Abstract
We report the construction of a 1.5 Mb resolution radiation hybrid map of the domestic cat genome. This new map includes novel microsatellite loci and markers derived from the 2X genome sequence that target previous gaps in the feline-human comparative map. Ninety-six percent of the 1793 cat markers we mapped have identifiable orthologues in the canine and human genome sequences. The updated autosomal and X chromosome comparative maps identify 152 cat-human and 134 cat-dog homologous synteny blocks. Comparative analysis shows the marked change in chromosomal evolution in the canid lineage relative to the felid lineage since divergence from their carnivoran ancestor. The canid lineage has a thirty-fold difference in the number of interchromosomal rearrangments relative to felids, while the felid lineage has primarily undergone intrachromosomal rearrangements. We have also refined the pseudoautosomal region and boundary in the cat and show that it is markedly longer than those of human or mouse. This improved RH comparative map provides a useful tool to facilitate positional cloning studies in the feline model.
Keywords: domestic cat, radiation hybrid map, canine genome, genome evolution, synteny, chromosome rearrangement
Introduction
Survey sequenced genomes (i.e., approximately 2X coverage) are useful for providing access to orthology-verified sequences for gene map construction, evolutionary genomic studies, and the annotation of the human genome sequence [1, 2]. Nearly two-dozen mammalian species are in the process of having their genome sequence determined by survey sequencing (http://www.genome.gov/11007951). The recent completion of a 2X domestic cat genome sequence provides a necessary boost for successful application of genome-based scans towards identifying genes of interest in this valuable animal model [3, 4]. However, navigating hundreds of thousands of sequence contigs and the remaining traces for a survey sequenced genome, and confidently establishing their orthology to regions of related mammalian genomes (such as human and dog for cat) using sequence similarity is a daunting task. This task is less challenging if higher coverage assemblies or high density comparative maps are available.
Radiation hybrid (RH) maps and linkage maps are important tools for both the long-range assembly and quality control of early genome-builds [1, 5, 6]. We present a new, denser RH map of the domestic cat (Felis catus). The marker development phase of this project focused on finding markers in the cat genome sequence traces that were located in holes of the previous cat-human comparative maps [7–9]. We further combined these new survey sequence-derived markers with a novel collection of feline microsatellite markers. By exploiting the feline genome survey sequence and the close evolutionary relationship with the dog genome, we were able to identify orthologous genome positions in the finished human and canine draft genome sequences for 96% of the markers. We constructed an enhanced comparative map relating the three genomes that provides 86% comparative coverage of the human genome and 85% comparative coverage of the canine genome. With an average spacing of 1 marker every 1.5 Mb in the feline euchromatic sequence, the map provides a solid framework and comparative tool to aid in the identification of genes controlling feline phenotypes and the chromosomal assignment of feline contigs and scaffolds during assembly. Alignment of the feline, canine and human chromosomes provides insight into the different chromosomal rearrangement characteristics that have occurred in the feline and canine lineages.
Results
Third Generation Cat-Human and First Generation Cat-Dog Ordered Comparative Maps
Combining new high-quality marker genotypes with previous RH data sets [7–9] produced a final set of 1845 markers that were evaluated and used to construct the RH map. After dropping 52 markers for quality control reasons the final computed RH map includes 1793 markers, with new data from 335 whole genome sequence trace-derived markers, 87 STS markers designed from ESTs and mRNAs, and 269 microsatellite markers. The map contains 1252 MLE-consensus framework markers (see Methods), 1680 markers that have assigned cR positions including 15 markers that had an identical vector with at least one other marker, and 113 markers that were binned (or placed) relative to the positioned markers. RH map linkage groups were initially established using a 2-point LOD score threshold of 8.0. For ten chromosomes the RH linkage groups correspond to separate arms of meta-or submetacentric chromosomes (A1, A2, B2, B4, C1, C2, D4, E2, E3, and X), and for eight chromosomes marker density was sufficient to produce single linkage groups for whole chromosomes (A3, B1, B3, D1, D2, D3, F1 and F2). Chromosome E1p formed two RH linkage groups because of the severe changes in retention frequency associated with the RH panel selectable marker (TK1) found on this chromosome. These RH groups were merged using evidence from the feline linkage map [8].
