SUMMARY
IFI16 is a member of the interferon-inducible HIN-200 family of nuclear proteins. It has been implicated in transcriptional regulation by modulating protein-protein interactions with p53 tumor suppressor protein and other transcription factors. However, the mechanisms of interaction remain unknown. Here, we report the crystal structures of both HIN-A and HIN-B domains of IFI16 determined at 2.0 and 2.35 Å resolution, respectively. Each HIN domain comprises a pair of tightly packed OB-fold subdomains that appear to act as a single unit. We show that both HIN domains of IFI16 are capable of enhancing p53-DNA complex formation and transcriptional activation via distinctive means. HIN-A domain binds to the basic C terminus of p53, whereas the HIN-B domain binds to the core DNA-binding region of p53. Both interactions are compatible with the DNA-bound state of p53 and together contribute to the effect of full-length IFI16 on p53-DNA complex formation and transcriptional activation.
INTRODUCTION
Interferons (IFNs) are cytokines involved in diverse biological functions including regulation of antiviral, antibacterial, immune, and inflammatory responses (Gutterman, 1994; Stark et al., 1998). The effects of IFN stimulation are often manifested through activation of IFN-inducible genes. Although many genes are known to be activated by this pathway, their biological and functional importance remains unclear.
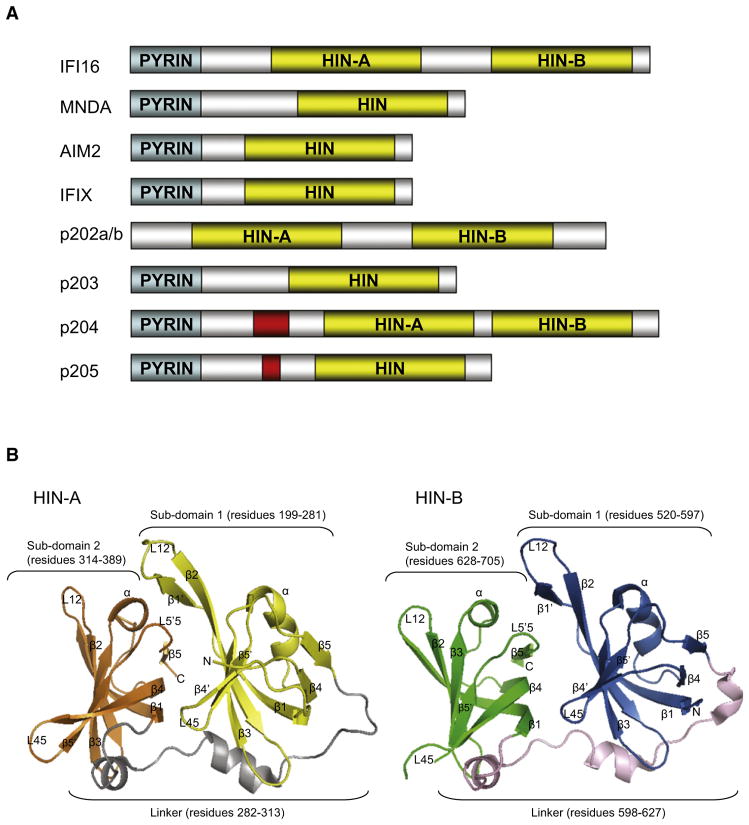
The IFN-inducible HIN-200 gene family encodes a class of homologous proteins that share a 200-amino acid signature motif (HIN) (Dawson and Trapani, 1996). Four human (IFI16, MNDA, AIM2, and IFIX) and five mouse (p202a, p202b, p203, p204, and p205) members of this family have been identified (Figure 1A). Most of the HIN-200 proteins possess two major protein domains. At the N terminus, there is a conserved α-helical PYRIN domain, which belongs to the death domain-containing protein superfamily involved in apoptosis, inflammation, and immune responses (Hiller et al., 2003; Stehlik and Reed, 2004). At the C terminus, all HIN-200 proteins possess either one or two copies of a conserved HIN domain, which has been implicated in DNA binding, as well as in mediating protein-protein interactions for transcriptional regulation (Koul et al., 1998; Xin et al., 2003; Ludlow et al., 2005; Yan et al., 2008).
Figure 1. The HIN Domains of IFI16.
(A) Overall domain architecture of the HIN-200 proteins in human (IFI16, MNDA, AIM2, IFIX) and mouse (p202a/b, p203, p204, p205). The PYRIN and HIN domains are indicated by the cyan and yellow shadings, respectively. Red shadings highlight S/T- rich region.
(B) Structure of HIN-A (left) or HIN-B (right) domain of IFI16 in cartoon representation. N and C termini are indicated.
See also Figure S1.
The mouse protein p202a, containing two HIN domains exclusively, is the most studied member of the HIN-200 family. The HIN repeats in p202a have been shown to interact with numerous transcription factors including pRB, p53, NFκB, AP-1, MyoD, and E2F, indicating that the HIN domain may serve as a scaffold to assemble large protein complexes to modulate the transcription of target genes (Choubey, 2000; Xin et al., 2003). The human HIN-200 homolog IFI16, whose primary sequence predicts a PYRIN domain and two tandem HIN domains, has also been implicated in binding to pRB, E2F1, p53, and BRCA1 (Johnstone et al., 2000; Aglipay et al., 2003; Xin et al., 2003). Recently, human AIM2 was shown to be essential for sensing cytoplasmic foreign DNA through its HIN domain and interacting with an apoptosis-associated protein, ASC, via its PYRIN domain, leading to activation of inflammatory responses (Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009).
The observation that these HIN-200 proteins interact with several cellular regulators involved in cell cycle control, proliferation, differentiation, and apoptosis hints that the physiological role of HIN-200 proteins may lie beyond the IFN system (Ludlow et al., 2005; Ding et al., 2006). Indeed, IFI16 is widely expressed in normal human endothelial and epithelial cells in addition to hematopoietic cells (Gariglio et al., 2002; Wei et al., 2003; Raffaella et al., 2004; Ludlow et al., 2005). Several studies have demonstrated that loss or reduced expression of IFI16 is often associated with various forms of human cancers, including those of the pancreas, prostate, and breast (Trapani et al., 1992; Xin et al., 2003; Fujiuchi et al., 2004); leading to the notion that IFI16 may play an important role in tumor suppression. However, the molecular mechanism by which IFI16 exerts its activity remains unknown. Understanding the IFI16-mediated protein-protein interaction will provide insight into its mechanism of action and function.
Inactivation of the p53 tumor suppressor gene occurs in more than half of human cancers, and loss of p53 function is a fundamental step in the pathogenesis of most human cancers (Vogelstein and Kinzler, 1992). The p53 protein functions as a transcription factor and can transactivate cellular genes through sequence-specific DNA binding to their promoters (Prives and Manley, 2001). Although the physiological significance of IFI16 in p53 biology is still under investigation, it has been reported to regulate p53-mediated transcriptional activation (Johnstone et al., 2000; Fujiuchi et al., 2004; Xin et al., 2004). In addition, IFI16 also plays a role in p53-mediated transmission of apoptosis signaling in response to DNA damage, by upregulating p53 and downregulating MDM2 (Fujiuchi et al., 2004). Furthermore, tetracycline-regulated IFI16 also induced apoptosis in the presence of ionizing radiation when coexpressed with p53 in human bladder carcinoma p53-deficient EJ cells (Fujiuchi et al., 2004). Together, these studies suggest that IFI16 is involved in the regulation of p53 stability, p53-mediated transcriptional activation, and apoptosis.
