 (641–746) using PCR-directed overlap extension (Heckman and Pease, 2007) and cloned into pGEX-6P-1 vector. The GST fusion protein was expressed in Escherichia coli BL21-Gold cells and purified using glutathione affinity chromatography with glutathione sepharose 4B beads (GE Healthcare) and eluted in elution buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 20 mM reduced glutathione and 0.1 % BME). GST-fusion protein of BRCT5 was cleaved with Thrombin protease (GE Healthcare) overnight at room temperature. BRCT5 was then separated from GST by cation exchange chromatography (buffer A: 50 mM HEPES pH 7, 0.1 % BME; buffer B: 50 mM HEPES pH 7, 1 M NaCl, 0.1 % BME). BRCT5 was further purified on a Superdex 75 column in storage buffer (10 mM Tris-HCl pH 8, 150 mM NaCl, 1 mM DTT). GST-TopBP1 BRCT4/5 was cleaved with PreScission protease overnight at 4 °.C. BRCT4/5 was purified by anion exchange chromatography (buffer A: 50 mM HEPES pH 7, 0.1 % BME; buffer B: 50 mM HEPES pH 7, 1 M NaCl, 0.1 % BME). Residual GST was removed by incubation with glutathione sepharose 4B beads (GE Healthcare) prior to a final purification step on a Superdex 75 column in storage buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT). The MBP-BRCT4/5 fusion protein was expressed in Escherichia coli BL21-DE3 cells and purified using amylose affinity chromatography with amylose resin (New England Biolabs) and eluted in elution buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 10 mM Maltose, 1 mM EDTA and 0.1 % BME). MBP fusion protein was purified by anion exchange chromatography (buffer A: 50 mM HEPES pH 7, 0.1 % BME; buffer B: 50 mM HEPES pH 7, 1 M NaCl, 0.1 % BME) and further purified on a Superdex 75 column in storage buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT). The size and oligomeric state of each of the proteins was confirmed by size exclusion chromatography on a Superdex 75 column calibrated with protein size standards.
Crystallization
TopBP1 BRCT5 was concentrated to 9 mg/mL for crystallization. Crystals were grown at 4 °C using hanging drop vapour diffusion by adding 2 µL protein with 1 µL reservoir consisting of 10 % PEG 1000 and 0.1 M Na/K phosphate pH 6.2. Crystals were flash-cooled in cryo-protectant containing reservoir solution and 26 % glycerol. TopBP1 BRCT4/5 was concentrated to 6.5 mg/mL for crystallization. Crystals were grown at room temperature in drops containing 1 µL protein and 1 µL reservoir (20 % PEG 3350 and 0.2 M NaSCN). Cryo-protectant used to flash-cool crystals contained reservoir solution supplemented with 15 % glycerol. For co-crystallization, TopBP1 BRCT4/5 concentrated to 12 mg/mL was incubated with 1:6 molar ratio of protein:peptide (Ac-GFIDpSDpTDVEEE-NH2) for 1 hr on ice. Co-crystals were grown at room temperature by adding 1 µL of protein:peptide mixture with 1 µL of reservoir solution containing 0.1 M ammonium acetate, 0.1 M bis-Tris pH 5.5 and 17 % PEG 10,000. Co-crystals were flash-cooled in reservoir solution supplemented with 20 % glycerol.
Data collection and structure determination
Data for BRCT5 and BRCT4/5 crystals were collected at the 8.3.1 beamline (Advanced Light Source, Berkeley). Intensity data from a BRCT5 crystal were processed using HKL2000 (Otwinowski and Minor, 1997) to the space group P6222 with unit cell dimensions a = 91.00 Å, b = 91.00 Å, c = 114.31 Å, α = 90 °, β = 90 °, γ = 120 °. A starting model consisting of an ensemble of N-terminal BRCT domains (PDB ID: 1JNX, 1R1Z and 2ADO) was used in molecular replacement with PHASER (McCoy, 2007). The solution was then partially built in COOT (Emsley and Cowtan, 2004) and refined to 3.3 Å resolution with TLS and restrained refinement in REFMAC5 (Murshudov et al., 1997) to a Rwork and Rfree of 0.3887 and 0.4256, respectively. Data from a BRCT4/5 crystal were scaled and reduced to the space group P2221 with unit cell dimensions a = 35.90 Å, b = 48.80 Å, c = 126.09 Å, α = 90 °, β = 90 °, γ = 90 °. The partially refined BRCT5 structure was used in molecular replacement to find one BRCT4/5 molecule in the asymmetric unit. Further refinement with rigid body and restrained refinement in REFMAC5 prior to automated model building using ARP/wARP (Cohen et al., 2008) successfully built 191 total residues with side chains. Further model building in COOT and refinement using PHENIX (Adams et al., 2010) at 1.9 Å resolution yielded a final Rwork and Rfree of 0.175 and 0.224, respectively. The final model lacks the N-terminal 549 residue and C-terminal 745–746 residues due to disorder in the crystal. The Ramachandran plot contains 100 % of all residues in favoured regions and 0 % in outlier regions.
