Abstract
Objective
A shift towards overall larger very low-density lipoprotein (VLDL), and smaller low-density lipoprotein and high-density lipoprotein (HDL) diameters occurs in insulin resistance (IR), which reflects shifts in the distribution of the subfraction concentrations. Fenofibrate, indicated for hypertriglyceridemia, simultaneously reduces IR and shifts in lipoprotein diameter. Individual responses to fenofibrate vary, and we conducted a genome-wide association study to identify genetic differences that could contribute to such differences.
Methods
Association analysis was conducted between single nucleotide polymorphisms (SNPs) on the Affymetrix 6.0 array and fasting particle diameter responses to a 12-week fenofibrate trial, in 817 related Caucasian participants of the Genetics of Lipid Lowering Drugs and Diet Network. Linear models were conducted, which adjusted for age, sex and study center as fixed effects, and pedigree as a random effect. The top three SNPs associated with each fraction were examined subsequently for associations with changes in subfraction concentrations.
Results
SNPs in AHCYL2 and CD36 genes reached, or closely approached, genome-wide levels of significance with VLDL and HDL diameter responses to fenofibrate, respectively (P=4 × 10−9 and 8 × 10−8). SNPs in AHCYL2 were associated with a decrease in the concentration of the large VLDL subfraction only (P = 0.002). SNPs associated with HDL diameter change were not associated with a single subfraction concentration change (P > 0.05) indicating small shifts across all subfractions.
Conclusion
We report novel associations between lipoprotein diameter responses to fenofibrate and the AHCYL2 and CD36 genes. Previous associations of these genes with IR emphasize the role of IR in mediating lipoprotein response to fenofibrate.
Keywords: AHCYL2, CD36, fenofibrate, inflammation, insulin resistance, insulin signaling, lipoprotein diameter, methylation, PPARγ, subclass
Introduction
Lipoproteins within the fractions of very low-density lipoprotein (VLDL), low-density lipoprotein (LDL) and high-density lipoprotein (HDL) are heterogenous in their composition. Within a given fraction, the constituent lipoprotein particles can be subdivided into several subfractions, based on size. Recent research indicates that the subfraction distribution within each fraction may be more informative in understanding lipoprotein metabolism and its associations with disease risk than traditional lipid measures [1]. As lipoprotein diameters are modifiable through drug, exercise, and dietary interventions [2–4], they may thus present an important epidemiological tool to understand the pathways to insulin resistance (IR) and atherosclerosis, and a clinical target for preventing the development of these conditions.
Increases in the number of small LDL and HDL particles are associated with the development of IR and atherosclerosis [5–7]. These changes in concentration are reflected as shifts to a smaller average LDL and HDL diameter [8], and research has supported similar associations between IR and smaller average LDL/HDL diameters, as well as between these conditions and larger average VLDL diameter [8,9]. Fenofibrate is an efficacious therapeutic agent indicated in hypertriglyceridemia, which reduces plasma triglyceride (TG) levels by 35–50% while conferring additional benefits on HDL-C concentrations and markers of inflammation [10–15]. In addition, fenofibrate is associated with shift towards larger more buoyant LDL particles in a variety of populations, which is associated with a decreased progression of coronary artery disease [16–19]. The effect of fenofibrate on changes in diameter in the other lipoprotein fractions is less well studied, although increases in the average HDL diameter have been reported in mice [20].
There is, however, a significant interindividual variation in the response to fenofibrate across a range of lipid phenotypes [21]. Although several genes have been associated with interindividual variations in the lipid-lowering effects of fenofibrate [22–24], studies have yet to examine which genetic variants may mediate the effects of fenofibrate on the change in the distribution of the lipoprotein subfractions, reflected as a change in the average particle diameter. Genome-wide association studies (GWAS) are advantageous in understanding individual variations as the method is hypothesis free and designed to generate new hypothesis regarding underlying biological pathways. In this study, we performed association tests between single nucleotide polymorphisms (SNPs) and lipoprotein particle diameters, for each fraction of lipoprotein, before and after 3 weeks of daily treatment with fenofibrate in genetically homogeneous Caucasian participants of the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study. In addition, we examined the association of the top three loci associated with the diameter response to fenofibrate for each of VLDL, LDL, and HDL with changes in the concentration of the various subtractions, to further examine the biological change driving the SNP–phenotype associations found.
Methods
Study population
GOLDN is part of the PROgram for GENetic Interaction Network, a group of family intervention studies focusing on gene–environment interactions. The participants in the GOLDN study were mainly rerecruited from two NHLBI Family Heart Study field centers: Minneapolis (Minnesota) and Salt Lake City (Utah). All participants were of European ancestry. Eligibility criteria were: (a) at least 18 years of age; (b) fasting TGs less than 1500mg/dl; (c) willing to participate in the study and attend the scheduled clinic exams; (d) member of a family with at least two members in a sibship; (e) ASTand ALTresults within normal range; and (f) creatinine less than or equal to 2.0mg/dl. Exclusion criteria were: (a) history of liver, kidney, pancreas, gall bladder disease, or malabsorption; (b) current pregnancy; (c) insulin use; (d) use of lipid lowering drugs (including prescription, OTC and nutraceuticals; participants taking these agents were withdrawn from them ≥4 weeks before the study with physician’s approval); (e) use of warfarin; (f) women of childbearing potential not using an acceptable form of contraception; (g) known hypersensitivity to fenofibrate; and (h) history of pancreatitis within 12 months before enrollment. Previous data on these conditions were available from the parent study, and individuals not fulfilling inclusion criteria were not invited to participate. A medication questionnaire was administered on the first visit, which confirmed eligibility for inclusion. A previous study demonstrated that Caucasians in Utah and Minnesota were homogeneous and pooling data across centers would not threaten the validity of this study [25]. From an initial sample size of 1 238 participants 817 agreed to undergo the fenofibrate trial and are included in the analysis.
