Abstract
New microbial genomes are constantly being sequenced, and it is crucial to accurately determine their taxonomic identities and evolutionary relationships. Here we report PhyloPhlAn, a new method to assign microbial phylogeny and putative taxonomy using >400 proteins optimized from among 3,737 genomes. This method measures the sequence diversity of all clades, classifies genomes from deep-branching candidate divisions through closely-related subspecies, and improves consistency between phylogenetic and taxonomic groupings. PhyloPhlAn improved taxonomic accuracy for existing and newly-sequenced genomes, detecting 157 erroneous labels, correcting 46, and placing or refining 130 new genomes. We provide examples of accurate classifications from subspecies (Sulfolobus spp.) to phyla, and of preliminary rooting of deep-branching candidate divisions, including consistent statistical support for Caldiserica (formerly candidate division OP5). PhyloPhlAn will thus be useful for both phylogenetic assessment and taxonomic quality control of newly-sequenced genomes. The final phylogenies, conserved protein sequences, and open-source implementation are available online.
The reconstruction of evolutionary relationships (phylogeny) from DNA sequences is one of the oldest challenges in bioinformatics. Microbial phylogenies in particular are crucial for comparative genomics and understanding selective pressures in rapidly-evolving single-celled organisms1; microbial systematics also relies on the precise definition of a comprehensive microbial tree of life2. Whole-genome phylogenies are needed for taxonomic assignment of newly-sequenced genomes, detection of horizontally-transferred genes3, and studying selection on genes, pathways4, and pathogen mutations during disease outbreaks5. Accurate phylogenetic trees are also vital for estimating the microbial biodiversity of entire communities and relating it with environmental factors or human disease6. Although a wide range of methods have been described for aligning and reconstructing trees from individual peptide sequences, none to date have scaled to produce a highly-resolved microbial phylogeny that takes full advantage of the millions of genes and thousands of genomes now sequenced.
The microbial tree of life has long been of particular interest; gene and protein sequence-based approaches at its reconstruction antedate modern genomics7. The 16S rRNA gene (subsequently abbreviated as 16S) is historically the most-adopted phylogenetic marker8, but other single genes have been used for the same task9. Although differences can certainly arise between gene-focused and genome-focused phylogenies for any one marker, this has generally not precluded the biological utility and overall accuracy of bifurcating whole-genome trees10. While 16S databases, and thus phylogenies, include millions of sequences covering a substantial fraction of all microbes11,12, relying on any single gene lacks phylogenetic resolution at evolutionarily short time scales, preventing differentiation of closely-related organisms13, and mutations or lateral transfers in any single gene may not be well-correlated with organismal evolution14.
As whole sequenced genomes have become more abundant, the most successful recent approach for selecting markers for whole-genome phylogenetic reconstruction is based on the concatenated sequences of 31 manually-curated conserved proteins15. This and other multi-gene methods have greatly improved the accuracy and resolution of the resulting microbial trees16 when appropriate alignment and tree reconstruction methods are combined with the target sequences17,18. Proteins previously selected for this process are mainly ribosomal (23 out of 31), making the method dependent both on manual curation and on a single (albeit critical) cellular machinery. This is not necessarily an ideal proxy for organismal evolution, particularly given different rates of evolution among gene lineages19–23, and it is thus of course desirable to select several markers as molecular clocks of differing rates (i.e. slow for resolving deep branches, rapid for placing recent divergences). Furthermore, the protein selection method was neither automated nor extended beyond the 191 genomes then available, and the implemented approaches, AMPHORA16 and AMPHORA224, have not been expanded to include more than 31 (in bacteria) or 104 (in archaea) proteins and ~1,000 complete genomes. In combination with the principle of statistical consistency25,26, this suggests that the quality of a reconstructed species tree likely correlates with the size of integrated sequence data, and small marker sets may thus not be representative of the threefold larger current catalog of draft and final genomes.
The converse problem, accurate placement of newly-sequenced genomes within a reconstructed phylogeny, presents a comparable challenge. Surprisingly, although well-studied methods for tree insertion are available for the 16S phylogeny27 and for arbitrary peptide sequences28, their accuracy for automated taxonomic assignment has been minimally studied. Classification approaches based on raw sequence similarity with a best-hit policy between new and labeled sequences typically neglect topology, and have not been assessed as a tool for recommending taxonomic labels for new genomes29. Moreover, genomes can themselves be occasionally taxonomically mislabeled or misplaced, leading to the propagation of errors if not properly considered. Since manual curation is impractical for the thousands of microbial genomes now being regularly sequenced, novel computational tools for taxonomic characterization are needed, incorporating accurate, highly resolved, and comprehensive phylogenies in order to guarantee reliable analyses.
In this work, we propose and validate a novel method for accurate microbial phylogeny reconstruction, detection of potentially mislabeled genomes, and taxonomy assignment using this phylogeny for newly sequenced genomes. The approach automatically and efficiently identifies hundreds of conserved proteins from the current catalog of >3,700 finished and draft microbial genomes and uses them to build a complete high-resolution phylogeny. We develop several measures for quantitatively assessing the quality of the resulting phylogenies, all of which indicate that sampling peptides from hundreds of proteins results in increased accuracy relative to available methods. As the phylogeny is able to resolve both very closely-related strains and deep- branching candidate divisions, newly-sequenced genomes can be automatically integrated and, in many cases, assigned taxonomy with high confidence. We thus determine taxonomy for 130 previously-unassigned genomes and have detected 157 sequenced microbes likely to be taxonomically misannotated, 46 with high-confidence corrections. The fully-automated pipeline is freely available and scales to thousands more sequenced genomes, thus remaining applicable to future genomic and metagenomic investigations.
Results
A high-resolution tree of life incorporating 400 markers
We present an automated, high-throughput method for generating high-resolution microbial phylogenies by automatically detecting and combining ubiquitously-conserved bacterial and archaeal proteins. Proteins are initially selected from among 2,780 bacterial and 107 archaeal genomes in IMG (version 3.4)31, and each are tested for conservation among over 10 million genes. We assess phylogenies built from up to the 500 proteins spanning the greatest diversity, as measured by a preliminary 16S-based phylogeny11. Phylogenetic trees are generated from subsequences of these proteins concatenating their most informative amino acid positions, each aligned separately (using MUSCLE32), and reconstructed into trees using FastTree33 and RAxML28 (see Methods).
The most accurate resulting tree of life is built using >4,600 aligned amino acid positions sampled from 400 proteins (Figure 1). This incorporates the original ~2,900 genomes, 848 more from IMG 3.5 and IMG-GEBA 3.5 (as of February 201231), and two additional genomes from candidate division OP134 and the Caldiserica phylum35. In addition to placing these genomes, taxonomic assignments are refined, flagged, or newly provided for a total of 262 genomes. PhyloPhlAn, the implementation of these methods, is generalizable to any set of genomes. The process can quickly re-identify the most conserved proteins in a genome set, although this is not needed for phylogenetic placement or taxonomy assignment for newly-sequenced genomes.
