Summary
MicroRNAs (miRNAs), small non-coding RNA molecules that post-transcriptionally regulate gene expression, are known to play key roles in regulating immune responses and autoimmunity. We investigated miR-146a expression in Sjögren's syndrome (SjS) patients as well as in the SjS-prone C57BL/6.NOD-Aec1Aec2 mouse model, to elucidate its involvement in SjS pathogenesis. Expression of miR-146a was examined in the peripheral blood mononuclear cells (PBMCs) of 25 SjS patients and 10 healthy donors, as well as in PBMCs, salivary and lacrimal glands in SjS-prone mice and wild-type C57BL/6J mice. Functional assays using THP-1 human monocytes were conducted to determine the biological roles of miR-146a in innate immunity. miR-146a expression was significantly increased in SjS patients compared to healthy controls, and was upregulated in the salivary glands and PBMCs of the SjS-prone mouse at both 8 weeks (prior to disease onset) and 20 weeks (full blown disease) of age. More importantly, functional analysis revealed roles for miR-146a in increasing phagocytic activity and suppressing inflammatory cytokine production while migration, nitric oxide production, and expression of antigen presenting/costimulatory molecules are not affected. Taken together, our data suggest that abnormal expression/regulation of miRNA in innate immunity may contribute to or be indicative of the initiation and progression of SjS.
Keywords: microRNA, Sjögren's syndrome, phagocytosis, innate immunity
Introduction
Sjögren's syndrome (SjS) is common systemic autoimmune disease mainly affecting the salivary and lacrimal glands resulting in dry mouth and dry eye, respectively. Despite extensive studies into the mechanisms which contribute to the development or pathogenesis of SjS, the events that trigger disease onset in the target exocrine glands remain unknown. Our previous studies examining the salivary glands of the non-obese diabetic (NOD) and more recently, the C57BL/6.NOD-Aec1Aec2 mouse models of SjS indicate alterations in the glandular environment even prior to disease onset, including apoptosis of acinar tissues, increase in caspase-1 activity, and altered cell proliferation [1-5].
It is becoming increasingly clear that epigenetic gene regulation may play an important role in a number of diseases, including autoimmune disorders. One example of epigenetic regulation of gene expression is small non-coding RNAs, including microRNAs (miRNAs), which are 18-22 nucleotides long and negatively regulate gene expression at the post-transcriptional level by binding to the 3′ untranslated region (UTR) of specific messenger RNAs (mRNAs) [6]. It is now known that miRNA regulation is critical for a variety of cellular processes such as apoptosis, differentiation, immune cell development and immune responses. Recent publications underscore the role of miRNAs in the regulation of innate immune responses in monocytes and macrophages [7-9]. Up-regulated miRNAs were identified in a monocytic cell line treated with the Toll-like receptor (TLR)-4 ligand LPS, specifically, miR-146a, miR-155, and miR-132 [9]. Transcription of miR-146a was shown to be regulated by NF-κB, and its target genes include IL-1 receptor associated kinase (IRAK-1) and TNF receptor-associated factor-6 (TRAF-6) [9]. Interestingly, these two genes were upregulated in the salivary glands of SjS-prone C57BL/6.NOD-Aec1Aec2 mice prior to disease onset, detected by microarray in our previous study [10]. Overall, miR-146a appears to function as the effector arm of a negative feedback mechanism regulating TLR signaling, suggesting its expression may be critical in preventing excess inflammation [11].
Aberrant miR-146a expression has been demonstrated in several immune-mediated diseases including psoriasis [12], rheumatoid arthritis (RA) and systemic lupus erythematosus patients (SLE). Two studies examined miRNA expression in RA synovial tissue and fibroblasts demonstrating increased miR-146a and miR-155 expression in RA synovial fibroblasts compared to those in osteoarthritis patients [13] and increased miR-146a expression in RA synovial tissue compared to that of osteoarthritis patients and normal controls [14]. Our group examined miRNA expression in the peripheral blood mononuclear cells (PBMCs) of RA patients and controls and demonstrated that miR-146a, miR-155, miR-132, and miR-16 were significantly upregulated in RA patients compared to controls, and that increased miR-146a and miR-16 expression correlated with disease activity [15]. In contrast, miR-146a was found to be underexpressed in SLE patients, and this underexpression negatively correlated with clinical disease activity [16]. Notably, miR-146a was also demonstrated to negatively regulate type I interferon induction in PBMCs [16].
Based on the emerging evidence for the role of miRNAs in autoimmune diseases, the recently dissected role of miRNAs in regulating innate immune signaling [8, 9], and elevated target genes of miRNA involving innate immunity in our SjS-prone mouse model prior to disease onset [10], we initiated our study to identify if abnormal miRNA expression/regulation would be present in a mouse model of SjS and patients with autoimmune SjS and what roles, if any, aberrant miRNA expression may play in SjS pathogenesis.
Results
miR-146a and miR-155 expression is increased in SjS patients compared to healthy controls
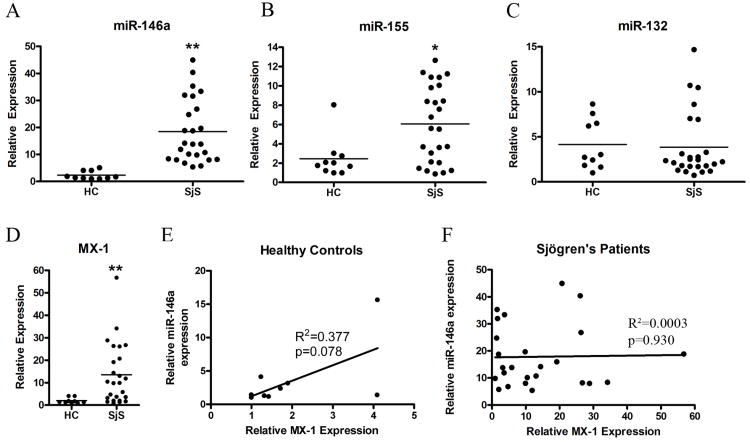
We began our study by examining miR-146a, miR-155, and miR-132 expression in PBMC samples from 25 SjS patients (Table 1) and 10 healthy controls as described in Methods. These miRNA were chosen based on previous studies demonstrating their differential expression in RA and SLE and their induction during innate immune responses [13-18]. As shown in Figure 1A, the average relative expression level of miR-146a was 8.0-fold higher for SjS patients than for healthy controls (p<0.0001 as determined by Mann Whitney test). miR-155 expression was 2.5-fold higher in SjS patients (Figure 1B, p<0.05), while miR-132 expression was similar between patients and controls (Figure 1C, p=0.55).