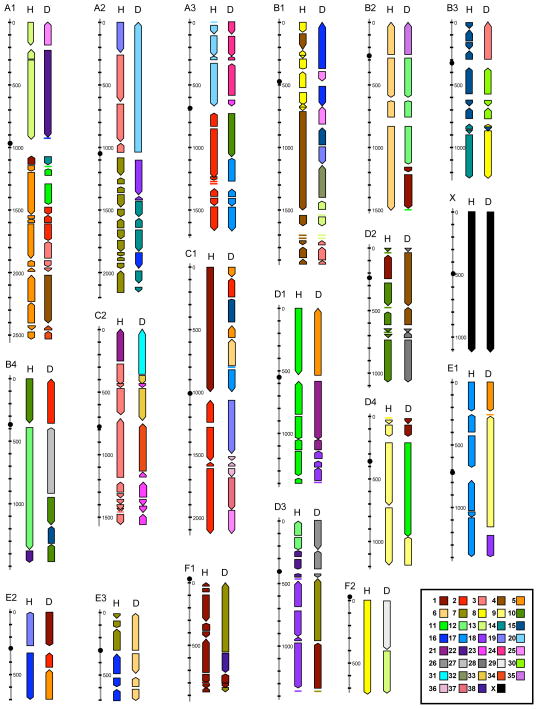
The 1793 markers cover all 19 feline autosomes and the X chromosome at an average spacing of 1.5 megabase-pairs (Mbp)(Table 1). Excluding the 113 binned markers would produce a marker density of 1 marker/1.6 Mbp. Marker density is also fairly uniform between chromosomes (Table 1), with chromosome F1 being the most marker dense at 1 marker/Mb, and chromosomes B1 and B2 being the most marker poor at 1 marker/1.7 Mb. Assuming a feline euchromatic genome size of 2.7 Mbp based on the assembly of genome survey sequence (Pontius et al. unpublished data), 1 centiray5000 corresponds to approximately 100 Kbp, or 10 cR5000/Mbp (Table 1). The final maps for each chromosome are available in tabular format as supplemental material (Supplemental Table 1), while a graphical display of each cat chromosome and blocks of conserved syntenic order with the human and canine genomes is presented in Figure 1.
Table 1.
Cat RH Map Marker Summary and Comparative Synteny Overview
| Feline Chromosome | Avg. Marker Density (Mb) | No. of MLE- consensus framework markers | Total markers on map | RH Length (cR5000) | Approx. Physical Lengtha (Mb) | cR/Mb | No. of Cat- Human Homologous Synteny Blocks | Orthologous Human Chromosomes | No. of Cat-Dog Homologous Synteny Blocks | Orthologous Dog Chromosomes |
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 1.5 | 111 | 168 | 2514.1 | 259 | 9.7 | 15 | 1b, 5, 13 | 16 | 2, 3, 4, 11, 14b, 16b, 22, 25, 34b |
| A2 | 1.4 | 107 | 138 | 2143.7 | 189 | 11.3 | 16 | 3, 7, 19 | 9 | 14, 16, 18, 20 |
| A3 | 1.5 | 73 | 99 | 1644.2 | 151 | 10.9 | 13 | 2, 20 | 10 | 10, 17, 23b, 24 |
| B1 | 1.7 | 93 | 120 | 1926.0 | 208 | 9.3 | 16 | 4, 8 | 15 | 3, 13, 15, 16, 19, 25, 32 |
| B2 | 1.7 | 64 | 90 | 1469.7 | 157 | 9.4 | 4 | 6 | 7 | 1, 12, 35 |
| B3 | 1.5 | 78 | 98 | 1221.5 | 151 | 8.1 | 10 | 14, 15 | 7 | 3, 8, 15b, 30 |
| B4 | 1.5 | 69 | 100 | 1443.3 | 146 | 9.9 | 3 | 10, 12, 22 | 5 | 2, 10, 15, 27 |
| C1 | 1.6 | 94 | 142 | 2111.0 | 232 | 9.1 | 5 | 1, 2 | 13 | 2, 5, 6, 15, 17, 19, 36, 37 |
| C2 | 1.4 | 81 | 112 | 1533.1 | 157 | 9.8 | 12 | 3, 21 | 10 | 23, 31, 33, 34 |
| D1 | 1.3 | 66 | 100 | 1394.9 | 130 | 10.8 | 6 | 11 | 7 | 5, 18, 21 |
| D2 | 1.4 | 51 | 76 | 1084.7 | 108 | 10.0 | 10 | 1b, 10 | 6 | 4, 26b, 28 |
| D3 | 1.4 | 65 | 78 | 1351.2 | 108 | 12.5 | 11 | 12, 18, 22 | 6 | 1, 7, 26 |
| D4 | 1.5 | 55 | 66 | 1184.9 | 100 | 11.9 | 5 | 9 | 4 | 1, 9, 17 |
| E1 | 1.3 | 46 | 77 | 1389.9 | 100 | 13.9 | 6 | 17 | 4 | 5, 9, 18b |
| E2 | 1.2 | 36 | 67 | 680.3 | 81 | 8.4 | 2 | 16, 19 | 3 | 1, 2, 5 |
| E3 | 1.4 | 35 | 45 | 697.2 | 62 | 11.2 | 6 | 7, 16 | 4 | 6 |
| F1 | 1.0 | 43 | 76 | 921.8 | 78 | 11.8 | 10 | 1 | 5 | 7, 38 |
| F2 | 1.5 | 33 | 54 | 717.8 | 78 | 9.2 | 1 | 8 | 2 | 13, 29 |
| X | 1.6 | 52 | 87 | 1081.1 | 135 | 8.0 | 1 | X | 1 | X |
| TOTAL | 1.5 | 1252 | 1793 | 26510.3 | 2630 | 10.1 | 152 | 134 |
Figure 1.
