Abstract
Salivary diagnostics has fascinated many researcheres and has been tested as a valuable tool in the diagnosis of many systemic conditions and for drug monitoring. Advances in the field of molecular biology, salivary genomics and proteomics have led to the discovery of new molecular markers for oral cancer diagnosis, therapeutics and prognosis. Oral cancer is a potentially fatal disease and the outcome of the treatment and prognosis largely depends on early diagnosis. Abnormal cellular products elucidated from malignant cells can be detected and measured in various body fluids including saliva and constitute tumor markers. This article discusses the various salivary tumor markers and their role in oral pre-cancer and cancer.
Keywords: DNA markers, oral cancer, protein markers, RNA markers, salivary markers, tumor markers
INTRODUCTION
Oral cancer is one of the major global public health problems and is the sixth most common human malignancy with a five year mortality rate of approximately 50%.[1] This has not changed significantly over the last 50 years.[2] A high prevalence of oral cancer in India and other Asian countries has been attributed to the influence of region specific epidemiological factors such as tobacco and betel quid chewing.[3] Another alarming scenario is the increasing incidence of oral squamous cell carcinoma (OSCC) in younger people owing to the heavy abuse of tobacco and tobacco related products.
The high morbidity rate in OSCC can be attributed to the delay in the diagnosis of the disease.[4] The diagnosis of oral cancer and/or the malignant potential of an oral lesion is based on various aspects such as (a) etiology-associated with the use of tobacco, presence of factors such as detection of HPV (b) clinical appearance of the lesion (leukoplakic, erythroplakic, nodular, ulcerative, verrucous) [Figures 1 and 2] (c) location of the lesion - the high risk sites being floor of the mouth, ventrolateral aspect of the tongue etc., (d) histopathological aspects - presence of epithelial dysplasia [Figure 3] and (e) molecular biological aspects of the lesion.[5] A detailed discussion on the clinical and histopathological diagnosis and the biological aspects of cancer development are not within the scope of this review.
Figure 1.
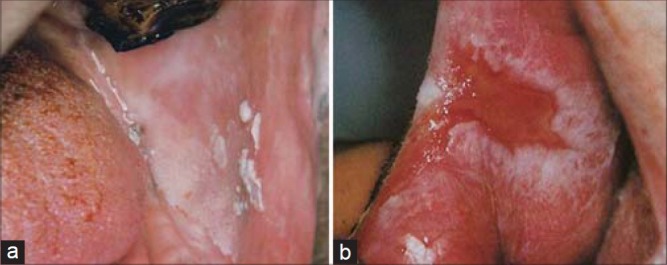
Potentially malignant lesions (a) Leukoplakia and (b) Erosive Lichen planus
Figure 2.
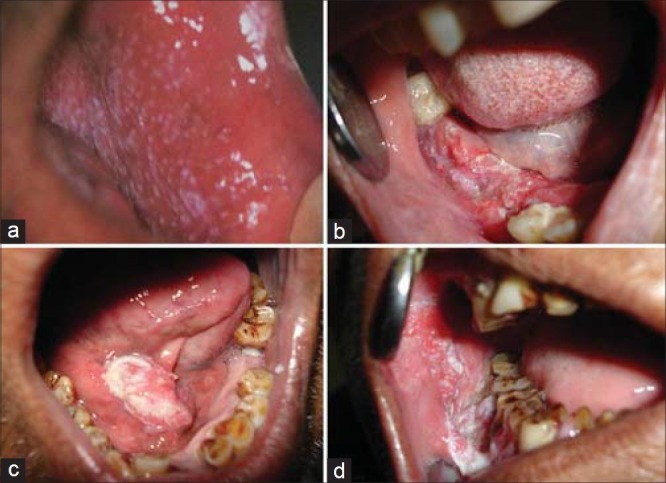
Various clinical presentations of oral cancer (a) Erythroleukoplakia (b) Ulcerative (c) Nodular/verruciod (d) Ulcero-proliferative
Figure 3.
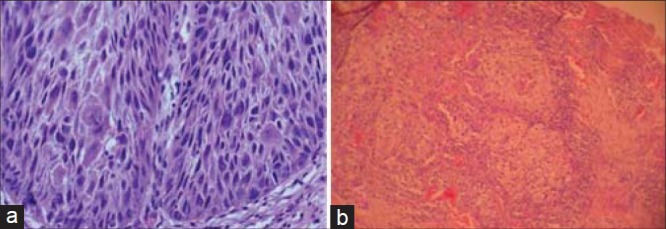
Photomicrograph showing (a) Severe dysplasia (b) Moderately differentiated OSCC
The diagnosis of epithelial dysplasia is based on a static picture though the histologic changes imply the possibility of a dynamic process. However, the problems in evaluating the significance of epithelial dysplasia arise from a lack of objectivity in the evaluation of established criteria, arbitrary division of the grading, and lack of sufficient knowledge of which criteria are important for the prediction of malignant potential.[6] A better understanding of the biological process of oral cancer have led to many studies in the field of molecular biological markers which help in assessment of cancer risk, potentiality of a lesion towards malignant transformation and also in predicting the prognosis.[7]
Neoplastic process gives/produces several abnormal cellular products which can be detected in various body fluids and on the surface of cancer cells either by biochemical methods or by immunohistochemistry; such products that are detected and measured are known as “tumor markers”.[8] These tumor markers aid the clinician greatly if the clinico-pathological picture is not accurately suggestive of malignancy and/or is indicative if the picture would soon change. Circulating tumor markers for OSCC have been investigated in various studies[9,10,11,12,13,14] and these have shown relatively moderate sensitivity and specificity values. Investigators over the years have used two approaches for the study of markers of malignant development: Epithelial dysplasia on one hand and oral cancer on the other are characterized by the presence/absence or the pattern of distribution of the marker in question. In general, the marker is characterized as a promising tool if the reaction pattern in epithelial dysplasias is similar to that in carcinomas and/or if the aberrant reaction pattern is positively related to the grade of epithelial dysplasia. These biological markers for determining future cancer development in oral pre-malignant lesions can be broadly divided into (a) genomic markers, including DNA content (ploidy) chromosome aberration, and changes in expression of oncogenes and tumor suppressor genes; (b) proliferation markers; and (c) differentiation markers, including keratins and carbohydrate antigens which will be discussed.