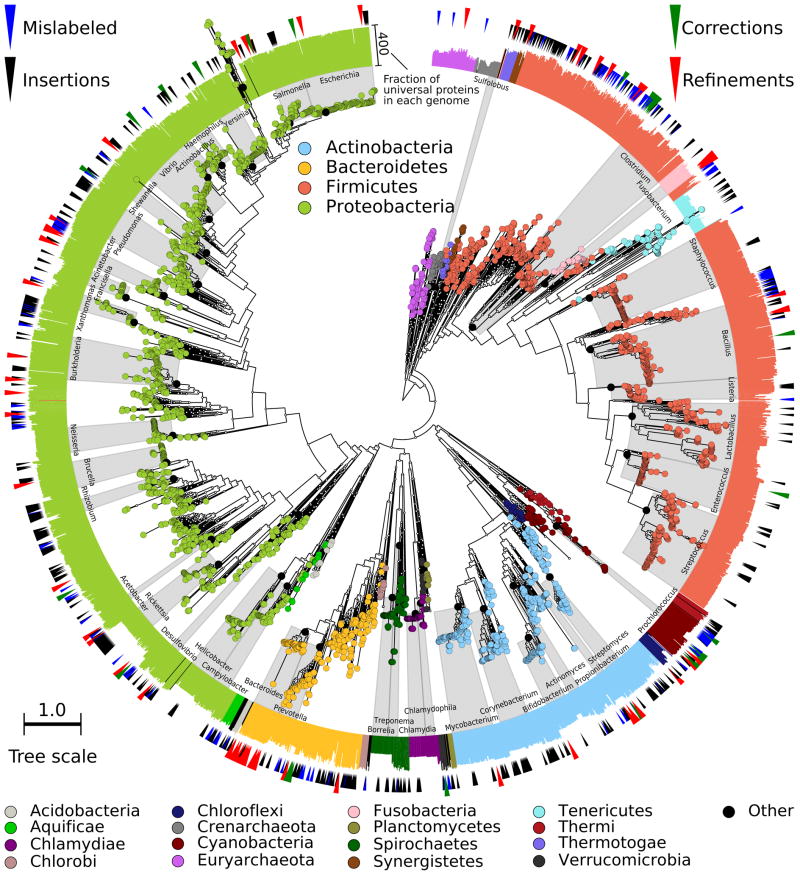
Figure 1. A high-resolution microbial tree of life with taxonomic annotations.
We reconstruct and validate a bacterial and archaeal phylogeny leveraging subsequences from 400 broadly-conserved proteins determined using 2,887 genomes and applied on a total of 3,737 genomes. The tree is built using RAxML28, with organisms colored based on phyla including at least 5 genomes. Scale indicates normalized fraction of total branch length. Gray labels indicate the lowest common ancestor of genera with at least 10 genomes (excluding predicted taxonomic mislabelings). External bar length represents the fraction of the 400 proteins contained in each genome. Red external triangles indicate genomes predicted by our method to be taxonomically mislabeled and confidently replaced; blue triangles indicate problematic labels that were refined but still did not fall within a fully consistent clade; green triangles indicate genomes whose incomplete taxonomic label we confidently refined; and black triangles indicate 566 genomes from IMG-GEBA that have been newly placed into the tree.
The new phylogenies have high accuracy and consistency
Unfortunately, no ground truth is available as a gold standard for assessing the topological accuracy of a phylogeny spanning billions of years of microbial evolution. As a surrogate, we quantitatively evaluate the consistency of our tree of life with respect to the IMG microbial taxonomy31. While no one taxonomy is perfect, this represents a well-accepted microbial categorization, as it has been extensively manually curated, and its genomic and phenotypic bases are well-established36. Likewise, while we do not expect any phylogeny and taxonomy to match perfectly, they are unlikely to match by chance; thus, greater relative similarity is a reliable measure of increased accuracy. The first measure of phylogenetic quality we derive is the consistency or precision of a clade, which for the purposes of this manuscript is defined as a systematic group or leaves of a subtree sharing taxonomic labeling. Its precision is defined as the fraction of genomes within a monophyletic subtree assigned to the same taxonomic group. For example, in our microbial tree of life (Figure 1), all Staphylococcus aureus genomes are contained in the same subtree without genomes from other organisms, thus achieving a precision of 1.0 (see Supplementary Methods equation (S1)). Unsurprisingly, most well-studied model organisms achieve near-perfect (>0.99) precision by our method, including Streptococcus pneumoniae, Salmonella enterica, Mycobacterium tuberculosis, and Vibrio cholerae, among others.
We next evaluate the recall of taxonomic clades based on the relative size of their largest taxonomically-consistent subtrees (LTCS). The LTCS of a clade is defined as the phylogenetic subtree containing only members of that clade and spanning the greatest distance. That phylogenetic diversity represents the diameter of the LTCS (longest distance between leaves; see Supplementary Methods), thus defining a clade’s recall as the fraction of its genomes within the LTCS diameter. Intuitively, a clade’s recall quantifies how many of its genomes occur “close together” within a phylogeny. All 20 phylum-level clades achieve perfect recall in our phylogeny, excluding Bacteroidetes at 0.99 due to an IMG 3.4 mislabeling discussed below. 33 of 36 (92%) class-level clades also have perfect recall, and 80 (82%) species-level clades possess recall >0.9.
Notably, the Tenericutes phylum was rooted within the Firmicutes. The placement of the Tenericutes has been controversial since the inception of the tree of life, and several early trees indeed placed the “phylum” within Firmicutes37,38. The recent concatenated proteins approach16 also supports this inner rooting, and some reconstructions, such as RAxML (Figure 1) and the All-Species Living Tree project39, root the Fusobacteria within Firmicutes as well. The PhyloPhlAn placement of these well-known yet challenging examples led us to examine the OP candidate divisions below and suggests that highly-diverged clades are sometimes better classified by high-resolution protein sequence analysis than by phenotypic traits, let alone by the single 16S gene sequence.