Feline chromosome maps (labeled above), and homologous synteny blocks (HSBs) in the human (H) and dog (D) genomes. HSBs are shown to the right of each cat chromosome map (only the map scale is shown). The dark cross-marks on each cat chromosome correspond to 100 cR5000 intervals. The inferred centromere positions are shown by dark circles. HSBs are color coded by human or dog chromosomes, defined by the key in the bottom right corner.
Comparative Synteny Analysis
We identified orthologous positions in the dog genome, and by proxy the human genome, for 96% of the 1793 markers placed on the final cat RH map (Supplemental Table 1). The resulting comparative maps identify all 32–34 cat-human conserved synteny blocks reported in previous Zoo-FISH studies [10, 11], in addition to two smaller blocks of orthology to human chromosome 1 on A1q-cen and D2p not detected by Zoo-FISH (Table 1). We also observed strong concordance with Zoo-FISH maps interrelating the cat and dog genomes, identifying all 68 synteny blocks observed in a previous study comparing cat and dog genomes [12]. The ordering of the canine synteny blocks along the cat chromosomes (Fig. 1) was also consistent between the two approaches, further supporting the long-range ordering of markers on each chromosome.
Taking into account marker order we identified 152 conserved segments (or homologous synteny blocks--HSBs [6, 13]) between the cat and human genomes, and 134 between the cat and dog genomes (Figure 1). These HSB counts include 13 and 9 comparative singletons in the cat-human and cat-dog comparative maps, respectively; these 22 HSBs are usually singletons in one species and part of a multimarker stretch of conserved gene order in the other species. These singletons likely represent lineage-specific rearrangements. By contrast, we found 18 other “interchromosomal” singletons that appear out of place with respect to all other markers on the same chromosome. Of these, 6 match gene exons that are members of multigene families, suggesting we may have mapped paralogs; the remainder are microsatellites or not gene-associated. For the time being, we do not include these singletons in the HSB counts until further mapping validation is performed.
We estimated comparative coverage for all human and dog autosomes and the X chromosomes (Table 2) following previously reported methods [6, 14]. Comparative coverage is defined as the sum of the spans of conserved chromosome segments in cat, divided by the size of the human or dog genomes after excluding centromere, telomere, and heterochromatic regions [6, 14], or regions lacking any cross-species homology in multi-species alignments [13]. Because we were targeting gaps in the cat-human comparative map, we find that our comparative coverage with human is slightly greater than the cat-dog comparative map (86% versus 85%), despite the higher number of breakpoints between cat and human. This may in part be due to the large number of canine-specific intrachromosomal rearrangement breakpoints relative to the more conserved cat chromosomes, for which marker density is low and for which we had no prior comparative mapping information to target. For the cat-human comparison, the mean and median remaining gap sizes are 2.8 and 2.3 Mbp, respectively, with 87% of the gaps being less than 5 Mbp. Larger gaps remain on several cat chromosomes, notably chromosome D4 where two gaps greater than 9 Mbp remain.
Table 2.