Table 1. Demographic and clinical data for patients.
| Subject | Sex | Age (years) | Diagnosis | Medications | SSA | SSB | Biopsy/Focus Score | Salivary Flow (ml/min) |
|---|---|---|---|---|---|---|---|---|
| SjS-1 | F | 65 | 1° SjS | Cevimeline | neg | neg | pos, 7 | 0.293 |
| SjS-2 | F | 65 | 1 °SjS | Cevimeline; Cyclosporine ophthalmic | neg | neg | pos, 4 | 0.035 |
| SjS-4 | F | 64 | 2° SjS, PBC | Cevimeline | neg | neg | pos, 12 | 0.065 |
| SjS-5 | F | 61 | 1° SjS | None | pos | pos | NA | 0.013 |
| SjS-6 | F | 72 | 1° SjS | Hydroxychloroquine; Cevimeline; Cyclosporine ophthalmic | pos | neg | pos | NA |
| SjS-7 | F | 22 | 1° SjS | Hydroxychloroquine | pos | pos | pos, 5 | 0.67 |
| SjS-8 | F | 58 | 1° SjS | Nabumetone; Cyclosporine ophthalmic | pos | neg | NA | 0.037 |
| SjS-9 | F | 77 | 1° SjS | None | pos | NA | NA | 0.03 |
| SjS-10 | F | 68 | 1° SjS | Cyclosporine opthalmic; Hydroxychloroquine | pos | neg | pos, 3 | 0.071 |
| SjS-11 | F | 59 | 1° SjS/nephritis | Cevimeline; Prednisone | pos | neg | pos, 9 | 0.001 |
| SjS-12 | F | 76 | 1° SjS | Hydroxychloroquine | pos | pos | pos, 2 | 0.045 |
| SjS-13 | F | 46 | 1° SjS | None | pos | neg | pos, 2 | 0.164 |
| SjS-14 | F | 47 | 1° SjS | Hydroxychloroquine | neg | neg | pos, 3 | 0.455 |
| SjS-15 | F | 65 | 1° SjS | Prednisone; Hydroxychloroquine; Methotrexate | pos | neg | NA | 0.011 |
| SjS-16 | F | 78 | 1° SjS | Hydroxychloroquine | pos | pos | pos, 5 | 0.042 |
| SjS-17 | F | 64 | 1° SjS | Hydroxychloroquine | pos | pos | pos, 9 | 0.002 |
| SjS-18 | F | 61 | 1° SjS | Hydroxychloroquine | pos | neg | NA | 0.385 |
| SjS-19 | F | 31 | 1° SjS/nephritis | Hydroxychloroquine; Mycophenolate motefil | pos | pos | NA | NA |
| SjS-21 | F | 53 | 1° SjS | Cevimeline; Hydroxychloroquine; Cyclosporine ophthalmic | pos | pos | NA | NA |
| SjS-22 | F | 72 | 1° SjS | None | pos | neg | NA | NA |
| SjS-24 | F | 65 | 1° SjS | None | pos | neg | NA | NA |
| SjS-25 | F | 57 | 1° SjS | Cyclosporine ophthalmic | pos | neg | NA | NA |
| SjS-26 | F | 32 | 1° SjS | Hydroxychloroquine; Cyclosporine ophthalmic | pos | neg | pos, 3 | 0.06 |
| SjS-27 | F | 60 | 1° SjS | None | pos | pos | NA | 0.012 |
| SjS-28 | F | 56 | 1° SjS | None | pos | pos | pos | 0.11 |
SjS, Sjögren's syndrome; PBC, primary biliary cirrhosis; NA, no data available; SSA, anti-Ro antibodies; SSB, anti-La antibodies
Figure 1. SjS patients exhibit increased miR-146a and miR-155 expression in PBMCs compared to healthy controls.
PBMC total RNA was isolated from healthy control individuals (n=10) and SjS-patients (n=25) and relative expression levels of A) miR-146a, B) miR-155 and C) miR-132 were analyzed by qRT-PCR using U44 RNA as an internal control. D) MX-1 gene expression was analyzed by qRT-PCR as a measure of the IFN signature in SjS patients compared to healthy controls. Bars indicate mean, asterisks (**) indicate p<0.0001, (*) p<0.05 as determined by Mann Whitney test for panels A-D. E) Relative expression of miR-146a versus MX-1 was plotted for healthy controls and SjS patients (F) to determine the presence of any correlations. No significant correlation was determined by linear regression with p=0.377 for healthy controls and p=0.930 for SjS patients.
Clinical and demographic information for SjS patients was examined for any correlations between increased miRNA expression and medications, SSA/SSB autoantibody reactivity, salivary flow rate, or salivary gland biopsy focal score (Table 1, Supplemental Figure 1). No significant correlations were found between miR-146a/155 expression and any of these parameters (Supplemental Figure 1). Although not statistically significant, there was a tendency for patients with upregulated miR-155 to be positive for anti-SSA, while those with lower miR-155 expression were SSA negative. However, larger patient cohorts need to be examined to determine if there is a true correlation.
The majority of SjS patients included in this study were being treated with secretagogues or immunosuppressant/anti-inflammatory medications at the time of sample collection. However, there were no clear correlations between these medications and miRNA expression.
The type 1 Interferon (IFN) signature of the SjS patients was assessed by examining the expression of IFN-inducible gene MX-1 (Figure 1D). As expected, MX-1 expression was significantly upregulated in SjS patients compared to healthy controls (p=0.002 as determined by Mann Whitney test). However, there was no correlation between MX-1 expression and miR-146a expression in healthy controls or SjS patients (Figure 1E and F).
To test if PBMC miR-146a expression correlated with target tissue miR-146a expression in SjS patients, in situ hybridization was used to examine miR-146a expression in minor salivary gland biopsies from 8 SjS patients and 4 non-SjS controls. As shown in Supplemental Figure 2, PBMC miR-146a expression does correlate with salivary gland miR-146a expression in SjS patients. Two representative patients are shown in Supplemental Figure 2A. miR-146a expression was blindly scored on a scale from 0-5, with 0 set as the absence of detectable expression and 5 set as expression intensity equal to U6 expression (Supplemental Figure 2B). Similarly to PBMC expression, miR-146a was significantly upregulated in SjS biopsy specimens compared to non-SjS controls (p=0.02 as determined by Mann Whitney test).
