Abstract
Serpins regulate various physiological reactions in humans and insects, including certain immune responses, primarily through inhibition of serine proteases. Six serpins have previously been identified and characterized in the tobacco hornworm Manduca sexta. In this study, we obtained a full-length cDNA sequence of another Manduca serpin, named serpin-7. The open reading frame of serpin-7 encodes a polypeptide of 400 amino acid residues with a predicted signal peptide of the first 15 residues. Multiple protein sequence alignment of the reactive center loop region of the M. sexta serpins indicated that serpin-7 contains Arg–Ile at the position of the predicted scissile bond cleaved by protease in the serpin inhibition mechanism. The same residues occur in the scissile bond of the reactive center loop in M. sexta serpin-4 and serpin-5, which are protease inhibitors that can block prophenoloxidase activation in plasma. Serpin-7 transcript was detected in hemocytes and fat body, and its expression increased in fat body after injection of larvae with Micrococcus luteus. Recombinant serpin-7 added to larval plasma inhibited spontaneous melanization and decreased prophenoloxidase activation stimulated by bacteria. Serpin-7 inhibited prophenoloxidase-activating protease-3 (PAP3), forming a stable serpin-protease complex. Considering that serpin-3 and serpin-6 are also efficient inhibitors of PAP3, it appears that multiple serpins present in plasma may have redundant or overlapping functions. We conclude that serpin-7 has serine protease inhibitory activity and is likely involved in regulation of proPO activation or other protease-mediated aspects of innate immunity in M. sexta.
Keywords: Hemolymph, Immunity, Melanization, Protease inhibitor, Serpin, Tobacco hornworm
1. Introduction
A key aspect of insect innate immunity is the ability of insects to melanize microbes and foreign objects (Ayres and Schneifer, 2008; Cerenius et al., 2008; Suwanchaichinda and Paskewitz, 1998). Melanization requires proteolytic activation of prophenoloxidase (proPO) to its active form (Kanost and Gorman, 2008; Vavricka et al., 2010). This activation is mediated by a cascade of multiple serine proteases, with proPO-activating proteases (PAPs) as a possible terminal component of the pathway. PAPs contain one or two regulatory clip domains at the amino terminus and a catalytic serine protease domain at the carboxyl terminus, which is activated by a specific proteolytic cleavage (Kanost et al., 2004). The activated proteases are regulated through their production as zymogens and rapid inhibition once they are activated, to prevent damage to the host’s physiological systems.
Serpins are a superfamily of proteins folding into a conserved tertiary structure with a reactive center loop (RCL) near the C-terminus, which acts as bait for a target protease (Gettins, 2002). After a protease cleaves a scissile bond between amino acid residues P1 and P1′ of the RCL, the serpin undergoes a conformational change and traps the protease in an inactive state, resulting in irreversible inhibition of the protease. Multiple serpin genes have been identified in insect genomes (Reichhart, 2005; Suwanchaichinda and Kanost, 2009; Zou et al., 2009). Several of these insect serpins regulate innate immune responses. In Manduca sexta, with the exception of serpin-2 (Gan et al., 2001), serpin-1, -3, -4, -5, and -6 have been characterized and shown to be inhibitory and to regulate proteases that function in cascades leading to activation of proPO and the cytokine spätzle (An and Kanost, 2010; An et al., 2011; Christen et al., 2012; Jiang and Kanost, 1997; Jiang et al., 2003; Ragan et al., 2010; Tong and Kanost, 2005; Tong et al., 2005; Wang and Jiang, 2004; Zhu et al., 2003).
In this study, we cloned a cDNA for an additional M. sexta serpin, serpin-7 (GenBank: HQ149330), and biochemically characterized the serpin-7 recombinant protein. Our findings indicated that serpin-7 can inhibit proPO activation in M. sexta plasma and it can inhibit PAP3.
2. Materials and methods
2.1. Insect rearing
Manduca sexta eggs were originally obtained from Carolina Biological Supply (Burlington, NC). The insect larvae were reared as described previously (Dunn and Drake, 1983).
2.2. cDNA cloning and sequencing
A partial 3′-end sequence obtained from an M. sexta EST (Gen- Bank: CA798822) was used to design primers for the oligo-capping rapid amplification of cDNA ends (RACE) method to obtain the remaining sequence of the serpin-7 transcript. The 5′-RACE reaction was performed according to the manufacturer’s protocol for a GeneRacer kit (Invitrogen). Fat body mRNA samples were prepared from day-2 fifth instar larvae 24 h after injection with 50 µg of Micrococcus luteus. Corresponding cDNA was produced and used as a template for amplification. The RACE products were cloned and sequenced, and the full-length cDNA was then amplified by RT-PCR. The product was cloned into a pGEM-T plasmid vector (Promega). Sequencing of plasmids and PCR products was carried out at the Iowa State University Sequencing Facility (Ames, IA). The deduced amino acid sequence was obtained by using the Translate tool provided by the Swiss Institute Bioinformatics (Gasteiger et al., 2003).