Saliva, a watery and frothy substance produced in the mouth of human and most other animals contains informative components that have been used as diagnostic markers for human disease. An increasing number of systemic diseases and conditions, amongst them oral cancer, have been shown to be reflected diagnostically in saliva. Saliva as a diagnostic fluid meets the demand for an inexpensive, non invasive and accessible in the diagnosis, prognosis prediction, as well as for monitoring the patients post therapy status in oral cancer.[14]
MATERIALS AND METHODS
Literature search in Medline, PubMed and Scholar Google was performed using the key words salivary markers, tumor markers in saliva, oral cancer, salivary diagnostics, DNA markers, Protein markers and RNA markers. Manual search from various printed articles was also performed.
DISCUSSION
Tumor marker
A tumor marker is a substance present in or produced by a tumor or tumors’ host in response to the tumor's presence that can be used to differentiate a tumor from normal tissue or determine the presence of a tumor based on measurement in blood or secretions.[15] Lehto, Ponten.[16] have defined tumor markers as “specific, novel or structurally altered cellular macromolecules or temporarily spatially or quantitatively altered normal molecules that are associated with malignant (and in some cases benign) neoplastic cells.” Tumor markers may be unique genes or their products that are formed only in tumor cells or they may be genes or gene products that are found in normal cells but are aberrantly expressed in unique locations in the tumor cells.[17] The distinguishing biological characteristics of tumor cells such as the capacity for invasion, metastasis, unlimited proliferation, evasion of apoptosis and angiogenesis are all mediated by complex molecular pathways; any one of these components are potential tumor markers.[18]
An ideal tumor marker
A biological marker should have certain characteristics that are applicable in all situations. Kaplan and Pesce[19] have suggested the following criteria for an ideal tumor marker:
Be easy and inexpensive to measure in readily available body fluids.
Be specific to the tumor being studied and commonly associated with it.
Have a stoichiometric relationship between plasma levels of the marker and the associated tumor mass.
Have an abnormal plasma level, urine level or both in the presence of micro-metastases, that is, at a stage when no clinical or presently available diagnostic methods reveal their presence.
Have plasma levels, urine levels or both, that are stable and not subject to wild fluctuations. If present in the plasma of healthy individuals, exist in a much lower concentration than that found in association with all stages of cancer.
In addition to the above, they[19] have stated that the ideal tumor marker should relate to the clinical setting and comply with the following:
They should prognosticate a higher or lower risk for eventual development of recurrence.
They should change as the current status of the tumor changes over time.
They should precede and predict recurrences before they are clinically detectable.
Saliva and its diagnostic potential
The ability to use saliva to monitor a patient's health and disease state is a highly desirable goal for health promotion and health care research. Saliva has long been proposed and used as a diagnostic medium[20] as it is easily accessible, collection is noninvasive, not time consuming, inexpensive and can be used for mass screening purposes.[21] A major drawback to using saliva as a diagnostic fluid has been the notion that informative analytes generally are present in lower amounts than in serum.[22] However with new techniques of detecting small quantities of salivary components including proteins and messenger Ribonucleic acid (mRNA), almost anything that can be measured in blood can be measured in saliva. Hence, saliva is considered as the blood stream of oral cavity. Alterations in the levels of certain mRNA molecules[23] and of certain proteins have been detected in several cancers,[24] including oral cancers indicating their possible use in cancer detection and follow-up.
Mechanism for the presence of biomarker in saliva
Though the exact mechanism for the presence of these tumor markers in saliva is not known, they may be either derived from serum or can be produced locally.[25] Figure 4[25,26,27,28,29,30,31,32,33,34,35,36,37] summarizes the possible mechanism that leads to the presence of biomarkers in saliva.
Figure 4.
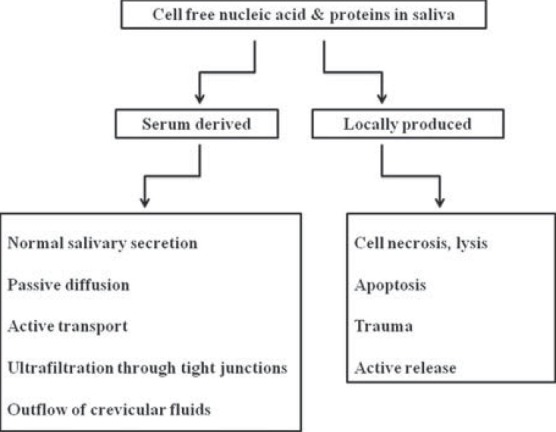
Mechanism that lead to the presence of molecular markers in saliva
Salivary biomarkers for oral cancer detection
Several salivary tumor markers are found to be significantly increased in the saliva of oral cancer patients.[14,38] Molecular markers for the diagnosis of oral cancer can be quested in 3 levels;[39] changes in the cellular Deoxyribonucleic acid (DNA) which results in altered mRNA transcripts leading to altered protein levels intracellularly, on the cell surface or extracellularly. Markopoulos et al.,[39] have summarized the molecular markers for the diagnosis of OSCC [Table 1].