SjS-prone mouse model exhibits elevated miR-146a expression in salivary glands and PBMCs
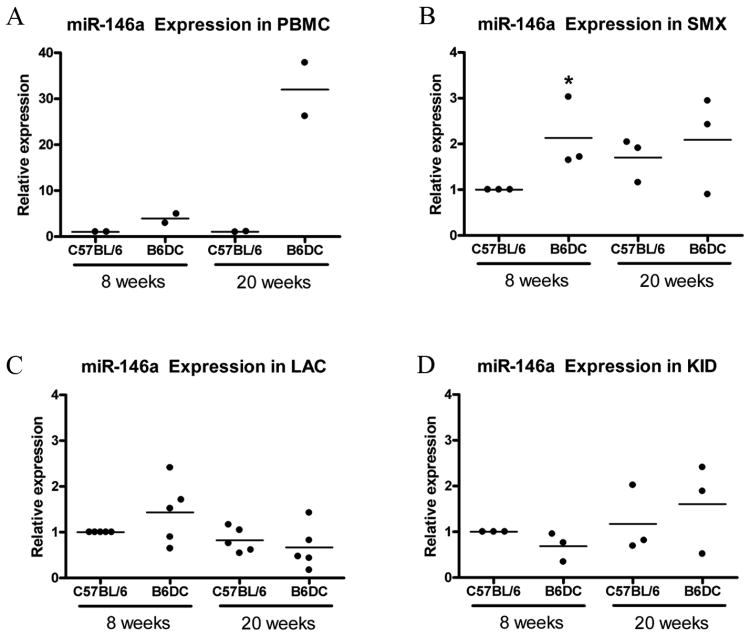
There are currently numerous SjS mouse models established, but none completely resemble human disease [19]. However, our model, the C57BL/6.NOD-Aec1Aec2 has distinct genetic advantages over other NOD derivatives while maintaining a SjS-like phenotype in the absence of the type 1 diabetic phenotype. To further investigate miR-146a expression in SjS-like disease in SjS-prone mouse model, miR-146a expression in the submandibular glands (SMX), lacrimal glands (LAC), PBMCs, and kidneys (KID) of female SjS-prone mice C57BL/6.NOD-Aec1Aec2 (B6DC) was examined. The B6DC mouse is a recently generated congenic strain with two genetic intervals derived from the NOD mouse on a C57BL/6J background manifesting primary SjS-like disease [2, 5, 10]. PBMCs were collected as described in Methods by pooling whole blood from 3-4 mice per group. Tissues were harvested from 8 week and 20 week old mice which enabled us to examine miRNA expression prior to disease onset and during advanced disease, respectively [2, 10].
As shown in Figure 2A, miR-146a in PBMCs was increased at 8 weeks (3.7 fold) in B6DC as well as at 20 weeks (30-fold), which is consistent with patient data. miR-146a expression was upregulated in SMX of 8 week old B6DC compared to C57BL/6 mice (2.1 fold, p<0.05 as determined by Mann Whitney). In 20 week old mice, miR-146a expression was similar between B6DC C57BL/6 mice (Figure 2B). Since SjS affects both salivary and lacrimal glands, miR-146a expression in LAC was also investigated. Our finding indicates that miR-146a expression in LAC was slightly altered in B6DC mice compared to wild-type mice, but these differences were not statistically significant (Figure 2C). This difference in miRNA expression between SMX and LAC is likely due to the differences in disease pathogenesis that occur in SMX versus LAC [2]. Kidney samples were also analyzed as a non-target tissue control, and as expected, miR-146a expression was similar between SjS-prone and wild-type mice (Figure 2D).
Figure 2. miR-146a expression is increased in SjS-prone mouse SMX and PBMCs.
qRT-PCR was used to examine miR-146a expression in SjS-prone (B6DC) and wild-type (C57BL/6) mice at 8 and 20 weeks old. A) PBMC (n=2 replicates of pooled sample), B) Submandibular gland (SMX, n=6), C) lacrimal gland (LAC, n=10), and D) kidney (KID, n=6) were analyzed. Data are presented as relative expression after normalization to U44 RNA and C57BL/6 8 week samples. Asterisks (*) indicate p<0.05 compared to age-matched wild-type control as determined by Mann Whitney, bars represent mean.
miR-155 and miR-132 expression was also examined in the SMX, LAC, KID, and PBMC of B6DC and C57BL/6 mice (Supplemental Figure 2). Interestingly, miR-155 was found to be significantly downregulated in 20 week B6DC SMX compared to C57BL/6 (p<0.05 as determined by Mann Whitney), and miR-132 was significantly upregulated in 8 week B6DC SMX compared to C57BL/6 (p<0.05 as determined by Mann Whitney, Supplementary Figure 2A). Similar to miR-146a, miR-155 and miR-132 expression was dramatically increased in 20 week B6DC PBMC compared to C57BL/6 (Supplementary Figure 2D). No significant differences were observed in miR-155 or miR-132 expression in LAC or KID (Supplementary Figure 2B and C).
These data may indicate that since upregulated miR-146a is detectable in SjS-prone mice prior to disease onset, alterations in miRNA expression in humans may be present even prior to disease onset in target tissues and PBMCs and could serve as biomarkers for SjS-prone individuals.
miR-146a has no significant effect on THP-1 cell migration or antigen presentation
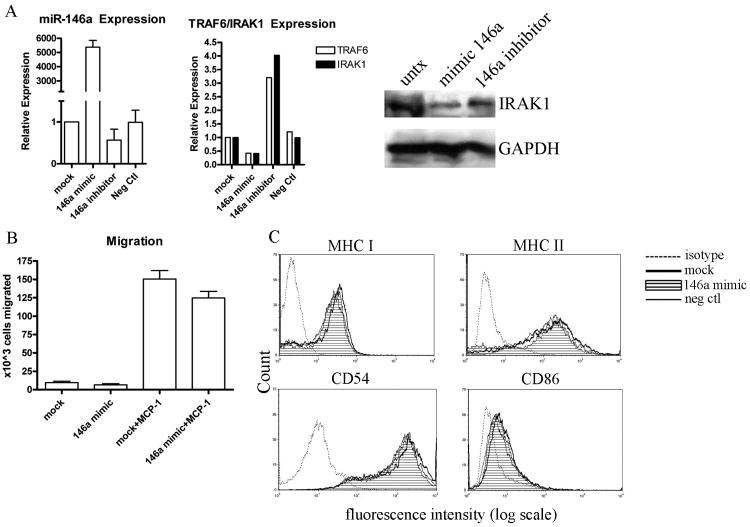
Since miR-146a expression is significantly upregulated in the PBMCs of both the mouse model of SjS and human SjS patients and miR-146a is known to have a regulatory role in the innate immune response, we wanted to determine the functional significance of miR-146a upregulation in SjS. To accomplish this, human monocytic THP-1 cells were transfected with synthetic miR-146a mimic in order to emulate the upregulated miR-146a that we have observed in SjS in vivo. Cells were also transfected with a miR-146a inhibitor to block the function of endogenous miR-146a in THP-1 cells. To verify successful transfection of these molecules, qRT-PCR was conducted 48 hours after transfection. As shown in Figure 3A, transfection with miR-146a mimic resulted in a dramatic increase in miR-146a expression, while transfection with miR-146a inhibitor resulted in a slight decrease in miR-146a expression, and transfection with negative control mimic had no effect on miR-146a expression. Confirmed target genes of miR-146a, TRAF6 and IRAK-1 [8, 15, 16, 20-22], were also analyzed by qRT-PCR after transfection with miR-146a mimic and were found to decrease 25% and 50%, respectively, and after transfection of miR-146a inhibitor were found to increase 3.2 and 4.0 fold, respectively (Figure 3A), as expected. Western blot data also demonstrate a significant reduction in IRAK-1 protein after transfection with miR-146a mimic (Figure 3A).