2.3. Expression and purification of recombinant serpin-7
Serpin-7 cDNA encoding the mature protein was amplified by PCR using high fidelity Platinum Taq DNA polymerase (Invitrogen), a forward primer (5′-ATCCATGGCGCCGAATGTGGGA-3′) containing an NcoI site, and a reverse primer (5′-ATGCGGCCGCTCACACAACTGAAG- 3′) containing an NotI site. After digestion with the restriction enzymes NcoI and NotI (New England BioLabs), the PCR product and the plasmid H6pQE60 (Lee et al., 1994) were purified by agarose gel electrophoresis. The purified PCR product was subsequently cloned into the plasmid vector at the corresponding restriction sites. The DNA construct was sequenced to confirm the correct reading frame, which included an amino-terminal hexahistidine tag.
The hexahistidine-tagged recombinant serpin-7 was expressed in Escherichia coli strain XL1-blue. The bacteria were grown in 1 L LB medium at 37 °C, 300 rpm. Carbenicillin was used as a selective agent, and isopropyl β-d-thiogalactoside (IPTG) was used to induce protein expression. The soluble portion of the expressed protein was efficiently purified under native conditions by Ni-NTA chromatography. Column elution fractions were dialyzed in Tris buffer (20 mM Tris HCl, 50 mM NaCl, pH 8.0) overnight at 4 °C and concentrated before use in the experiments.
2.4. Multiple sequence alignments
The amino acid sequence of serpin-7 was aligned with other M. sexta serpins previously identified by using the ClustalW program. The prediction of the P1-P1′ residues and the reactive center loop region was manually identified. The three-dimensional structure of Manduca serpin-1K has been previously characterized (Li et al., 1999), and was used for comparison in this study.
2.5. Inhibition of spontaneous melanization of larval plasma
Hemolymph of day-1 fifth-instar larvae was collected into a chilled microcentrifuge tube from a cut in the dorsal horn. Hemocytes were removed by centrifugation at 5,000× g for 10 min. Six µl of the plasma was immediately mixed with 1, 2 and 3 µg of the purified recombinant serpin-7 in a total volume of 10 µl, adjusted with Tris buffer (0.1 M Tris HCl, pH 8.0/0.1 M NaCl). The mixtures were incubated at room temperature and photographed after 30 and 60 min.
2.6. Detection of serpin-protease complexes
Active recombinant M. sexta PAP3 was kindly provided by Dr. Maureen Gorman at Kansas State University. Reaction mixtures of PAP3 with serpin-7 were prepared according to Michel et al. (2006) and separated by electrophoresis using gradient 4–20% Tris · HCl Ready gels (Bio-Rad, Hercules, CA). The resolved proteins were transferred onto a nitrocellulose membrane and subjected to immunoblot analysis using antisera against either PAP3 or Histag (Qiagen) as primary antibodies (1:2500 dilution). Alkaline phosphatase-conjugated goat anti-rabbit IgG (1:3000 dilution) was used as the secondary antibody, and the antibody binding was visualized by using the alkaline phosphatase-conjugate substrate kit (Bio-Rad).
2.7. Inhibition of protease activities
Inhibition assays of PAP3 activity and larval prophenoloxidase activation were carried out as described previously (Michel et al., 2006). Briefly, for inhibition of PAP3 activity, recombinant serpin-7 at various concentrations was mixed with active PAP3 for 15 min at room temperature. After adding the N-acetyl-Ile-Glu-Ala- Arg-p-nitroanilide substrate solution, protease activity was monitored at 405 nm in a PowerWave340 microplate reader (Bio-Tek, Winooski, VT). Three replicates were performed (1 Unit = 0.001 ΔA405/min). For inhibition of prophenoloxidase activation, recombinant serpin-7 at different concentrations was incubated with 1 µl of day-1 fifth instar larval plasma. After incubation at room temperature for 10 min, M. luteus suspension was added to the mixtures and further incubated for 5 min. After adding the dopamine substrate solution, PO activity was measured by monitoring absorbance at 470 nm. Three replicates with different larval plasma samples were performed.
2.8. Serpin-7 gene expression
Collections of hemocytes and fat body from larvae injected with either 0.85% NaCl (control samples) or M. luteus (immune-challenged samples) were prepared according to Tong and Kanost (2005). Total RNAs of the hemocytes and fat body samples were prepared as described. RNA samples in the amount of 2.5 µg were used as templates for cDNA synthesis by using the SuperScript II First-Strand Synthesis System for RT-PCR (Invitrogen). One µl of each cDNA sample was used as a template for PCR. The primers were: 5′-GCAGAGAGCGGAATTACCAG-3′ and 5′- ACACCAGCAAAGAGGACGAT-3′ for serpin-7, and 5′-TCAGGCCGAGTCTTTGAGAT- 3′ and 5′-AGCACTCCTTGCCTGAGAAG-3′for ribosomal protein S3 (rpS3). PCR products of serpin-7 were amplified with 25 cycles for hemocyte samples and 28 cycles for fat body samples (94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s). The rpS3 was used as an internal control (19 cycles; 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s). All PCR products were analyzed by agarose gel electrophoresis and detected by ethidium bromide staining.