Comparative Coverage of Cat HSBs on Dog and Human Genomes
| Human Chr. | Comparative human length (Mbp) | Comparative Coverage by Cat (%) | Dog Chr. | Comparative canine length (Mbp) | Comparative Coverage by Cat (%) |
|---|---|---|---|---|---|
| 1 | 224 | 89 | 1 | 122 | 80 |
| 2 | 238 | 86 | 2 | 85 | 85 |
| 3 | 195 | 89 | 3 | 92 | 73 |
| 4 | 188 | 85 | 4 | 88 | 97 |
| 5 | 178 | 86 | 5 | 89 | 84 |
| 6 | 167 | 96 | 6 | 76 | 88 |
| 7 | 155 | 76 | 7 | 80 | 94 |
| 8 | 143 | 80 | 8 | 75 | 78 |
| 9 | 114 | 75 | 9 | 51 | 98 |
| 10 | 132 | 79 | 10 | 70 | 92 |
| 11 | 131 | 88 | 11 | 73 | 87 |
| 12 | 131 | 96 | 12 | 72 | 73 |
| 13 | 96 | 90 | 13 | 63 | 72 |
| 14 | 88 | 79 | 14 | 61 | 86 |
| 15 | 82 | 70 | 15 | 64 | 73 |
| 16 | 76 | 91 | 16 | 57 | 89 |
| 17 | 78 | 84 | 17 | 64 | 83 |
| 18 | 75 | 82 | 18 | 63 | 77 |
| 19 | 51 | 92 | 19 | 54 | 89 |
| 20 | 59 | 82 | 20 | 58 | 99 |
| 21 | 33 | 96 | 21 | 50 | 95 |
| 22 | 35 | 74 | 22 | 61 | 96 |
| X | 152 | 99 | 23 | 53 | 76 |
| Total | 2821 | 86 | 24 | 48 | 78 |
| 25 | 51 | 93 | |||
| 26 | 38 | 68 | |||
| 27 | 46 | 94 | |||
| 28 | 39 | 87 | |||
| 29 | 42 | 86 | |||
| 30 | 40 | 71 | |||
| 31 | 38 | 97 | |||
| 32 | 39 | 94 | |||
| 33 | 31 | 80 | |||
| 34 | 42 | 72 | |||
| 35 | 27 | 95 | |||
| 36 | 31 | 66 | |||
| 37 | 31 | 90 | |||
| 38 | 23 | 65 | |||
| X | 124 | 98 | |||
| Total | 2311 | 85 |
Pairwise counts of cat-human and cat-dog breakpoints reveal slightly more breakpoints between the former (133) compared with the latter (115) (Table 3). However, when adjusted for divergence time, the rate of chromosome breakage is notably higher in the canine lineage (1 breakpoint/million year [My]) than the human lineage (0.70 breakpoints/My). Further discrimination between intrachromosomal and interchromosomal breakpoints reveals that within carnivores, interchromosomal rearrangements have predominated in the canid lineage (89% of all rearrangements), whereas inversions have been the primary mechanism remodeling felid chromosomes (95% of all rearrangements) since divergence from a common carnivoran ancestor 55 My ago (Table 3).
Table 3.
Chromosomal breakpoint statisitics
| Taxon | interchromosomal breakpoints | intrachromosomal breakpoints | Total breakpoints | breakpoints/Myra | interchromosomal breakpoints/Myra | intrachromosomal breakpoints/Myra |
|---|---|---|---|---|---|---|
| cat vs. human | 19 | 114 | 133 | 0.70 | 0.10 | 0.60 |
| cat vs. dog | 65 | 50 | 115 | 1.00 | 0.59 | 0.45 |
| canid-specificb | 63 | 8 | 71 | 1.29 | 1.15 | 0.15 |
| felid-specificb | 2 | 42 | 44 | 0.80 | 0.04 | 0.76 |
Refinement of the Cat Pseudoautosomal Boundary
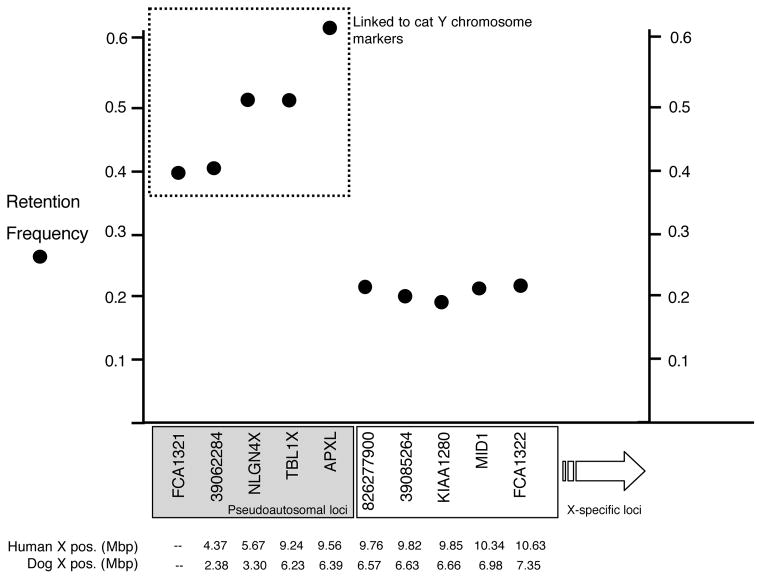
RH mapping of the most terminal Xp markers produced markedly higher retention frequencies when compared to other Xp markers (Fig. 2). Two-point linkage analysis revealed that these markers were strongly linked to existing Y chromosome markers [15], (LOD scores ranging between 9 and 13 for markers 39062284, TBL1X, NLGN4X, and APXL) while weakly linked to X chromosome markers, despite their best BLAST hits and detectable orthology to the X chromosomes of human, mouse, rat and dog. Inspection of RH vectors revealed that these five markers scored positive (by PCR) for all or nearly all RH panel DNAs containing fragments of the adjacent terminal Xp markers (e.g. KIAA1280, 39085624, MID1, see Supplementary Table 1) but differed from these X chromosome RH vectors only by being positive for clone DNAs that were also positive for feline Y chromosome fragments [15]. Thus, the co-amplification of X and Y chromosome fragment RH cell lines is responsible for the anomalously high retention frequencies, and we conclude that these feline markers reside in a pseudoautosomal region (PAR) that is expanded compared to the primate PAR.
