Abstract
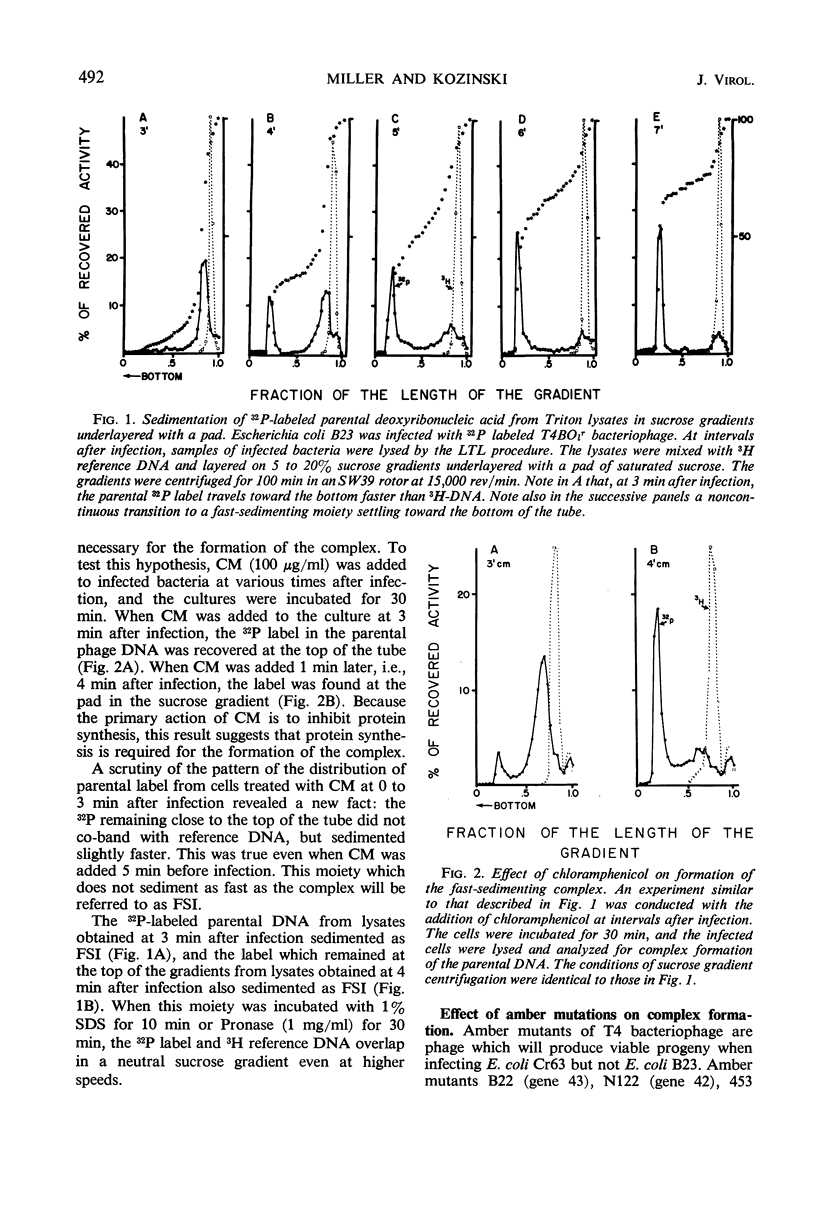
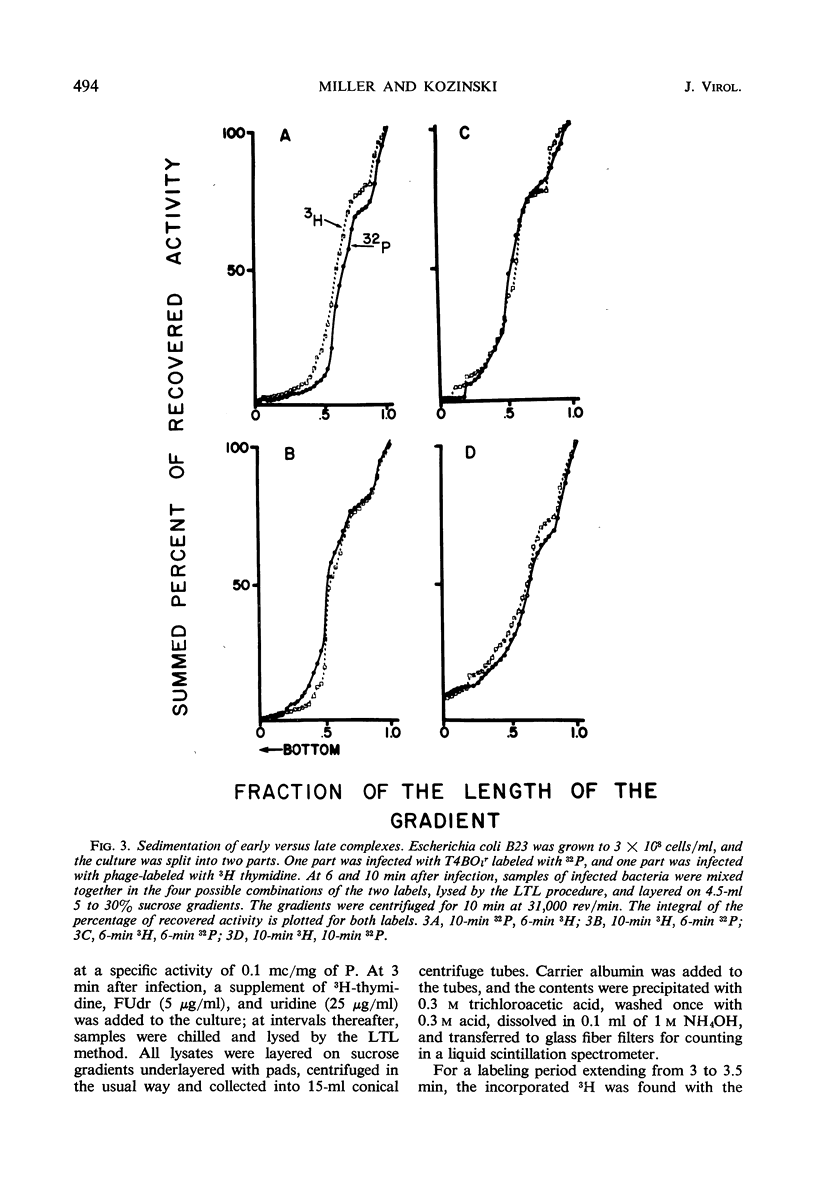
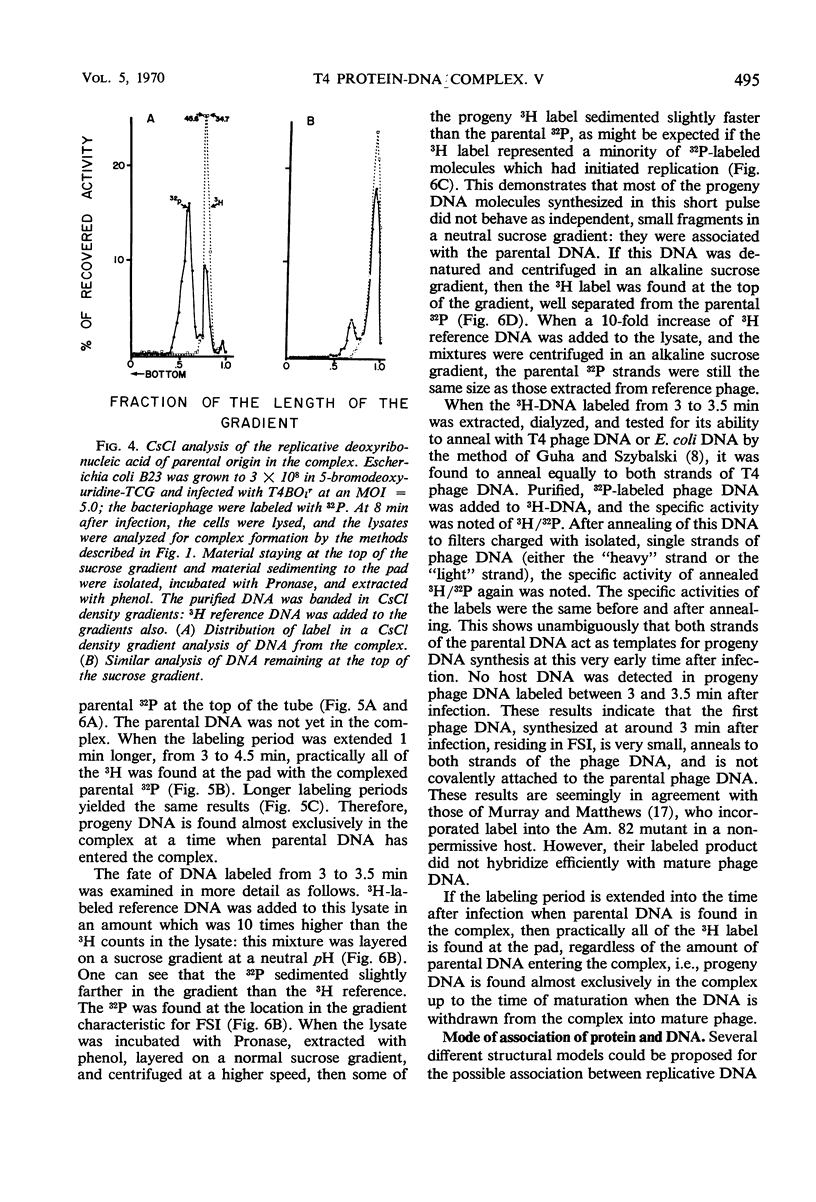
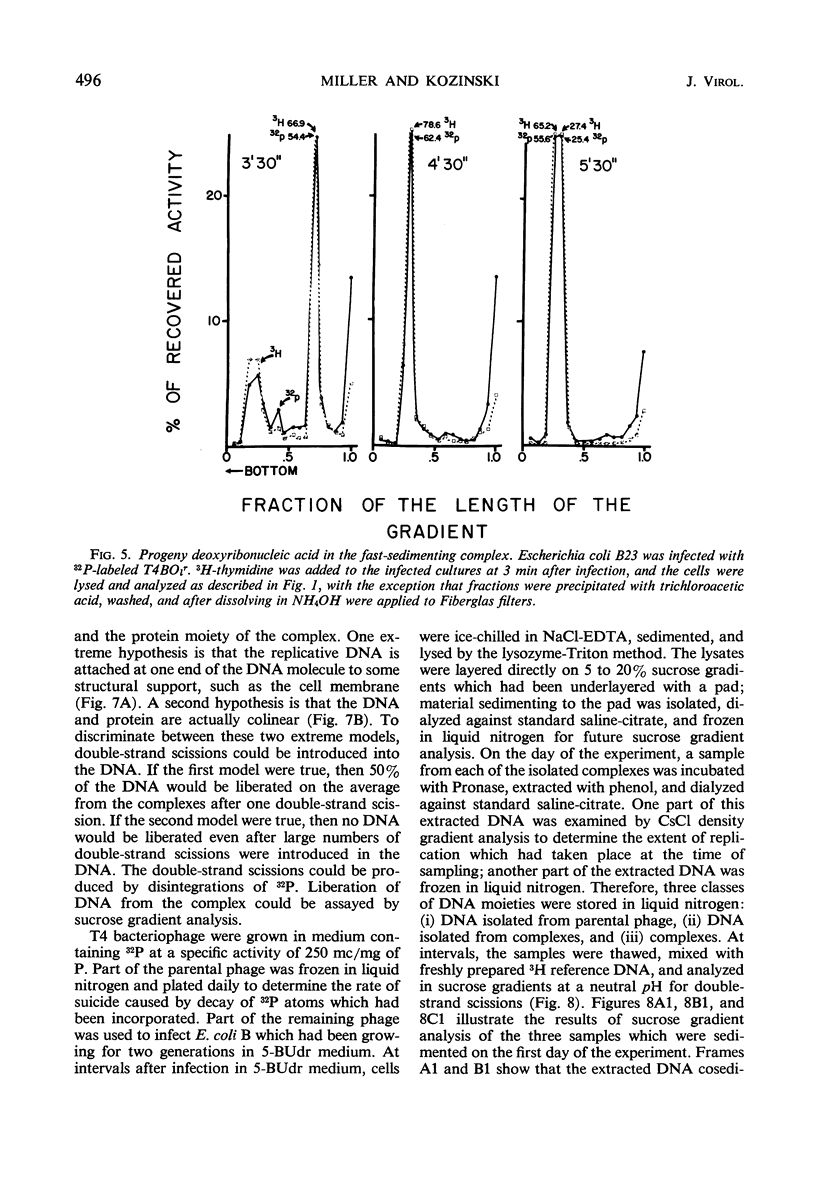
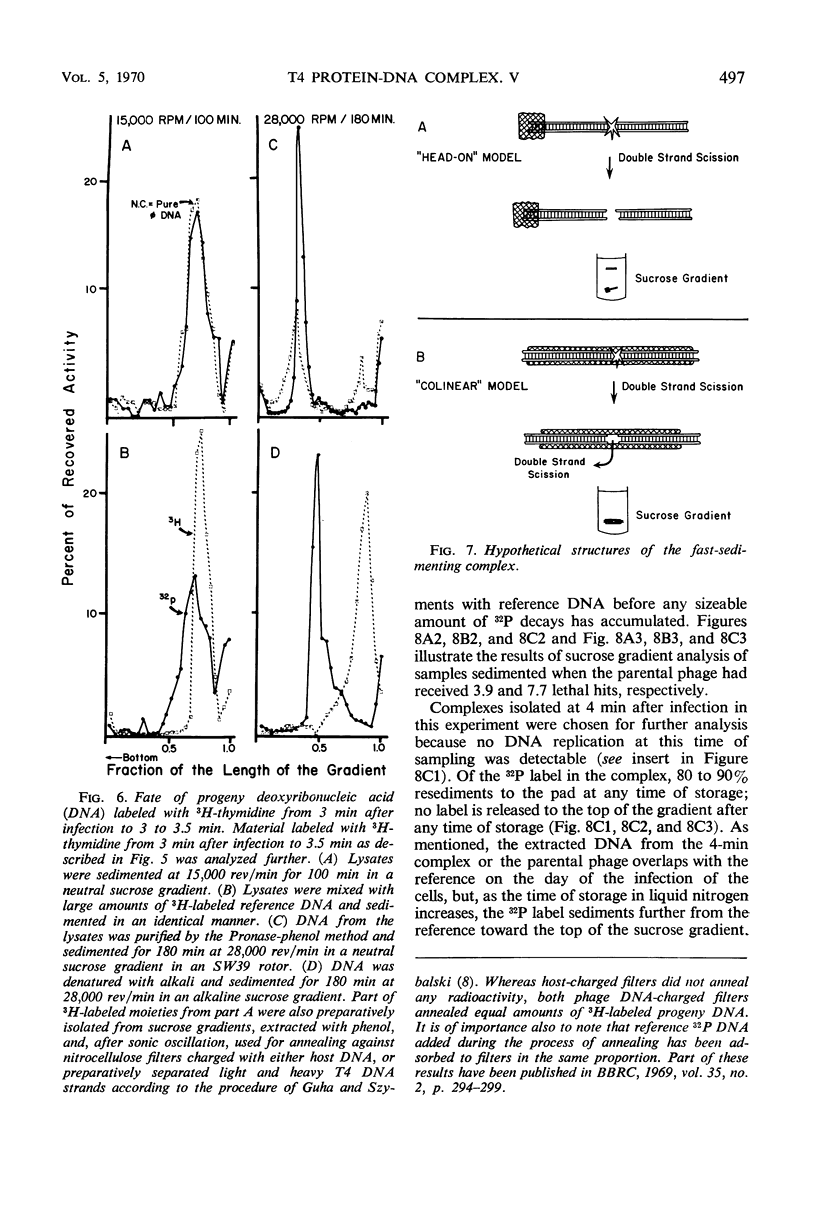
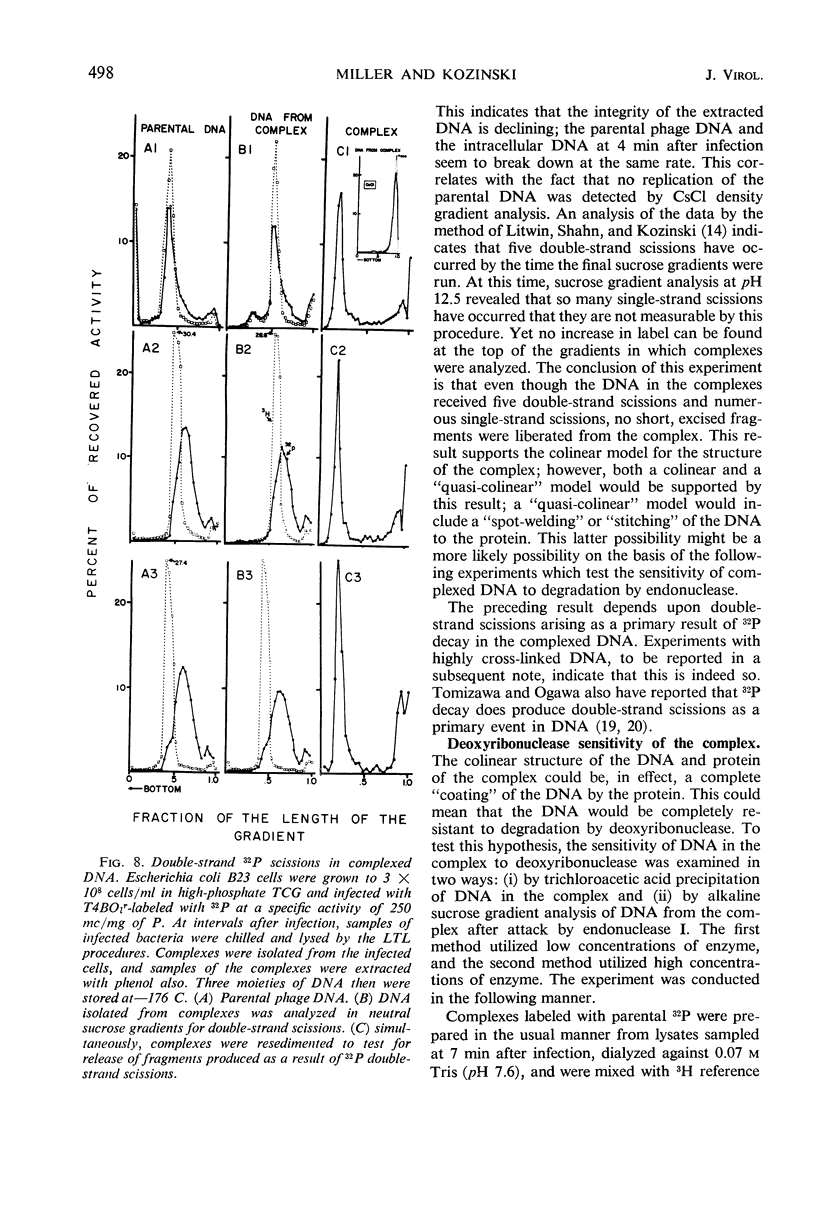
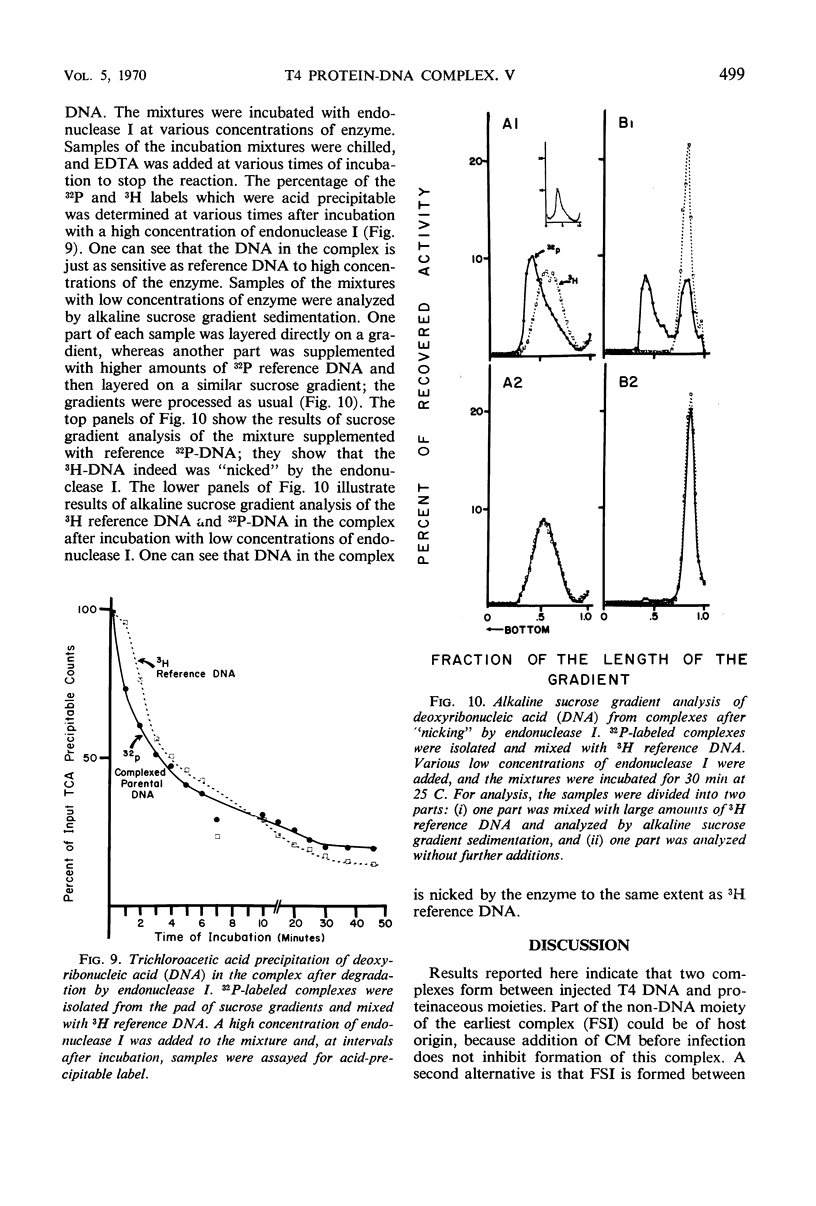
Soon after infection parental deoxyribonucleic acid (DNA) enters a structure sedimenting fast to the bottom of a sucrose gradient. The addition of chloramphenicol (CM) prevents formation of this structure, whereas treatment with Pronase releases DNA which sediments thereafter with the speed characteristic of phenol-extracted replicative DNA. It is assumed therefore that the structure responsible for fast sedimentation of replicative DNA is a newly synthesized protein. Those fast-sedimenting complexes contain preferentially the replicative form of parental DNA; this was proven by density labeling experiments. Progeny DNA labeled with 3H-thymidine added after