3. Results
3.1. Sequence and structural features of serpin-7
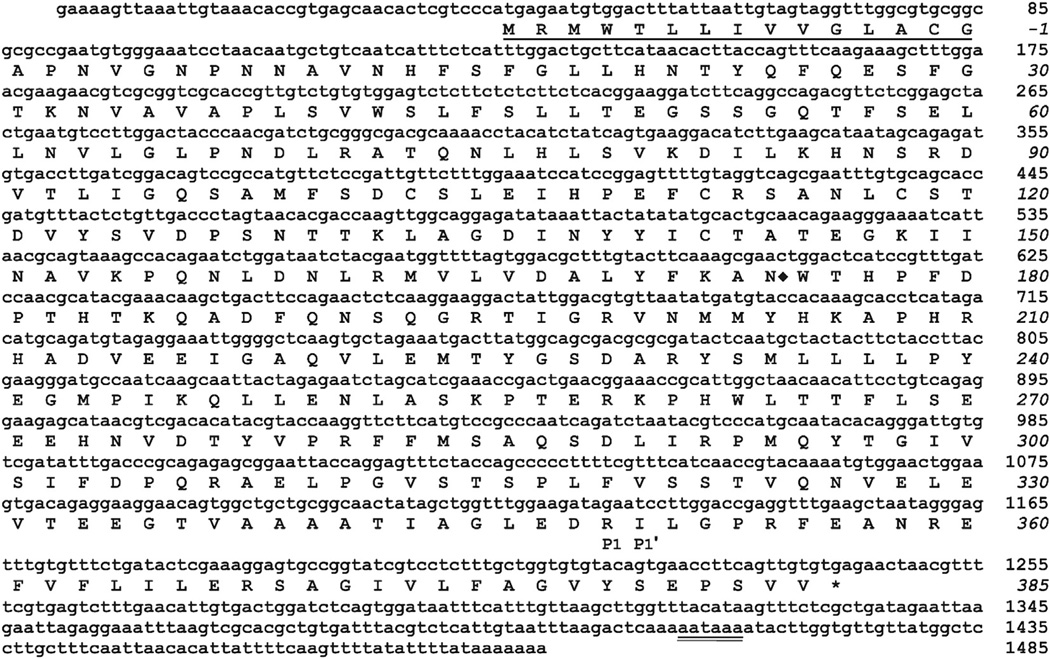
A M. sexta EST (GenBank: CA798822) encoded a partial amino acid sequence that exhibited the putative conserved domain of the serpin superfamily. Based on the cDNA sequence of this EST, which represented an incomplete 3′ end of a serpin transcript, we performed RACE and obtained a full-length nucleotide sequence, named serpin-7. The open reading frame of the serpin-7 cDNA encodes a polypeptide of 400 amino acid residues, with a predicted signal peptide consisting of the first 15 residues (Fig.1). There is one potential N-linked glycosylation site at Asn174. The calculated molecular mass and isoelectric point of the mature protein are 42.78 kDa and 5.29, respectively. The predicted residues at P1-P1′ positions of the predicted scissile bond in the RCL region are Arg349 and Ile350, similar to M. sexta serpin-4 and serpin-5 (Fig. 2), which have an Arg–Ile scissile bond and are known to be inhibitory and to regulate proPO activation (An and Kanost, 2010; Tong and Kanost, 2005; Tong et al., 2005).
Fig 1.
Nucleotide and amino acid sequence of M. sexta serpin-7. The deduced amino acid sequence is shown below the nucleotide sequence of the cDNA. The one-letter code for each amino acid is aligned with the second nucleotide of each codon. The signal peptide is underlined. The P1 and P1′ residues of the predicted scissile peptide bond in the RCL are indicated underneath the corresponding residues. A polyadenylation signal near the 3′ end is double underlined. One possible N-linked glycosylation site is marked with ◆ after the Asn residue.
Fig 2.
Multiple sequence alignment of M. sexta serpins. The translated amino acid sequence of serpin-7 was aligned with the six serpins from M. sexta, which have been characterized previously. The predicted P1-P1′ residues are highlighted. The square box represents the reactive center loop region.
A database search of the NCBI nonredundant protein sequences showed that the serpin-7 mature protein was most similar in amino acid sequence to Bombyx mori serpin-32 (Zou et al., 2009) with 63% identity (E-value = 0.0) (Table 1). Among M. sexta serpins, serpin-7 was most similar to serpin-4 with 32% identity. Serpin-7 also showed similarity with serpin-5 but at a lower score than that with serpin-4. In comparisons with Anopheles gambiae serpins, serpin-7 was most similar in amino acid sequence to serpin-8 (Suwanchaichinda and Kanost, 2009) with 30% identity.
Table 1.