In order to better understand the molecular details and mechanism of p53 regulation by IFI16, here, we present the crystal structures of the HIN-A and HIN-B domains of IFI16 determined at 2.0 and 2.35 Å resolution, respectively. These structures represent the first experimentally determined HIN domain structures reported in the HIN-200 protein family. Although they are structurally similar, HIN-A recognizes the C terminus of p53, whereas HIN-B binds to the DNA-binding region of p53, highlighting important functional differences between the two HIN domains. Furthermore, the two HIN domains of IFI16 may work cooperatively to stimulate the transcriptional activity of p53. Together, our structures provide a molecular description of the conserved HIN domain in this family, and offer insight into the mechanism by which IFI16 recognizes, binds, and potentially regulates p53.
RESULTS AND DISCUSSION
Overall Structures of IFI16 HIN-A and HIN-B Domains
The crystallized proteins of the first (HIN-A) and second (HIN-B) IFI16 HIN domains span residues 192–393 and 515–710, respectively. Data collection and refinement statistics are summarized in Table 1. As shown in Figure 1B, each HIN domain adopts an overall α/β fold that is further organized as two subdomains oriented in tandem. The first subdomain (residues 199–281 in HIN-A; residues 520–597 in HIN-B) is composed of eight β strands and one α helix, forming a globular barrel with the approximate dimensions 13 × 18 × 36 Å 3 for HIN-A and 14 × 18 × 35 Å 3 for HIN-B. Correspondingly, the second subdomain (residues 314–389 in HIN-A; residues 628–705 in HIN-B) consists of six β strands and one α helix, forming a second globular barrel with the approximate dimensions 11 × 19 × 21 Å 3 for HIN-A and 10 × 20 × 22 Å 3 for HIN-B. In both cases the two subdomains are connected by an extended interdomain linker (residues 282–313 in HIN-A; residues 598–627 in HIN-B) with two α helices in between for HIN-A, and three α helices for HIN-B.
Table 1.
Summary of Data Collection and Refinement Statistics
| HIN-A (Se-SAD) | HIN-B (Native) | |
|---|---|---|
| Data Collection | ||
| Space group | P21 | P212121 |
| Unit cell parameters | ||
| a, b, c (Å) | 43.3, 88.9, 112.8 | 43.0, 92.9, 100.3 |
| α, β, γ (°) | 90.0, 99.4, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength λ(Å) | 0.97918 | 1.0 |
| Rmerge (%) | 7.2 (38.9) | 6.6 (37.9) |
| I/σ (I) | 31.4 (2.7) | 26.4 (4.8) |
| Completeness (%) | 96.4 (85.7) | 99.9 (100) |
| Redundancy | 4.1 (3.6) | 7.2 (7.3) |
| Refinement | ||
| Resolution (Å) | 41.3–2.0 (2.07–2.00) | 42.1–2.35 (2.43–2.35) |
| Number of unique reflections | 55105 | 17421 |
| Rwork/Rfree (%) | 20.8/26.0 | 22.9/29.2 |
| Number of atoms | ||
| Protein | 6160 | 2938 |
| Water | 159 | 17 |
| Mean B factor (Å2) | 47.7 | 55.6 |
| Rms deviation from ideal geometry | ||
| Bond length (Å) | 0.011 | 0.016 |
| Bond angle (°) | 1.332 | 1.466 |
| PDB accession code | 2OQ0 | 3B6Y |
Values in parentheses are for the highest resolution shell.
Structural comparison of the individual IFI16 HIN-A or HIN-B subdomain with known folds using the DALI algorithm (Holm and Sander, 1993) revealed that each subdomain of HIN-A and HIN-B has high homology to the oligonucleotide/oligosaccharide binding (OB) fold found in many proteins (see Figure S1A available online), including replication protein A (RPA) (Bochkarev et al., 1997; Deng et al., 2007), single-stranded DNA-binding protein (SSB) (Kerr et al., 2003), aspartyl-tRNA-synthetase (AspRS) (Charron et al., 2003; Poterszman et al., 1994), BRCA2 (Yang et al., 2002), and telomere-binding protein α subunit (α-TEBP) (Classen et al., 2001; Theobald and Schultz, 2003; Buczek and Horvath, 2006), with Z-scores ranging from 7.1 to 11.3. This finding is consistent with prediction based on comparative sequence analysis that HIN domain contains two OB folds (Albrecht et al., 2005). A canonical OB fold consists of two three-stranded antiparallel β sheets packed orthogonally, forming a closed β-barrel in a 1-2-3-5-4-1 topology, in which the β1 strand is shared by both sheets (Theobald et al., 2003). Comparison of the IFI16 HIN subdomains with these well-characterized OB folds reveals significant variability in the length and arrangement of the β strands in forming the barrel, as well as the L12 and L45 loops, which are known to be important for recognizing and binding to oligonucleotide or peptide substrates (Theobald et al., 2003). For example the L12 and L45 loops of each HIN subdomain consist of less than five residues, whereas for other proteins these corresponding loops, especially L45, can be greater than 15 residues (e.g., AspRS, RPA70, α-TEBP). In addition, helical turn variations in the α helix connecting strands β3 and β4 are also observed among the HIN subdomains and other OB-fold proteins. This α helix is present in all four HIN subdomains and is another conserved feature of the OB-fold template. This helix packs against the open edge of the barrel and may be essential for structural integrity of the fold (Theobald et al., 2003; Kerr et al., 2003). In the case of SSB and α-TEBP, the OB fold of these two proteins uses a distorted helical turn to mimic the α helix and, thus, accomplish the same effect (Kerr et al., 2003). In total, over 50 structural homologs of IFI16 HIN subdomains were identified, reflecting the widespread occurrence of the OB fold. Surprisingly, when a DALI search was performed using the complete HIN-A or HIN-B domain (rather than the individual subdomain), the closest match was the structure of the DNA-bound form of RPA 70 kDa subunit (RPA70, PDB: 1JMC) (Bochkarev et al., 1997), which contains two consecutive OB folds but is only weakly superimposable with root-mean-square deviation (rmsd) of 5.8 Å (over 173 Cα atoms) and 5.6 Å (over 170 Cα atoms), respectively (Figure S1B). Such a large deviation is primarily seen in the size and conformation of various secondary structural elements and variable loops, as well as in the interdomain linker, which could possibly influence the subdomain/subdomain orientation. In fact the interdomain linker bridging the two OB folds of RPA70 is highly flexible and can adopt different conformations in the absence of DNA. Moreover, unlike the HIN domain of IFI16, the two OB folds of RPA70 do not come close enough to interact with each other without DNA (Bochkareva et al., 2001).
Despite a moderate sequence identity of ~40% between the HIN-A and HIN-B domains, their crystal structures can be superimposed readily with rmsd of 1.3 Å for all the main chain Cα atoms (Figure S1C). However, differences between the HIN-A and HIN-B domains are seen in various loop regions and the orientation of nearby secondary structural elements. For example the L12 loops in subdomain 1 and subdomain 2 have maximal shifts of 8.1 and 3.1 Å, respectively, when HIN-A and HIN-B are superimposed. Other substantial differences between HIN-A and HIN-B are observed in the conformations of the L45 and L3α loops in subdomain 2, with deviations of 2.0 and 2.9 Å, respectively.
Subdomain/Subdomain Interactions of IFI16 HIN Domains
For each HIN domain of IFI16, there appears to be extensive interactions at the interface between its two OB-fold subdomains. First, recombinant proteins comprising only one of the subdomains were insoluble (data not shown), suggesting that both subdomains may be required to provide proper structural stability for the HIN domain. Second, several key intramolecular interactions are evident between residues from each subdomain (Figure 2). The observation that various aromatic interactions, in addition to other noncovalent contacts, are present at the subdomain/subdomain interface of each HIN domain may explain why disrupting these interactions was detrimental to the overall fold of the structure. Furthermore, these aromatic residues are mostly conserved across the HIN-200 protein family, further suggesting that they are likely the key components of the HIN domain.
Figure 2. Subdomain/Subdomain Interactions of IFI16 HIN Domains.