Figure 3. miR-146a has no effect on migration or expression of antigen presenting molecules.
THP-1 cells were transfected with miR-146a mimic, miR-146a inhibitor, or negative control as described in Methods. Two days after transfection, migration and expression of antigen presenting and costimulatory moleucles were monitored and compared to mock transfected cells. A) miR-146a, TRAF6, and IRAK-1 expression after transfection of mimic/inhibitor/negative control was determined by qRT-PCR. B) Migration assay was conducted using 8 μm transwell membranes as described in Methods (n=5 independent experiments, bars represent mean with standard error). C) The surface expression of MHC class I, MHC class II, CD54, and CD86 was detected by flow cytometry after 48 hours of stimulation with 50ng/ml of IFNγ. The THP-1 cells were transfected with either miR-146a mimic (filled peak), negative control miRNA (solid line), or mock transfected (bold solid line). The mouse K isotype moAb controls for FITC, PE, and APC (dotted lines) are also shown. Data are representative of 3 independent experiments.
After transfection with miR-146a mimic or inhibitor, cells were monitored in functional assays as described in Materials and Methods. THP-1 cell migration was monitored after transfection using an 8 μm transwell membrane. Cells were placed in the top chamber, and MCP-1 was added to the bottom chamber, creating a chemokine gradient which induces the cells to migrate across the permeable membrane. No significant difference in cell migration were observed between miR-146a mimic transfected and mock transfected cells, indicating that miR-146a is not involved in directly regulating MCP-1 induced migration (Figure 3B). LPS-induced nitric oxide production was also monitored after transfection with 146a mimic, but no significant differences were observed (data not shown).
To characterize any changes in MHC antigen presentation in response to elevated levels of miR-146a, we examined cell surface expression of MHC class I, MHC class II, the crucial costimulatory molecule, CD86, and the adhesion molecule CD54 on IFNγ-stimulated THP-1 cells by flow cytometry. We found no detectable differences in expression of MHC I, MHC II, CD86 or CD54 between 146a mimic-transfected and mock transfected cells (Figure 3C). This suggests that miR-146a may not be directly involved in the alteration of antigen presentation and the immune response to IFNγ stimulation. However, we cannot rule out that our analysis by flow cytometry may not be sensitive enough to detect minute/intricate changes in expression.
miR-146a significantly upregulates phagocytic activity in THP-1 cells
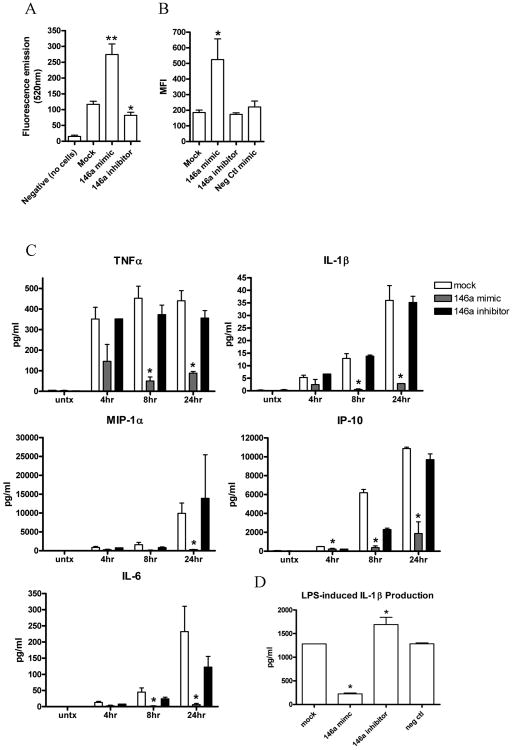
After transfection with miR-146a mimic or inhibitor, cells were monitored for phagocytic activity as described in Materials and Methods. Briefly, transfected and mock-transfected cells were incubated with fluorescein-labeled Escherichia coli for 2 hours, quenched with trypan blue, and then the engulfed bacteria were quantitatively detected using a fluorescent plate reader (Figure 4A) or flow cytometer (Figure 4B). Interestingly, cells transfected with miR-146a mimic showed a significant increase in phagocytic activity compared with mock transfected cells (p<0.0001 as determined by t test, Figure 4A & 4B), and cells transfected with 146a inhibitor showed a significant decrease in phagocytic activity compared with mock transfected cells (Figure 4A, p<0.05 as determined by t test). These data suggest that miR-146a plays a role in upregulating phagocytic activity in human monocytes.
Figure 4. miR-146a upregulates phagocytic activity and suppresses cytokine production in human monocytic THP-1 cells.