Amino acid sequence identity between Manduca sexta serpin-7 (GenBank:ADM86478.1) and some other insect serpins.
| GenBank accession | Name | Species | Length (residues) | Total score | Max identity (%) |
|---|---|---|---|---|---|
| NP_001139723.1 | Serpin-32 | Bombyx mori | 395 | 544 | 63a |
| AEW46895.1 | Serpin-001 | Chilo suppressalis | 401 | 484 | 61 |
| EHJ68469.1 | Serpin-32 | Danaus plexippus | 523 | 427 | 62 |
| BAM19106.1 | Serpin-77Ba | Papilio polytes | 396 | 213 | 34 |
| AAS68504.1 | Serpin-4B | Manduca sexta | 407 | 210 | 32 |
| AAS68503.1 | Serpin-4A | Manduca sexta | 407 | 209 | 32 |
| XP_001863329.1 | Serpin | Culex quinquefasciatus | 418 | 190 | 31 |
| ABJ52807.1 | Serpin-8 | Anopheles gambiae | 434 | 184 | 30 |
| AAS68507.1 andAAS68508.1 | Serpin-5A and Serpin-5B | Manduca sexta | 396 | 181 | 32 |
| EFA11609.1 | Serpin-30 | Tribolium castaneum | 402 | 178 | 28 |
| AFG28186.1 | Serpin-4 | Glossina morsitans | 437 | 177 | 30 |
| XP_001661905.1 | Serpin | Aedes aegypti | 454 | 167 | 31 |
| NP_649205.3 | Serpin-77Ba isoform A | Drosophila melanogaster | 450 | 152 | 28 |
E value = 0.0.
3.2. Serpin-7 gene expression
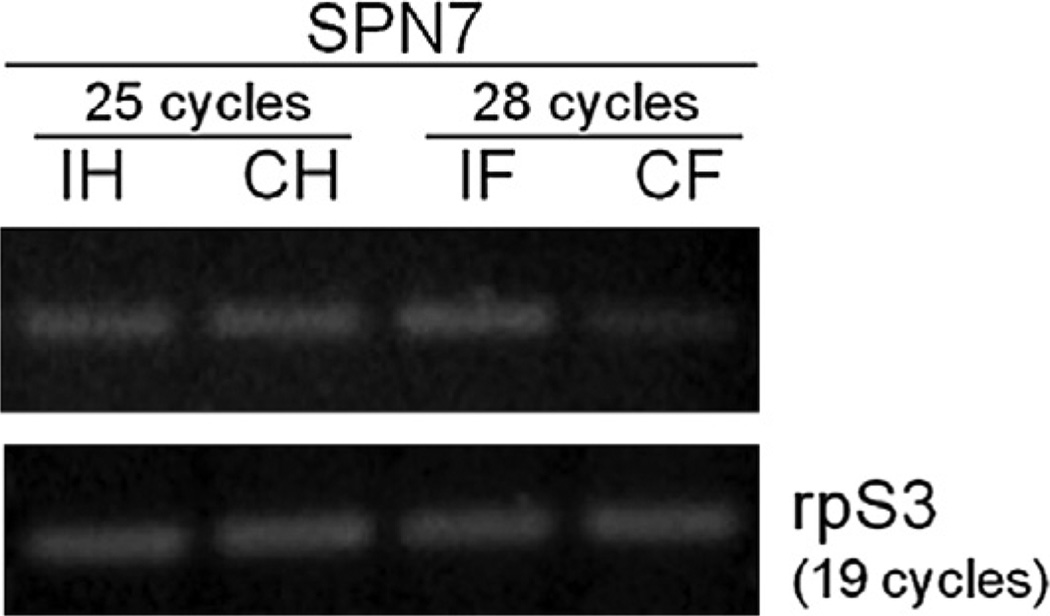
Hemocytes and fat body are the major tissues involved in both humoral and cellular immune responses in insects (Lavine and Strand, 2002; Tsakas and Marmaras, 2010). To determine tissue specificity of serpin-7 in the insect immune system, transcriptional regulation of serpin-7 upon immune challenge was examined by RT-PCR. Indeed, constitutive levels of serpin-7 transcripts were detected in both hemocytes and fat body of M. sexta larvae (Fig. 3). However, the transcript level of serpin-7 increased significantly in fat body after larvae were immune-challenged with bacteria. No significant difference was found in the transcript level in hemocytes between the treated and control larvae (Fig. 3).
Fig 3.
Serpin-7 gene expression in hemocytes and fat body. RT-PCR reactions were performed using specific primers for serpin-7, and ribosomal protein S3 (rpS3) was used as an internal control. cDNA samples of hemocytes (H) and fat body (F) from control (C) and Micrococcus-injected larvae (I) were used as templates.