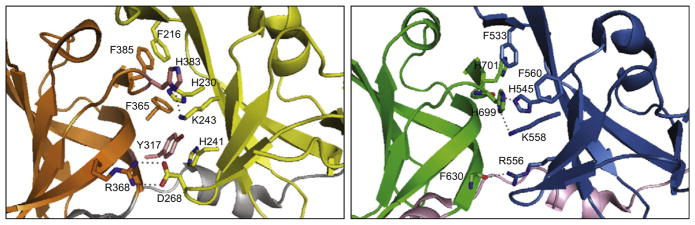
Close-up view of residues involved in subdomain/subdomain interaction for IFI16 HIN-A (left) and HIN-B (right).
To further evaluate if the interface formed by the OB-fold subdomains is interaction specific and not an artifact of crystal packing, the Protein Interfaces, Surfaces and Assemblies service PISA at European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) was utilized. As presented in Table 2, various interaction and thermodynamic parameters associated with interface analysis were acquired for HIN-A and HIN-B, and in comparison with that of other consecutive OB fold-containing proteins RPA70 (DNA bound; PDB: 1JMC; or unbound, PDB: 1FGU), α-TEBP (PDB: 2I0Q), and BRCA2 (PDB: 1MIU). The ΔiG P value is a measure of interface specificity. It indicates the probability of getting a smaller than obtained solvation free-energy gain ΔiG upon interface formation, if interface atoms were selected randomly from a protein surface. A P value of 0.5 implies that the formed interface is not unique, whereas P value >0.5 indicates that the interface is less hydrophobic than it could be and is likely to be an artifact of crystal packing. Finally, P value <0.5 signifies that the interface formed has surprising hydrophobicity, implying it is interaction specific (Krissinel and Henrick, 2007). Comparing these ΔiG P values, we can easily observe that the interface formed by the tandem OB-fold subdomains of HIN-A (P value 0.179) or HIN-B (P value 0.232) is the most interaction specific. Interestingly, unlike HIN-A or HIN-B, the interface formed by the OB-fold subdomains of RPA70 or α-TEBP appears likely to be the result of crystal packing because the ΔiG P value for each was above 0.5. Taken together, this suggests that the HIN domains of IFI16 may constitute a unique arrangement of closely interacting tandem OB folds that are stabilized by an extensive array of hydrogen bonding and aromatic interactions.
p53 C Terminus Binds to IFI16 HIN-A Domain
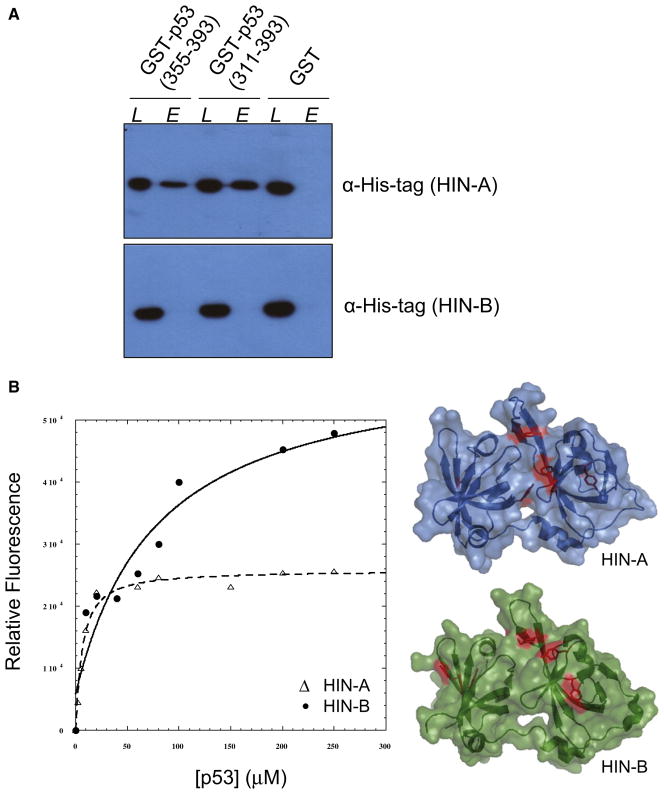
The role of IFI16 in p53-mediated transcriptional activation and apoptosis was first suggested based on its physical association with p53. Johnstone et al. (2000) mapped the interaction of the C terminus of p53 to a large region of IFI16 encompassing the first HIN-A domain (residues 155–476), but not the region containing the second HIN-B domain (residues 477–729). To evaluate whether the HIN-A domain itself is sufficient for interacting with p53, we performed a protein-protein interaction assay using recombinant His-tagged IFI16 HIN-A domain (residues 192–393) with GST-p53 C terminus (residues 355–393), GST-p53 C terminus with the tetramerization domain (residues 311–393), or GST alone. A similar assay was also conducted using His-tagged IFI16 HIN-B domain (residues 515–710). As shown in Figure 3A, the HIN-A domain was able to bind both constructs of p53 C terminus, indicating that residues outside of the structured HIN-A domain are not required for p53 interaction. Secondary structural analyses on these regions (residues 155–191 and 394–476) indicate that they are primarily unstructured (data not shown) and, thus, are not likely to participate in binding p53 C terminus, which also lacks secondary structural features. Despite having a similar fold to HIN-A, the HIN-B domain was not able to bind any of the p53 C terminus constructs.
Figure 3. Interaction of p53 C Terminus with IFI16 HIN-A or HIN-B Domain.
(A) An equimolar mixture of His-tagged IFI16 HIN-A or HIN-B domain and a GST fusion protein containing the indicated p53 construct was mixed with glutathione-sepharose (L). After washing, protein was eluted with glutathione and visualized by western (E) using antibody against His-tag.
(B) Increasing amounts of p53 (355–393) were incubated with IFI16 HIN-A (open triangle) or HIN-B (filled circle) domain, and binding was quantified by changes in tyrosine fluorescence. Tyrosine side chains are shown in red on the surface representation of HIN-A and HIN-B crystal structures.
To determine the strength of the interaction, we next measured quenching of the intrinsic tyrosine fluorescence of HIN-A or HIN-B domain upon addition of the p53 C terminus (residues 355–393), which does not possess any tyrosine residue. HIN-A domain contains five tyrosine residues (Y218, Y246, Y267, Y317, Y324), two of which (Y218, Y267) are surface exposed. Similarly, HIN-B domain contains five tyrosine residues (Y535, Y579, Y589, Y648, Y649) with three being surface exposed (Y535, Y589, Y649). Two of the three surface-exposed tyrosine residues of HIN-B (Y535, Y589) are situated near the same region on the first OB-fold subdomain as that of HIN-A. As demonstrated by the titration curve (Figure 3B), p53 C terminus binds IFI16 HIN-A domain with an apparent affinity of Kd ~20 μM. However, the same fragment of p53 C terminus did not show saturated binding to the HIN-B domain (Kd > 200 μM). This result further supports our findings that HIN-A is the binding site for the C terminus of p53, suggesting that distinct features present in HIN-A may be essential for p53 recognition.
IFI16 HIN-A Domain Enhances p53-DNA Complex Formation
The C terminus of p53 (residues 355–393) has been implicated in several important activities of p53, including autoregulation, protein-protein interactions, and nonspecific DNA binding (Hupp et al., 1992; Gu and Roeder, 1997; Anderson et al., 1997; Brazda et al., 2000; Ayed et al., 2001; Liu et al., 2003; Weinberg et al., 2004). It has been suggested that the binding of IFI16 to p53 C terminus may be important for p53 binding to DNA. This was initially shown by polyacrylamide electrophoretic mobility shift assays (EMSAs) in which the nuclear lysates of Mol-4 T cells, containing constitutively expressed IFI16 and p53, could bind p53-consensus oligonucleotides, and that binding was reduced in the presence of an IFI16 antibody (Johnstone et al., 2000). However, it was not clear whether IFI16 was associating with p53 directly.