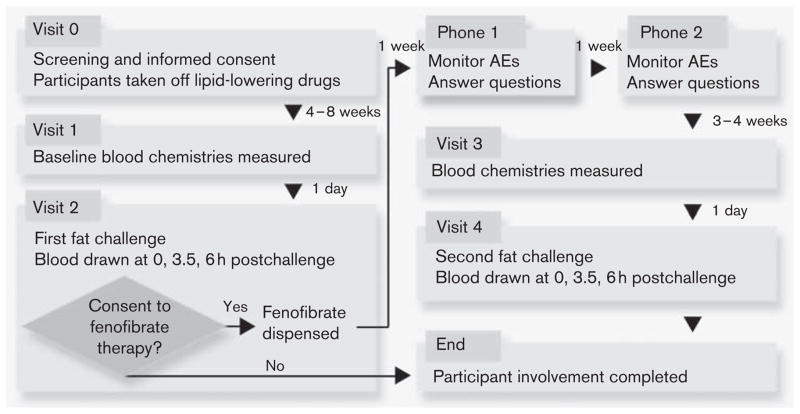
The details of the GOLDN visits are published elsewhere [26] and depicted in Fig. 1. After granting informed consent, participants underwent a baseline screening visit. This visit included a fasting blood draw and pregnancy test, if applicable. The day before the first clinical exam, participants came to the clinic for a fasting blood draw. The fenofibrate intervention consisted of a 3-week treatment period, in which participants took fenofibrate (160mg) daily. Lipoproteins were measured twice on the last 2 days of the treatment period after a minimum 8-h fast.
Fig. 1.
Details of the GOLDN study. Reproduced with permission from Frazier-Wood et al. [26]. AE, adverse events; GOLDN, Genetics of Lipid Lowering Drugs and Diet Network.
Biochemical measurements
All plasma samples used for this analysis were collected after an 8-h fast. All samples were analyzed for lipoprotein profiles once all collections were carried out in each study. Measurements of VLDL, LDL, and HDL diameter were determined by nuclear magnetic resonance (NMR) spectroscopy. NMR detects the signal emitted by lipoprotein methyl-group protons when in the field of a magnet charged at 400MHz. The NMR signal is decoded to obtain estimates of particle numbers for each of several lipoprotein fractions. The weighted average particle diameter for each lipoprotein fraction (VLDL, LDL, and HDL) is calculated as the sum of the average lipoprotein particle diameters multiplied by the relative mass percentage, based on the amplitude of the methyl NMR signal (nm). NMR groups intermediate-density lipoproteins as a subclass of LDL [27,28]. Details of the range of diameters within each subfraction are given in Table 1.
Table 1.
Diameter ranges of lipoprotein subclasses when measured by NMR
| NMR lipoprotein parameter | Diameter range (nm) |
|---|---|
| VLDL | |
| Large VLDL/chylomicrons | > 60 |
| Medium VLDL | 35–60 |
| Small VLDL | 27–35 |
| LDL | |
| Large LDL | 21.2–23 |
| Small LDL | 18–21.2 |
| Medium–small LDL | 19.8–21.2 |
| Very small LDL | 18–19.8 |
| HDL | |
| Large HDL | 8.8–13 |
| Medium HDL | 8.2–8.8 |
| Small HDL | 7.3–8.2 |
Adapted from Jeyarajah et al. [28].
HDL, high-density lipoprotein; LDL, low-density lipoprotein; NMR, nuclear magnetic resonance; VLDL, very low-density lipoprotein.
Genotyping
DNA extraction and purification in the GOLDN study has been described in detail by Irvin et al. [29]. A total of 9 06 600 SNPs were genotyped using the Affymetrix Genome-Wide Human 6.0 array (Affymetrix Inc., Santa Clara, California, USA) and the Birdseed calling algorithm (Broad Institute, Cambridge, Massachusetts, USA) [30]. The samples were processed in two different batches by two different technicians. After the imputation, we created a hybrid dataset that included 2 543 887 SNPs, of which 584 029 were initially genotyped in the GOLDN population. SNPs that were monomorphic (55 530) or had a call rate less than 96% (82 462) were removed from the analysis. In addition, SNPs were excluded from the analysis based on the number of families with Mendelian errors as follows: for minor allele frequency (MAF) of at least 20%, removed if errors were present in more than three families (1486 SNPs); for 20%>MAF≥10%, removed if errors were present in more than two families (1338 SNPs); for 10%>MAF≥5%, removed if errors were present in more than one family (1767 SNPs); for MAF less than 5%, removed if any errors were present (9592 SNPs). In families with remaining errors, the SNPs that showed a Mendelian error were considered as missing (31 595 SNPs). Furthermore, 16 participants with call rates less than 96% were also removed from any subsequent analyses. Subsequently, 748 SNPs failing the Hardy–Weinberg equilibrium test at P-value less than 10−6 were excluded from association analyses. Finally, after excluding markers with MAF less than 1%, Hardy–Weinberg equilibrium (P<10−6), missing strand information, or discrepancies with the mlinfo file, we used the MACH software (version 1.0.16) to impute untyped SNPs using Human Genome Build 36, CEU population, as the reference [31,32]. Missing typed data were considered as missing in the final genotype dataset.