THP-1 cells were transfected with miR-146a mimic or miR-146a inhibitor as described in Methods, and 48 hours after transfection, phagocytic activity and LPS-induced cytokine production were monitored and compared to mock transfected cells. A) Phagocytosis was quantitatively measured as described in Methods. Cells were transfected with miR-146a mimic (n=23), miR-146a inhibitor (n=18), or mock transfected (n=27). Asterisk (**) indicates p<0.0001 or (*) p<0.05 compared to mock transfected cells as determined by t test. Bars represent mean with standard error. Data shown from at least 5 independent experiments. B) Phagocytic activity was also monitored by flow cytometry as described in Methods. Cells were transfected with miR-146a mimic (n=6 independent experiments), miR-146a inhibitor (n=6 independent experiments), negative control miRNA mimic (n=6 independent experiments), or mock transfected (n=6 independent experiments). Mean fluorescence intensity (MFI) shown, asterisk (*) indicates p<0.05 compared to mock as determined by one-way ANOVA. Bars represent mean with standard error. C) After transfection, cells were stimulated with 1 μg/ml LPS for 4, 8, or 24 hours or left untreated. Culture supernatants were collected and cytokine production was quantitatively measured using multiplex analysis as described in Methods (n=3, asterisk indicates p<0.05 as determined by t test). Bars represent mean with standard error. D) LPS-induced IL-1β production was detected by ELISA after transfection of miR-146a mimic/inhibitor or negative control mimic (n=3 independent experiments, asterisk indicates p<0.05 as determined by one-way ANOVA). Bars represent mean with standard error.
miR-146a negatively regulates pro-inflammatory cytokine production
Next, we monitored the effect of miR-146a on cytokine production by stimulating transfected and mock transfected cells with 1 μg/ml LPS for 4, 8, or 24 hours. Culture supernatants were harvested, and cytokine production was quantitatively measured using multiplex analysis as described in Methods. Production of TNF-α, IL-1β, MIP-1α, IP-10, and IL-6 was significantly reduced in cells transfected with miR-146a mimic compared to mock transfected cells (Figure 4C, p<0.05 as determined by t test). However, cytokine production was not affected by transfection with miR-146a inhibitor, which could be due to relatively low basal levels of miR-146a in THP-1 cells. ELISA to detect IL-1β production was also performed to confirm these data, and as shown in Figure 4D, similar results were obtained with significant reduction after transfection with miR-146a mimic (p<0.05 as determined by one way ANOVA) and significantly increased production after transfection with miR-146a inhibitor (p<0.05 as determined by one-way ANOVA). Transfection with negative control mimic showed no effect. These data indicate that miR-146a negatively regulates pro-inflammatory cytokine production, which is consistent with previous studies demonstrating the ability of miR-146a to negatively regulate some pro-inflammatory cytokines/chemokines [8, 20, 23-25].
Taken together, these data indicate a role for miR-146a regulation of phagocytosis and pro-inflammatory cytokine production in THP-1 human monocytes, but antigen presentation and migration do not appear to be affected by miR-146a expression.
Discussion
SjS affects not only the salivary and lacrimal glands but also other exocrine glands or organs potentially via a process called epitope spreading. Complex disease pathogenesis of SjS has hindered the advancement of our understanding on disease initiation, thus delaying the identification of susceptible individuals. Recent evidence showing miRNA as a micromanager of various stages of immune regulations has generated interest in the involvement of miRNAs in autoimmune disorders. In an effort to elucidate early disease pathogenesis of SjS, we investigated miRNAs that are involved in innate immune responses in our study to examine if miRNAs are involved in prolonged and persistent inflammation characterized in SjS.
Our data have demonstrated that the SjS-prone mouse model (B6DC) exhibits increased miR-146a expression in SMX tissue and PBMCs prior to disease onset (8 weeks old) and during full blown disease (20 weeks old). Interestingly, the expression of these miRNA was increased in the target SMX tissue even prior to disease onset. In PBMCs, miRNA expression was higher during full blown disease, but was still elevated prior to disease onset, potentially due to prolonged inflammatory process in SjS. These data suggest that miR-146a is involved in early disease pathogenesis, which coincides with initial observations by Taganov et al. suggesting [9] its induction as a result of innate immune response. RNA expression in PBMC may be indicative of the chronic inflammation present before disease onset in SjS.
In our cohort of SjS patients, miR-146a expression was significantly increased compared to healthy donors. In our previous work studying miRNA expression in RA patients, we also found that miR-146a expression was significantly increased compared to healthy controls [15]. However, the extent to which we observed miR-146a upregulation in SjS patients was much greater at 7.9-fold higher compared to 2.6-fold higher in RA patients. This was somewhat surprising and particularly interesting given that SjS is often considered to be closely related to SLE as an interferon-driven disease, and in SLE patients, miR-146a was found to be downregulated [16]. Interestingly, there was no correlation detected between elevated miR-146a expression and MX-1 expression in the SjS cohort, as has been previously demonstrated in SLE [16]. Despite the evidence of miR-146a's general regulatory role over the inflammatory response, this difference could be utilized in better determining disease specific miRNAs in the development of a diagnostic miRNA profile. These data also indicate that there may be a defect in miR-146a regulation of the type 1 IFN response. Interestingly, a recent study demonstrates the association between a polymorphism in the 3′ UTR of miR-146a target IRAK1 with RA susceptibility [21], indicating that a possible defect in miR-146a regulation of IRAK1 could contribute to RA. Further studies are needed to determine if such a regulatory defect contributes to SjS.
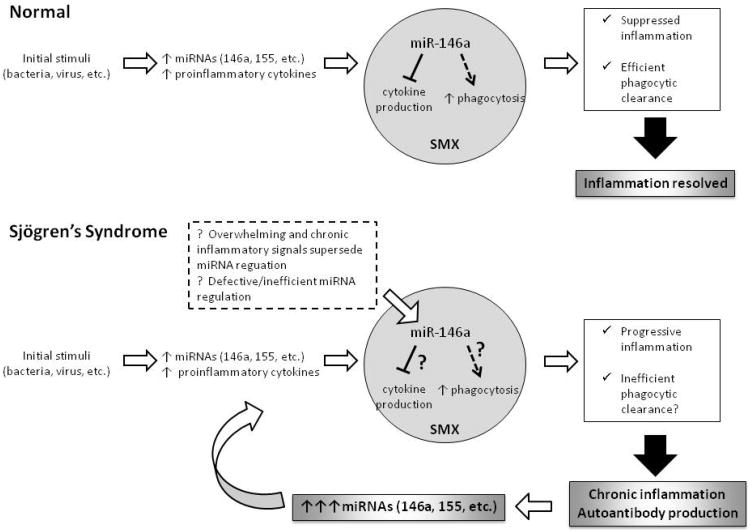
Our in vitro functional analysis of miR-146a suggests a role for miR-146a in up-regulating phagocytosis and downregulating inflammatory cytokine/chemokine production in monocytes. In an in vivo situation, this data would suggest that miR-146a upregulation in normal individuals should contribute to efficient phagocytic clearance and the reduction of inflammatory cytokine production resulting in the resolution of the inflammatory environment (Figure 5). However, in SjS patients there is chronic inflammation in the target tissues due in part to apoptotic cellular debri and other unknown triggers. In this scenario, upregulated phagocytic activity (due to increased miR-146a expression) in the presence of autoantibodies could contribute to the pro-inflammatory response as has been reported in SLE ([26], Figure 5). Additionally, despite increased levels of miR-146a in these patients (which should help decrease inflammatory cytokine production), in pre-disease state SjS-prone mice and in human patients, elevated IL-1β and IL-18 levels have been observed in the salivary glands as well as in sera [1, 27-30]. This discrepancy could indicate the possibility that miR-146a is unable to properly regulate its targets in SjS due to unknown mechanisms, such as other over-riding signals or inability to bind target genes, illustrated in Figure 5. Therefore, it is speculated that the inability of miRNA to suppress cytokines and increase phagocytic activity would lead to prolonged inflammation and inefficient clearance of self antigens in vivo, resulting in chronic inflammation and activation of adaptive immune response. Further studies are underway to test if the effect of miR-146a upregulation on monocytic functions that we observed in vitro is altered in SjS-prone mice or in SjS patients.