Here, we performed EMSA using purified recombinant proteins of IFI16 HIN-A domain (residues 192–393) and p53 (residues 82–393) to assess whether the direct interaction between the two proteins can influence p53’s sequence-specific DNA-binding function. This p53 variant contains the central core sequence-specific DNA-binding domain, the tetramerization domain, and the C-terminal regulatory domain. As shown in Figure 4A, addition of IFI16 HIN-A protein was able to augment p53 (residues 82–393)’s binding to its consensus double-stranded DNA sequence 5′-GGACATGCCCGGGCATGTCC-3′, forming a stable protein-DNA complex in a dose-dependent manner (compare lane 6: 0 μM IFI16 to lane 10: 80 μM IFI16). Increasing amounts of IFI16 HIN-A protein by itself on the other hand, did not show any binding to the same DNA molecules (lanes 1–5).
Figure 4. EMSA of p53 (82–393) with p53-Consensus DNA Sequence in the Presence of IFI16 HIN-A Domain.
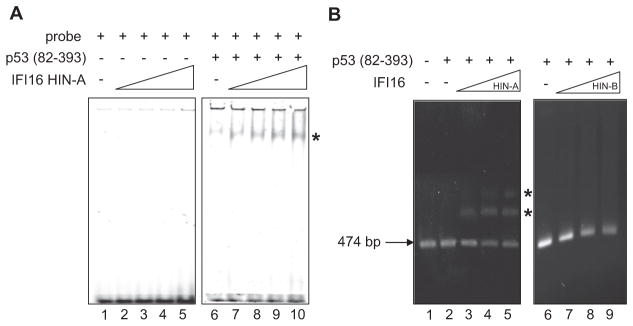
(A) Polyacrylamide EMSA reactions without p53 (lanes 1–5) or with constant amount (20 μM) of p53 (lanes 6–10), in the presence of increasing protein concentration (0, 10, 20, 40, 80 μM) of HIN-A.
(B) Agarose EMSA of constant amount (0.2 μM) of p53 in the presence of increasing protein concentration (0, 0.07, 0.2, 0.33 μM) of HIN-A (lanes 2–5) or HIN-B (lanes 6–9).
We also performed EMSA using agarose gel to determine the effect of IFI16 HIN-A domain on p53 (residues 82–393) binding to larger DNA molecules (474 base pairs [bp]) containing p53-consensus sequence (Figure 4B). In the absence of HIN-A, binding of p53 to its DNA target was not evident at low concentration (lane 2). However, in the presence of increasing amounts of HIN-A, p53 was able to bind to the probe, as indicated by the formation of retarded bands with corresponding disappearance of free DNA (lanes 3–5). Notably, a more slowly migrating band was also observed in lanes 4 and 5, indicative of higher molecular weight protein-DNA complexes. Using agarose EMSA, we also showed that IFI16 HIN-B domain, which does not interact with p53 C terminus, was not capable of activating p53 (residues 82–393)’s DNA-binding activity (Figure 4B, lanes 6–9). These results are consistent with previously reported data in which proteins that bind to the C terminus of p53 have been shown to promote stable formation of sequence-specific p53-consensus DNA complexes in EMSA experiments (Hupp et al., 1995; Anderson et al., 1997; Brazda et al., 2000; Sarkari et al., 2010). Thus, IFI16 shows biochemical features of other C-terminal p53-binding proteins and can influence p53’s DNA-binding properties.
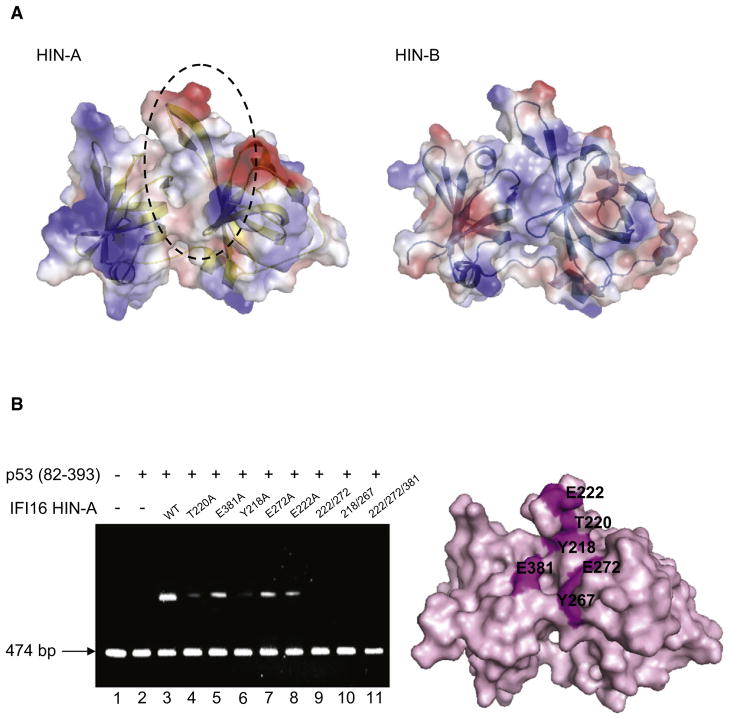
p53-Binding Surface of IFI16 HIN-A Domain
Given the overall structural similarities of HIN-A and HIN-B domains of IFI16, we investigated the structural features that are unique to HIN-A domain and mediate interaction with p53 C terminus. Comparing the electrostatic potential surfaces of the two HIN domains based on our crystal structures (Figure 5A), we hypothesized that the interaction with the basic C terminus of p53 may be mediated by an overt acidic/hydrophobic patch observed on the surface of HIN-A (formed by residues Y218, T220, E222, Y267, E272, E381), but not HIN-B domain of IFI16. This surface encompasses the two exposed tyrosine residues that presumably gave rise to the changes in fluorescence signal upon p53 binding (Figure 3B) and is in an area of the OB fold commonly found to interact with binding partners (Theobald et al., 2003). To further elucidate the molecular basis of p53 binding to HIN-A, we performed mutagenesis studies examining the roles of the aforementioned surface-exposed residues in mediating p53 interaction and activation (Figure 5B). Compared to activation of p53 (residues 82–393) by wild-type HIN-A domain (lane 3), T220A, E381A, Y218A, E272A, E222A single mutants (lanes 5–8) were less effective at activation of p53, especially T220A and Y218A. On the other hand, double mutants 222/272 (lane 9) and 218/267 (lane 10), and triple mutant 222/272/381 (lane 11) all abrogated p53 activation completely, suggesting that these residues contribute to the p53 C terminus-binding surface on HIN-A. Circular dichroism measurements indicated that all the mutants were folded and had levels of secondary structure similar to that of wild-type IFI16 (data not shown). Together these six residues form a cluster between strands β2 and β5′ of subdomain 1 and loop L5′5 of subdomain 2, creating the likely p53 C terminus-interacting surface.
Figure 5. p53 C Terminus-Binding Surface of IFI16 HIN-A Domain.
(A) Transparent electrostatic surface representation of IFI16 HIN-A (left) and HIN-B (right) crystal structures. White, red, and blue colors correspond to neutral, negatively, and positively charged surfaces, respectively.
(B) Agarose EMSA of p53 (82–393) with wild-type or mutant IFI16 HIN-A (left). Molar ratio of p53 to HIN-A is 1:3. Surface representation of IFI16 HIN-A (right). Residues involved in binding are colored in magenta.