Data for crystals of the BRCT4/5-peptide complex were collected at the CMCF 08ID-1 beamline (Canadian Light Source, Saskatoon). Intensity data were scaled and reduced using the XDS package (Kabsch, 2010) to the space group P1 with unit cell dimensions a = 58.81 Å, b = 59.10 Å, c = 78.31 Å, α = 102.05 °, (β = 98.04 °, γ = 114.34 °. The apo BRCT4/5 structure was used in PHASER to successfully find 4 copies in the asymmetric unit. Model building was carried out in COOT and refined using TLS refinement (1 group/chain) and 2-fold NCS restraints in PHENIX. The BRCT4/5 molecules are arranged as two dimers (designated AB and CD) related by 2-fold non-crystallographic symmetry (Fig. S2B). BRCT4/5 molecules A and C lack the N-terminal 549–550 residues, C-terminal 742–746 residues and loop residues 584–588. Molecules B and D lack the N-terminal 549–550 residues, C-terminal 743–746 residues and loop residues 584–589. The BRCT4/5 molecules were fully refined before building of the 2 peptides. Peptide A lacks the N-terminal −6 residue and C-terminal +5 residue and peptide B lacks the N-terminal −6 to −4 residues and C-terminal +5 residue (Fig. S3A). Since there are slight deviations in the two peptide chain conformations (Fig. S3B), 2-fold NCS restraints were not imposed for the peptide chains during refinement, which also yielded the lowest Rfree statistics. The wxu_scale was set to 0.1 to reduce the X-ray/ADP weight. The final model was refined in Phenix at 2.6 Å resolution to a Rwork and Rfree of 0.190 and 0.234, respectively. The Ramachandran plot contained 96.8 % of all residues in favoured regions, 3.0 % in allowed regions and 0.3 % in outlier regions.
Data collection and refinement statistics for the apo and peptide-bound structures are listed in Table 1. Models were validated with MolProbity (Chen et al., 2010). Secondary structure prediction of the models was performed with DSSP (Kabsch and Sander, 1983) and converted using DSSP2PDB (http://structure.usc.edu/dssp2pdb/). Hydrogen bonding was verified using HBPLUS (McDonald and Thornton, 1994). Structure figures were prepared with PyMOL (Version 1.4, Schrödinger, LLC.).
Fluorescence polarization
FP measurements were carried out using an Envision multi-label plate reader (Perkin Elmer) on a 384-well OptiPlate (Perkin Elmer). All peptides were synthesized and purified by Biomatik. FP assays were performed by mixing 10 nM FITC-labeled MDC1 phospho-peptide (FITC-GFIDpSDpTDVEEE-NH2; FITC-GFIDpSDpTDDEEE-NH2; FITC-GFIDSDpTDVEEE-NH2; FITC-GFIDpSDTDVEEE-NH2; FITC-GFIDSDTDVEEE-NH2) with freshly concentrated TopBP1 in FP assay buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT, 0.05 % Tween-20). The wells were incubated for 15 min at room temperature prior to taking FP measurements at an excitation wavelength of 485 nm and emission wavelength of 538 nm. Curve fitting and Kd calculations were obtained using PRISM software (GraphPad). Kd values presented in Table 2 are the averages from at least three independent titrations.
Immunofluorescence staining
Cells grown on coverslips were treated with HU (2 mM) for 3 hours. Cells were fixed in 3% paraformaldehyde for 10 minutes, and then permeabilized in 0.5% Triton X-100 containing solution for 5 minutes. Cells were incubated with primary antibodies diluted in 5% goat serum at room temperature for 30 minutes. Cells were washed twice with PBS and then incubated with either FITC-conjugated or rhodamine-conjugated secondary antibodies at room temperature for 30 minutes. Nuclei were counterstained with DAPI. The coverslips were mounted onto glass slides with anti-fade solution and visualized using a Nikon Eclipse E800 fluorescence microscope with a 60 × NA 1.3 oil objective lens. Images were photographed and analyzed using a Spot 2 Megasample camera and Photoshop software (Adobe).