Statistical methods
Outcomes were defined using predicted values from linear regression models (slopes). Where necessary, data were monotonically transformed prefenofibrate and postfenofibrate to normalize the distribution (further details found in Supplementary Table 1, http://links.lww.com/FPC/A496). For raw or normalized data, fasting data from visit 4 (prefenofibrate; Fig. 1) was the outcome, and fasting data from visit 2 (postfenofibrate; Fig. 1) along with age, sex, number of fenofibrate tablets taken per day, and data collection center were predictors.
Tests of genome-wide association
For the initial GWAS, the associations of interest were assessed using linear mixed models, adjusted for sex, age, and center as fixed effects, and phenotypic dependence among family members as a function of their kinship (R software, kinship package [33]). The additive assumption was used to model genotypes. Population substructure was assessed using principal components generated using EIGENSOFT 3.0 software (http://www.genepath.med.harvard.edu/~reich/Software.htm). As the first 10 principal components did not show a significant association with any outcome (P<0.001), they were not included in the mixed models that tested for genotype–phenotype associations. For the initial GWAS, the Bonferroni correction was used to establish genome-wide significance, with the threshold of P less than 2 × 10−8. Genome-wide Manhattan plots were generated to visualize the results (Supplementary Fig. 1, http://links.lww.com/FPC/A498). Quantile–quantile plots were constructed to evaluate deviations from the expected test statistic distribution (Supplementary Fig. 2, http://links.lww.com/FPC/A499). Plots of the top three hits for each phenotype were completed using LocusZoom [34] (Supplementary Fig. 3, http://links.lww.com/FPC/A500).
Post-hoc associations with subfractions
The same models were used in R, with the kinship package. Because of the strong a priori hypothesis a false discovery rate (FDR) correction was used on all significant results (P<0.05) within each fraction and corrected Q values are additionally presented [35].
Gene set-based analysis
A pathway analysis on the results of the GWAS were analyzed using the program gene set-based analysis of polymorphisms (GeSBAP [36]). GeSBAP maps SNPs onto gene (±5 kb) using HapMap data. A gene set-based test is conducted [37] whereby the SNPs that fall within a category defined by Gene Ontology (GO [38]) are combined into a single P-value for that category, corrected for multiple testing using an FDR correction [35]. In addition, the percentage of genes within the GO category that are significantly associated with the phenotype in a gene-based test is given (Supplementary Fig. 4, http://links.lww.com/FPC/A502). To avoid overinterpreting the data, although all results are presented, we discuss only those where the P-value for the biological pathway was significant at an FDR corrected P less than 0.01 and 100% of the genes within the GO biological pathway were significantly associated with the phenotype at an FDR-corrected P less than 0.01.
Results
General characteristics of the GOLDN study population are summarized in Table 2. Half of the participants were female (50.8%). All participants were of European ancestry. In both the prefenofibrate and postfenofibrate conditions, there were no significant differences between men and women for demographic variables or fasting VLDL diameter; however, fasting LDL and HDL diameters varied significantly by sex (both P<0.001). Change in lipoprotein diameter from prefenofibrate to postfenofibrate was significantly different for LDL (Δ=0.12±0.80 nm; P<0.0001) and HDL (Δ=0.12±0.23nm; P<0.0001) diameter, but not VLDL diameter (Δ=0.49±9.17nm; P=0.12).
Table 2.
Means (±SD) or percentages for demographic characteristics and lipoprotein diameters for the GOLDN study participants
| Men | Women | Pa | ||
|---|---|---|---|---|
| Age (years) | 48.3 (15.6) | 48.1 (15.9) | 0.57 | |
| Field center (% from Minnesota) | 49.6 | 49.6 | 0.89 | |
| Age (years) | 48.3 (15.6) | 48.1 (15.9) | 0.57 | |
| Men and women
|
||||
| Prefenofibrate | Postfenofibrate | Δ | ||
| VLDL diameter (nm) | 51.39 (7.83) | 51.87 (8.73) | 0.49 (9.17) | 0.12 |
| LDL diameter (nm) | 20.80 (0.88) | 20.89 (0.58) | 0.12 (0.80) | < 0.0001 |
| HDL diameter (nm) | 8.85 (0.85) | 8.73 (0.40) | 0.12 (0.23) | < 0.0001 |
GOLDN, Genetics of Lipid Lowering Drugs and Diet Network; HDL, high-density lipoprotein; LDL, low-density lipoprotein; VLDL, very low-density lipoprotein.
P-values examining differences in diameters were derived from t-tests.