Choosing an optimal number of universal protein markers
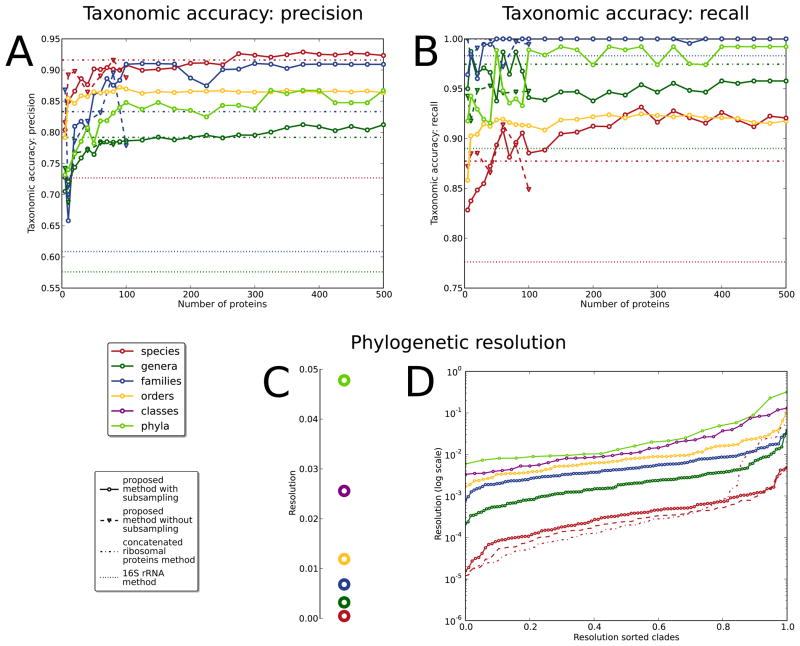
We next compare the accuracy of our approach with 16S and AMPHORA-based phylogenies across a wide range of parameters (Figure 2, and Supplementary Figure S1). Specifically, we vary the number of conserved proteins considered from 5 to 500 (Figures 2A–B) and the number of amino acids sampled from within these proteins from 30 to 4. Remarkably, we find that both phylogenetic precision and recall continue to increase at all taxonomic levels for reconstructions using up to 300 conserved proteins. Small improvements at higher phylogenetic levels continue as amino acids from up to 500 proteins are included, showing that it is indeed beneficial to employ as many conserved proteins as possible in the phylogenetic reconstruction procedure. This observation is most striking at the species level due to the fact that less universally-conserved proteins provide conversely better resolution over short evolutionary timescales. PhyloPhlAn thus leverages this behavior by combining many proteins with a core of highly-conserved universal sequences, allowing both broad (phylum-level) and detailed (species-level) accuracy.
Figure 2. Selecting informative subsequences improves the accuracy of phylogenetic tree reconstruction.
As compared to a gold standard derived from the IMG taxonomy, both precision (A) and recall (B) of inferred phylogenies increase at all taxonomic levels as up to the 500 most-conserved proteins are sampled (values averaged across all clades at each level). Comparison with full-length protein sequence phylogenies (up to 100 proteins) confirms that alignments subsampled at the most discriminative amino acids are both more accurate and more efficient. This approach outperforms single 16S rRNA gene phylogenies at all taxonomic levels, as well as trees based on curated ribosomal protein concatenation15,16 for all but the most specific clades. (C) The relative phylogenetic diversity of all taxonomic levels is consistent across varying protein numbers and is on average remarkably logarithmic, providing quantitative support for the existing multi-level microbial taxonomy. (D) Relative phylogenetic diversity among individual clades at each taxonomic level, however, shows a tremendous range of diversities, with some underrepresented phyla comprising only as much sequence divergence among available genomes as some species. This suggests that while taxonomic levels are consistent on average, clade-specific diversity thresholds should be employed when linking phylogenetic divergence with individual taxonomic labels. Again, even the most diverse species reconstructed by this method are better resolved than those using the 16S rRNA gene alone, for which many demonstrate improbably high putative phylogenetic diversity.
Based on these results, we select the 400 most ubiquitous proteins to build our recommended tree of life, as gains in accuracy at any taxonomic level beyond this point were modest. For up to 100 proteins, the approach is also feasible using full-length protein sequences without any subsampling of alignment positions (Figures 2A–B), but this provides no apparent benefit. It is also of note that the taxonomic levels achieving highest precision are the broadest (phyla) and the most specific (species), suggesting that those categories are both the most phylogenetically and taxonomically well defined. Difficulties in microbial taxonomic assignments at intermediate (class through genus) levels are well-known and reflected by the substantial number of provisional clades among these levels (e.g. 31 genomes in incertae sedis genera).
We additionally compare our phylogenetic reconstructions with state-of-the-art methods based on the 16S gene and on 31 concatenated protein alignments15,16. While concatenating ribosomal proteins greatly outperforms the 16S-based phylogeny as expected (Figure 2A, B), both are outperformed at most phylogenetic levels by our methodology. The 31 ribosomal proteins15 are remarkably precise and consistent at the species level, but performance decreases among all higher-level taxonomic clades. This may reflect the ambiguity of manual curation-based methods in assigning microbial taxonomy at intermediate levels, and emphasizes the need for automated approaches. Despite its ubiquity, the single 16S gene sequence proves highly noise-prone as a sole marker for phylogeny (Figure 2A, B). Specifically on precision, our tree outperforms that of 16S-based approaches at every taxonomic level from species to phylum, averaging for clades with at least four genomes 92.5% for species, 86.6% for genera, 80.9% for families and orders, 86.8% for classes, and 90.9% for phyla. Of 98 total species-level clades with at least four genomes, our final tree groups 80 (82%) of them with >0.9 precision, in contrast to only 58 (59%) in previous 16S gene-based approaches.
We additionally assess that the reconstructed phylogeny is robust to horizontal gene transfer (HGT) by means of systematic gene transfer simulation (Supplementary Figure S2 and Supplementary Methods). Even at extreme levels of synthetic HGT, this had a limited impact on inferred phylogenetic relationships as compared to the HGT-free PhyloPhlAn reference tree. Distances among leaf nodes in the HGT trees remained highly correlated with those in the original tree (correlation coefficients from 0.998 to 0.976 for 5% and 50% synthetic HGT, respectively). The strategy of including up to several hundred markers as diverse, repeated measures of divergence thus appears robust even to high levels of HGT, although the magnitude of branch lengths in the reconstructed trees could be underestimated for clades with extensive HGT.
Phylogenetic diversity at different taxonomic levels
We next determine what the reconstructed phylogeny reveals about the diversity of taxonomic clades. We define a third quantitative measure, the relative phylogenetic diversity of a taxonomic clade within the phylogeny, as the total branch length spanned by the placement of all genomes within the clade. Averaging this over all species, genera, and so forth, determines the typical “diameter” of each taxonomic level (Figure 2C), which proves to be remarkably logarithmic. Classes typically capture half the sequence diversity of phyla (2.5% vs. 4.8% of total diversity), orders half that of classes (1.2% vs. 2.5%), and so forth. PhyloPhlAn again produces generally more discriminative phylogenies than existing methods, regardless of the number of proteins considered. The 16S gene alone, for example, places almost 15% of species in such a manner as to include inconsistently high diversity.