Table 1.
Molecular markers for oral cancer[39]
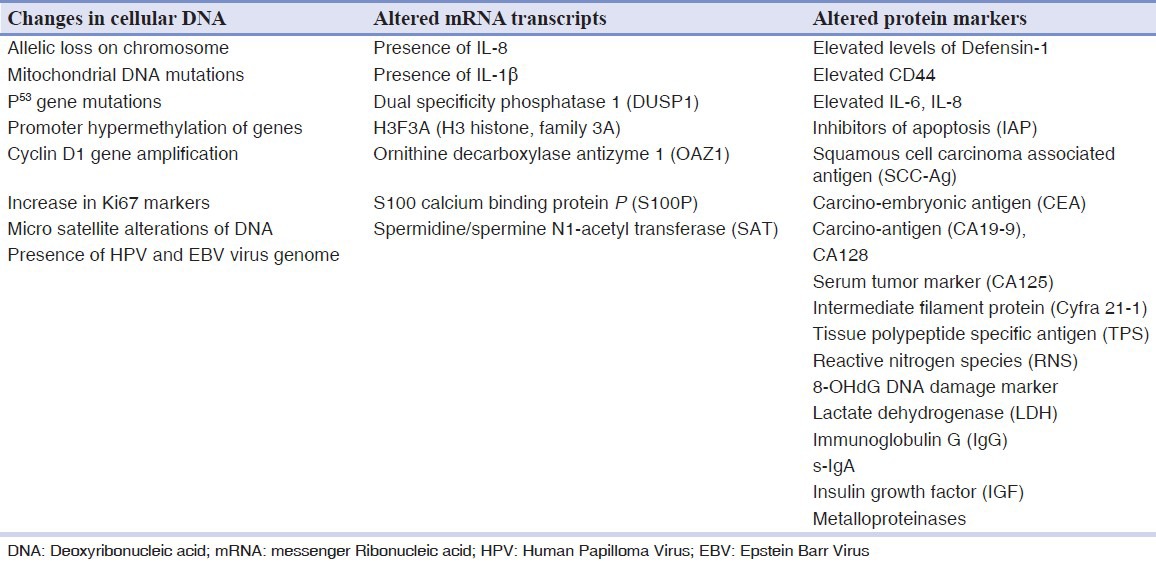
Alterations in host cellular DNA
DNA markers are universal i.e., there is not a single tumor cell with in the lesion which does not contain its genetic material. They originate from dead cells, are detected in the early stage of tumorigenesis and are absolutely oncospecific; showing a direct cause-and-effect relationship with tumorigenesis. However, tissue specificity of DNA markers is very low.[40] There are several changes in the host DNA of dysplastic or cancer cells which include point mutation, deletion, translocation, amplification, methylation, cyclin D1, epidermal growth factor (EGFR), microsatellite instability and presence of HPV. Sudbo, et al.,[41] and Femiano, et al.,[42] have found that premalignant lesions with aneuploidy transform into malignancy more frequently than lesions with normal DNA content irrespective of the histopathological grade of dysplasia. DNA aneuploidy appears to be associated with advanced stage carcinomas, perineural invasion and lymph node metastasis.[43] Hence, DNA content of a tumor may help in predicting the aggressiveness of the tumor.
Loss of heterozygosity (LOH) is defined as loss of genomic material in one of the chromosomal pair. Studies have shown that LOH in regions that contain a known human suppressor gene is an early predictor of malignant transformation of precancerous lesion.[44] This helps the clinician to identify high and low risk lesions in the context of management. Studies[45,46,47] have demonstrated frequent LOH in chromosome 3p, 9q, 13q and 17p as an early event in oral carcinogenesis. Rosin, et al.,[48] in their study found that allelic loss at 3p and 9q increase the risk of malignant transformation by 3.8 fold and the risk of further increase to 33 fold when LOH occurs at chromosomes 4q, 8p, 11q, 13q and 17p in addition to the former.
Mitochondrial DNA mutations have also been useful to detect exfoliated OSCC cells in saliva.[39] Such mutations have been identified in 46% of head and neck cancer and in 67% of saliva samples from OSCC patients by direct sequencing.[49]
p53 gene located on chromosome 17p13.1 exhibits mutation in 50-70% of epithelial tumors[50,51] and LOH of p53 allele has been reported in 22% of pre-cancer and 20% of oral cancer. However, the prognostic significance of p53 in oral cancer is yet to be established although there are multiple studies comparing the expression of p53 in premalignant lesions and malignancies.[52,53] Boyle et al.,[51] using plaque hybridization identified tumor specific p53 mutations in 71% saliva samples from patients with head and neck cancer. Other genes such as p16, p27, p63, p73 related to p53 and cell cycle have been found to be altered in varying degrees in oral cancer.[50]
Promoter methylation, an alternate form of gene silencing, which depends on the epigenetic factor has been described to be involved in OSCC.[50] The main genes to be methylated are CDKN2A, CDH1, MGMT, DAPK1.[50,54] CDKN2A which are involved in the retinoblastoma pathway of the cell cycle appear to be methylated in 23-67% of primary OSCC's. CDH1 a gene responsible for cell adhesion, promotes metastasis when mutated and shows promoter methylation in up to 85% of tumors.[54] Rosas, et al.,[55] identified aberrant methylation of at least one of these genes (p16, MGMT, DAP-K) in OSSC and detected promoter hyper methylation in 65% of matched saliva samples in OSCC patients.