Initial GWAS
The top three loci for SNP associations with the prefenofibrate to postfenofibrate treatment differences for each lipoprotein diameter are described in Table 3. Variants in the S-adenosylhomocysteine hydrolase-like 2 isoform (AHCYL2) gene, and the CD36 antigen (CD36) genes reached (or nearly approached) genome-wide levels of significance with VLDL and HDL diameter responses to fenofibrate, respectively (P=3.95 × 10−9 and 7.52 × 10−8). The next top two loci reported for each of the fraction are within the cytokinesis 4 (DOCK4) gene with VLDL diameter response (P=1.39 × 10−7 and 1.45 × 10−7), and within the peroxisome proliferator-activated receptor (PPARγC1B) gene for HDL diameter response to fenofibrate (P=2.87 × 10−7). Although there were no associations reaching genome-wide levels of significance with LDL diameter response to fenofibrate, the top three loci were in the jumonji domain containing 1C isoform a (JMJD1C) gene (P=1.81 × 10−6), the phosphodiesterase 10A (PDE10) gene (P=2.01 × 10−6) and near the neurexophilin-1 (NXPH1) gene (P=1.45 × 10−06). As the two SNPs in the DOCK4 gene were in high LD (r>0.8) conditional analysis was run whereby both SNPs were simultaneously modeled as predictors. In this model, neither SNP remained significant (rs10428959: P=0.32; rs6466397: P=0.34), suggesting that either could be causal or they could be tagging the causal variant. There were no other SNPs in LD>r=0.03 in our data, and therefore this question remains open.
Table 3.
Top three genetic loci in SNP-phenotype associations with VLDL, LDL and HDL particle diameter changes in response to fenofibrate in GOLDN study participants
| Marker | Chromosome | Position | HWE | Minor allele | β (SE) | Gene | P |
|---|---|---|---|---|---|---|---|
| VLDL diameter | |||||||
| rs11766298 | 7 | 128792083 | – | T | 4.41 (0.74) | AHCLY2 | 3.95 × 10−9 |
| rs10428959 | 7 | 111465142 | – | T | 6.00 (1.13) | DOCK4 | 1.39 × 10−7 |
| rs6466397 | 7 | 111454471 | – | G | −5.98 (1.13) | DOCK4 | 1.45 × 10−7 |
| LDL diameter | |||||||
| rs10952132 | 7 | 9033345 | > 0.99 | T | 0.31 (0.64) | NXPH1 | 1.45 × 10−6 |
| rs10995485 | 10 | 64696864 | – | G | −1.18 (0.25) | JMJ1C | 1.81 × 10−6 |
| rs519595 | 6 | 165855411 | – | G | −0.60 (0.12) | PDE10 | 2.01 × 10−6 |
| HDL diameter | |||||||
| rs11574703 | 7 | 80124844 | – | T | −3.72 (0.68) | CD36 | 7.52 × 10−8 |
| rs5001812 | 2 | 166412540 | – | T | −4.67 (0.89) | – | 2.17 × 10−7 |
| rs9285640 | 5 | 149122260 | – | G | −29.95 (5.78) | PPARγC1B | 2.87 × 10−7 |
GOLDN, Genetics of Lipid Lowering Drugs and Diet Network; HDL, high-density lipoprotein; HWE, Hardy–Weinberg equilibrium; LDL, low-density lipoprotein; VLDL, very low-density lipoprotein.
Post-hoc associations with subfractions
The decrease in VLDL diameter, associated with variants in the AHCYL2 and DOCK4 genes reflected an association of these variants with a decrease in the concentration of large VLDL particles (P=0.002–0.02; Q=0.01–0.06); these variants were not associated with changes in the concentration of medium or small VLDL particles (P>0.05; Table 4). The top three loci associated with an increase in LDL diameter reflected similar associations between these loci and changes across all five subfractions of the LDL fraction (all P<0.02; Q<0.02; Table 5). Finally, the associations of HDL diameter response to fenofibrate with variants in the CD36 and PPARγC1B genes seem to reflect small changes across all subfraction concentrations, and are not driven by significant changes in the concentration of any of the HDL subfractions (all P>0.05; Table 6).
Table 4.
The association of the top three genetic loci in SNP-phenotype associations VLDL particle diameters response to fenofibrate with VLDL subfraction concentration responses to fenofibrate
| Marker | Large VLDL
|
Medium VLDL
|
Small VLDL
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P | Q | β (SE) | P | Q | β (SE) | P | Q | |
| rs11766298 | 1.86 (0.78) | 0.02 | 0.06 | −1.40 (0.77) | 0.07 | – | −1.12 (0.90) | 0.21 | – |
| rs10428959 | 3.58 (1.14) | 0.002 | 0.01 | −1.32 (1.16) | 0.26 | – | −1.02 (1.39) | 0.46 | – |
| rs6466397 | −3.58 (1.14) | 0.002 | 0.01 | 1.31 (1.16) | 0.26 | – | 1.02 (1.39) | 0.46 | – |
SNPs, single nucleotide polymorphisms; VLDL, very low-density lipoprotein.
Table 5.