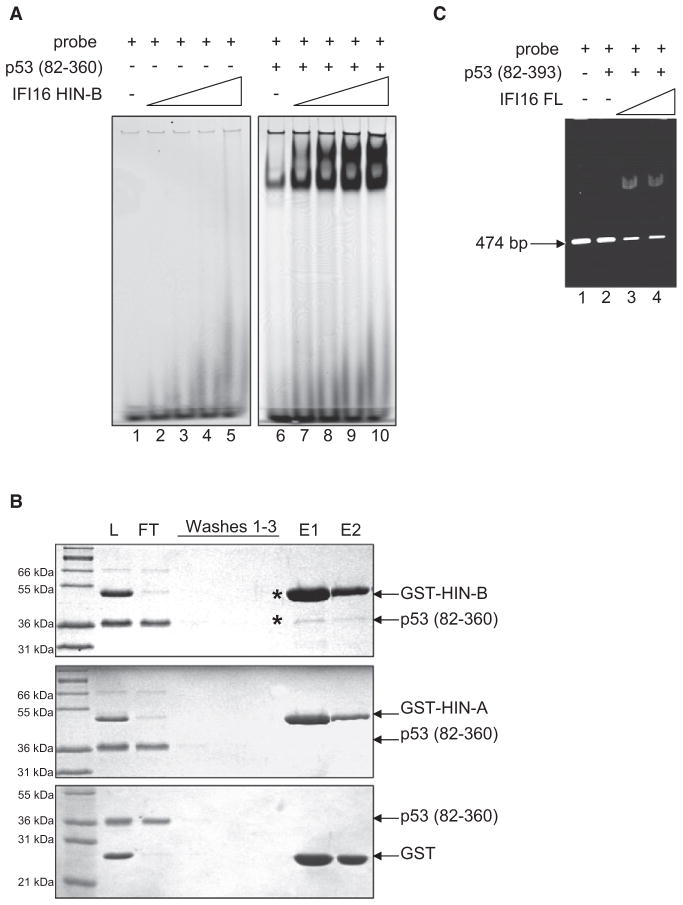
IFI16 HIN-B Domain Enhances p53-DNA Binding by Associating with p53 Core Domain
Although HIN-B domain of IFI16 does not interact with p53 C terminus, we turned our effort next to investigate if HIN-B domain could influence instead p53 (residues 82–360) binding to consensus DNA. Employing the polyacrylamide EMSA shown in Figure 6A, increasing amounts (0–80 μM) of HIN-B protein was observed to greatly enhance p53-DNA binding as well as forming higher molecular weight protein-DNA complexes. Next, we asked if HIN-B could associate with p53 (residues 82–360) directly using a protein-protein interaction assay. As shown in Figure 6B, p53 (residues 82–360) can interact weakly with GST-tagged HIN-B, but not GST-tagged HIN-A or GST alone. As presented above, IFI16 HIN-B domain does not interact with the C terminus of p53 (residues 311–393) (Figure 3A), nor does it bind significantly to the 474 bp DNA containing p53-consensus sequence (Figure 4B). Therefore, our data suggest that binding of IFI16 HIN-B domain to p53 may be mediated through the core domain (residues 82–310) of p53, especially when it is already bound to DNA.
Figure 6. IFI16 HIN-B Domain Enhances p53-DNA Binding by Associating with p53 Core Domain.
(A) Polyacrylamide EMSA reactions without p53 (82–360) (lanes 1–5) or with constant amount (2 μM) of p53 (lanes 6–10), in the presence of increasing protein concentration (0, 10, 20, 40, 80 μM) of HIN-B.
(B) GST-pull-down of p53 and HIN-B domain. An equimolar mixture of His-tagged p53 (82–360) and GST-tagged IFI16 HIN-B, GST-tagged IFI16 HIN-A, or GST protein alone was mixed with glutathione-sepharose (L). After washing (Washes 1–3), protein was eluted (E1, E2) with reduced glutathione and visualized by Coomassie blue staining.
(C) Agarose EMSA of constant amount (0.2 μM) of p53(82–393) in the presence of increasing protein concentration (0, 0.07, 0.2 μM) of full-length IFI16.
Cooperative Effect of Full-Length IFI16 on p53-DNA Binding
Having studied the effects of individual HIN-A and HIN-B domains of IFI16 on p53 sequence-specific DNA binding, it is tempting to ask if the presence of both HIN domains together enhances the activity cooperatively. To address such a question, we purified the 82 kDa full-length IFI16 (residues 1–729) and performed EMSA analysis with p53 (residues 82–393). As seen in Figure 6C, increasing amounts of full-length IFI16 were incubated with a constant amount of p53 and p53-consensus DNA probe. Notably, addition of lowest concentration (0.07 μM) of full-length IFI16 was able to better enhance p53-DNA binding (compare lane 3 of Figure 6C with lane 3 of Figure 4B). This suggests that the presence of both HIN-A and HIN-B domains in the full-length IFI16 protein may cooperatively elevate the effect of individual HIN domains on p53 binding to its consensus sequence. Although full-length IFI16 also possesses a conserved PYRIN domain at its N terminus, this domain was shown by Johnstone et al. (2000) not to interact with p53.
IFI16 Enhances p53-Mediated Transcription Activity
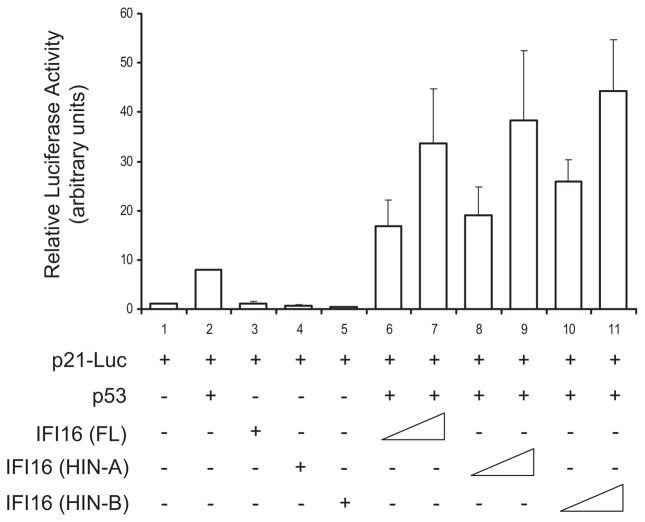
We next examined whether this IFI16-mediated enhancement of p53 sequence-specific DNA binding has an effect on p53 transcriptional activation. We performed luciferase reporter assays to examine the effect of IFI16 on p53-mediated p21 promoter activation. p21 is a functionally important p53-target gene involved in cell cycle control and transcriptional regulation. H1299, a human nonsmall cell lung carcinoma cell line, is used for this study because it is known not to express endogenous p53. In addition we have observed that H1299 cells also did not express endogenous IFI16 (Figure S2). The cells were cotransfected with p53 and/or IFI16 (full-length, HIN-A or HIN-B) together with a luciferase reporter gene containing the 20 bp 5′response element of p21. As shown in Figure 7, p53 alone, but not IFI16 (full-length, HIN-A or HIN-B), increased the relative luciferase signal by 8-fold. A greater than 15-fold dose-dependent increase in luciferase activity was observed when increasing amounts of full-length IFI16 were cotransfected with p53. Interestingly, when coexpressed with p53, the HIN-A or HIN-B domain of IFI16 was sufficient to promote p53-mediated luciferase activity in a dose-dependent manner, comparable to that of the full-length IFI16. This finding indicates that IFI16, via either HIN-A or HIN-B domain, is capable of upregulating p53-mediated transactivation function.
Figure 7. IFI16 Enhances p53-Mediated Transcription of the p21 Promoter.
H1299 cells were cotransfected with the luciferase reporter vector containing the full-length p21 promoter, p21-Luc, and with pcDNA3-p53 and/or pcDNA3-IFI116 (full-length [FL], HIN-A or HIN-B). The cells were lysed and assayed for luciferase activity 48 hr after transfection. Standard error was calculated using three independent experimental readings. See also Figure S2.