These behaviors represent averages, however, and are by no means consistent among clades (Figure 2D). The most diverse taxonomic level, phyla, range from a maximum in Proteobacteria (one third of total diversity) to a minimum in Acidobacteria. At lower taxonomic levels, only a weak relationship between diversity and number of available genomes can be observed for families, genera, and species (Supplementary Figure S3, respective R2 0.04, 0.22, and 0.30). E. coli, for example, is the most-sequenced species, but it is only the 18th most diverse. More surprisingly, genus and species-level clades span more than two orders of magnitude in relative phylogenetic diversity: species with the highest diversity (>0.25%) included Buchnera, Prochlorococcus, Clostridium and Bacillus, whereas several Brucella species span very low relative phylogenetic diversity (<0.005%). All of these examples contain at least 12 sequenced genomes, and it is important to underscore that the true biological diversity of some clades may be underestimated if the genomes are insufficiently representative of the diversity within those clades.
Analyzing specific subclades of Bacteria and Archaea
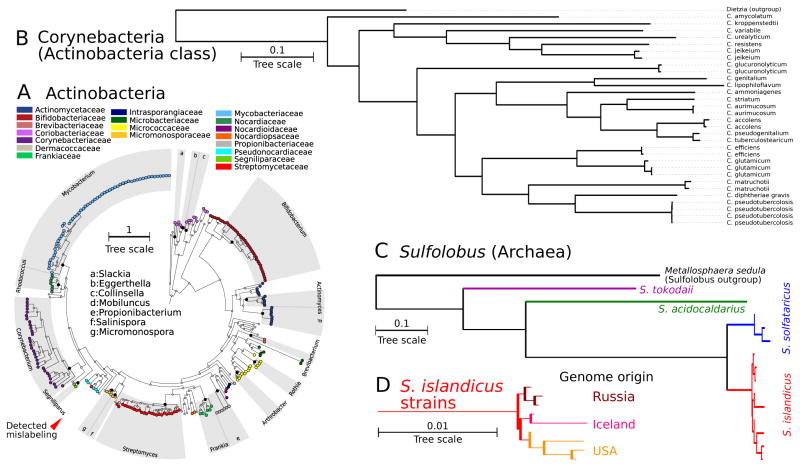
A recent study of 45 genomes in the Actinobacteria phylum40 showed that a consistent and resolved phylogeny of this clade could not be achieved with rRNA genes (5S, 16S, or 23S); one was inferred by combining several (155) concatenated genes together with features such as synteny and phylogenetic profiling. Our Actinobacteria reconstruction (Figure 3A) achieves a fully consistent grouping of all 19 families, with the sole exception a Streptomyces genome phylogenetically included in Pseudonocardiaceae due to a known mislabeling31. Our method automatically flags this as a likely misannotation (red triangle in Figure 3A). All other genera and species included in the existing phylogeny40 are correctly inferred here, confirming that this catalog of 400 proteins allows accurate phylogenetic reconstruction without the need for additional genomic information. We then specifically investigate the Corynebacterium genus, as this clade has recent been phylogenetically characterized with conflicting topologies using the single 16S and rpoB genes9. In the PhyloPhlan tree of the 31 Corynebacterium genomes (Figure 3B), multiple strains in the same species always cluster together with relatively little divergence. Interestingly, intra-species distances between complete and draft genomes are as small as those between complete genomes (see C. efficiens and C. aurimucosum), confirming the PhyloPhlAn consistency in processing partially assembled genomes.
Figure 3. Inferred phylum, genus, and species phylogenetic trees.
(A) The inferred Actinobacteria phylum subtree, with genomes colored by family and genera annotated by root node. All 19 families are grouped consistently, which cannot be achieved by 16S gene sequences alone40. (B) The Corynebacterium genus subtree, with highly concordant species and strain grouping not achieved by previous analyses9. (C) Archaeal genomes of genus Sulfolobus, and (D) for S. islandicus, an inset of the inferred strain-level tree. For this particular organism, all 9 genomes group consistently according to the geography of their site of origin.
The Archaea show similar accuracy (Figure 3C); Sulfolobus is the genus with the greatest number of sequenced organisms (14), all representing thermoacidophiles isolated mainly from volcanic springs. Its phylogenetic tree is again monophyletic with respect to species and highlights inter-species diversity more than 20x higher than intra-clade diversity for S. solfataricus and S. islandicus. However, the low diversity in these two species does not affect the discriminative power of the subtree, as confirmed by the inset S. islandicus genomes (Figure 3D). This strain-level phylogeny further exactly matches the distribution of these species, reflecting the geography of genomes sequenced from acid environments in American, Russian (7 genomes41), and Icelandic (2 genomes) locations42.
Phylogenetic placement of deep branching organisms
We next analyze the integration of two genomes from candidate divisions OP1 and Caldiserica (formerly OP5), which are particularly challenging cases lacking close relatives in the existing phylogeny, and the placement of similarly deep-branching representatives of divisions TM7 and OP11. Based on 16S data, OP1 was previously assessed as most closely related to Thermatoga; TM7 was closest to Chloroflexi (specifically Chloroflexus auranticus43), and OP11 was very deep-branching44. A later study concatenated 44 highly-conserved proteins and concluded that Coprothermobacter (family Thermodesulfobiaceae) was most closely related to the Dictyoglomi and Thermatoga, as well as confirming the Actinobacteria / Deinococcus-Thermus / Cyanobacteria / Chloroflexi grouping45.
Our final phylogeny (Figure 1 and Supplementary Figure S4) concurs with the confident subset of these previous placements and introduces a potentially deeper branching for Caldiserica. The description of the only current Caldiserica isolate, Caldisericum exile35, places it between the Proteobacteria and Acidobacteria phyla based on 16S data. Our results (Supplementary Figure S4) suggest a much deeper phylogeny, with Dictyoglomus and Thermotoga as sister phyla and consistent bootstrapping support (85%) for the combined Caldiserica-Coprothermobacter subclade; this placement should of course be explored by further targeted analyses.
The placement of OP11 agrees with previous placements external to all major bacterial phyla44, but was based on a reduced core of 30 genes from a single draft genome of 417 proteins. Its placement is particularly challenging due to an unusually large fraction of short peptides (41.2% of proteins <100AA versus an average of 13.3% s.d. 5.7% for all genomes) and, as a result, the corresponding subtrees have low bootstrap support. Our phylogeny supports deep-branching of Candidatus acetothermum autotrophum (OP1), in agreement with 16S and protein-based studies, but between Thermotoga and Archaea rather than Deinococcus-Thermus34,44. We group Actinobacteria, Deinococcus-Thermus, Cyanobacteria, and Chloroflexi as sister phyla and propose that TM7, represented by only one genome, is not only closely related to Chloroflexi but possibly rooted within them, although the limited bootstrapping support (40%) suggests that this hypothesis needs independent validation.