Figure 2.
Putative boundary of the feline pseudoautosomal region as defined by RH STS mapping. The retention frequency is plotted for the most terminal Xp chromosome markers in the cat RH map. The cat markers are listed based on the inferred HSB order with dog and human (Fig. 1). The physical coordinates for each marker (where known) in the dog and human genome sequences are shown below the x-axis. The pseudoautosomal markers are boxed in gray, and are also indicated by their linkage to Y chromosome STS markers (dashed box). X-specific markers have an average retention frequency around 0.20, very similar to the X chromosome-wide average.
Discussion
We report an updated 1.5 Mb resolution, RH-based, physical map of the feline genome. This increased marker density resulted in a considerably more detailed cat-human comparative map, and a first generation ordered cat-dog comparative map. Notable improvements include several hundred new markers that more evenly cover previous gaps in the cat-human comparative map, made possible due to access and annotation of domestic cat trace archive sequences. In addition, the close evolutionary relationship between the cat and dog genomes allowed for the determination of microsatellite marker orthology between the two carnivoran genomes and the human genome, with a final total of 96% of all mapped feline markers finding positions in both genomes. This is a significant advance over prior versions of the cat-human comparative map where no feline microsatellite markers were assigned to orthologous regions in the human genome, and only 500–700 gene-based comparative anchors connected the two species’ chromosomes [8–9].
With this nearly threefold increase in comparative alignment, we present the first fine-scale syntenic comparison of the cat, dog and human genomes beyond previous chromosome painting studies [10–12, 16]. Classification of breakpoints to different lineages showed a clear increase in the number of breakpoints assigned to the dog lineage relative to cat (Table 3). As was observed by Lindblad-Toh et al. [5], most of the canid-lineage rearrangements are interchromosomal, while most of the rearrangements between the human and feline genomes are intrachromosomal (inversions). Otherwise our counts and classifications of breakpoints are incomparable to those of Lindblad-Toh et al.in part because they used sequence-based comparison, and because some reuse breakpoints will be necessarily classified as human-specific rearrangements due to limited taxon sampling.
A striking example of the accelerated rate of evolution in the canid lineage is observed on cat chromosome C1 (Fig. 1; Table 1), where cat and human share five HSBs corresponding to two human chromosomes (1p and 2q), most of the rearrangements being found on C1q (human 2q) due to inversions. By contrast, cat C1 and dog are distinguished by thirteen HSBs, most of these due to translocations involving nine different canine chromosomes. Similar examples of this pattern are seen on two of the other largest cat chromosomes, A1 and B1, where there are three times the number of cat-dog HSBs compared to cat-human HSBs. Most of this increase is the result of canid-specific interchromosomal rearrangements (Table 1).
After adjusting for evolutionary divergence time, these ordered mapping comparisons confirm the radically rearranged nature of canid chromosome evolution seen by chromosome painting data [12, 16]. While felid chromosomes are quite conserved at the syntenic level, even with species from other mammalian orders like human, they have still undergone a fair amount of intrachromosomal rearrangement (0.76/My) not appreciated by chromosome painting studies [10, 11]. The overall rate of rearrangement within the felid lineage is roughly similar to that between the felid and human lineage (0.80 versus 0.70). By contrast, canid chromosomes have undergone more than 1.5 times as much total breakage as felid chromosomes, with nearly thirty times as many interchromosomal rearrangements. At present it is not clear what genomic/structural features might explain the different pattern of rearrangement between felids and canids. Full genome comparisons of sequence features at chromosome breakpoints in dog and cat genomes remain a fertile area for exploration in the future once better quality genome assemblies become available for both species.
The identification and mapping of PAR markers in the RH panel allowed us to refine the boundary of this region to less than 200 kilobases between the APXL and KIAA1280 genes. Compared to the current human PAR1 boundary near 3 Mbp on the X chromosome, the feline (and probably canine) PAR extends as far as 9 Mbp on the human X chromosome, though is still within the limits of an ancient eutherian PAR originally defined by the AMELX/Y genes [17]. These findings are consistent with comparative FISH mapping studies of human Xp genes in carnivores and artiodactyls [18] that show the gene content of a formerly larger ancestral PAR1 extended as least as far as the steroid sulfatase (STS) gene (~7Mbp on human X), which is still conserved in other eutherian mammals but notably reduced in primates, indicating the reduction of the ancestral PAR has occurred variably among mammalian lineages [17–19]. Some of the genes in the ancient PAR between 3 and 9 Mbp on the human X have evolved Y-specific counterparts, such as TBL1Y and NLGN4Y [20]. Similarly, the X chromosome counterpart of a recently discovered carnivore-specific Y chromosome gene, TETY2, resides just within the APXL-KIAA1280 gene interval that currently defines the extent of the felid PAR boundary [15]. This suggests the present felid PAR boundary is a recent carnivore-specific reduction of the formerly larger eutherian PAR [17] that may be shared with the dog. Though not part of the current canine genome annotation [5], further definition of the dog PAR boundary and comparison to cat would be of interest.