The association of the top three genetic loci in SNP-phenotype associations LDL particle diameters response to fenofibrate with LDL subfraction concentration responses to fenofibrate
| Marker | Large LDL
|
Medium–small LDL
|
Small LDL
|
Very small LDL
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P | Q | β (SE) | P | Q | β (SE) | P | Q | β (SE) | P | Q | |
| rs10952132 | 0.29 (0.65) | 7.31 × 10−6 | 3.00 × 10−5 | −0.16 (0.69) | 0.02 | 0.02 | −0.29 (0.07) | 2.26 × 10−5 | 4.8 × 10−5 | −0.29 (0.07) | 4.13 × 10−5 | 6.85 × 10−5 |
| rs10995485 | −1.04 (0.25) | 3.35 × 10−5 | 6.00 × 10−5 | 1.12 (0.25) | 1.16 × 10−5 | 3.00 × 10−5 | 0.82 (0.26) | 0.002 | 0.002 | 0.75 (0.27) | 0.006 | 0.007 |
| rs519595 | −0.58 (0.13) | 5.02 × 10−6 | 3.00 × 10−5 | 0.44 (0.14) | 0.001 | 0.001 | 0.58 (0.13) | 1.17 × 10−5 | 3.00 × 10−5 | 0.52 (0.14) | 0.0002 | 3.00 × 10−4 |
LDL, low-density lipoprotein; SNPs, single nucleotide polymorphisms.
Table 6.
The association of the top three genetic loci in SNP-phenotype associations HDL particle diameters response to fenofibrate with HDL subfraction concentration responses to fenofibrate
| Marker | Large HDL
|
Medium HDL
|
Small HDL
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P | Q | β (SE) | P | Q | β (SE) | P | Q | |
| rs11574703 | −0.62 (0.70) | 0.38 | – | 1.85 (1.23) | 0.13 | – | 0.24 (0.69) | 0.73 | – |
| rs5001812 | −1.09 (0.91) | 0.23 | – | −0.66 (1.99 | 0.75 | – | 2.77 (0.90) | 0.002 | 0.01 |
| rs9285640 | 3.45 (5.88) | 0.56 | – | −13.27 (15.73) | 0.39 | – | 7.33 (5.85) | 0.21 | – |
HDL, high-density lipoprotein; SNPs, single nucleotide polymorphisms.
There were no associations (FDR corrected Q>0.10) between any of our lipoprotein diameter responses to fenofibrate with the 95 SNPs recently reported as being associated with fasting HDL-cholesterol, LDL-cholesterol, total cholesterol, or TG levels in a recent meta-analysis of 46 GWAS of these traits ([39]; Supplementary Table 2, http://links.lww.com/FPC/A503).
Gene-based analysis
GeSBAP identified a number of biological pathways implicated in lipoprotein diameter response to fenofibrate (Supplementary Fig. 4, http://links.lww.com/FPC/A502). Gene sets where 100% of the GO biological pathways were significantly associated with one of the phenotypes showed that neuron adhesion pathways (P=4.1 × 10−2) were associated with the response of LDL diameter to fenofibrate, and pathways regulating insulin receptor signaling (P=1.9 × 10−2) and interleukin-6 (IL-6; P=4.6 × 10−3) were associated with HDL diameter responses to fenofibrate. There were no gene categories where 100% of the included genes were significantly associated with VLDL diameter response to fenofibrate.
Discussion
This is, to our knowledge, the first study to look at genetic associations with lipoprotein subfraction responses to fenofibrate. We used the GWAS data as a discovery mechanism to reveal associations between a variant in the AHCYL2 gene with VLDL diameter response to fenofibrate, which reached genome-wide levels of significance. In addition, variants in the CD36, PPARγC1B, and JMJD1C genes were associated with HDL (CD36, PPARγC1B) and LDL (JMJD1C) diameter responses to fenofibrate, at levels approaching genome-wide significance, and may have biological relevance through previous associations with NMR and IR phenotypes.
We saw one locus that was associated with particle diameter responses to fenofibrate at genome-wide levels of significance. The response of VLDL diameter to fenofibrate was significantly associated with a single variant (rs11766298) in the AHCYL2 gene on chromosome 7. The association of AHCYL2 with VLDL diameter change results from decreases in the concentration of large VLDL particles also associated with AHCYL2. The protein encoded by AHCYL2 acts as a homotetramer and may be involved in the conversion of S-adenosylhomocysteine to L-homocysteine and adenosine. High levels of S-adenosylhomocysteine inhibit methylation [40], are associated with IR [41] and may be a sensitive indicator of CVD [42].
An intronic variant of the CD36 gene (rs11574703) was suggestively associated with a change in diameter for HDL particles, in that the association approached genome-wide levels of significance. The various subfractions of lipoprotein compete to bind in their oxidized state to the CD36 receptor, which thus may contribute to the regulation of lipid metabolism, and to the pathogenesis of atherosclerosis [43]. Null mutations in the CD36 gene, in mice, are associated with an increase in cholesterol, mainly within the HDL fraction, and additionally an increase in triacylglycerol within the very small LDL fraction [44]. Our study is the first study to suggest that CD36 mutations may affect the response of the content of HDL lipoproteins to fenofibrate, in humans.