Efficiently expanding the tree of life with new genomes
The identification of the 400 most-conserved proteins in microbial genomes is made computationally tractable by our approach, but is still an expensive operation that is avoidable when creating or updating a phylogeny with new genomes. The PhyloPhlAn implementation stores a non-redundant database of the 400 proteins and their variants that is used for identifying them in new genomes by translated mapping with USEARCH46. Without any other prior information, a full tree of the 3,174 genomes in IMG version 3.5 is produced in under two hours on a 16-CPU system using the FastTree application33.
Even this step is typically unnecessary, however, as the pipeline also allows new genomes to be incorporated directly into an existing tree. IMG-GEBA 3.5 provides 566 new genomes without species labels, for example, and they have been integrated in this way and are indicated with black triangles in Figure 1 and detailed in Supplementary Figure S4. This most current phylogenetic tree is reconstructed with RAxML version 7.3.228 in place of FastTree33 to increase accuracy at the expense of computational time (620 instead of 2 CPU-hours).
Refining taxonomic assignments for new genomes
Since microbial genomes are now being sequenced by the hundreds, an important application of automated phylogenetic reconstruction is to suggest taxonomic labels for newly-sequenced genomes. This is possible using guilt-by-association to transfer nearby taxonomic labels, and it is most straightforward when the genome is inserted within a monophyletic clade at any taxonomic level. Transfer is particularly confident when the genome’s distance from the lowest common ancestor of a monophyletic subtree is consistent with the subtree’s diameter (see Supplementary Methods). Of the 566 IMG-GEBA genomes inserted above, when taxonomic information is stripped before PhyloPhlAn assignment, the pipeline assigns 56 to the species level, 164 at genus, 250 at family, 350 at order, and 414 at class using the most stringent confidence threshold (Supplementary Data 1).
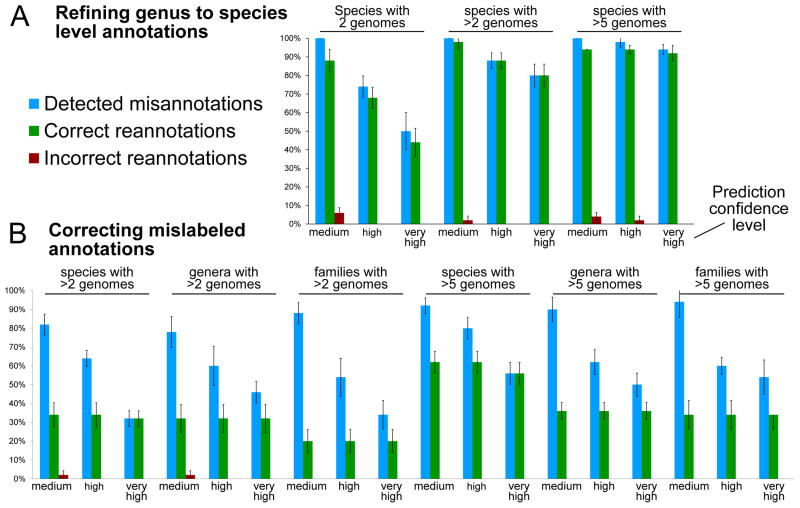
In many cases, newly-sequenced genomes can be assigned at least partial taxonomy by a depositor, e.g. to the family or genus level. This can be incorporated as additional information and either refined to a more specific level or flagged as suspect. Of the 566 partially-labeled IMG-GEBA genomes, 51 are confidently refined to a species-level taxonomic assignment, whereas 20 of them are flagged as potentially misplaced and relabeled (Supplementary Data 2). 36 additional genomes are flagged as suspect without further confident refinement. These results are again obtained with the most stringent confidence threshold; results at more lenient thresholds are still informative but might require manual review. The accuracies of PhyloPhlAn’s three confidence levels are quantified on artificial datasets obtained by removing species-level labels from known genomes and re-imputing their taxonomy, with precision exceeding 80% for well-represented clades and zero false positives at high confidence (Figure 4A).
Figure 4. Accuracy of correctly re-inferred taxonomic labels for artificially-mislabeled organisms.
Barplots report the percentages (with s.d.) of successfully-recovered cases. (A) For 5 iterations, 10 taxa are selected at random from species with 2, more than 2, or more than 5 genomes, and their species-level label removed. The PhyloPhlAn phylogenetic tree (which is built without any taxonomic information) is then used to re-impute the removed labels at medium, high, and very high confidence thresholds. No incorrect refinements are produced at the highest confidence threshold, and average recall rates for species with at least three taxa exceed 90% at high confidence. (B) We repeat this procedure by mislabeling (rather than removing labels for) species, genus, or family-level assignments. No false positives are produced at high or very high confidence, and only 2 over all experiments (<1%).
Detection of taxonomic mislabeling and label assignment
Our phylogenetic reconstruction method also suggests corrections to incomplete or misannotated entries in the current microbial taxonomy. Potential misclassifications are automatically flagged for inspection by checking whether a taxon is outside its largest consistent monophyletic subtree (see Methods); when possible, refinements are provided for genomes missing species-level labels by removing and re-imputing partial taxonomy. When applied to all 2,726 annotated genomes, potential corrections to the current microbial taxonomy range from simple typos to apparent phenotypic misclassifications. More than 5% (157) are detected as potentially misplaced, in addition to 410 genomes with incomplete taxonomy (totaling 17.8%). 26 of the 157 flagged cases could be confidently reassigned to an equally-specific taxonomic level, as well as a further 20 of the 114 genomes flagged during the IMG-GEBA insertions (58 more at less- strict confidence thresholds). Finally, between both genome sets, 71 of 445 genomes lacking detailed taxonomy are very confidently refined, resulting in the red (corrections), green (refinements), and blue (flags) annotations in Figure 1 and the complete list of taxonomic corrections in Supplementary Data 3.
A striking example of misannotation in the existing taxonomy is the strain ATCC 43243 of Bacteroides pectinophilus, which we place well outside the otherwise fully-consistent Bacteroidetes phylum. It instead falls within Clostridia, as verified by a manual phylogenetic analysis of the bacteroides47. This is fortunately the only phylum-level misclassification, with several of the others occurring at lower taxonomic levels associated with independent external confirmations. A representative subset of inferred taxonomic changes is reported in Table 1.
Table 1.