Finally, this enhanced mapping resource, coupled with the forthcoming assembly and annotation of the feline genome sequence (Pontius et al., in prep.) will stimulate and facilitate the identification of feline genes of interest using positional cloning approaches. In the past three years twelve feline mutations in nine genes associated with coat color and disease phenotypes have been discovered [21–29]; six of these utilized cat genome mapping resources to assess linkage in candidate genes, while the remainder were identified by directly sequencing candidate genes. More recently, the first feline genome scan was used to identify a novel disease gene through positional mapping efforts [4]. With the availability of a detailed comparative map, and integration with developing linkage maps and the 2X sequence, we anticipate that the identification of causative mutations for many feline phenotypes will accelerate, as disease gene mapping has done so recently in the canine model system [30].
Materials and Methods
Marker and Primer Design
We examined approximately 40, 000 random cat 2x traces generated by Agencourt Biosciences, and performed BLAST searches with the human and canine genomes. We then chose traces with best reciprocal hits to orthologous regions of both dog and human genomes and used these to design primers for radiation hybrid mapping. Novel microsatellite markers were isolated from a (dG•dT)n (dC•dA)n enriched microsatellite library as described [31]. Finally, we designed primers for feline ESTs and mRNAs from GenBank not present on the previous map. All primers were designed with Primer3 [32]. We tested each primer pair using PCR in cat, hamster, and a 10:1 hamster: cat mixture of DNA, to identify those that produced a single bright band in cat that was absent or of differing mobility compared to hamster.
Radiation Hybrid Genotyping
RH genotyping for all new gene-based or trace-archive derived markers was performed using previously described methods [7–9]. Genotyping was performed on the 5,000-rad feline whole genome radiation hybrid panel [33], and resolved on 2% agarose gels stained with ethidium bromide, or were scored using a Taq-man-based assay. Markers were dropped before map computation for one of the following reasons: weak amplification, high hamster background amplification, or excessively high retention frequency (>70% and not predicted to reside on the selectable locus chromosome or near a centromere) or excessively low retention frequency compared to other markers on the same chromosome. These new genotypes were merged with vectors from Refs 7–9 to compile a novel data set. In this process, 24 markers were dropped from eligibility for the new map due to suspect genotypes.
Map Construction
Two-point linkage groups were initially computed at a LOD score of 8.0, though a small number of markers were included that fell below this threshold due to a number of reasons, such as being close to a centromere which tends to inflate retention frequencies. These were assigned to chromosomes based on previous physical mapping, FISH and chromosome painting studies [7–11]. Most metacentric and submetacentric chromosome arms comprised a single linkage group. Gaps generally resulted from high marker retention frequencies near centromeres [7–9]. Three chromosomes were comprised of multiple linkage groups at LOD=8; these groups were oriented and merged into single groups using best pairwise lod scores and orientation from linkage maps. Markers within each chromosome arm or linkage group were ordered using a reduction from the problem of RH mapping to the traveling salesman problem (TSP) [34], as implemented in the software rh_tsp_map [35]. The computations to construct the map were done using programs from the software package rh_tsp_map (ftp://ftp.ncbi.nih.gov/pub/agarwala/rhmapping/rh_tsp_map.tar.gz) and using the package CONCORDE (http://www.isye.gatech.edu/~wcook/rh) linked with QSopt (http://www.isye.gatech.edu/~wcook/qsopt) to solve the TSP instances to guaranteed optimality. We followed the multistep procedure used to construct some horse chromosome maps, described in detail in [36]. As in [9, 36], we call the first and most reliable map the “MLE-consensus map” because the markers on that map are required to have the same optimal order under three different formulations of the maximum likelihood (MLE) criterion [35]. In addition, we required that in a flips test, the MLE-consensus map be at least 0.5 LOD units better than the second best alternative map. One other difference from the procedure in [36] is that for the map herein, ten markers binned with LOD score <0.1 (comparing best placement to second best placement) were placed in a larger interval that combined their best interval with their second best interval.