Amplification and over expression of c-MYCIN-MYC has been observed in 20-40% of oral cancers. Das, et al.,[56] have reported amplification of 11q13, containing 1NT2, HST1 and Cyclin D oncogenes in 30-50% of patients with oral cancer. Cyclin D1 gene amplification has been found to be associated with poor prognosis in OSCC.[57] STAT 3 expression and activation was found in 82% of oral cancers related to chewing tobacco, while there was no expression in normal mucosa and premalignant lesions.[58]
Levels of Ki67 marker were increased while 8-oxoguanine DNA glycosylase, phosphorylated-Src and mammary serine protease inhibitor (Maspin) were found decreased in the saliva of patients with OSCC.[59]
The presence of HPV and EBV virus genomic sequence have been identified as possible DNA markers in detecting OSCC and tumor progression.[60]
RNA as a biomarker
RNA has been found to be a robust and informative marker and salivary RNA signatures have been identified for oral cancer. RNA for years was thought to degrade in saliva due to the various RNAases that is present in saliva.[61] However, cell free RNA is present in saliva both in intact and fragmented forms.[62] It has been speculated that salivary mRNA is contained in apoptotic bodies[29,30] or actively released in exosomes or micro vesicles.[32,34,35] Researchers[63] compared the clinical accuracy of saliva with that of blood RNA biomarker for oral cancer detection and found four RNA biomarkers that have a sensitivity and specificity of 91% and 71% and a collective receiver operator characteristic (roc) value of 0.95.[18]
Studies have found seven mRNA molecules; (1) IL-8,[63,64] (2) IL1β (interleukin 1)[63,64] which take part in signal transduction, proliferation, inflammation and apoptosis, (3) DUSP1[64] (dual specificity phosphatase 1) with a role in protein modification, signal transduction and oxidative stress, (4) H3F3A[64] (H3 histone, family 3A) having a DNA binding activity, (5) OAZ1[63,64] (ornithine decarboxylase antizyme 1) taking part in polyamine biosynthesis, (6) S100P[64] (S100 calcium binding protein P) with a role in protein binding and calcium ion binding and (7) SAT[63,64] (spermidine/spermine N1-acetyltransferase) which takes part in enzyme and transferase activity, to be significantly elevated in OSCC patients rather than in healthy controls.
Protein markers
Protein markers are differentiation antigens of corresponding normal tissue and characterize a certain stage of its maturation. They originate from live cells and show high tissue specificity. However, they may be detected in other pathologies as well.[40] Salivary protein markers have shown moderate sensitivity and specificity as prognostic markers.[39] A group of leading researchers[64,65,66] using new and sophisticated approaches, such as Luminex Multianalyte Profiling (xMAP) technology, shotgun proteomics, capillary reversed-phase liquid chromatography with quadruple time off light mass spectrometry and matrix-assisted laser desorption/ionization-mass spectrometry (MALDI-MS) has contributed significantly in recent years to the research in saliva for cancer diagnosis. Their studies have shown that saliva contains proteins that may serve as biomarkers for OSCC, since 46 peptides/proteins were found at significantly different levels between the OSCC and control groups.
Carbonylation (indicative of oxidative damage to proteins) has attracted a great deal of attention in cancer research because of its irreversible and irreparable nature, which becomes cytotoxic and is associated with cancer.[67] It is currently reported that a substantial increase in salivary carbonyls (246%) is seen in OSCC patients and points to the fact that there is a significant free radical attack to which the epithelial cells are exposed.[59]
Metalloproteinases such as MMP-9,[59] MMP-11,[68] MMP-2[68] are significantly altered in OSCC. MMP 9 are shown to degrade type IV collagen, a major type in the basement membrane, other types of collagen (V, VII, X), elastin and fibronectin.[59] Studies[68,69] have shown high MMP-9 expression in stromal cells surrounding the invading front of metastasizing tumors.[69,70] MMP-9 polymorphism was shown to have a strong association with increased risk for developing OSCC.[71] Shpitzer, et al.,[59] found a 39% increase in MMP-9 with a sensitivity of 100% and specificity of 79% in OSCC patients.
IL-6, IL-8 are post inflammatory cytokines that play a prominent role in immune host defense responses to infection, they are found to stimulate angiogenesis, influence tissue remodeling and take part in regulation of cell proliferation and differentiation. They are identified as important mediators of cancer development and powerful activators of not only apoptosis but also anti apoptotic signaling cascade[72] and hence play a role in early detection of oral pre-malignancies and OSCC. Rajkumar, et al.,[38] in their study found concentration of IL-6 and Il-8 in saliva to be significantly increased. St John, et al.,[73] detected higher levels of IL-8 in saliva of OSCC patients. Studies have found significant increase in the levels of tumor marker in patients with OSCC compared to dysplastic oral lesions and controls, thus suggesting its diagnostic value as a marker of malignant transformation in oral premalignant lesions.[74] Arllano-Garcia, et al.,[75] using Luminex MAP technology showed that both IL-8 and IL-1β were expressed at significantly higher levels in OSCC patients.