Full-Length IFI16 Model with p53
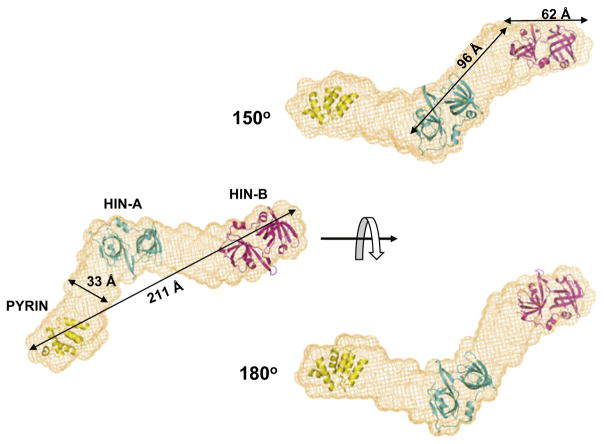
IFI16 is involved in transcriptional regulation by modulating protein-DNA and protein-protein interactions with transcription factors such as p53. Using small-angle X-ray scattering (SAXS), an averaged bead model/molecular envelope of the full-length IFI16 protein was derived (Figure 8), with a putative arrangement of its three folded domains, PYRIN (based on homology modeling of PYRIN domain of MNDA, PDB: 2DBG), HIN-A (PDB: 2OQ0), and HIN-B (PDB: 3B6Y) in the bead model. The model adopts a zigzag, elongated overall shape, which is in accordance with the noncompact conformation indicated by the Kratky plot (Figure S3B) and the relatively large Rg value 56 ± 2 Å (Figure S3A). The cross section throughout the bead model is approximately 30–50 Å. In addition the volume of the averaged bead model is 1.95 × 105 Å 3, which is in a good agreement with the Porod volume 1.8 ± 0.2 × 105 Å 3, estimated directly from the scattering profile. The three domains of IFI16 are oriented relatively independent of each other. In this arrangement, kinks also occur at the junctions between PYRIN/HIN-A and HIN-A/HIN-B at the nonstructured linkers. This model of full-length IFI16 is consistent with its potential function as a scaffold for multiple protein-protein interactions including p53, in which HIN-A and HIN-B domains of IFI16 interact with the C terminus and core domain of p53, respectively. HIN-A may relieve unproductive nonspecific DNA interactions of p53, or otherwise guide the C terminus to facilitate sequence-specific DNA binding by the core domain, whereas HIN-B is likely to interact with DNA-bound core domain stabilizing the p53-DNA complex.
Figure 8. Model of Full-Length IFI16.
An averaged bead model/molecular envelope of the full-length IFI16 calculated from SAXS data, with dimension measurements and putative arrangement of its three folded domains: PYRIN, HIN-A, and HIN-B. See also Figure S3.
Conclusion
IFI16 plays an important role in regulating cell proliferation and transcription through different protein-protein interactions. We have solved the crystal structures of the HIN-A and HIN-B domains of IFI16, representing the first experimentally determined HIN domain structures of the HIN-200 family. Despite having low-sequence identity to the OB folds commonly found in various proteins, both HIN domains of IFI16 possess tandem OB folds joined by an α-helical interdomain linker. It is worth noting that the majority of the OB-fold structures reported to date in the Protein Data Bank (PDB) are solitary. Thus, IFI16 joins mouse BRCA2 (PDB: 1MIU), Oxytricha nova α-TEBP (PDB: 2I0Q), and human RPA70 (PDB: 1JMC) to be the only proteins with structural information showing consecutive OB folds. Structural analysis of the human RPA70 has revealed a similar domain organization such that it has four domains (RPA70N, DBD-A, DBD-B, DBD-C) each with an OB fold. The N-terminal OB fold of RPA70 has a role in mediating protein-protein interaction with p53 (Bochkareva et al., 2005), whereas the remaining three OB folds all participate in single-stranded DNA binding (Bochkarev et al., 1997, 2000, 2001). Although the HIN domain of IFI16 constitutes a unique arrangement of tandem OB folds with specific interdomain interactions, it is possible that the four OB folds are also involved in a modular and concerted fashion for multiple substrate recognition analogous to those of RPA70.
The work presented here extends previous studies demonstrating the association of IFI16 HIN-A domain with the C terminus of p53 and further defines the mechanism by which IFI16 stimulates p53-mediated transcriptional activation. Although HIN-A and HIN-B domains of IFI16 are structurally similar and both modulate p53 sequence-specific DNA binding, they interact with p53 differently. IFI16 recognizes the basic p53 C terminus through an acidic/hydrophobic surface present on and unique to HIN-A domain, whereas HIN-B domain associates instead with core domain of p53. It is likely that HIN-A domain prevents p53 from nonspecific DNA interaction via its C terminus, whereas HIN-B domain stabilizes p53-DNA binding. Given its putative role as a scaffold molecule, full-length IFI16 is also likely involved in recruitment of additional factors to the p53-binding sites via its PYRIN domain. Indeed, the BRCA1 tumor suppressor protein is known to interact with IFI16 PYRIN domain and promote p53-dependent apoptosis (Aglipay et al., 2003). Interestingly, we and others have shown that p53 can also associate directly with BRCA1 (Zhang et al., 1998; Mark et al., 2005), thus suggesting intricate crosstalk among the three proteins and also adding another level of complexity in the function and regulation of IFI16.
EXPERIMENTAL PROCEDURES
Expression and Purification
IFI16 gene fragments coding for HIN-A domain (residues 192–393) and HIN-B domain (515–710) were amplified from human IFI16 cDNA (Mammalian Gene Collection, Structural Genomic Consortium) by PCR and subcloned into pET15b (Novagen) expression vector. Both IFI16 domains HIN-A and HIN-B proteins were expressed in E. coli BL21-CondonPlus cells (Strata-gene) with N-terminal His6-tag in selenomethionine (Se-Met)-containing media. Protein purification was performed by Co2+ affinity column (TALON).
Recombinant human p53 protein (residues 355–393, 311–393, 82–360, 82–393) was subcloned into pET15b (Novagen) or pGEX-2TK (Amersham) vector and expressed in and purified from E. coli BL21-pLysS cells (Stratagene) as previously described (Ayed et al., 2001; Mark et al., 2005).
Crystallization, Data Collection, and Structure Determination of IFI16 HIN-A
Crystals of the Se-Met-labeled IFI16 HIN-A (35 mg/ml) were obtained by the sitting drop vapor diffusion method by mixing the protein with an equal volume of reservoir solution containing 26% PEG 6000, 0.2 M MgCl2, and 0.1 M Tris-HCl at pH 8.5. The crystals belong to the mono-clinic space group P21 with unit cell parameters of a = 43.3 Å, b = 88.9 Å, c = 112.8 Å, and β = 99.4°. For data collection and structure determination details, please refer to Supplemental Experimental Procedures.
Crystallization, Data Collection, and Structure Determination of IFI16 HIN-B
Native crystals of IFI16 HIN-B (23 mg/ml) were obtained by the sitting drop vapor diffusion method by mixing the protein with an equal volume of reservoir solution containing 26% PEG 4000, 0.2 M NH4SO4, and 0.1 M sodium cacodylate buffer at pH 6.5. The crystals belong to the orthorhombic space group P212121 with unit cell parameters of a = 43.0 Å, b = 92.9 Å, and c = 100.3 Å with two molecules in the asymmetric unit. For data collection and structure determination details, please refer to Supplemental Experimental Procedures.