infection can also be detected preferentially in this fast-sedimenting moiety. The association of the DNA with the complexing protein is of a colinear or quasi-colinear type. This was proven by introducing double-strand scissions into DNA embedded in the replicative complex; double-strand scissions do not liberate DNA from the fast-sedimenting complex. Despite the apparent intimate relation between protein and DNA, DNA residing in complexes is fully sensitive to the action of nucleases. Shortly prior to the appearance of the fast-sedimenting complex, parental DNA displays still another characteristic: at about 3 min after infection, it sediments faster than reference, but sizeably slower than the complex which appears at roughly 4 to 5 min after infection. The transition between these two fast-sedimenting types of moieties is not continuous. This fast-sedimenting intermediate, which appears at 3 min after infection, cannot be inhibited by the addition of CM either at the moment or prior to infection. Fast-sedimenting intermediate can be destroyed by sodium dodecyl sulfate, Pronase, or phenol extraction. The progeny DNA labeled with 3H-thymidine between 3 and 3.5 min after infection can be recovered in fast-sedimenting intermediate. The contribution of newly synthesized progeny DNA is so small that it cannot be detected as a shift of the parental density in a density labeling experiment. Small fragments of progeny DNA recovered in fast-sedimenting intermediate are not covalentlv attached to parental molecules and represent both strands of T4 DNA.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botstein D., Levine M. Intermediates in the synthesis of phage P22 DNA. Cold Spring Harb Symp Quant Biol. 1968;33:659–667. doi: 10.1101/sqb.1968.033.01.075. [DOI] [PubMed] [Google Scholar]