Figure 5. Summary of potential role of miRNA regulation in SjS.
Based on our data showing increased miR-146a expression in SjS and miR-146a's role in upregulating phagocytosis and downregulating pro-inflammatory cytokine production, we propose this model. In normal individuals, an initial stimuli triggers and inflammatory response and increased miR-146a expression which then acts to negatively regulate cytokine production and enhance phagocytic activity. This suppressed inflammation and efficient phagocytic clearance results in the resolution of inflammation. However, in SjS, it is unclear whether miR-146a can properly function which could potentially lead to chronic inflammation, autoimmunity, and prolonged miR-146a elevation. It is also speculated that enhanced phagocytic clearance of apoptotic debri in SjS target tissues could contribute to loss of tolerance and activation of the adaptive immune response. Question marks indicate speculated points.
In conclusion, our study provides interesting insight into abnormal regulation of miRNA present in autoimmune SjS and the similarities of miRNA expression in the SjS-mouse model compared with human SjS patients. In addition, our mouse model and human data suggest that the increased expression of miR-146a observed is SjS-specific since B6DC mice exhibit no other autoimmune conditions and human SjS patients presented a distinct profile of miR-146a expression compared to RA and SLE patients. Our future goal is to obtain a larger cohort of SjS patients, disease controls, and healthy donors and utilize miRNA microarray technology to analyze a large population of miRNAs within these individuals. From this data we hope to be able to create a diagnostic miRNA profile specific for SjS that can be utilized for early SjS screening and is less invasive and more cost efficient than biopsy procedures and miRNA targeted therapeutics. More importantly, we plan to further elucidate any pathological roles that miRNA dysregulation may play in SjS initiation and/or progression.
Materials and Methods
Animals
C57BL/6J and C57BL/6.NOD-Aec1Aec2 mice were bred and maintained under SPF conditions within the Animal Care Services at the University of Florida, Gainesville. C57BL/6.NOD-Aec1Aec2 exhibits dry mouth and dry eye condition, resembling clinical manifestations of primary SjS [2, 5, 10]. The animals were maintained on a 12 h light-dark schedule and provided water and food ad libitum. For this study, female mice were utilized at 8 or 20 weeks of age, prior to disease onset and during advanced disease, respectively. Both breeding and use of these animals were approved by the University of Florida IACUC. The mice were sacrificed using American Veterinary Medical Association approved procedures.
Patients and control subjects
Twenty-five patients who were diagnosed as SjS according to the modified European-American diagnostic criteria for SjS (24 primary SjS, 1 secondary SjS) were included in this study [31]. Their demographic, clinical, and laboratory characteristics are summarized in Table 1. Ten gender-matched healthy donors with no history of autoimmune disease were included as control individuals. This study was approved by the University of Florida Institutional Review Board, and written permission was obtained from all who participated in the study.
PBMC collection and qRT-PCR
Whole blood samples were collected in EDTA-treated tubes and PBMCs were isolated using the Leukolock filter system (Ambion, Austin, TX). Total RNA was extracted using TRI reagent (Ambion) according to Ambion's alternative protocol for extraction of RNA from cells captured on Leukolock filters. Total RNA concentrations were determined and 100ng of each RNA sample were used for quantitative real-time RT-PCR (qRT-PCR) as previously described [15]. miRNA qRT-PCR was performed using the TaqMan MicroRNA Reverse Transcription Kit, TaqMan Universal PCR Master Mix, and TaqMan MicroRNA Assay primers for human miR-146a, human and murine miR-155, and human miR-132 (Applied Biosystems, Foster City, CA, USA). All reactions were analyzed using StepOne Real-Time PCR System (Applied Biosystems). The levels of miRNA were normalized to U44 controls for human samples and U6 for mouse samples, and the cycle threshold (Ct) values, corresponding to the PCR cycle number at which fluorescence emission reaches a threshold above baseline emission, were determined and the relative miRNA expression was calculated using the 2-ΔΔCt method [32].
miRNA in situ hybridization
Minor salivary gland biopsies were harvested and fixed in 10% neutral buffered formalin, embedded in paraffin, and 5 μm sections were cut and placed on glass slides. In situ hybridizations were performed using mercury LNA microRNA ISH Optimization Kit (Exiqon, Denmark) according to the manufacturer's suggested protocol. Hybridization was performed at 55°C with double-DIG labeled LNA probes for hsa-miR-146a and U6 small RNA (Exiqon). Following hybridization and stringent washes, blocking and DIG detection was carried out using the Roche DIG Wash and Block Buffer set (Roche Applied Science, Indianapolis, IN) and NBT/BCIP (Roche Applied Science). The slides were mounted and images were taken by light microscopy at 400× magnification.
Cell culture and transfection
THP-1 human monocytes obtained from American Type Culture Collection (Manassas, VA) were cultured in RPMI 1640 medium with 10% fetal bovine serum. THP-1 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Pre-miR miRNA Precursor molecules (miR-146a mimic), Anti-miR miRNA Inhibitors (miR-146a inhibitor), and negative control miRNA precursors were purchased from Ambion. THP-1 cells were transfected with miR-146a mimic, inhibitor, or negative control at a final concentration of 40 nM and cells were incubated 48 hours before use in functional assays. Transfection efficiency was monitored by qRT-PCR detection of miR-146a.