3.3. Purification of recombinant serpin-7
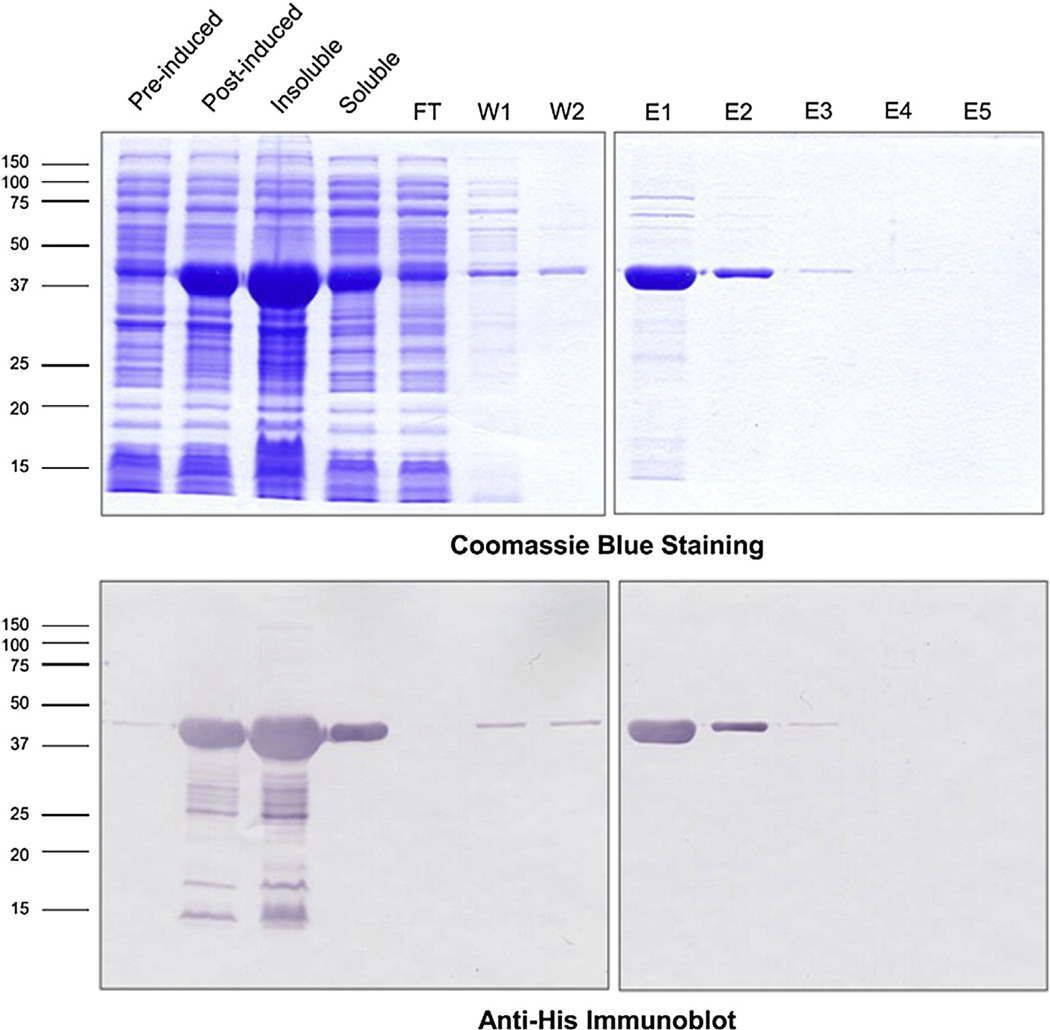
Serpin-7 with an amino-terminal hexahistidine tag was expressed in E. coli and purified by nickel-affinity chromatography. The recombinant protein in the soluble fraction of the bacterial cells was used for purification. Fig. 4 illustrates the purity of the recombinant protein as shown in the SDS gels with Coomassie blue staining. The immunoblots using the anti-Histag antibody confirms that the recombinant protein is about the expected size (calculated mass, ~44 kDa).
Fig 4.
Purification of recombinant serpin-7. The hexahistidine-tagged recombinant serpin-7 was expressed in E. coli strain XL1-blue. IPTG was used to induce protein expression. The soluble fraction was purified by nickel-affinity chromatography (FT, flow-through fraction; W, wash fraction; E, elution fraction). Samples were subjected to SDS-PAGE and stained with Coomassie blue (upper panel), or analyzed by immunoblotting with an antibody against the hexahistidine tag (lower panel). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Inhibition activities of serpin-7
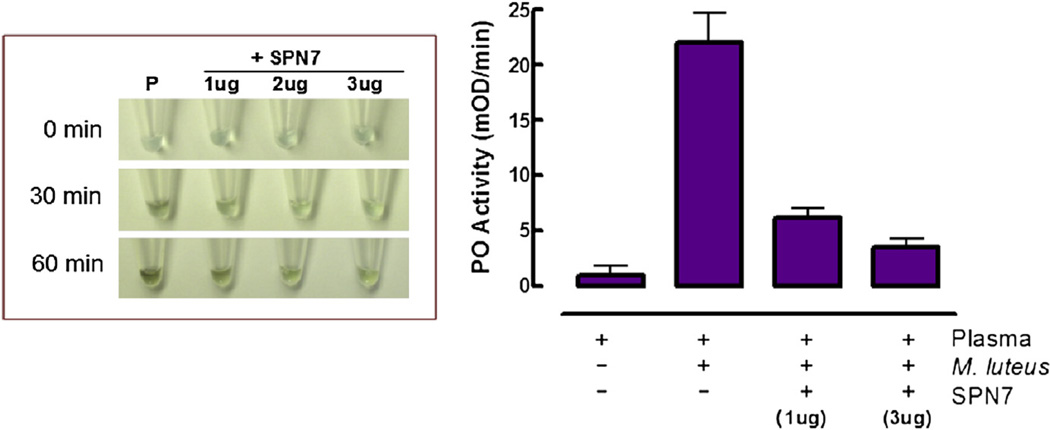
In order to test whether serpin-7 may be involved in regulating immunity in M. sexta, we examined inhibition activities of the purified recombinant serpin-7. Spontaneous melanization of larval plasma typically occurs with increasing time after hemolymph is collected and exposed to air. The color of plasma changed to dark brown or blackish within 30 min and continued to darken up to 60 min. However, addition of serpin-7 to plasma samples substantially slowed the melanization process, and apparently stopped it at higher concentrations (Fig. 5, left panel). In addition, serpin-7 significantly blocked proPO activation stimulated by exposure of plasma to M. luteus (Fig. 5, right panel).