- Earhart C. F., Tremblay G. Y., Daniels M. J., Schaechter M. DNA replication studied by a new method for the isolation of cell membrane-DNA complexes. Cold Spring Harb Symp Quant Biol. 1968;33:707–710. doi: 10.1101/sqb.1968.033.01.079. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. Studies on the nature of replicating DNA in T4-infected Escherichia coli. J Mol Biol. 1966 Jun;18(1):127–143. doi: 10.1016/s0022-2836(66)80081-5. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. The absence of mature phage DNA molecules from the replicating pool of T-even-infected Escherichia coli. J Mol Biol. 1966 Jun;18(1):109–126. doi: 10.1016/s0022-2836(66)80080-3. [DOI] [PubMed] [Google Scholar]
- Guha A., Szybalski W. Fractionation of the complementary strands of coliphage T4 DNA based on the asymmetric distribution of the poly U and poly U,G binding sites. Virology. 1968 Apr;34(4):608–616. doi: 10.1016/0042-6822(68)90082-2. [DOI] [PubMed] [Google Scholar]
- KOZINSKI A. W., KOZINSKI P. B. Fragmentary transfer of P32-labeled parental DNA to progeny phage. II. The average size of the transferred parental fragment. Two-cycletransfer. Repair of the polynucleotide chain after fragmentation. Virology. 1963 Jun;20:213–229. doi: 10.1016/0042-6822(63)90109-0. [DOI] [PubMed] [Google Scholar]
- Knippers R., Sinsheimer R. L. Process of infection with bacteriophage phiX174. XX. Attachment of the parental DNA of bacteriophage phiX174 to a fast-sedimenting cell component. J Mol Biol. 1968 May 28;34(1):17–29. doi: 10.1016/0022-2836(68)90231-3. [DOI] [PubMed] [Google Scholar]
- Kozinski A. W., Felgenhauer Z. Z. Molecular recombination in T4 bacteriophage deoxyribonucleic acid. II. Single-strand breaks and exposure of uncomplemented areas as a prerequisite for recombination. J Virol. 1967 Dec;1(6):1193–1202. doi: 10.1128/jvi.1.6.1193-1202.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozinski A. W., Kozinski P. B. Early intracellular events in the replication T4 phage DNA. II. Partially replicated DNA. Proc Natl Acad Sci U S A. 1965 Aug;54(2):634–640. doi: 10.1073/pnas.54.