Representative examples of taxonomic assignments inferred by PhyloPhlAn.
| Name | Genome ID | Type | Most accurate taxonomy | |
|---|---|---|---|---|
| Original | New | |||
| Plautia stali symbiont | IMG 651324086 | detection (refinement) | kingdom: Bacteria | genus: Pantoea |
|
| ||||
| Burkholderiales bacterium 1_1_47 | IMG 648861006 | detection (refinement) | order: Burkholderiales | species: Parasutterella excrementihominis |
|
| ||||
| Enterococcus faecalis PC4. 1 | IMG 647000238 | detection (correction) | species: Enterococcus faecalis | species: Enterococcus faecium |
|
| ||||
| Bacteroides sp. 3_1_19 | IMG 648861002 | detection (correction) | genus: Bacteroides | genus: Parabacteroides |
|
| ||||
| Porphyra umbilicalis endophyte | IMG-GEBA 2511231155 | imputation (refinement) | class: Planctomycetia | family: Pirellulaceae |
|
| ||||
| Shewanella sp. W3-18–1 | IMG-GEBA 2511231030 | imputation (refinement) | genus: Shewanella | species: Shewanella baltica |
|
| ||||
| Sediminibacterium sp OR43 | IMG-GEBA 2509887033 | imputation (correction) | genus: Sediminibacterium | family: Chitinophagaceae |
|
| ||||
| Citromicrobium sp. JLT1363 | IMG-GEBA 2512047056 | imputation (correction) | genus: Citromicrobium | family: Erythrobacteraceae |
We evaluate this approach by repeatedly artificially mislabeling 10 currently correct genomes and re-imputing them at increasingly strict confidence thresholds (Figure 4B). This procedure is run five times each for randomly selected genomes in clades at increasing levels of specificity and with decreasing amounts of existing evidence (>5 to 2 supporting genomes). No false positive imputations occur at the highest PhyloPhlAn confidence threshold among all 45 runs, and only 2 at medium confidence. Corresponding recall rates range from 78% and 82% in the most difficult cases to 94% in the best.
Discussion
We developed and validated an automated method for generating a highly-resolved microbial tree of life that can be applied to taxonomically label newly-sequenced microbial genomes. The method scales efficiently to incorporate all available finished and draft bacterial and archaeal genomes and leverages phylogenetic information from hundreds of proteins well-conserved among microbial organisms. In the first comprehensive evaluation of the taxonomic precision and recall in microbial trees of life, the final phylogenetic trees produced by this method outperformed both the commonly employed single 16S gene8 and state-of-the-art curated multiple protein approaches15,16. Total achievable accuracy continued to increase as informatively varying peptides were sampled from up to 500 total proteins, thus addressing potential pitfalls of single gene and manually-curated methods and allowing the rapid taxonomic assignment of any newly-sequenced microbial genome and the detection of 157 genomes likely to be currently misannotated.
New microbial genomes are now being sequenced by the hundreds; thus, it is increasingly important to provide an accurate, high-resolution, automated framework for phylogenetic placement and recommended taxonomy. Long-standing phenotyping and biochemistry are vital for taxonomic validation, but it is impractical to perform these for all isolates in a high-throughput environment. PhyloPhlAn provides a systematic check for the thousands of genomes already sequenced and is compatible with draft genomes missing individual markers such as the 16S gene, microbes sequenced from uncultured samples48, partial assemblies from metagenomic data49, and genomes with extensive HGT. Such genomes are often fragmentary, uncultured, or phenotypically uncharacterized prior to sequencing and classification. Computational efficiency is also crucial for the increasing size of microbial genomics, and PhyloPhlAn scales at best (and typically) linearly and at worst quadratically with total genomes, making it suitable for much larger sequence compendia (Supplementary Figure S5). Finally, there are many theoretical motivations for improving large phylogenetic reconstructions by including many well-distributed genomes50,51, and PhyloPhlAn thus represents a means to efficiently employ the increasing catalog of microbial genomes. Variations on this method have already been useful for efficient and high-throughput taxonomic assessment of whole uncultured communities52 and can be crucial for mining meta’omic datasets53.
Opportunities exist to further refine all of PhyloPhlAn’s three primary steps: identifying informative conserved residues, reconstructing a species tree, and inferring new genomes’ putative taxonomy. We currently favor conserved proteins by drawing proportionally more residues from them, tending to identify residues that are “slightly saturated”54. The parameters or method used to select these residues, and the numbers of loci included, could be optimized using holdout sequences or new genomes. Different tree reconstruction algorithms can be used with the selected residues, and this raises the possibility of evaluating additional evolutional models and tree-combining55 or alignment-free56 reconstructions. Likewise, different genome placement classifiers could be used to assign putative taxonomy. This is particularly of interest, since more advanced ortholog/paralog detection methods and annotation-free identification of conserved target sequences may further improve accuracy on partial genomes derived from metagenomes or single cell sequencing (Supplementary Tables S1 and S2).
The tasks enabled by a microbial tree of life with phylum-to-species accuracy include additional evolutionary and comparative genomic applications not covered in our initial analyses. For example, some microbial clades have very broad pan-genomes (e.g. Prochlorococcus marinus with a pan-genome 4x larger than its median genome), for which we could detect enrichments for specific genes, pathways, or functionality within phylogenetically well-defined sub-trees. When considering the entire tree of life, the relationship between functional and evolutionary distances can be compared for investigating convergent functional specialization of unrelated bacteria57 or, conversely, divergence in recent speciation58. Overall, the high consistency achieved by this phylogenetic tree built using hundreds of well-conserved proteins provides a solid foundation for future high-throughput studies of taxonomy (see Supplementary Discussion), comparative genomics, systematics, and taxonomic classification relying on an accurate and comprehensive microbial tree of life.
Methods
PhyloPhlAn reconstruction pipeline and availability
The developed open source software, documentation, tutorial, resulting data, and supplemental information are available online and for download (PhyloPhlAn website30, with a copy in Supplementary Software). PhyloPhlAn implements all phylogenetic reconstruction steps (conserved protein detection, tree building, and integration of new genomes into the tree, all described in the Supplementary Methods) and taxonomic curation strategies (mislabeling detection, label imputation/refinement for new genomes, and label imputation/refinement for detected mislabeling, described below). The PhyloPhlAn approach is based on the 400 most universal proteins that have been identified by off-line preprocessing of all available microbial genomes. The pre-processing steps include core gene identification13 and merging core genes into universal protein families (described below), and ranking each protein family for ubiquitous conservation and covered diversity in the microbial domain (Supplementary Methods).
Genomic input data
All 2,887 sequenced microbial genomes were retrieved from the Integrated Microbial Genomes31 (IMG) version 3.4 with corresponding coding sequence (CDS) calls, translated protein sequences, and taxonomic assignments. Genomes were screened for length >50,000 nt, at least 50 CDSs, and at least 75% of the genome coding sequences. 51 of the remaining genomes lacked a taxonomic label below the family level and were considered to be taxonomically uncharacterized. 1,221 16S gene sequences representing IMG species were retrieved from Greengenes11. CDS to COGs assignments and 16S rRNA gene annotations were downloaded from IMG and used only for identifying the 31 ribosomal proteins for re-performing the corresponding method15,16 with these genomes as described in the Supplementary Methods (“Building phylogenetic trees using 16S and ribosomal proteins”). The PhyloPhlAn pipeline was further tested on the 3,171 genomes from IMG 3.5 as of February 2012; 566 additional genomes not contained in IMG 3.5 were downloaded from IMG-GEBA31 as of March 2012, and the genomes of candidate division OP134 and Caldiserica35 were retrieved from the GOLD database59 (GOLD ids Gc02183 and Gi17125 respectively).