Functional assays
Phagocytosis assay
The Vybrant Phagocytosis Assay Kit (Invitrogen, Carlsbad, CA) was used according to the manufacturer's protocol. Briefly, transfected and mock-transfected cells were incubated with fluorescein-labeled Escherichia coli for 2 hours, quenched with trypan blue, and a Spectramax M5 fluorescence plate reader (Molecular Devices) was used to quantitatively detect the engulfed bacteria within the cells. Phagocytosis was also monitored by flow cytometry by incubating transfected/mock-transfected cells with the fluorescein-labeled E. coli from the above mentioned kit for 4 hours, washing 3 times with PBS, and resuspended in 500ul FACS buffer (PBS with 1% FBS, 2mM EDTA). The cells were then analyzed with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA), and data anaylsis was performed using CellQuest Pro software (BD Biosciences).
Migration Assay
The migration assay was performed using a 6.5mm Transwell with 8.0μm pore polycarbonate membrane insert (Corning Life Sciences, Acton, MA). THP-1 cells (1×106) (transfected with pre-miR-146a or mock transfected) were added to the top chamber in 0.1ml RPMI with 0.5% FBS and allowed to incubate 30 minutes. MCP-1 (monocyte chemotactic protein-1) was then added to the bottom chamber at a concentration of 25ng/ml, and the cells are incubated at 37°C for 3 hours. Cells that migrated to the bottom chamber were then harvested and counted.
LPS-induced cytokine production
THP-1 cells were transfected as previously described, and then treated with 1 ug/ml LPS (Salmonella enteric serotype minnesota; Sigma, St. Louis, MO) for 4, 8, or 24 hours in culture medium. The human cytokine LINCOplex premixed kit (LINCO Research, St. Charles, MO) was used according to the manufacturer's protocol to quantitatively detect the following human cytokines in the supernatant: IL-1β, IL-6, IP-10, MIP-1α, MCP-1, IFN-γ, IL-8, IL-12p70, IL-10, and TNF-α. Human IL-1β ELISA Ready-SET-Go kit (eBiosciences, San Diego, CA) was also utilized to measure IL-1β production.
Expression of antigen presenting/costimulatory molecules
THP-1 cells were transfected as described above, incubated for 48 hours, and then stimulated with 50 ng/ml IFNγ (BD Biosciences) for 48 hours. Cell surface expression of MHC class I, MHC class II, CD54, and CD86 molecules was then examined by flow cytometry using the following antibodies: FITC-conjugated anti-HLA-ABC (eBiosciences), PE-conjugated anti-HLA-DR, PE-conjugated anti-CD86, APC-conjugated anti-CD54 (BD Biosciences).
Statistical analysis
Statistical analyses were performed using Prism 4.0 (GraphPad Software, Inc., San Diego, CA). Mann Whitney was used to determine significant differences in miRNA expression between patients and controls and wild-type and disease mice, and P < 0.05 was considered significant. Student's t test or one way ANOVA were used for all other experiments unless indicated otherwise.
Supplementary Material
Supplemental Figure 1. miR-146a and miR-155 expression is not correlated with clinical parameters. miR-146a and miR-155 expression was analyzed by qRT-PCR in PBMCs of SjS patients and correlated with salivary flow rate (A), salivary gland biopsy focal scores (B), and presence/absence of anti-SSA/SSB autoantibodies (C-E). R-squared and p values determined by linear regression for A and B, p values determined by Mann Whitney test.
Supplemental Figure 2. miR-146a expression in salivary gland biopsies of SjS patients corresponds with PBMC expression. Eight SjS patient and four non-SjS control salivary gland biopsy samples were selected to examine miR-146a expression by in situ hybridization. A) SjS-2, which exhibited low miR-146a expression in PBMCs and SjS-4, which exhibited high miR-146a expression in PBMCs are shown as representative images. U6 RNA was used as an internal control and was uniformly expressed. Arrows indicate positive staining. 400× magnification shown. B) miR-146a expression was blindly scored on a scale from 0-5, with 0=absence of detectable expression and 5=expression intensity equal to U6 expression (p=0.02 as determined by Mann Whitney test). Bars represent mean.
Supplemental Figure 3. miR-155 and miR-132 are differentially expressed in SjS-prone mouse tissues and PBMCs. qRT-PCR was used to examine miR-155 (white bars) and miR-132 (black bars) expression in SjS-prone (B6DC) and wild-type (C57BL/6) mice at 8 and 20 weeks old. A) PBMC (n=2 replicates of pooled sample), B) Submandibular gland (SMX, n=6), C) lacrimal gland (LAC, n=10), and D) kidney (KID, n=6), were analyzed. Data are presented as average relative expression after normalization to U44 RNA and C57BL/6 8 week samples. Asterisks (*) indicate p<0.05 compared to age-matched wild-type control as determined by Mann Whitney, bars represent mean with standard error.
Acknowledgments
This work was supported by NIH/NIDCR grant DE019644(SC). KMP is supported by NIH/NIDCR training grant T32DE007200.
List of Abbreviations Used
- B6DC
C57BL/6.NOD-Aec1Aec2 mouse model
- LAC
lacrimal gland
- miRNA
microRNA
- PBMCs
peripheral blood mononuclear cells
- SjS
Sjögren's syndrome
- SLE
systemic lupus erythematosus
- SMX
submandibular salivary gland
- RA
rheumatoid arthritis
Footnotes
Conflict of Interest: The authors declare they have no conflicts of interest.