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozinski A. W., Lin T. H. Early intracellular events in the replication of T4 phage DNA. I. Complex formation of replicative DNA. Proc Natl Acad Sci U S A. 1965 Jul;54(1):273–278. doi: 10.1073/pnas.54.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin S., Shahn E., Kozinski A. W. Interpretation of sucrose gradient sedimentation pattern of deoxyribonucleic acid fragments resulting from random breaks. J Virol. 1969 Jul;4(1):24–30. doi: 10.1128/jvi.4.1.24-30.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. C., Kozinski A. W., Litwin S. Molecular Recombination in T4 Bacteriophage Deoxyribonucleic Acid: III. Formation of Long Single Strands During Recombination. J Virol. 1970 Mar;5(3):368–380. doi: 10.1128/jvi.5.3.368-380.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray R. E., Mathews C. K. Addition of nucleotides to parental DNA early in infection by bacteriophage T4. J Mol Biol. 1969 Sep 14;44(2):233–248. doi: 10.1016/0022-2836(69)90172-7. [DOI] [PubMed] [Google Scholar]
- Salivar W. O., Sinsheimer R. L. Intracellular location and number of replicating parental DNA molecules of bacteriophages lambda and phi-X174. J Mol Biol. 1969 Apr 14;41(1):39–65. doi: 10.1016/0022-2836(69)90124-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Ogawa H. Breakage of DNA in rec+ and Rec- bacteria by disintegration of radiophosphorus atoms in DNA and possible cause of pleiotropic effects of RecA mutation. Cold Spring Harb Symp Quant Biol. 1968;33:243–251. doi: 10.1101/sqb.1968.033.01.028. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Ogawa H. Breakage of polynucleotide strands by disintegration of radiophosphorus atoms in DNA molecules and their repair. II. Simultaneous breakage of both strands. J Mol Biol. 1967 Nov 28;30(1):7–15. doi: 10.1016/0022-2836(67)90239-2. [DOI] [PubMed] [Google Scholar]