Identification of core genes
Our unsupervised pipeline identifies the most ubiquitous proteins in thousands of genomes while avoiding computationally infeasible brute-force pairwise sequence comparisons between all >10M microbial CDSs. The three main steps of the method are (i) identifying nucleotide level core genes, i.e. those consistently present in at least one low-diversity clade (approximately from species to family levels), (ii) finding strong amino acid homologs between core genes to detect universal proteins conserved in multiple lineages, and (iii) ranking these universal proteins based on the number of genomes containing them and the total diversity they span.
For identifying core genes, we employed and expanded a previous method for hierarchically identifying CDS homologs by means of recursive clustering on a guide tree13,52. Each genome was first transformed into a collection of clustered CDSs. From each cluster, a single representative (seed) was selected. Seeds from all strain-level genomes in each species were compared using UCLUST32 at 75% nucleotide identity to identify species-level core genes. We introduced several refinements to this step to make the identification procedure robust to missing genomic regions and errors in CDS calls and taxonomic assignment. To capture missing or unannotated CDSs, each seed was aligned by BLASTN against every raw genome, and high-confidence matches were added to the corresponding gene family clusters. To address draft genomes and misannotated open reading frames, we generalized the definition of core gene using a probabilistic model. The presence/absence of a gene family across a group of genomes was modeled as a beta function of expected posterior probability density. We selected gene families with a >95% probability of being core in each clade given a 5% missing gene rate from annotation and assembly errors (measured from missing 16S annotations), and propagated them to the next level of identification. Genomes assigned by IMG directly to the genus (or higher) level were not considered during species-level core identification, but were included subsequently. Once species core genes were determined, clustering and comparing gene families was recursively applied to successively higher taxonomic levels (from genus to phylum).
Merging core genes into universal protein families
To detect proteins with homologs in a large fraction of genomes, we performed a translated nucleotide search against the microbial proteomes for a reduced set of conserved core genes. Specifically, we selected the 50 most conserved core genes in each lineage at the highest level of the taxonomic guide tree covering a maximum of genomes, resulting in a catalog of 39,000 CDSs. NCBI Blastx60 (e-value <1e-50) generated a bipartite graph between core genes and amino acid sequences. The proteomes of the 51 organisms without clear taxonomy (excluded from core gene identification above) were included in this translated search to permit downstream phylogenetic profiling.
Unsurprisingly, several very similar sets of proteins were targeted by more than one core gene, when several amino acid sequences were conserved in multiple lineages but were missing or substituted by functionally related proteins in specific clades. We thus binned together each set of proteins targeted by approximately the same set of core genes, initially evaluating three different approaches: intersection of the overlapping sets of target proteins, union, or selection of the largest of those sets, in all cases thresholded at a minimum overlap of 95%, and maximum disjoint fraction at 5%. Preliminary evaluation showed that the final maximum cardinality approach was the most accurate, and we thus selected it for downstream analyses. The resulting fully disjoint catalog of protein families comprised 513 sets of proteins each present in at least 1,000 genomes. It is worth mentioning that the use of core genes rather than all gene calls for alignment against proteomes does not cause the misdetection of any ubiquitous proteins; any protein present in at least 1,000 genomes must be core in at least one genus-level or higher clade, and the process takes possibly missing or misannotated CDSs into account13. Universal proteins are then ranked for ubiquitous conservation as described in the Supplementary Methods.
Detecting potentially misannotated genomes
Genomes that are phylogenetically rooted well outside the largest monophyletic subtree (i.e. the LTCS) of their putative taxonomic clades are flagged as potentially mislabeled. This test is performed all genomes within any clades (at any taxonomic level) containing at least four representative genomes; a reliable LTCS cannot be defined when fewer sequences are available. LTCS calculation and all subsequent taxonomic label comparisons are performed using only genomes for which a fully defined curated taxonomy is available. Let D be the ratio between the genome’s distance from the LTCS and the 75th percentile of all within-clade distances. If D<1.0, the genome is not flagged. Similarly, let R′ be the ratio of two additional distances from the genome of interest, first to the closest genome in the LCA sharing the target level’s taxonomic label, and second to the closest of any genome in the LTCS. R′<1.0 indicates at least one other genome outside the LTCS supports the current label, in which case the genome is again not flagged.
Genomes meeting neither of these criteria are flagged as potential misannotations at one of three different confidence levels. Let R″ be the fraction of the clade of interest included in the LTCS; high values of R″ reflect subclades that are consistent except for the genome under consideration. Genomes with D 2.0, R′≥ 2, and R″≥ 0.8 are flagged with very high confidence; genomes with 1.25<D<2.0, 1.5<R′<2, and 0.7<R″<0.8 receive high confidence; and flagged genomes not meeting these thresholds are annotated as medium-confidence misannotations. Medium-confidence assignments are thus still unlikely to be correct due to the lack of strong phylogenetic evidence supporting a putative taxonomic label.
Inferring taxonomy for unlabeled or misannotated genomes
Genomes with incomplete or absent manually-assigned taxonomy, or whose taxonomic label has been flagged as inconsistent (as described above), can be provided with a putative improved label based on evidence from the surrounding phylogeny. By default, the pipeline will taxonomically re-profile only very high-confidence predictions. First, we identify the largest otherwise fully- monophyletic subtree containing the genome of interest. If such a subtree exists (i.e. if it consists of more than one taxon in addition to the target), the taxonomic label of this subtree is initially assigned to the genome with medium confidence. This confidence score is increased if the distance of the target genome from the other genomes inside the subtree is consistent with the overall distribution of intra-clade distances. Specifically, the distance between the target and the closest other within-clade genome is compared to the distribution of all minimum pairwise distances between leaves in the subtree. If the target rank is within the 95th percentile, the relabeling is increased to very high confidence, and high confidence is assigned at the 90th percentile.
If no such monophyletic subtree exists for a flagged or unannotated target, we reassign taxonomy only if the genome under investigation is extremely close to a well-defined genome. In particular, very high confidence is assigned for cases in which the distance of the closest fully characterized taxon is below the median of all closest pairwise distances in the same clade for the taxonomic level of interest or smaller than 0.001% of the total diversity in the tree. High confidence is assigned if the distance ranks between the 50th and 75th percentile, and medium confidence between the 75th and 90th percentiles. Genomes falling above the 90th percentile remain flagged, but no new putative taxonomy is automatically provided.