Supplementary Material
HIGHLIGHTS.
TopBP1 BRCT4/5 adopts a novel BRCT-BRCT packing interface
MDC1 phospho-peptide binds via an extended positively charged. surface of BRCT5
Mutation of the binding surface of BRCT5 abrogates recruitment to DNA damage foci
ACKNOWLEDGEMENTS
We thank the staff at the ALS beamline 8.3.1 and Dr. Pawel Grochulski and the staff at the Canadian Light Source CMCF beamline 08ID-1 for assistance with synchrotron data collection. This work was supported by grants from the Canadian Cancer Society [to J.N.M.G]; and the National Institutes of Health [CA92584 to J.N.M.G., CA092312 to J.C.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Coordinates for the TopBP1 BRCT4/5 (RCSB accession: 3UEN) and peptide-bound complex (RCSB accession: 3UEO) are in the Protein Data Bank.
SUPPLEMENTAL INFORMATION
Supplemental figures 1–3 can be found with this article online.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel OJ, Jakob B, Cherry AL, Gueven N, Fusser M, Kijas AW, Peng C, Katyal S, McKinnon PJ, Chen J, et al. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2010;38:1489–1503. doi: 10.1093/nar/gkp1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescutti R, Negrini S, Kohzaki M, Halazonetis TD. TopBP1 functions with 53BP1 in the G1 DNA damage checkpoint. EMBO J. 2010;29:3723–3732. doi: 10.1038/emboj.2010.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- Cohen SX, Ben Jelloul M, Long F, Vagin A, Knipscheer P, Lebbink J, Sixma TK, Lamzin VS, Murshudov GN, Perrakis A. ARP/wARP and molecular replacement: the next generation. Acta Crystallogr D Biol Crystallogr. 2008;64:49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Huen MS, Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci. 2010;35:101–108. doi: 10.1016/j.tibs.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowsey P, Morrice NA, Hastie CJ, McLauchlan H, Toth R, Rouse J. Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR. DNA Repair (Amst) 2007;6:1536–1544. doi: 10.1016/j.dnarep.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kilkenny ML, Dore AS, Roe SM, Nestoras K, Ho JC, Watts FZ, Pearl LH. Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev. 2008;22:2034–2047. doi: 10.1101/gad.472808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Lee MS, Green R, Marsillac SM, Coquelle N, Williams RS, Yeung T, Foo D, Hau DD, Hui B, Monteiro AN, Glover JN. Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res. 2010;70:4880–4890. doi: 10.1158/0008-5472.CAN-09-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CC, Glover JN. BRCT domains: Easy as one, two, three. Cell Cycle. 2011;10:2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CC, Gong Z, Chen J, Glover JN. Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. J Biol Chem. 2011;286:4292–4301. doi: 10.1074/jbc.M110.189555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Paik JC, Wang B, Lin FT, Lin WC. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 2006;25:4795–4807. doi: 10.1038/sj.emboj.7601355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Rappas M, Oliver AW, Pearl LH. Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res. 2011;39:313–324. doi: 10.1093/nar/gkq743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng ZZ, Zhao YQ, Huang JF. Functional Evolution of BRCT Domains from Binding DNA to Protein. Evol Bioinform Online. 2011;7:87–97. doi: 10.4137/EBO.S7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell. 2004;14:405–412. doi: 10.1016/s1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- Singh N, Basnet H, Wiltshire TD, Mohammad DH, Thompson JR, Heroux A, Botuyan MV, Yaffe MB, Couch FJ, Rosenfeld MG, Mer G. Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1. Proc Natl Acad Sci U S A. 2012;109:14381–14386. doi: 10.1073/pnas.1212366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Wang J, Gong Z, Chen J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol. 2011;193:267–273. doi: 10.1083/jcb.201010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Williams RS, Dovey CL, Guenther G, Tainer JA, Russell P. gammaH2A binds Brc1 to maintain genome integrity during S-phase. EMBO J. 2010;29:1136–1148. doi: 10.1038/emboj.2009.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci U S A. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wu L, Cui G, Botuyan MV, Chen J, Mer G. Structure of a second BRCT domain identified in the nijmegen breakage syndrome protein Nbs1 and its function in an MDC1-dependent localization of Nbs1 to DNA damage sites. J Mol Biol. 2008;381:361–372. doi: 10.1016/j.jmb.2008.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Michael WM. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle. 2009;8:2877–2884. doi: 10.4161/cc.8.18.9485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.