References
- 1.Bulosan M, Pauley KM, Yo K, Chan EK, Katz J, Peck AB, Cha S. Inflammatory caspases are critical for enhanced cell death in the target tissue of Sjogren's syndrome before disease onset. Immunol Cell Biol. 2008 doi: 10.1038/icb.2008.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cha S, Brayer J, Gao J, Brown V, Killedar S, Yasunari U, Peck AB. A dual role for interferon-gamma in the pathogenesis of Sjogren's syndrome-like autoimmune exocrinopathy in the nonobese diabetic mouse. Scand J Immunol. 2004;60:552–565. doi: 10.1111/j.0300-9475.2004.01508.x. [DOI] [PubMed] [Google Scholar]
- 3.Cha S, van Blockland SC, Versnel MA, Homo-Delarche F, Nagashima H, Brayer J, Peck AB, Humphreys-Beher MG. Abnormal organogenesis in salivary gland development may initiate adult onset of autoimmune exocrinopathy. Exp Clin Immunogenet. 2001;18:143–160. doi: 10.1159/000049194. [DOI] [PubMed] [Google Scholar]
- 4.Robinson CP, Yamamoto H, Peck AB, Humphreys-Beher MG. Genetically programmed development of salivary gland abnormalities in the NOD (nonobese diabetic)-scid mouse in the absence of detectable lymphocytic infiltration: a potential trigger for sialoadenitis of NOD mice. Clin Immunol Immunopathol. 1996;79:50–59. doi: 10.1006/clin.1996.0050. [DOI] [PubMed] [Google Scholar]
- 5.Cha S, Nagashima H, Brown VB, Peck AB, Humphreys-Beher MG. Two NOD Idd-associated intervals contribute synergistically to the development of autoimmune exocrinopathy (Sjogren's syndrome) on a healthy murine background. Arthritis Rheum. 2002;46:1390–1398. doi: 10.1002/art.10258. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Killedar SJ, Eckenrode SE, McIndoe RA, She JX, Nguyen CQ, Peck AB, Cha S. Early pathogenic events associated with Sjogren's syndrome (SjS)-like disease of the NOD mouse using microarray analysis. Lab Invest. 2006;86:1243–1260. doi: 10.1038/labinvest.3700487. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Chen XP, Li YJ. MicroRNA-146a and human disease. Scand J Immunol. 2010;71:227–231. doi: 10.1111/j.1365-3083.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- 12.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Stahle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 14.Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 17.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, Xu ZM, Yin YB. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 18.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int. 2008;29:749–754. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 19.Chiorini JA, Cihakova D, Ouellette CE, Caturegli P. Sjogren syndrome: advances in the pathogenesis from animal models. J Autoimmun. 2009;33:190–196. doi: 10.1016/j.jaut.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nahid MA, Pauley KM, Satoh M, Chan EK. miR-146a is critical for endotoxin-induced tolerance: Implication in Innate Immunity. J Biol Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. A polymorphism in the 3′-UTR of interleukin-1 receptor-associated kinase (IRAK1), a target gene of miR-146a, is associated with rheumatoid arthritis susceptibility. Joint Bone Spine. 2010;77:411–413. doi: 10.1016/j.jbspin.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. The role of microRNA-146a (miR-146a) and its target IL-1R-associated kinase (IRAK1) in psoriatic arthritis susceptibility. Scand J Immunol. 2010;71:382–385. doi: 10.1111/j.1365-3083.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 23.Curtale G, Citarella F, Carissimi C, Goldoni M, Carucci N, Fulci V, Franceschini D, Meloni F, Barnaba V, Macino G. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 2010;115:265–273. doi: 10.1182/blood-2009-06-225987. [DOI] [PubMed] [Google Scholar]
- 24.Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 25.Pauley KM, Satoh M, Pauley BA, Dominguez-Gutierrez PR, Wallet SM, Holliday LS, Cha S, Reeves WH, Chan EK. Formation of GW/P bodies as marker for microRNA-mediated regulation of innate immune signaling in THP-1 cells. Immunol Cell Biol. 2009;88:205–212. doi: 10.1038/icb.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz LE, van Bavel C, Franz S, Berden J, Herrmann M, van der Vlag J. Apoptosis in the pathogenesis of systemic lupus erythematosus. Lupus. 2008;17:371–375. doi: 10.1177/0961203308089990. [DOI] [PubMed] [Google Scholar]
- 27.Bombardieri M, Barone F, Pittoni V, Alessandri C, Conigliaro P, Blades MC, Priori R, McInnes IB, Valesini G, Pitzalis C. Increased circulating levels and salivary gland expression of interleukin-18 in patients with Sjogren's syndrome: relationship with autoantibody production and lymphoid organization of the periductal inflammatory infiltrate. Arthritis Res Ther. 2004;6:R447–456. doi: 10.1186/ar1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eriksson P, Andersson C, Ekerfelt C, Ernerudh J, Skogh T. Relationship between serum levels of IL-18 and IgG1 in patients with primary Sjogren's syndrome, rheumatoid arthritis and healthy controls. Clin Exp Immunol. 2004;137:617–620. doi: 10.1111/j.1365-2249.2004.02562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szodoray P, Alex P, Brun JG, Centola M, Jonsson R. Circulating cytokines in primary Sjogren's syndrome determined by a multiplex cytokine array system. Scand J Immunol. 2004;59:592–599. doi: 10.1111/j.0300-9475.2004.01432.x. [DOI] [PubMed] [Google Scholar]
- 30.Yamano S, Atkinson JC, Baum BJ, Fox PC. Salivary gland cytokine expression in NOD and normal BALB/c mice. Clin Immunol. 1999;92:265–275. doi: 10.1006/clim.1999.4759. [DOI] [PubMed] [Google Scholar]
- 31.Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, Daniels TE, Fox PC, Fox RI, Kassan SS, Pillemer SR, Talal N, Weisman MH. Classification criteria for Sjogren's syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61:554–558. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. miR-146a and miR-155 expression is not correlated with clinical parameters. miR-146a and miR-155 expression was analyzed by qRT-PCR in PBMCs of SjS patients and correlated with salivary flow rate (A), salivary gland biopsy focal scores (B), and presence/absence of anti-SSA/SSB autoantibodies (C-E). R-squared and p values determined by linear regression for A and B, p values determined by Mann Whitney test.
Supplemental Figure 2. miR-146a expression in salivary gland biopsies of SjS patients corresponds with PBMC expression. Eight SjS patient and four non-SjS control salivary gland biopsy samples were selected to examine miR-146a expression by in situ hybridization. A) SjS-2, which exhibited low miR-146a expression in PBMCs and SjS-4, which exhibited high miR-146a expression in PBMCs are shown as representative images. U6 RNA was used as an internal control and was uniformly expressed. Arrows indicate positive staining. 400× magnification shown. B) miR-146a expression was blindly scored on a scale from 0-5, with 0=absence of detectable expression and 5=expression intensity equal to U6 expression (p=0.02 as determined by Mann Whitney test). Bars represent mean.
Supplemental Figure 3. miR-155 and miR-132 are differentially expressed in SjS-prone mouse tissues and PBMCs. qRT-PCR was used to examine miR-155 (white bars) and miR-132 (black bars) expression in SjS-prone (B6DC) and wild-type (C57BL/6) mice at 8 and 20 weeks old. A) PBMC (n=2 replicates of pooled sample), B) Submandibular gland (SMX, n=6), C) lacrimal gland (LAC, n=10), and D) kidney (KID, n=6), were analyzed. Data are presented as average relative expression after normalization to U44 RNA and C57BL/6 8 week samples. Asterisks (*) indicate p<0.05 compared to age-matched wild-type control as determined by Mann Whitney, bars represent mean with standard error.