Abstract
13–197 is a novel NF-ĸB inhibitor that shows promising in-vitro efficacy data against pancreatic cancer. In this study, we characterized the pharmacokinetics (PK), tissue distribution, protein binding, and metabolism of 13–197 in mice and rats. A valid, sensitive, and selective LC-MS/MS method was developed. This method was validated for the quantification of 13–197, in the range of 0.1–500 ng/ml in mouse plasma, liver, kidney, lung, heart, spleen, brain, urine and feces. 13–197 has low bioavailability of 3 and 16% in mice, and rats respectively. 13–197 has faster absorption in mice with 12-fold shorter Tmax than in rats. Tissue concentrations were 1.3–69.2 folds higher in mice than in rats at 72h after intravenous administration. 13–197 is well distributed to the peripheral tissues and has relatively high tissue: plasma concentration ratios, ranging from 1.8 to 3634, in both mice and rats. 13–197 also demonstrated more than 99% binding to plasma proteins in both mice and rats. Finally, less than 1% of 13–197 is excreted unchanged in urine and feces, and metabolite profiling studies detected more than 20 metabolites in mice and rats plasma, urine, and feces, which indicates 13–197 is extensively metabolized and primarily eliminated by metabolism rather than by excretion.
Keywords: LC-MS/MS, NF-кB inhibitor 13–197, Pharmacokinetics, Protein binding, Metabolism
Introduction
Pancreatic cancer (PC) is one of the most lethal human diseases, with a 5-year survival rate of < 4% and a median survival of less than 6 months (Ji et al., 2007). PC is resistant to most treatments, including chemotherapy, radiation, and several combination therapies. Gemcitabine, a nucleoside analog, is the current chemotherapeutic agent of choice for PC. However, gemcitabine has a very low response rate and the initial responders to gemcitabine, typically develop chemoresistance rapidly (el-Kamar et al., 2003). Recent studies have shown that NF-кB is up regulated in chemoresistant PC. NF-кB is constitutively over expressed in most tumor cell lines and in many tumor tissues derived from patients, but not in normal tissues (Aggarwal et al., 2004). In addition, the clinically silent onset of PC has been attributed to the upregulation of pro-inflammatory pathways including NF-кB (Farrow and Evers, 2002). NF-кB has been shown to regulate the expression of over 200 immune, growth, and inflammation genes (Sen and Baltimore, 1986; Baldwin, 1996). This knowledge triggered extensive efforts to investigate the potential of NF-кB inhibitors for PC therapy.
Currently used drugs for the treatment of advanced PC’s such as curcumin, parthenolide, and genistein have been shown to inhibit NF-кB (Kwok et al., 2001; Kunnumakkara et al., 2007; Holcomb et al., 2008). Although many compounds were identified as NF-кB inhibitors, only a handful has progressed to clinical stages of drug development. This high attrition rate is attributed to the pleiotropic nature of NF-кB, which makes it both an attractive and a challenging target. The extensive efforts in this area have led to identifying a quinoxaline urea analog (13–197), which inhibits the activation of NF-кB via κB kinase β (IKKβ), in PC cell lines. IKKβ is a key kinase activator in the NF-κB pathway. In addition, 13–197 induces caspace-mediated apoptosis and can inhibit tumor growth in vivo as well as the growth of a panel of PC cell lines with low (1–5µM) IC50. Preliminary data demonstrated that 13–197 has comparable or stronger NF-ĸB inhibition compared to curcumin and parthenolide.
Pharmacokinetics (PK)-related issues account for more than 50% of drug development failures, preventing new chemical entities (NCEs) from reaching the market (Cheng et al., 2002). As a result, in addition to paying attention to the traditional concern of attaining potency and selectivity towards the biological targets of interest, PK considerations have been moved to early stages of drug discovery, which represents a significant paradigm shift in the approach of drug discovery and development in pharmaceutical industry (Clark and Grootenhuis, 2002). Therefore, the promising in vitro efficacy data of 13–197 has triggered our efforts to characterize its preclinical PK profile in mice and rats. To characterize the PK profile of 13–197, a valid, sensitive, and selective bioanalytical method with high sensitivity, simple sample preparation, and short run time was developed to quantify 13–179 in biological tissues and fluids. This liquid chromatography-tandem mass spectrometry (LC-MS/MS) method utilized ultraperformance liquid chromatography (UPLC) and hybrid ion trap-triple quadrupole (Q-Trap) MS. The method was validated to ensure precise and accurate measurements according to FDA guidelines. In addition, the metabolic profile was investigated to identify major metabolites in rats and mice plasma, urine, and feces using information-dependent acquisition MS/MS methods. Finally, binding to rat and mouse plasma proteins was also determined using the blood/plasma partitioning method.