Protein-Protein Interaction Assays
Purified GST, GST-p53 (residues 355–393), or GST-p53 (residues 311–393) fusion protein was first mixed with His-tagged IFI16 (residues 192–393) or His-tagged IFI16 (residues 515–710) protein in assay buffer (20 mM HEPES [pH 7.6], 150 mM NaCl) and then incubated with the glutathione beads (Amersham) at 4°C for 2 hr. After extensive washing with assay buffer, bound proteins were eluted with 30 mM reduced glutathione and detected after SDS-PAGE by western analysis using antibody against His-tag (QIAGEN). Similar assays were conducted for purified GST, GST-HIN-A (residues 192–393), or GST-HIN-B (residues 515–710) fusion protein with His-tagged p53 protein (residues 82–360).
Intrinsic Tyrosine Fluorescence Assay
Direct binding of p53 C terminus (residues 355–393) to IFI16 HIN-A domain (residues 192–393) or HIN-B domain (residues 515–710) was characterized based on principles employed by Leclerc et al. (1993), Gupta et al. (2008), and Yan et al. (2008). A p53-binding titration was performed manually by mixing a solution of IFI16 protein (final concentration 1 μM) with increasing amounts of p53 (0–250 μM) in a buffer containing 25 mM Tris-HCl (pH 8.0) and 75 mM NaCl in a total volume of 150 μl at room temperature. After mixing, the intrinsic tyrosine fluorescence intensity (I) was measured using a time-based experimental module, which averages the fluorescence intensity from 30 readings with each reading for 1 s and sets the excitation wavelength at 278 nm, emission wavelength at 307 nm, and bandwidth at 4 nm. The quenched fluorescence (Iq) was calculated by Io−I, in which Io was the fluorescence intensity of IFI16 protein without p53. Background signals from the buffer and ligand were corrected. The dissociation constants (Kd) were determined by fitting the binding curve to Equation 1, using KaleidaGraph Version 3.0 (Abelbeck Software):
| (1) |
where Iq is the fluorescence intensity at a given total concentration of target ligand, [L]. [P] is the total concentration of protein in the reaction. Kd is the dissociation constant. IN is the fluorescence intensity at saturation, and Io is the fluorescence intensity in the absence of ligand.
Polyacrylamide EMSA
p53 (2–20 μM) was incubated with double-stranded DNA probe 5′-GGACATGCCCGGGCATGTCC-3′ (1–2 pmol) tagged with the monoreactive fluorescent reagent Cy5-Dye (Sigma) in EMSA buffer (20 mM Tris-HCl [pH 6.8], 150 mM NaCl, 10% [v/v] glycerol, 5 mM DTT) in the presence of IFI16 HIN-A or HIN-B protein (at final concentrations of 0–80 μM). Complexes were resolved by electrophoresis at 100 V at room temperature using a pre-run 5% polyacrylamide gel containing Tris-Borate-EDTA buffer and then visualized by red fluorescence emission measured at 635 nm using a STORM860 scanner (Molecular Dynamics).
Agarose EMSA
DNA (600 ng of PvuII fragments of pPGM2) (Brazda et al., 2006; Jagelska et al., 2008), p53, and IFI16 were mixed at various ratios in 20 μl of the DNA-binding buffer (5 mM Tris-HCl [pH 7.0], 1 mM EDTA, 50 mM KCl, and 0.01% Triton X-100). The samples were incubated for 10 min at 4°C and loaded onto a 1% agarose gel (SERVA) containing 0.33× Tris-Borate-EDTA buffer. Agarose electrophoresis was performed for 4 hr at 100 V at 4°C. The gels were stained with ethidium bromide and photographed.
Cell Culture and Luciferase Assay
Human nonsmall cell lung carcinoma cell line H1299 was obtained from the Benchimol laboratory and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). pcDNA3-p53 and the luciferase reporter construct carrying the p21 promoter (p21-Luc) were also obtained from the Benchimol laboratory. An amplified fragment of full-length IFI16, HIN-A, or HIN-B from human IFI16 cDNA (Mammalian Gene Collection, Structural Genomic Consortium) was subcloned into pcDNA3 (Invitrogen). H1299 cells at ~80% confluence in a 6-well plate were transfected using Lipofectamine 2000 (Invitrogen). At 48 hr after transfection, luciferase assay was performed using the Luciferase Assay Kit (Promega) and measured using the SpectraMax M5 luminometer (Molecular Devices). Standard error was calculated using three independent experimental readings.
SAXS Experiments, Data Analysis, and Bead Model Reconstruction
Both SAX and wide-angle X-ray scattering (WAXS) were performed at beam-line 12-ID of the Advanced Photon Sources (APS) at the Argonne National Laboratory. A detailed description about experimental procedure, processing and analysis of scattering data, and bead model reconstruction can be found in Supplemental Experimental Procedures.
Supplementary Material
Table 2.
Summary of PISA Analysis of OB-Fold Subdomain/Subdomain Interaction
| Subdomain 1 | Subdomain 2 | Interface | ΔiG | ΔiG | NHB | NSB | NDS | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Nat | Nres | Nat | Nres | area (Å2) | kcal/mol | P Value | ||||
| HIN-A: 2OQ0 | 65 | 20 | 53 | 11 | 567.4 | −8.8 | 0.179 | 4 | 4 | 0 |
| HIN-B: 3B6Y | 49 | 15 | 45 | 11 | 484.4 | −5.2 | 0.232 | 3 | 0 | 0 |
| RPA70A: 1JMC | 34 | 12 | 25 | 11 | 278.9 | 0.1 | 0.650 | 5 | 2 | 0 |
| RPA70A: 1FGU | 6 | 1 | 7 | 2 | 52.5 | −0.2 | 0.576 | 0 | 0 | 0 |
| TEBP: 2I0Q | 85 | 19 | 75 | 17 | 689.0 | −4.5 | 0.556 | 12 | 5 | 0 |
| BRCA2: 1MIU | 48 | 14 | 37 | 10 | 394.1 | −4.5 | 0.415 | 3 | 1 | 0 |
Nat, number of interfacing atoms in the corresponding subdomain; Nres, number of interfacing residues in the corresponding subdomain; NHB, number of hydrogen bonds across the interface; NSB, number of salt bridges across the interface; NDS, number of disulfide bonds across the interface.
Acknowledgments
We thank the Benchimol laboratory for the H1299 cells and p21-Luc reporter plasmid. We also thank Yi Sheng and Rob Laister for critical revision of the manuscript. This work was supported by grants from Canadian Cancer Society (to C.H.A.), the Ontario Ministry of Health and Long Term Care, the US National Institute of Health Protein Structure Initiative (P50-GM62413-01 and GM67965) through the Northeast Structural Genomics Consortium, a Canadian Institute of Health Research (CIHR) Canada Graduate Scholarship (2005–2008), and Terry Fox Foundation Studentship (2008–2010) (to J.C.C.L.). V.B. is supported by the ASCR (Grants M200040904, AV0Z50040507, and AV0Z50040702) and by the GACR (301/10/1211). The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the CIHR, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust. C.H.A. holds a Canada Research Chair in Structural Proteomics.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors for the reported crystal structures have been deposited in the PDB under the accession codes 2OQ0 and 3B6Y.
Supplemental Information includes Supplemental Experimental Procedures and three figures and can be found with this article online at doi:10.1016/j. str.2010.12.015.