Fig 5.
Inhibition of spontaneous melanization and bacteria-stimulated prophenoloxidase (proPO) activation by serpin-7. Left: Day-1 fifth-instar larval plasma samples (P) were mixed with the recombinant serpin-7 at different concentrations. Photographs were taken at the indicated times after mixing. Right: Larval plasma samples were mixed with or without serpin-7. Micrococcus luteus was added as indicated. PO activity was measured by using dopamine as a substrate (Mean ± SE, n = 3).
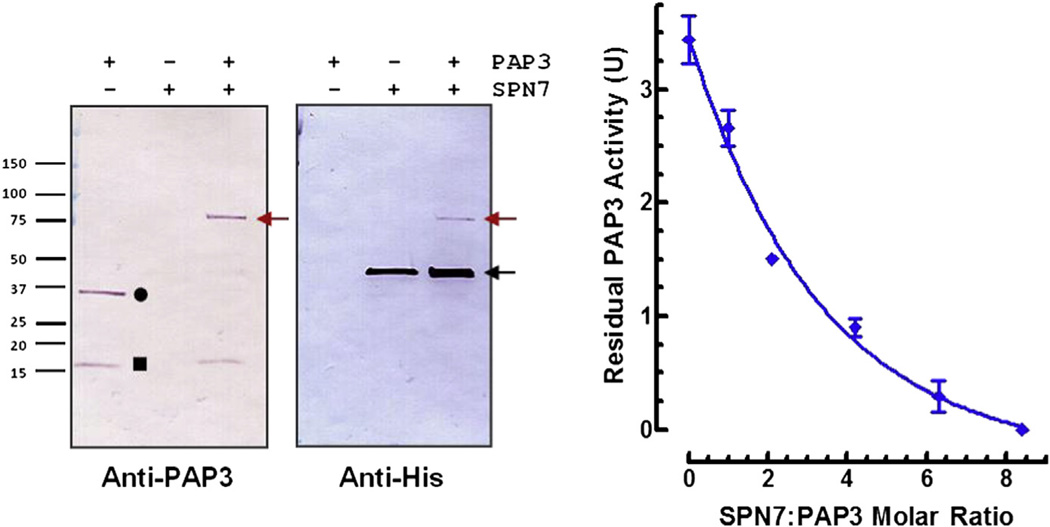
We examined the ability of serpin-7 to inhibit PAP3, a protease that directly activates proPO (Jiang et al., 2003). Serpin-7 inhibited PAP3 in a concentration-dependent manner (Fig. 6, right panel). Fifty percent inhibition occurred at an approximately 2:1 molar ratio of serpin-7:PAP3. Active PAP3 is composed of two chains, a clip domain and a catalytic (serine protease) domain, connected by a disulfide bond (Jiang and Kanost, 2000; Jiang et al., 2003). After incubation of PAP3 with serpin-7, the immunoreactive band representing the PAP3 catalytic domain (~35 kDa) disappeared, and a new band at a higher molecular mass appeared at ~75 kDa (Fig. 6, left panel), which corresponds to the combined molecular masses of PAP3 catalytic domain and cleaved serpin (calculated mass of the predicted N-terminal fragment of cleaved recombinant serpin-7, ~40 kDa). The identity of this serpin-protease complex was also confirmed by detection with a specific antibody against the hexahistidine tag of the recombinant serpin-7. The immunoreactive band representing serpin-7 after interaction with the active PAP3 (Fig. 6, left panel, anti-His immunoblot, far right lane, black arrow) appeared to be thicker than the band representing serpin-7 alone. The changed appearance of the band was possibly due to the combination of the uncleaved and cleaved serpin-7 as two bands locating very close to each other. These results confirm that serpin-7 can inhibit PAP3, which likely contributes to its effect on proPO activation in plasma.
Fig 6.