In addition, two variants approached genome-wide levels of significance (P<2 × 10−6) and warrant further examination in their association with the fenofibrate responses of particle diameters, because of the previous associations with related phenotypes. We observed associations with markers in the PPARγC1B (rs9285640) gene with HDL response to fenofibrate and in the intronic region of JMJ1C (rs10995485) gene with LDL diameter response to fenofibrate. PPARγC1B has been associated previously with the homeostatic model assessment of IR, lipid-induced IR and type 2 diabetes [45–48]. As our analysis suggests that PPARγC1B mediates the effect of fenofibrate, the role of IR, similarly associated with PPARγC1B, in mediating the responses of lipoproteins to fenofibrate should be further examined. The preliminary evidence that polymorphisms associated IR may mediate lipoprotein diameter responses to fenofibrate is strengthened by our gene set-based analysis which showed that the insulin signaling receptor biological pathway, as well as the IL-6 inflammation pathway, were associated with HDL diameter response to fenofibrate. The nominal association of JMJ1C with VLDL diameter response to fenofibrate is interesting as JMJC1 has been associated previously with total number of VLDL parameters (particle concentration) in a recent GWAS [49]. This GWAS report represented first lipoprotein-related association with this gene, and here we additionally report that JMJ1C may also associate with LDL subfraction response to fenofibrate, whereby the changes in concentration of all subfractions of LDL are mediated by individual differences in variants on the JMJ1C gene.
Our study should be seen in the light of a number of possible limitations. First, we limited our analysis to Caucasians of European descent at an a priori lower risk of cardiovascular disease, thus generalizations from our findings may be limited. Second, we assumed an additive genetic model, and thus may have missed loci that associate with fenofibrate response through other modes of inheritance, and any epistatic genetic effects. Third, information on covariates that may affect fenofibrate response, such as renal function, was limited in our cohort; therefore, we are unable to more finely understand the biology underlying our top hits. Fourth, while we consider our sample size excellent for a clinical trial, it is more modest for a current day GWAS; therefore lack of power may explain why only one hit reached genome-wide significance, and we hope the data here may inform more targeted follow-up studies. Finally, as this is the first study to examine genome-wide predictors of NMR responses to fenofibrate, replication remains a key issue. However, this is especially challenging in the context of both a clinical trial, and with the use of the newer phenotypes revealed through NMR data, which may be unavailable in existing datasets. We are unaware of any be other studies holding NMR, GWAS, and fenofibrate response data. Thus, as the results from the gene-based analysis support our GWAS findings and fit with previous biological findings from independent groups, they are exploratory and should be considered suggestive only, and a basis for future research paradigms.
Nonetheless, we provide here important evidence that genetic variants in the AHCLY2, PPARγC1B, and JMJ1C genes are suggestively associated with the responses of particle diameters to fenofibrate. In particular, a variant associated previously with the conversion of S-adenosylhomocysteine to L-homocysteine and adenosine, and variants involved in the insulin receptor signaling pathway and the IL-6 inflammation related pathways may be an important mediator of responses to fenofibrate. As lipoprotein diameters could present a therapeutic target in clinical interventions for IR and atherosclerosis, the associations of these genes, and the role these biological pathways may play in preventing incident disease under therapeutic interventions, warrants further research.
Supplementary Material
Acknowledgments
GOLDN is funded by National Heart, Lung, and Blood Institute grant U01HL072524-04.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Website (www.pharmacogeneticsandgenomics.com).
Conflicts of interest
There are no conflicts of interest.
References
- 1.Petersen AK, Stark K, Musameh MD, Nelson CP, Römisch-Margl W, Kremer W, et al. Genetic associations with lipoprotein subfractions provide information on their biological nature. Hum Mol Genet. 2012;21:1433–1443. doi: 10.1093/hmg/ddr580. [DOI] [PubMed] [Google Scholar]
- 2.Lemieux I, Laperrière L, Dzavik V, Tremblay G, Bourgeois J, Després JP, et al. A 16-week fenofibrate treatment increases LDL particle size in type IIA dyslipidemic patients. Atherosclerosis. 2002;162:363–371. doi: 10.1016/s0021-9150(01)00711-0. [DOI] [PubMed] [Google Scholar]
- 3.Dreon DM, Fernstrom HA, Williams PT, Tremblay G, Bourgeois J, Després JP. Reduced LDL particle size in children consuming a very-low-fat diet is related to parental LDL-subclass patterns. Am J Clin Nutr. 2000;71:1611–1616. doi: 10.1093/ajcn/71.6.1611. [DOI] [PubMed] [Google Scholar]