Cytokeratins are cytoskeletal intermediate filaments present in almost all normal and malignant epithelial cells.[76] In malignant epithelial cells, the activated protease increases degradation of cytokeratin free filaments into the blood. Increased levels of Cyfra 21-1 in saliva has been found in OSCC.[10,11,14]
Defensins are peptides which process antimicrobials and cytotoxic properties. They are found in azurophil granules of polymorphonuclear leukocytes.[77,78] Mizukawa, et al.,[79] found elevated levels of salivary Defensin 1 in OSCC patients compared with healthy controls. Some of the other salivary biomarkers which are significantly altered in OSCC patients as compared with healthy controls are inhibitors of apoptosis (IAP),[80] squamous cell carcinoma associated antigen (SCC-Ag),[13,14] carcino-embryonic antigen (CEA),[12,14] carcino-antigen (CA19-9),[13,14] CA128,[13] serum tumor marker (CA125),[14,81] tissue polypeptide specific antigen (PPS),[14,82] reactive nitrogen species (RNS),[9] lactate dehydrogenase (LDH)[11] and immunoglobulin G (IgG),[11] s-Ig A,[68] insulin growth factor (IGF).[68]
Uses of tumor markers
Recent technological advances in molecular biology has led to discovery of new tumor markers and thus increased its scope of use in oral biology. Tumor markers have a role in the diagnosis, prognosis, formulating the treatment and in the detection of recurrence of cancer. Chan and Sell.[15] have summarized the potential uses of tumor markers as follows:
(a) screening in general population (b) differential diagnosis in symptomatic patients (c) clinical staging of cancer (d) estimating tumor volume (e) prognostic indicators for disease progression (f) evaluating the success of treatment (g) detecting recurrences (h) Monitoring responses to therapy (i) radioimmunolocalization of tumor masses and (j) Determining direction for immunotherapy.
CONCLUSION
Present day screening tests by visual examination, or other well established and time tested diagnostic tools such as vital staining with toluidine blue, brush biopsy, chemiluminescence, tissue autofluorescence and the diagnostic gold standard being tissue biopsy followed by histopathological evaluation for OSCC are still critical for the diagnosis of potentially malignant lesions and malignancies. The field of salivary diagnostics is a broad, complex and crosscutting area of scientific research with enormous potential to impact the diagnosis of various diseases including oral cancer. The significant increase in tumor markers in saliva is quite encouraging as saliva offers many advantages such as saliva harvesting is noninvasive thus making it an effective alternative to serum testing. Though at present no single tumor marker can validate the presence or prognosis of disease, a panel of biomarkers would be more helpful. Recent advances and emerging technologies such as nanotechnology, proteomic and genomics make salivary diagnostics a highly sensitive tool. The present methods are not ready for immediate direct use and much needs to be done to come up with a fast, simple, cost effective clinical diagnostic system. Saliva will possibly outperform other media in the diagnosis of not only cancer but other diseases as well.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Myers JN, Elkins T, Roberts D, Byers RM. Squamous cell carcinoma of the tongue in young adults: Increasing incidence and factors that predict treatment outcomes. Otolaryngol Head Neck Surg. 2000;122:44–51. doi: 10.1016/S0194-5998(00)70142-2. [DOI] [PubMed] [Google Scholar]
- 2.Sparano A, Weinstein G, Chalian A, Yodul M, Weber R. Multivariate predictors of occult neck metastasis in early oral tongue cancer. Otolaryngol Head Neck Surg. 2004;131:472–6. doi: 10.1016/j.otohns.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Tsantoulis PK, Kastrinakis NG, Tourvas AD, Laskaris G, Gorgoulis VG. Advances in the biology of Oral cancer. Oral Oncol. 2007;43:523–34. doi: 10.1016/j.oraloncology.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Peacock S, Pogrel A, Schmidt BL. Exploring the reasons for delay in treatment of oral cancer. Am Dent Assoc. 2008;139:1346–52. doi: 10.14219/jada.archive.2008.0046. [DOI] [PubMed] [Google Scholar]
- 5.Reibel J. Prognosis of oral pre-malignant lesions: Significance of clinical, histopathological, and molecular biological characteristics. Crit Rev Oral Biol Med. 2003;14:47–62. doi: 10.1177/154411130301400105. [DOI] [PubMed] [Google Scholar]
- 6.Warnakulasuriya S. Histological grading of oral epithelial dysplasia: Revisited. J Pathol. 2001;194:294–7. doi: 10.1002/1096-9896(200107)194:3<294::AID-PATH911>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz JL. Biomarkers and molecular epidemiology and chemoprevention of oral carcinogenesis. Crit Rev Oral Biol Med. 2000;11:92–122. doi: 10.1177/10454411000110010501. [DOI] [PubMed] [Google Scholar]
- 8.Nayak AG, Chatra L. Tumor Markers: An overview. JIAOMR. 2010;22:147–50. [Google Scholar]