Comparative Analysis
For each domestic cat locus, physical positions for orthologous genes were obtained from human sequence (Build 35). Sequence traces, ESTs and microsatellites were assigned orthologous positions based upon nucleotide discontiguous MegaBLAST (http://www.ncbi.nlm.nih.gov/blast/) [37] searches to the reference assembly of the dog genome (CanFam1) and the human genome (Build 35), using an E-value threshold of e−10. In cases where the cat marker found a match only in the dog genome, we identified the corresponding stretch of orthology in the human genome using the dog-human alignment net of the UCSC Genome Browser. Homologous synteny blocks were defined per Ref. 13. Specifically, we searched for runs of two or more uninterrupted markers on the same chromosome between two species. Inverted segments were defined by runs of three or more markers each separated by 1 Mbp. Some out of place markers were expected due to mapping/genotyping errors or limitations of RH mapping resolution. These were assigned to their closest HSB if the intervening markers did not span more than a few Mbp. Markers that were binned or placed with a LOD score <0.5 were not used in determining marker order, though they could be used to determine the extent of coverage of a HSB.
Supplementary Material
Acknowledgments
We thank N. Coomber, S. Corriveau, G. Pei, and T. Raudsepp for technical assistance and/or insightful discussion on these topics. This work was supported by funds from the Winn Feline Foundation and Texas A&M University (WJM). This research was supported in part by the intramural research program of the NIH, NLM (RA, AAS). This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400 (SJO, MMR). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Hitte C, et al. Facilitating genome navigation: survey sequencing and dense radiation-hybrid gene mapping. Nat Rev Genet. 2005;6:643–648. doi: 10.1038/nrg1658. [DOI] [PubMed] [Google Scholar]
- 2.Margulies EH, et al. An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc Natl Acad Sci USA. 2005;102:4795–4800. doi: 10.1073/pnas.0409882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien SJ, Menotti-Raymond M, Murphy WJ, Yuhki N. The feline genome project. Ann Rev Genet. 2002;36:657–686. doi: 10.1146/annurev.genet.36.060602.145553. [DOI] [PubMed] [Google Scholar]
- 4.Fyfe JC, et al. An ~140kb deletion associated with feline spinal muscular atrophy implies an essential LIX1 function for motor neuron survival. Genome Res. 2006 doi: 10.1101/gr.5268806. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 6.Everts-van der Wind A, et al. A high-resolution whole-genome cattle-human comparative map reveals details of mammalian chromosome evolution. Proc Natl Acad Sci USA. 2005;102:18526–18531. doi: 10.1073/pnas.0509285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy WJ, et al. A radiation hybrid map of the cat genome: implications for comparative mapping. Genome Res. 2000;10:691–702. doi: 10.1101/gr.10.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menotti-Raymond M, et al. Second-generation integrated genetic linkage/radiation hybrid maps of the domestic cat (Felis catus) J Hered. 2003;94:95–104. doi: 10.1093/jhered/esg008. [DOI] [PubMed] [Google Scholar]
- 9.Menotti-Raymond M, et al. Radiation hybrid mapping of 304 novel microsatellites in the domestic cat genome. Cytogenet Genome Res. 2003;102:272–276. doi: 10.1159/000075762. [DOI] [PubMed] [Google Scholar]
- 10.Rettenberger G, et al. ZOO-FISH analysis: cat and human karyotypes closely resemble the putative ancestral mammalian karyotype. Chrom Res. 1995;3:479–486. doi: 10.1007/BF00713962. [DOI] [PubMed] [Google Scholar]
- 11.Wienberg J, et al. Conservation of human versus feline genome organization revealed by reciprocal chromosome painting. Cytogenet Cell Genet. 1997;77:211–217. doi: 10.1159/000134579. [DOI] [PubMed] [Google Scholar]
- 12.Yang F, et al. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chrom Res. 2000;8:393–404. doi: 10.1023/a:1009210803123. [DOI] [PubMed] [Google Scholar]
- 13.Murphy WJ, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 14.Meyers SN, et al. Piggy-BACing the human genome II. A high-resolution, physically anchored, comparative map of the porcine autosomes. Genomics. 2005;86:739–752. doi: 10.1016/j.ygeno.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Murphy WJ, et al. Novel gene acquisition on carnivore Y chromosomes. PLoS Genet. 2006;2:353–363. doi: 10.1371/journal.pgen.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nash WG, Menninger JC, Wienberg J, Padilla-Nash HM, O’Brien SJ. The pattern of phylogenomic evolution of the Canidae. Cytogenet Cell Genet. 2001;95:210–224. doi: 10.1159/000059348. [DOI] [PubMed] [Google Scholar]