References
- Aglipay JA, Lee SW, Okada S, Fujiuchi N, Ohtsuka T, Kwak JC, Wang Y, Johnstone RW, Deng C, Qin J, Ouchi T. A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene. 2003;22:8931–8938. doi: 10.1038/sj.onc.1207057. [DOI] [PubMed] [Google Scholar]
- Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–687. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- Anderson ME, Woelker B, Reed M, Wang P, Tegtmeyer P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol Cell Biol. 1997;17:6255–6264. doi: 10.1128/mcb.17.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Korolev S, Bochkarev A. The role for zinc in replication protein A. J Biol Chem. 2000;275:27332–27338. doi: 10.1074/jbc.M000620200. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Belegu V, Korolev S, Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, Bochkarev A. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazda V, Palecek J, Pospisilova S, Vojtesek B, Palecek E. Specific modulation of p53 binding to consensus sequence within supercoiled DNA by monoclonal antibodies. Biochem Biophys Res Commun. 2000;267:934–939. doi: 10.1006/bbrc.1999.2056. [DOI] [PubMed] [Google Scholar]
- Brazda V, Jagelska EB, Fojta M, Palecek E. Searching for target sequences by p53 protein is influenced by DNA length. Biochem Biophys Res Commun. 2006;341:470–477. doi: 10.1016/j.bbrc.2005.12.202. [DOI] [PubMed] [Google Scholar]
- Buczek P, Horvath MP. Structural reorganization and the cooperative binding of single-stranded telomere DNA in Sterkiella nova. J Biol Chem. 2006;281:40124–40134. doi: 10.1074/jbc.M607749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron C, Roy H, Blaise M, Giege R, Kern D. Non-discriminating and discriminating aspartyl-tRNA synthetases differ in the anticodon-binding domain. EMBO J. 2003;22:1632–1643. doi: 10.1093/emboj/cdg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D. P202: an interferon-inducible negative regulator of cell growth. J Biol Regul Homeost Agents. 2000;14:187–192. [PubMed] [Google Scholar]
- Classen S, Ruggles JA, Schultz SC. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J Mol Biol. 2001;314:1113–1125. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- Dawson MJ, Trapani JA. HIN-200: a novel family of IFN-inducible nuclear proteins expressed in leukocytes. J Leukoc Biol. 1996;60:310–316. doi: 10.1002/jlb.60.3.310. [DOI] [PubMed] [Google Scholar]
- Deng X, Habel JE, Kabaleeswaran V, Snell EH, Wold MS, Borgstahl GE. Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation. J Mol Biol. 2007;374:865–876. doi: 10.1016/j.jmb.2007.09.074. [DOI] [PubMed] [Google Scholar]
- Ding Y, Lee JF, Lu H, Lee MH, Yan DH. Interferon-inducible protein IFIXalpha1 functions as a negative regulator of HDM2. Mol Cell Biol. 2006;26:1979–1996. doi: 10.1128/MCB.26.5.1979-1996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiuchi N, Aglipay JA, Ohtsuka T, Maehara N, Sahin F, Su GH, Lee SW, Ouchi T. Requirement of IFI16 for the maximal activation of p53 induced by ionizing radiation. J Biol Chem. 2004;279:20339–20344. doi: 10.1074/jbc.M400344200. [DOI] [PubMed] [Google Scholar]
- Gariglio M, Azzimonti B, Pagano M, Palestro G, De Andrea M, Valente G, Voglino G, Navino L, Landolfo S. Immunohistochemical expression analysis of the human interferon-inducible gene IFI16, a member of the HIN200 family, not restricted to hematopoietic cells. J Interferon Cytokine Res. 2002;22:815–821. doi: 10.1089/107999002320271413. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Gupta VB, Indi SS, Rao KSJ. Studies on the role of amino acid stereospecificity in amyloid beta aggregation. J Mol Neurosci. 2008;34:35–43. doi: 10.1007/s12031-007-0070-z. [DOI] [PubMed] [Google Scholar]
- Gutterman JU. Cytokine therapeutics: lessons from interferon alpha. Proc Natl Acad Sci USA. 1994;91:1198–1205. doi: 10.1073/pnas.91.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller S, Kohl A, Fiorito F, Herrmann T, Wider G, Tschopp J, Grutter MG, Wuthrich K. NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure. 2003;11:1199–1205. doi: 10.1016/j.str.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Sparks A, Lane DP. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- Jagelska EB, Brazda V, Pecinka P, Palecek E, Fojta M. DNA topology influences p53 sequence-specific DNA binding through structural transitions within the target sites. Biochem J. 2008;412:57–63. doi: 10.1042/BJ20071648. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Wei W, Greenway A, Trapani JA. Functional interaction between p53 and the interferon-inducible nucleoprotein IFI 16. Oncogene. 2000;19:6033–6042. doi: 10.1038/sj.onc.1204005. [DOI] [PubMed] [Google Scholar]
- Kerr ID, Wadsworth RI, Cubeddu L, Blankenfeldt W, Naismith JH, White MF. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul D, Obeyesekere NU, Gutterman JU, Mills GB, Choubey D. p202 self-associates through a sequence conserved among the members of the 200-family proteins. FEBS Lett. 1998;438:21–24. doi: 10.1016/s0014-5793(98)01263-0. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Leclerc E, Leclerc L, Marden MC. Asymmetry of calmodulin revealed by peptide binding. J Fluoresc. 1993;3:45–49. doi: 10.1007/BF00865288. [DOI] [PubMed] [Google Scholar]
- Liu G, Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J Biol Chem. 2003;278:17557–17565. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
- Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Mark WY, Liao JCC, Lu Y, Ayed A, Laister R, Szymczyna B, Chakrabartty A, Arrowsmith CH. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein-protein and protein-DNA interactions? J Mol Biol. 2005;345:275–287. doi: 10.1016/j.jmb.2004.10.045. [DOI] [PubMed] [Google Scholar]
- Poterszman A, Delarue M, Thierry JC, Moras D. Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase. J Mol Biol. 1994;244:158–167. doi: 10.1006/jmbi.1994.1716. [DOI] [PubMed] [Google Scholar]
- Prives C, Manley JL. Why is p53 acetylated? Cell. 2001;107:815–818. doi: 10.1016/s0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- Raffaella R, Gioia D, De Andrea M, Cappello P, Giovarelli M, Marconi P, Manservigi R, Gariglio M, Landolfo S. The interferon-inducible IFI16 gene inhibits tube morphogenesis and proliferation of primary, but not HPV16 E6/E7-immortalized human endothelial cells. Exp Cell Res. 2004;293:331–345. doi: 10.1016/j.yexcr.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Sarkari F, Sheng Y, Frappier L. USP7/HAUSP promotes the sequence-specific DNA binding activity of p53. PLoS ONE. 2010;5:e13040. doi: 10.1371/journal.pone.0013040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Stehlik C, Reed JC. The PYRIN connection: novel players in innate immunity and inflammation. J Exp Med. 2004;200:551–558. doi: 10.1084/jem.20032234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani JA, Browne KA, Dawson MJ, Ramsay RG, Eddy RL, Show TB, White PC, Dupont B. A novel gene constitutively expressed in human lymphoid cells is inducible with interferon-gamma in myeloid cells. Immunogenetics. 1992;36:369–376. doi: 10.1007/BF00218044. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. p53 function and dysfunction. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- Wei W, Clarke CJ, Somers GR, Cresswell KS, Loveland KA, Trapani JA, Johnstone RW. Expression of IFI 16 in epithelial cells and lymphoid tissues. Histochem Cell Biol. 2003;119:45–54. doi: 10.1007/s00418-002-0485-0. [DOI] [PubMed] [Google Scholar]
- Weinberg RL, Freund SM, Veprintsev DB, Bycroft M, Fersht AR. J Mol Biol. 2004;342:801–811. doi: 10.1016/j.jmb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- Xin H, Pereira-Smith OM, Choubey D. Role of IFI 16 in cellular senescence of human fibroblasts. Oncogene. 2004;23:6209–6217. doi: 10.1038/sj.onc.1207836. [DOI] [PubMed] [Google Scholar]
- Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22:4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- Yan H, Dalal K, Hon BK, Youkharibache P, Lau D, Pio F. RPA nucleic acid-binding properties of IFI16-HIN200. Biochim Biophys Acta. 2008;1784:1087–1097. doi: 10.1016/j.bbapap.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Zhang H, Somasundaram K, Peng Y, Tian H, Bi D, Weber BL, El-Deiry WS. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.