- 9.Nagler RM, Barak M, Ben-Aryeh H, Peled M, Filatov M, Laufer D. Early diagnostic and treatment monitoring role of Cyfra 21-1 and TPS in oral squamous cell carcinoma. Cancer. 1999;35:1018–25. [PubMed] [Google Scholar]
- 10.Bhatavdekar JM, Patel DD, Vora HH, Balar DB. Circulating prolactin and TPS in monitoring the clinical course of male patients with metastatic tongue cancer: A preliminary study. Anticancer Res. 1993;13:237–40. [PubMed] [Google Scholar]
- 11.Yen TC, Lin WY, Kao CH, Cheng KY, Wang SJ. A study of a new tumour marker, CYFRA 21-1, in squamous cell carcinoma of the head and neck, and comparison with squamous cell carcinoma antigen. Clin Otolaryngol. 1998;23:82–6. doi: 10.1046/j.1365-2273.1998.00101.x. [DOI] [PubMed] [Google Scholar]
- 12.Krimmel M, Hoffmann J, Krimmel C, Cornelius CP, Schwenzer N. Relevance of SCC-Ag, CEA, CA 19.9, and CA125 for diagnosis and follow-up in oral cancer. J Craniomaxillofac Surg. 1998;26:243–8. doi: 10.1016/s1010-5182(98)80020-6. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann J, Munz A, Krimmel M, Alfter G. Intraoperative and postoperative kinetics of serum tumor markers in patients with oral carcinoma. J Oral Maxillofac Surg. 1998;56:1390–3. doi: 10.1016/s0278-2391(98)90400-1. [DOI] [PubMed] [Google Scholar]
- 14.Nagler R, Bahar G, Shpitzer T, Feinmesser R. Concomitant analysis of salivary tumor markers: A new diagnostic tool for oral cancer. Clin Cancer Res. 2006;12:3979–84. doi: 10.1158/1078-0432.CCR-05-2412. [DOI] [PubMed] [Google Scholar]
- 15.Chan DW, Sell S. Tumor markers. In: Burtis CA, Ashwood ER, editors. Tietz's textbook of clinical chemistry. 2nd ed. Philadelphia: WB Saunders; 1994. pp. 897–925. [Google Scholar]
- 16.Lehto VP, Ponten J. Tumor markers in human biopsy material. Rev Oncol. 1989;28:743–7. doi: 10.3109/02841868909092305. [DOI] [PubMed] [Google Scholar]
- 17.Rassekh CH, Johnson JT, Eibling DE. Circulating markers in squamous cell carcinoma of the head and neck: A review. Eur J Cancer B Oral Oncol. 1994;30:23–8. doi: 10.1016/0964-1955(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 18.Speight PM, Morgan PR. The natural history and pathology of oral cancer and precancer. Comm Dent Health. 1993;10(Suppl 1):31–41. [PubMed] [Google Scholar]
- 19.William B, Ronald BH. 6th ed. Hamilton (ON): BC Decker; 2003. Characteristics of the ideal ztumor marker in holland-frei cancer medicine. [Google Scholar]
- 20.Kaufman E, Lamster I. The diagnostic applications of saliva: A review. Crit Rev Oral Biol Med. 2002;13:197–212. doi: 10.1177/154411130201300209. [DOI] [PubMed] [Google Scholar]
- 21.Samaranayake L. Saliva as a diagnostic fluid. Int Dent J. 2007;57:295–9. doi: 10.1111/j.1875-595x.2007.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller SM. Saliva testing: A nontraditional diagnostic tool. Clin Lab Sci. 1994;7:39–44. [PubMed] [Google Scholar]
- 23.Hu S, Li Y, Wang J, Xie Y, Tjon K, Wolinsky L, et al. Human saliva proteome and transcriptome. J Dent Res. 2006;85:1129–33. doi: 10.1177/154405910608501212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigler LR, Streckfus CF, Dubinsky WP. Salivary biomarkers for the detection of malignant tumors that are remote from the oral cavity. Clin Lab Med. 2009;29:71–85. doi: 10.1016/j.cll.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Baum BJ. Principles of saliva secretion. Ann NY Acad Sci. 1993;694:17–23. doi: 10.1111/j.1749-6632.1993.tb18338.x. [DOI] [PubMed] [Google Scholar]
- 26.Haeckel R, Hanecke P. Application of saliva for drug monitoring: An in vivo model for transmembrane transport. Eur J Clin Chem Clin Biochem. 1996;34:171–91. [PubMed] [Google Scholar]
- 27.Aps JK, Martens LC. Review: The physiology of saliva and transfer of drugs into saliva. Forensic Sci Int. 2005;150:119–31. doi: 10.1016/j.forsciint.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Halicka HD, Bedner E, Darzynkiewicz Z. Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res. 2000;260:248–56. doi: 10.1006/excr.2000.5027. [DOI] [PubMed] [Google Scholar]
- 29.Hasselmann D, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem. 2001;47:1488–9. [PubMed] [Google Scholar]
- 30.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–95. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 31.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–83. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 32.García JM, García V, Peña C, Domínguez G, Silva J, Diaz R, et al. Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched. RNA. 2008;14:1424–32. doi: 10.1261/rna.755908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, et al. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One. 2009;4:e4722. doi: 10.1371/journal.pone.0004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: Current perspectives. Proteomics. 2008;8:4083–99. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 36.Al-Nedawi K, Meehan B, Rak J. Microvesicles: Messengers and mediators of tumor progression. Cell Cycle. 2009;8:2014–8. doi: 10.4161/cc.8.13.8988. [DOI] [PubMed] [Google Scholar]