- 4.Williams PT, Krauss RM, Vranizan KM, Albers JJ, Wood PD. Effects of weight-loss by exercise and by diet on apolipoproteins A-I and A-II and the particle-size distribution of high-density lipoproteins in men. Metabolism. 1992;41:441–449. doi: 10.1016/0026-0495(92)90082-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hulthe J, Bokemark L, Wikstrand J, Fagerberg B. The metabolic syndrome, LDL particle size, and atherosclerosis: the Atherosclerosis and Insulin Resistance (AIR) study. Arterioscler Thromb Vasc Biol. 2000;20:2140–2147. doi: 10.1161/01.atv.20.9.2140. [DOI] [PubMed] [Google Scholar]
- 6.Gray RS, Robbins DC, Wang W, Yeh JL, Fabsitz RR, Cowan LD, et al. Relation of LDL size to the insulin resistance syndrome and coronary heart disease in American Indians. The Strong Heart Study. Arterioscler Thromb Vasc Biol. 1997;17:2713–2720. doi: 10.1161/01.atv.17.11.2713. [DOI] [PubMed] [Google Scholar]
- 7.Mora S, Szklo M, Otvos JD, Greenland P, Psaty BM, Goff DC, Jr, et al. LDL particle subclasses, LDL particle size, and carotid atherosclerosis in the Multi-Ethnic Study of Atherosclerosis (MESA) Atherosclerosis. 2007;192:211–217. doi: 10.1016/j.atherosclerosis.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Garvey WT, Kwon S, Zheng D, Shaughnessy S, Wallace P, Hutto A, et al. Effects of insulin resistance and type 2 diabetes on lipoprotein subclass particle size and concentration determined by nuclear magnetic resonance. Diabetes. 2003;52:453–462. doi: 10.2337/diabetes.52.2.453. [DOI] [PubMed] [Google Scholar]
- 9.Wood AC, Glasser S, Garvey WT, Kabagambe EK, Borecki IB, Tiwari HK, et al. Lipoprotein lipase S447X variant associated with VLDL, LDL and HDL diameter clustering in the metabolic syndrome. Lipids Health Dis. 2011;10:143. doi: 10.1186/1476-511X-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebestjen M, Keber I, Zegura B, Simcic S, Bozic M, Fressart MM, Stegnar M. Statin and fibrate treatment of combined hyperlipidemia: the effects on some novel risk factors. Thromb Haemost. 2004;92:1129–1135. doi: 10.1160/TH03-04-0250. [DOI] [PubMed] [Google Scholar]
- 11.Rosenson RS. Effect of fenofibrate on adiponectin and inflammatory biomarkers in metabolic syndrome patients. Obesity. 2009;17:504–509. doi: 10.1038/oby.2008.530. [DOI] [PubMed] [Google Scholar]
- 12.Rosenson RS, Huskin AL, Wolff DA, Helenowski IB, Rademaker AW. Fenofibrate reduces fasting and postprandial inflammatory responses among hypertriglyceridemia patients with the metabolic syndrome. Atherosclerosis. 2008;198:381–388. doi: 10.1016/j.atherosclerosis.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Jonkers IJ, Mohrschladt MF, Westendorp RG, van der Laarse A, Smelt AH. Severe hypertriglyceridemia with insulin resistance is associated with systemic inflammation: reversal with bezafibrate therapy in a randomized controlled trial. Am J Med. 2002;112:275–280. doi: 10.1016/s0002-9343(01)01123-8. [DOI] [PubMed] [Google Scholar]
- 14.Okopien B, Krysiak R, Kowalski J, van der Laarse A, Smelt AH. The effect of statins and fibrates on interferon-gamma and interleukin-2 release in patients with primary type II dyslipidemia. Atherosclerosis. 2004;176:327–335. doi: 10.1016/j.atherosclerosis.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Knopp RH, Walden CE, Warnick GR, Albers JJ, Ginsberg J, McGinnis BM. Effect of fenofibrate treatment on plasma lipoprotein lipids, high-density lipoprotein cholesterol subfractions, and apolipoproteins B, AI, AII, and E. Am J Med. 1987;83:75–84. doi: 10.1016/0002-9343(87)90875-8. [DOI] [PubMed] [Google Scholar]
- 16.Diabetes Atherosclerosis Intervention Study Investigators. Effect of fenofibrate on progression of coronary-artery disease in type 2 diabetes: the Diabetes Atherosclerosis Intervention Study, a randomised study. Lancet. 2001;357:905–910. [PubMed] [Google Scholar]
- 17.Feher MD, Caslake M, Foxton J, Cox A, Packard CJ. Atherogenic lipoprotein phenotype in type 2 diabetes: reversal with micronised fenofibrate. Diabetes Metab Res Rev. 1999;15:395–399. doi: 10.1002/(sici)1520-7560(199911/12)15:6<395::aid-dmrr65>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 18.Filippatos TD, Gazi IF, Liberopoulos EN, Athyros VG, Elisaf MS, Tselepis AD, Kiortsis DN. The effect of orlistat and fenofibrate, alone or in combination, on small dense LDL and lipoprotein-associated phospholipase A2 in obese patients with metabolic syndrome. Atherosclerosis. 2007;193:428–437. doi: 10.1016/j.atherosclerosis.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Vakkilainen J, Steiner G, Ansquer J-C, Perttunen-Nio H, Taskinen MR. Fenofibrate lowers plasma triglycerides and increases LDL particle diameter in subjects with type 2 diabetes. Diabetes Care. 2002;25:627–628. doi: 10.2337/diacare.25.3.627. [DOI] [PubMed] [Google Scholar]
- 20.Bouly M, Masson D, Gross B, Perttunen-Nio H, Taskinen MR. Induction of the phospholipid transfer protein gene accounts for the high density lipoprotein enlargement in mice treated with fenofibrate. J Biol Chem. 2001;276:25841–25847. doi: 10.1074/jbc.M101160200. [DOI] [PubMed] [Google Scholar]
- 21.Keating GM, Ormrod D. Micronised fenofibrate: an updated review of its clinical efficacy in the management of dyslipidaemia. Drugs. 2002;62:1909–1944. doi: 10.2165/00003495-200262130-00013. [DOI] [PubMed] [Google Scholar]