Formation of serpin-protease complex and inhibition of PAP3 activity by serpin-7. Left: Purified active PAP3 was incubated with serpin-7 (~1:3 protease:serpin molar ratio) for 15 min. Samples were subjected to SDS-PAGE and immunoblotting. Proteins were visualized by using antibodies against PAP3 or hexahistidine tag of the recombinant serpin-7. Symbols represent: PAP3-Serpin-7 complex, red arrow; non-complexed intact form of serpin-7, black arrow; catalytic domain of PAP3, circle; clip domain of PAP3, square. Right: The reaction mixtures were incubated at room temperature for 15 min before adding IEAR-pNA as a substrate. The residual PAP3 activity was measured as the rate of increase in absorbance at 405 nm (Mean ± SD, n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
A full-length cDNA for M. sexta serpin-7 was cloned and sequenced, and we began characterization of serpin-7 biochemical properties. The predicted P1-P1′ residues of serpin-7 (Arg–Ile), which are expected to be cleaved by a target protease, are the same as those in M. sexta serpin-4 and serpin-5, which can each block proPO activation in plasma (Tong and Kanost, 2005). Serpin- 4 can inhibit hemolymph protease-21 (HP21) (Tong et al., 2005), and serpin-5 can inhibit hemolymph protease-6 (HP6) (An and Kanost, 2010). Our preliminary experiment shows that HP6 can also form a protease-serpin covalent complex with serpin-7 (Supplementary materials, Fig. S1). Nonetheless, further work is needed to characterize the inhibitory selectivity of serpin-7 for M. sexta hemolymph proteases. The amino acid sequence of serpin-7 is most similar to B. mori serpin-32 (Zou et al., 2009), and these two proteins are identical in the P5-P5′ positions of the predicted RCL (Gly-Leu-Glu-Asp-Arg–Ile-Leu-Gly-Pro-Arg) (Supplementary materials, Fig. S2). It seems likely that M. sexta serpin-7 and B. mori serpin-32 are orthologs, with common inhibitory functions. In our previous study, A. gambiae serpin-8 (AgSRPN8) was predicted to be part of a phylogenetic clade with M. sexta serpin-4 and serpin-5 (Suwanchaichinda and Kanost, 2009). Serpin-7 was also most similar to AgSRPN8 among the A. gambiae serpins, suggesting that this group of M. sexta serpins may form a cluster with structural and/or functional similarity to AgSRPN8.
Hemocytes and fat body are important tissues involved in immunity in insects. We found that serpin-7 mRNA was detected in both hemocytes and fat body in M. sexta larvae. Upon immune stimulation by bacteria, the serpin-7 transcript level increased in fat body. A similar up-regulation pattern occurs for M. sexta serpin- 3, serpin-4 and serpin-5 (Tong and Kanost, 2005; Zhu et al., 2003). However, serpin-6 expression increases in both hemocytes and fat body after the exposure to bacteria (Zou and Jiang, 2005). The B. mori serpin-32, which was identified as an ortholog of serpin-7 in this study, was also up-regulated in larvae after bacteria injection (Zou et al., 2009). It is likely that some specific protease inhibition reactions catalyzed by serpin-7 occur in plasma, and thus up-regulation of serpin-7 in the fat body and its secretion into hemolymph may be critical in controlling protease activation pathways that are involved in immune responses in the insect, particularly to bacterial challenge in this case. We speculate that serpin-7 may have a key function in immunity.
Serpin-7 inhibited PAP3 activity, with 50% inhibition at a serpin: protease molar ratio of ~ 2:1. Serpin-7 can form a conventional serpin-protease covalent complex with the PAP3 catalytic domain. Serpin-4 and serpin-5 cannot efficiently inhibit PAP3 activity (50% inhibition at serpin:protease molar ratios of ~ 40:1 and ~ 17:1 by serpin-4 and serpin-5, respectively) (Tong and Kanost, 2005). On the other hand, serpin-3 (Zhu et al., 2003) and serpin-6 (Zou and Jiang, 2005) can form serpin-protease complexes with PAP3, and their inhibition of PAP3 activity is more efficient than serpin-7 with serpin:protease molar ratio of ~ 2:1 for 100% inhibition. It appears that some M. sexta serpins may have partially redundant functions, with multiple serpins able to inhibit a single protease. It is not yet known what proteases serve as natural targets of serpin-7, but it is likely that serpin-7 inhibits exclusively trypsin-type proteases (Supplementary materials, Fig. S3). Our results, demonstrating recombinant serpin-7 can inhibit spontaneous melanization of larval plasma as well as activation of proPO stimulated by exposure to bacteria, indicate that serpin-7 may have a role in regulating the proPO activation cascade and immune responses in M. sexta.
Supplementary Material
Acknowledgments
We thank Sandi Yungeberg for maintaining the insect colony. This work was supported by NIH grant GM41247. Contribution number 13-306-J from the Kansas Agricultural Experiment Station.
Abbreviations
- HP
hemolymph protease
- PAP
prophenoloxidase-activating protease
- proPO
prophenoloxidase
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ibmb.2013.03.015.