- 37.Aharon A, Brenner B. Microparticles, thrombosis and cancer. Best Pract Res Clin Haematol. 2009;22:61–9. doi: 10.1016/j.beha.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Rajkumar K, Kumar AR, Ramyamalini V, Nandhini G, Kumar TD, Ashwini BK, et al. Estimation of serological and salivary biomarkers in patients with Oral Squamous cell carcinoma, premalignant lesions and conditions. SRM Univ J Dent Sci. 2010;1:14–19. [Google Scholar]
- 39.Markopoulos AK, Michailidou EZ, Tzimagiorgis G. Salivary markers for oral cancer detection. The Open Dent J. 2010;4:171–8. doi: 10.2174/1874210601004010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lichtenstein AV, Potapova GI. Genetic defects as tumor markers. Mol Biol. 2003;37:159–69. [PubMed] [Google Scholar]
- 41.Sudbo J, Kildal W, Risberg B, Koppang HS, Danielsen HE, Reith A. DNA content as a prognostic marker in patients with oral leukoplakia. N Engl J Med. 2001;344:1270–8. doi: 10.1056/NEJM200104263441702. [DOI] [PubMed] [Google Scholar]
- 42.Femiano F, Scully C. DNA cytometry of oral leukoplakia and oral lichen planus. Med Oral Patol Oral Cir Bucal. 2005;10(Suppl 1):E9–14. [PubMed] [Google Scholar]
- 43.Rubio Bueno P, Naval Gias L, García Delgado R, Domingo Cebollada J, Díaz González FJ. Tumor DNA content as a prognostic indicator in squamous cell carcinoma of the oral cavity and tongue base. Head Neck. 1998;20:232–9. doi: 10.1002/(sici)1097-0347(199805)20:3<232::aid-hed8>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Rosin MP. Loss of heterozygosity: A potential tool in management of oral premalignant lesions? J Oral Pathol Med. 2001;30:513–20. doi: 10.1034/j.1600-0714.2001.300901.x. [DOI] [PubMed] [Google Scholar]
- 45.Califano J, Van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, et al. Genetic progression model for head and neck cancer: Implications for field cancerization. Cancer Res. 1996;56:2488–92. [PubMed] [Google Scholar]
- 46.Lee JJ, Hong WK, Hittelman WN, Mao L, Lotan R, Shin DM, et al. Predicting cancer development in oral leukoplakia: Ten years of translational research. Clin Cancer Res. 2000;6:1702–10. [PubMed] [Google Scholar]
- 47.Partridge M, Pateromichelakis S, Phillips E, Emilion GG, A’Hern RP, Langdon JD. A case-control study confirms that microsatellite assay can identify patients at risk of developing oral squamous cell carcinoma within a field of cancerization. Cancer Res. 2000;60:3893–8. [PubMed] [Google Scholar]
- 48.Rosin MP, Cheng X, Poh C, Lam WL, Huang Y, Lovas J, et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin Cancer Res. 2000;6:357–62. [PubMed] [Google Scholar]
- 49.Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–9. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 50.Wanninayake M Tilakaratne. the cancer handbook. 2nd ed. United States: John Wiley and Sons Ltd; 2007. Oral cavity and major and minor salivary glands; pp. 1–15. [Google Scholar]
- 51.Boyle JO, Hakim J, Koch W, Van der Riet P, Hruban RH, Roa RA, et al. The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res. 1993;53:4477–80. [PubMed] [Google Scholar]
- 52.Murti PR, Warnakulasuriya KA, Johnson NW, Bhonsle RB, Gupta PC, Daftary DK, et al. p53 expression in oral precancer as a marker for malignant potential. J Oral Pathol Med. 1998;27:191–6. doi: 10.1111/j.1600-0714.1998.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 53.Shahnavaz SA, Regezi JA, Bradley G, Dubé ID, Jordan RC. p53 gene mutations in sequential oral epithelial dysplasias and squamous cell carcinomas. J Pathol. 2000;190:417–22. doi: 10.1002/(SICI)1096-9896(200003)190:4<417::AID-PATH544>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 54.Ha PK, Califano JA. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006;7:77–82. doi: 10.1016/S1470-2045(05)70540-4. [DOI] [PubMed] [Google Scholar]
- 55.Rosas SL, Koch W, Da Costa Carvalho MG, Wu L, Califano J, Westra W, et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–42. [PubMed] [Google Scholar]
- 56.Das BR, Nagpal JK. Understanding the biology of oral cancer. Med Sci Monit. 2002;8:RA258–67. [PubMed] [Google Scholar]
- 57.Vielba R, Bilbao J, Ispizua A, Zabalza I, Alfaro J, Rezola R, et al. p53 and cyclin D1 as prognostic factors in squamous cell carcinoma of the larynx. Laryngoscope. 2003;113:167–72. doi: 10.1097/00005537-200301000-00031. [DOI] [PubMed] [Google Scholar]
- 58.Nagpal JK, Mishra R, Das BR. Activation of Stat-3 as one of the early events in tobacco chewing-mediated oral carcinogenesis. Cancer. 2002;94:2393–400. doi: 10.1002/cncr.10499. [DOI] [PubMed] [Google Scholar]
- 59.Shpitzer T, Hamzany Y, Bahar G, Feinmesser R, Savulescu D, Borovoi I, et al. Salivary analysis of oral cancer biomarkers. Br J Cancer. 2009;101:1194–8. doi: 10.1038/sj.bjc.6605290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimakage M, Horii K, Tempaku A, Kakudo K, Shirasaka T, Sasagawa T. Association of Epstein-Barr virus with oral cancers. Hum Pathol. 2002;33:608–14. doi: 10.1053/hupa.2002.129786. [DOI] [PubMed] [Google Scholar]