- 22.Cesari M, Penninx BW, Newman AB, Kritchevsky SB, Nicklas BJ, Sutton-Tyrrell K. Inflammatory markers and onset of cardiovascular events: results from the Health ABC study. Circulation. 2003;108:2317–2322. doi: 10.1161/01.CIR.0000097109.90783.FC. [DOI] [PubMed] [Google Scholar]
- 23.Cullen P. Evidence that triglycerides are an independent coronary heart disease risk factor. Am J Cardiol. 2000;86:943–949. doi: 10.1016/s0002-9149(00)01127-9. [DOI] [PubMed] [Google Scholar]
- 24.Hausenloy DJ, Yellon DM. Targeting residual cardiovascular risk: raising high-density lipoprotein cholesterol levels. Postgrad Med J. 2008;84:590–598. doi: 10.1136/hrt.2007.125401. [DOI] [PubMed] [Google Scholar]
- 25.Pankow JS, Province MA, Hunt SC, Arnett DK. Regarding ‘testing for population subdivision and association in four case-control studies’. Am J Hum Genet. 2002;71:1478–1480. doi: 10.1086/344582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frazier-Wood AC, Ordovas JM, Straka RJ, Hixson JE, Borecki IB, Tiwari HK, Arnett DK. The PPAR alpha gene is associated with triglyceride, low-density cholesterol and inflammation marker response to fenofibrate intervention: the GOLDN study. Pharmacogenomics J. 2012 doi: 10.1038/tpj.2012.9. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otvos JD, Jeyarajah EJ, Bennett DW, Krauss RM. Development of a proton nuclear magnetic resonance spectroscopic method for determining plasma lipoprotein concentrations and subspecies distributions from a single, rapid measurement. Clin Chem. 1992;38:1632–1638. [PubMed] [Google Scholar]
- 28.Jeyarajah EJ, Cromwell WC, Otvos JD. Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clin Lab Med. 2006;26:847–870. doi: 10.1016/j.cll.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Irvin MR, Kabagambe EK, Tiwari HK, Parnell LD, Straka RJ, Tsai M, et al. Apolipoprotein E polymorphisms and postprandial triglyceridemia before and after fenofibrate treatment in the Genetics of Lipid Lowering and Diet Network (GOLDN) Study. Circ Cardiovasc Genet. 2010;3:462–467. doi: 10.1161/CIRCGENETICS.110.950667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Willer CJ, Sanna S, Abecasis GR. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atkinson B, Thereau T. Kinship: mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees. R package version; 1(1.0–17) 2007 Available at: http://www.crantastic.org/packages/kinship.
- 34.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 36.Medina I, Montaner D, Bonifaci N, Pujana MA, Carbonell J, Tarraga J, et al. Gene set-based analysis of polymorphisms: finding pathways or biological processes associated to traits in genome-wide association studies. Nucleic Acids Res. 2009;37:W340–W344. doi: 10.1093/nar/gkp481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Shahrour F, Arbiza L, Dopazo H, Huerta-Cepas J, Mínguez P, Montaner D, Dopazo J. From genes to functional classes in the study of biological systems. BMC Bioinformatics. 2007;8:114. doi: 10.1186/1471-2105-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang PK, Gordon RK, Tal J, Zeng GC. S-adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- 41.Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci USA. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerins DM, Koury MJ, Capdevila A, Rana S, Wagner C. Plasma Sadenosylhomocysteine is a more sensitive indicator of cardiovascular disease than plasma homocysteine. Am J Clin Nutr. 2001;74:723–729. doi: 10.1093/ajcn/74.6.723. [DOI] [PubMed] [Google Scholar]
- 43.Calvo D, Gomez-Coronado D, Suarez Y, Lasunción MA, Vega MA. Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL. J Lipid Res. 1998;39:777–788. [PubMed] [Google Scholar]
- 44.Febbraio MA. Null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem. 1999;274:19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 45.Oberkofler H, Pfeifenberger A, Soyal S, Felder T, Hahne P, Miller K, et al. Aberrant hepatic TRIB3 gene expression in insulin-resistant obese humans. Diabetologia. 2010;53:1971–1975. doi: 10.1007/s00125-010-1772-2. [DOI] [PubMed] [Google Scholar]
- 46.Hoeks J, Hesselink MKC, Russell AP, Mensink M, Saris WH, Mensink RP, Schrauwen P. Peroxisome proliferator-activated receptor-gamma coactivator-1 and insulin resistance: acute effect of fatty acids. Diabetologia. 2006;49:2419–2426. doi: 10.1007/s00125-006-0369-2. [DOI] [PubMed] [Google Scholar]
- 47.Park KS, Shin HD, Park BL, Cheong HS, Cho YM, Lee HK, et al. Putative association of peroxisome proliferator-activated receptor gamma coactivator 1beta (PPARGC1B) polymorphism with type 2 diabetes mellitus. Diabet Med. 2006;23:635–642. doi: 10.1111/j.1464-5491.2006.01882.x. [DOI] [PubMed] [Google Scholar]
- 48.Sun L, Yang Z, Jin F, Zhu XQ, Qu YC, Shi XH, Wang L. The Gly482Ser variant of the PPARGC1 gene is associated with type 2 diabetes mellitus in northern Chinese, especially men. Diabet Med. 2006;23:1085–1092. doi: 10.1111/j.1464-5491.2006.01949.x. [DOI] [PubMed] [Google Scholar]
- 49.Chasman DI, Paré G, Mora S, Hopewell JC, Peloso G, Clarke R, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.