- 17.Iwase M, et al. The amelogenin loci span an ancient pseudoautosomal boundary in diverse mammalian species. Proc Natl Acad Sci U S A. 2003;100:5258–5263. doi: 10.1073/pnas.0635848100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toder R, et al. Genes located in and near the human pseudoautosomal region are located in the X–Y pairing region in dog and sheep. Chrom Res. 1997;5:301–306. doi: 10.1023/B:CHRO.0000038760.84605.0d. [DOI] [PubMed] [Google Scholar]
- 19.Park SH, et al. Rapid divergency of rodent CD99 orthologs: implications for the evolution of the pseudoautosomal region. Gene. 2005;353:177–188. doi: 10.1016/j.gene.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eizirik E, et al. Molecular genetics and evolution of melanism in the cat family. Curr Biol. 2003;13:448–453. doi: 10.1016/s0960-9822(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 22.Somers KL, et al. Mutation analysis of feline Niemann-Pick C1 disease. Mol Genet Metab. 2003;79:99–103. doi: 10.1016/s1096-7192(03)00074-x. [DOI] [PubMed] [Google Scholar]
- 23.Lyons LA, et al. Feline polycystic kidney disease mutation identified in PKD1. J Am Soc Nephrol. 2004;15:2548–2555. doi: 10.1097/01.ASN.0000141776.38527.BB. [DOI] [PubMed] [Google Scholar]
- 24.Lyons LA, Imes DL, Rah HC, Grahn RA. Tyrosinase mutations associated with Siamese and Burmese patterns in the domestic cat (Felis catus) Anim Genet. 2005;36:119–126. doi: 10.1111/j.1365-2052.2005.01253.x. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt-Küntzel A, Eizirik E, O’Brien SJ, Menotti-Raymond M. Tyrosinase and tyrosinase related protein 1 alleles specify domestic cat coat color phenotypes of the albino and brown loci. J Hered. 2005;96:289–301. doi: 10.1093/jhered/esi066. [DOI] [PubMed] [Google Scholar]
- 26.Lyons LA, Foe IT, Rah HC, Grahn RA. Chocolate coated cats: TYRP1 mutations for brown color in domestic cats. Mamm Genome. 2005;16:356–366. doi: 10.1007/s00335-004-2455-4. [DOI] [PubMed] [Google Scholar]
- 27.Goree M, Catalfamo JLJL, Aber S, Boudreaux MK. Characterization of the mutations causing hemophilia B in 2 domestic cats. J Vet Intern Med. 2005;19:200–204. doi: 10.1892/0891-6640(2005)19<200:cotmch>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Meurs KM, et al. A cardiac myosin binding protein C mutation in the Maine Coon cat with familial hypertrophic cardiomyopathy. Hum Mol Genet. 2005;14:3587–3593. doi: 10.1093/hmg/ddi386. [DOI] [PubMed] [Google Scholar]
- 29.Imes DL, Geary LA, Grahn RA, Lyons LA. Albinism in the domestic cat (Felis catus) is associated with a tyrosinase (TYR) mutation. Anim Genet. 2006;37:175–178. doi: 10.1111/j.1365-2052.2005.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker HG, Ostrander EA. Canine genomics and genetics: running with the pack. PLoS Genet. 2005;1:507–513. doi: 10.1371/journal.pgen.0010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarno R, David VA, Franklin WL, O’Brien SJ, Johnson WE. Development of microsatellite markers in the guanaco, Lama guanicoe: utility for South American camelids. Mol Ecol. 2000;9:1922–1924. doi: 10.1046/j.1365-294x.2000.01077-3.x. [DOI] [PubMed] [Google Scholar]
- 32.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 33.Murphy WJ, Menotti-Raymond M, Lyons LA, Thompson MA, O’Brien SJ. Development of a feline whole genome radiation hybrid panel and comparative mapping of human chromosome 12 and 22 loci. Genomics. 1999;57:1–8. doi: 10.1006/geno.1998.5695. [DOI] [PubMed] [Google Scholar]
- 34.Applegate D, Bixby R, Chvátal V, Cook W. On the solution of traveling salesman problems. Documenta mathematica, extra volume International Congress of Mathematics; 1998. pp. 645–656. [Google Scholar]
- 35.Agarwala R, Applegate DL, Maglott D, Schuler DGD, Schäffer AA. A fast and scalable radiation hybrid map construction and integration strategy. Genome Res. 2000;10:350–364. doi: 10.1101/gr.10.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brinkmeyer-Langford C, et al. A high-resolution physical map of equine homologues of HSA19 shows divergent evolution compared to other mammals. Mamm Genome. 2005;16:631–649. doi: 10.1007/s00335-005-0023-1. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comp Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 38.Springer MS, Murphy WJ, Eizirik E, O’Brien SJ. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy WJ, Stanyon R, O’Brien SJ. Evolution of mammalian genome organization inferred through comparative gene mapping. Genome Biol. 2001;2:0005.1–0005.8. doi: 10.1186/gb-2001-2-6-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.