- 61.Eichel HJ, Conger N, Chernick WS. Acid and alkaline ribonucleases of human parotid, submaxillary, and whole saliva. Arch Biochem Biophys. 1964;107:197–208. doi: 10.1016/0003-9861(64)90322-4. [DOI] [PubMed] [Google Scholar]
- 62.Hu Z, Zimmermann BG, Zhou H, Wang J, Henson BS, Yu W, et al. Exon-level expression profiling: A comprehensive transcriptome analysis of oral fluids. Clin Chem. 2008;54:824–32. doi: 10.1373/clinchem.2007.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and Genomics. JADA. 2006;137:313–21. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 64.Zimmermann BG, Wong DT. Salivary mRNA targets for cancer diagnostics. Oral Oncol. 2008;44:425–9. doi: 10.1016/j.oraloncology.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–52. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu S, Yen Y, Ann D, Wong DT. Implications of salivary proteomics in drug discovery and development: A focus on cancer drug discovery. Drug Discov Today. 2007;12:911–6. doi: 10.1016/j.drudis.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 67.Nyström T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24:1311–7. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shpitzer T, Bahar G, Feinmesser R, Nagler RM. A comprehensive salivary analysis for oral cancer diagnosis. Cancer Res Clin Oncol. 2007;133:613–7. doi: 10.1007/s00432-007-0207-z. [DOI] [PubMed] [Google Scholar]
- 69.Pories SE, Zurakowski D, Roy R, Lamb CC, Raza S, Exarhopoulos A, et al. Urinary metalloproteinases: Noninvasive biomarkers for breast cancer risk assessment. Cancer Epidemiol Biomarkers Prev. 2008;17:1034–42. doi: 10.1158/1055-9965.EPI-07-0365. [DOI] [PubMed] [Google Scholar]
- 70.Chen L, Sun B, Zhang S, Zhao X, He Y, Zhao S, et al. Influence of microenvironments on microcirculation patterns and tumor invasion-related protein expression in melanoma. Oncol Rep. 2009;21:917–23. doi: 10.3892/or_00000304. [DOI] [PubMed] [Google Scholar]
- 71.Vairaktaris E, Vassiliou S, Nkenke E, Serefoglou Z, Derka S, Tsigris C, et al. A metalloproteinase-9 polymorphism which affects its expression is associated with increased risk for oral squamous cell carcinoma. Eur J Surg Oncol. 2008;34:450–5. doi: 10.1016/j.ejso.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 72.Pezelj-Ribaric S, Prso IB, Abram M, Glazar I, Brumini G, Simunovic-Soskic M. Salivary levels of tumor necrosis factor-alpha in oral lichen planus. Mediators Inflamm. 2004;13:131–3. doi: 10.1080/09629350410001688530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.St John MA, Li Y, Zhou X, Denny P, Ho CM, Montemagno C, et al. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130:929–35. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- 74.Rhodus NL, Ho V, Miller CS, Myers S, Ondrey F. NF-kappa B dependent cytokine levels in saliva of patients with oral preneoplastic lesions and oral squamous cell carcinoma. Cancer Detect Prev. 2005;29:42–5. doi: 10.1016/j.cdp.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 75.Arellano-Garcia ME, Hu S, Wang J, Henson B, Zhou H, Chia D, et al. Multiplexed immunobead-based assay for detection of oral cancer protein biomarkers in saliva. Oral Dis. 2008;14:705–12. doi: 10.1111/j.1601-0825.2008.01488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moll R, Franke WW, Schiller DI. The catalog of human cytokeratins: Patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–21. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 77.Lichtenstein A, Ganz T, Selsted ME, Lehrer RI. In vitro tumor cell cytolysis mediated by peptide defensins of human and rabbit granulocytes. Blood. 1986;68:1407–10. [PubMed] [Google Scholar]
- 78.Lehrer RI, Ganz T, Selsted ME. Defensins: Endogenous antibiotic peptides of animal cells. Cell. 1991;64:229–30. doi: 10.1016/0092-8674(91)90632-9. [DOI] [PubMed] [Google Scholar]
- 79.Mizukawa N, Sugiyama K, Fukunaga J, Ueno T, Mishima K, Takagi S, et al. Defensin-1, a peptide detected in the saliva of oral squamous cell carcinoma patients. Anticancer Res. 1998;18:4645–9. [PubMed] [Google Scholar]
- 80.Kurokawa H, Tsuru S, Okada M, Nakamura T, Kajiyama M. Evaluation of tumor markers in patients with squamous cell carcinoma in the oral cavity. Int J Oral Maxillofac Surg. 1993;22:35–8. doi: 10.1016/s0901-5027(05)80353-4. [DOI] [PubMed] [Google Scholar]
- 81.Kurokawa H, Yamashita Y, Tokudome S, Kajiyama M. Combination assay for tumor markers in oral squamous cell carcinoma. J Oral Maxillofac Surg. 1997;55:964–6. doi: 10.1016/s0278-2391(97)90071-9. [DOI] [PubMed] [Google Scholar]
- 82.Bahar G, Feinmesser R, Shpitzer T, Popovtzer A, Nagler RM. Salivary analysis in oral cancer patients DNA and protein oxidation, reactive nitrogen species and antioxidant profile. Cancer. 2007;109:54–9. doi: 10.1002/cncr.22386. [DOI] [PubMed] [Google Scholar]