References
- An C, Kanost MR. Manduca sexta serpin-5 regulates prophenoloxidase activation and the toll signaling pathway by inhibiting hemolymph proteinase HP6. Insect Biochem. Mol. Biol. 2010;40:683–689. doi: 10.1016/j.ibmb.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Ragan EJ, Kanost MR. Serpin-1 splicing isoform J inhibits the pro-Spätzle-activating proteinase HP8 to regulate expression of antimicrobial hemolymph proteins in Manduca sexta. Dev. Comp. Immunol. 2011;35:135–141. doi: 10.1016/j.dci.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Schneider DS. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLOS. Biol. 2008;6:2764–2773. doi: 10.1371/journal.pbio.0060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerenius L, Lee BL, Söderhäll K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–271. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Christen JM, Hiromasa Y, An C, Kanost MR. Identification of plasma proteinase complexes with serpin-3 in Manduca sexta. Insect Biochem. Mol. Biol. 2012;42:946–955. doi: 10.1016/j.ibmb.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn PE, Drake DR. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm Manduca sexta. J Invertebr. Pathol. 1983;41:77–85. [Google Scholar]
- Gan H, Wang Y, Jiang H, Mita K, Kanost MR. A bacterial-induced, intracellular serpin in granular hemocytes of Manduca sexta. Insect Biochem. Mol. Biol. 2001;31:887–898. doi: 10.1016/s0965-1748(01)00034-0. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettins PGW. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4803. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. Characterization and functional analysis of 12 naturally occurring reactive site variants of serpin-1 from Manduca sexta. J. Biol. Chem. 1997;272:1082–1087. doi: 10.1074/jbc.272.2.1082. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem. Mol. Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu X, Zhu Y, Kanost MR. Prophenoloxidase-activating proteinase-3 (PAP3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem. Mol. Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Gorman MG. Phenoloxidases in insect immunity. In: Beckage N, editor. Insect Immunology. San Diego: Academic Press/Elsevier; 2008. pp. 69–96. [Google Scholar]
- Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 2002;32:1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- Lee E, Linder ME, Gilman AG. Expression of G-protein α subunits in Escherichia coli. Methods Enzymol. 1994;236:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- Li J, Wang Z, Canagarajah B, Jiang H, Kanost M, Goldsmith EJ. The structure of active serpin 1K from Manduca sexta. Structure. 1999;7:103–109. doi: 10.1016/s0969-2126(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Michel K, Suwanchaichinda C, Morlais I, Lambrechts L, Cohuet A, Awono-Ambene PH, Simard F, Fontenille D, Kanost MR, Kafatos FC. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc. Natl. Acad. Sci. U S A. 2006;103:16858–16863. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan EJ, An C, Yang C, Kanost MR. Analysis of mutually-exclusive alternatively spliced serpin-1 isoforms and identification of serpin-1 proteinase complexes in Manduca sexta hemolymph. J. Biol. Chem. 2010;285:29642–29650. doi: 10.1074/jbc.M110.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhart J. Tip of another iceberg: Drosophila serpins. Trends Cell. Biol. 2005;15:659–665. doi: 10.1016/j.tcb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Suwanchaichinda C, Kanost MR. The serpin gene family in Anopheles gambiae. Gene. 2009;442:47–54. doi: 10.1016/j.gene.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanchaichinda C, Paskewitz SM. Effects of larval nutrition, adult body size, and adult temperature on the ability of Anopheles gambiae (Diptera: Culicidae) to melanize sephadex beads. J. Med. Entomol. 1998;35:157–161. doi: 10.1093/jmedent/35.2.157. [DOI] [PubMed] [Google Scholar]
- Tong Y, Jiang H, Kanost MR. Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activation pathway. J. Biol. Chem. 2005;280:14932–14942. doi: 10.1074/jbc.M500532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Kanost MR. Manduca sexta serpin-4 and serpin-5 inhibit the prophenol oxidase activation pathway: cDNA cloning, protein expression, and characterization. J. Biol. Chem. 2005;280:14923–14931. doi: 10.1074/jbc.M500531200. [DOI] [PubMed] [Google Scholar]
- Tsakas S, Marmaras VJ. Insect immunity and its signaling: an overview. Invert. Surviv. J. 2010;7:228–238. [Google Scholar]
- Vavricka CJ, Christensen BM, Li J. Melanization in living organisms: a perspective of species evolution. Protein Cell. 2010;1:830–841. doi: 10.1007/s13238-010-0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Purification and characterization of Manduca sexta serpin-6: a serine proteinase inhibitor that selectively inhibits prophenoloxidase-activating proteinase-3. Insect Biochem. Mol. Biol. 2004;34:387–395. doi: 10.1016/j.ibmb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang Y, Gorman MJ, Jiang H, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J. Biol. Chem. 2003;278:46556–46564. doi: 10.1074/jbc.M309682200. [DOI] [PubMed] [Google Scholar]
- Zou Z, Jiang H. Manduca sexta serpin-6 regulates immune serine proteases PAP-3 and HP8. J. Biol. Chem. 2005;280:14341–14348. doi: 10.1074/jbc.M500570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Picheng Z, Weng H, Mita K, Jiang H. A comparative analysis of serpin genes in the silkworm genome. Genomics. 2009;93:367–375. doi: 10.1016/j.ygeno.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.