Supplementary Material
Acknowledgments
We would like to thank Ashlee Earl and the Human Microbiome Project Strains Working Group for insightful suggestions, Morgan Price for his helpful comments on applying FastTree, and Katherine Huang, Levi Waldron, Joshua Reyes and Timothy Tickle for their suggestions on methodology and tree visualization. This work was supported in part by NIH 1R01HG005969 and NSF DBI-1053486 to CH and by Danone research grant PLF-5972-GD.
Footnotes
Author contributions
N.S and C.H conceived the method; N.S. implemented the software and performed the experiments; N.S, D.B, X.M., and C.H. analyzed the data; N.S. and C.H. wrote the manuscript.
Conflict of interest
The authors declare no competing interests.
References
- 1.Ochman H, Wilson Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 2.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki W, Takagi T. Rapid pathway evolution facilitated by horizontal gene transfers across prokaryotic lineages. PLoS Genetics. 2009;5:e1000402. doi: 10.1371/journal.pgen.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardy JL, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. New Engl J Med. 2011;364:730. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 6.Manichan C, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 8.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. PNAS. 1977;74:5088. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khamis A, Raoult D, La Scola B. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J Clin Microbiol. 2005;43:1934. doi: 10.1128/JCM.43.4.1934-1936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge F, Wang LS, Kim J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS biology. 2005;3:e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole J, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segata N, Huttenhower C. Toward an Efficient Method of Identifying Core Genes for Evolutionary and Functional Microbial Phylogenies. PloS one. 2011;6:e24704. doi: 10.1371/journal.pone.0024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguileta G, et al. Assessing the performance of single-copy genes for recovering phylogenies. Syst Biol. 2008;57:613–627. doi: 10.1080/10635150802306527. [DOI] [PubMed] [Google Scholar]
- 15.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 16.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson JD, Linard B, Lecompte O, Poch O. A comprehensive benchmark study of multiple sequence alignment methods: current challenges and future perspectives. PloS one. 2011;6:e18093. doi: 10.1371/journal.pone.0018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Warnow T. Fast and accurate methods for phylogenomic analyses. BMC bioinformatics. 2011;12:S4. doi: 10.1186/1471-2105-12-S9-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome research. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocha EP, Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Molecular biology and evolution. 2004;21:108–116. doi: 10.1093/molbev/msh004. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. Journal of molecular evolution. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 22.Huelsenbeck JP, Bull J, Cunningham CW. Combining data in phylogenetic analysis. Trends in Ecology & Evolution. 1996;11:152–158. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 23.Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- 24.Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 25.Penny D, Hendy MD, Steel MA. Progress with methods for constructing evolutionary trees. Trends in Ecology & Evolution. 1992;7:73–79. doi: 10.1016/0169-5347(92)90244-6. [DOI] [PubMed] [Google Scholar]
- 26.Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Biology. 1978;27:401–410. [Google Scholar]
- 27.Ludwig W, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 29.Gao B, Gupta RS. Microbial systematics in the post-genomics era. Antonie van Leeuwenhoek. 2011:1–10. doi: 10.1007/s10482-011-9663-1. [DOI] [PubMed] [Google Scholar]
- 30.Segata N, Boernigen D, Morgan X, Huttenhower C. PhyloPhlAn. 2012 doi: 10.1038/ncomms3304. http://huttenhower.sph.harvard.edu/phylophlan. [DOI] [PMC free article] [PubMed]
- 31.Markowitz VM, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS one. 2010;5 doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takami H, et al. A Deeply Branching Thermophilic Bacterium with an Ancient Acetyl-CoA Pathway Dominates a Subsurface Ecosystem. PloS one. 2012;7:e30559. doi: 10.1371/journal.pone.0030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori K, Yamaguchi K, Sakiyama Y, Urabe T, Suzuki K. Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. nov. and Caldisericia classis nov. International journal of systematic and evolutionary microbiology. 2009;59:2894–2898. doi: 10.1099/ijs.0.010033-0. [DOI] [PubMed] [Google Scholar]
- 36.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludwig W, Schleifer KH. Microbial phylogeny and evolution, concepts and controversies. Oxford University Press; New York: 2005. Molecular phylogeny of bacteria based on comparative sequence analysis of conserved genes; pp. 70–98. [Google Scholar]
- 38.Woese C, Maniloff J, Zablen L. Phylogenetic analysis of the mycoplasmas. PNAS. 1980;77:494. doi: 10.1073/pnas.77.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yarza P, et al. The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Systematic and applied microbiology. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Alam MT, Merlo ME, Takano E, Breitling R. Genome-based phylogenetic analysis of Streptomyces and its relatives. Mol Phyl Evol. 2010;54:763–772. doi: 10.1016/j.ympev.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 41.Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. Biogeography of the Sulfolobus islandicus pan-genome. PNAS. 2009;106:8605. doi: 10.1073/pnas.0808945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo L, et al. Genome Analyses of Icelandic Strains of Sulfolobus islandicus, Model Organisms for Genetic and Virus-Host Interaction Studies. J Bacteriol. 2011;193:1672. doi: 10.1128/JB.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. PNAS. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishida H, Beppu T, Ueda K. Whole-genome comparison clarifies close phylogenetic relationships between the phyla Dictyoglomi and Thermotogae. Genomics. 2011;98:370–375. doi: 10.1016/j.ygeno.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 47.Karlsson FH, Ussery DW, Nielsen J, Nookaew I. A closer look at Bacteroides: phylogenetic relationship and genomic implications of a life in the human gut. Microbial ecology. 2011:1–13. doi: 10.1007/s00248-010-9796-1. [DOI] [PubMed] [Google Scholar]
- 48.Sczesnak A, et al. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host & Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 50.Hillis DM. Inferring complex phylogenies. Nature. 1996;383:130. doi: 10.1038/383130a0. [DOI] [PubMed] [Google Scholar]
- 51.Rannala B, Huelsenbeck JP, Yang Z, Nielsen R. Taxon sampling and the accuracy of large phylogenies. Systematic Biology. 1998;47:702–710. doi: 10.1080/106351598260680. [DOI] [PubMed] [Google Scholar]
- 52.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012 doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed]
- 53.Segata N, et al. Computational meta’omics for microbial community studies. Molecular Systems Biology. 2013;9:1–15. doi: 10.1038/msb.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philippe H, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS biology. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science (New York, NY) 2009;324:1561–1564. doi: 10.1126/science.1171243. [DOI] [PubMed] [Google Scholar]
- 56.Nelesen S, Liu K, Wang LS, Linder CR, Warnow T. DACTAL: divide-and-conquer trees (almost) without alignments. Bioinformatics. 2012;28:i274–i282. doi: 10.1093/bioinformatics/bts218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. PNAS. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zdziarski J, et al. Host Imprints on Bacterial Genomes—Rapid, Divergent Evolution in Individual Patients. PLoS pathogens. 2010;6 doi: 10.1371/journal.ppat.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pagani I, et al. The Genomes OnLine Database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.