Material and methods
Chemicals and reagents
13–197 was synthesized and purified (> 99%) in Dr’s Amarnath Natarjan’s Laboratory. Efavirenz (EFV) was obtained from Hetero Labs Ltd. (Hyderabad, India). HPLC-grade methanol, acetonitrile, ammonium acetate, ammonium formate, ammonium hydroxide, formic acid, and acetic acid were obtained from Fisher Scientific (Fair Lawn, NJ, USA).
Liquid chromatographic and mass spectrometric conditions for 13–97 quantification
A Waters ACQUITY ultra-performance liquid chromatography (UPLC) system (Waters, Milford, MA) coupled to a 4000 Q TRAP® quadrupole linear ion trap hybrid mass spectrometer with an electrospray ionization (ESI) source (Applied Biosystems, MDS Sciex, Foster City, CA) was used throughout. All chromatographic separations were performed with an ACQUITY UPLC® BEH Shield RP18 column (2.1×100mm, 1.7µm; Waters) equipped with an ACQUITY UPLC C18 guard column (Waters, Milford, MA).
Mobile phase A consisted of 7.5 mM ammonium formate (pH-3.0) and mobile phase B comprised of 5% acetonitrile (ACN) in methanol (MeOH). The initial mobile phase composition was 82.5% B for the first 3.5 min and was gradually increased to 90% B in 0.1 min and held constant for 2 min. Mobile phase B was then reset to 82.5% in 0.15 min and the column was equilibrated for 2.25 min before the next injection. A flow rate of 0.3 ml/min was used and the injection volume of all samples was 10 µl. MS/MS analyses were performed with negative ESI mode and using the following parameters: ion spray voltage, −4500 V; source temperature, 550 °C; curtain gas (nitrogen), 10 arbitrary units; and collision gas (nitrogen), “High”. Specific detection was performed by monitoring the transition 473.1→276 m/z for 13–197 and 313→243 m/z for IS.
Chromatographic and mass spectrometric conditions for metabolite identification
Mass shift analyses were performed using the same LC-MS/MS system. Data analyses were performed using Analyst 1.5.2 and LightSight 2.2.1 software (Applied Biosystems, Foster City, CA). The same mobile phase as described above was also used. The gradient profile was held at 10% mobile phase B for 1 min, increased to 95% mobile phase B from 1 to 25 min. Mobile Phase B was then held at 95% for 2 min and brought back to 10% in 1 min, followed by 7 min equilibration. The flow rate was 0.25 ml/min and the injection volume was 10 µl.
MRM-information dependent acquisition-enhanced product ion (MRM-IDA-EPI) scans were used for the identification of the metabolites, which resulted in the identification of a higher number of metabolites compared to neutral loss (NL), precursor ion (PI), and enhanced mass spectrometry (EMS) scans. The details of our approach in metabolite identification was previously described (Huang et al., 2010; Huang et al., 2012; Zheng et al., 2012). Briefly, EPI spectra of analytes were collected to identify the three most abundant fragments for each analyte. MRM channels of the parent compound and MRM channels for ”guessed" metabolites were used as survey scans. The MRM channels for guessed metabolites based on predicted biotransformation pathways were generated by LightSight 2.2.1 software.
Sample preparation
Simple protein precipitation using ice-cold ACN was used for sample clean up. One ml of ice-cold ACN was added to 100 µl plasma samples pre-spiked with 10 µl of 4.0 µg/ml IS (EFV). Samples were then vortexed, and centrifuged at 16,000 × g for 10 min. The supernatant was aspirated, evaporated under vacuum, and reconstituted in a 100 µl 75% methanol. After centrifugation at 16,000 × g for 10 min, 10 µl of each sample was used for LC-MS/MS analysis. Liver, kidney, lung, heart spleen, and brain tissues were homogenized in deionized water (1:2 w/v). 100 µl aliquots of tissue homogenates were spiked with 10 µl of 4.0 µg/ml IS (EFV) and were extracted and analyzed similarly to plasma samples. Urine samples were prepared by solid phase extraction (SPE) using 1 cc Oasis HLB cartridges. 100 µl aliquots of urine samples were spiked with 10 µl IS, vortexed, and loaded onto SPE cartridges pre-conditioned with 1ml MeOH, followed by 1ml H2O. Loaded cartridges were washed with 1ml H2O and eluted with 1ml MeOH. The eluate was evaporated under vacuum at room temperature and reconstituted in a 100 µl of 75% MeOH. Feces were homogenized in deionized water (1:8 w/v), and a 100 µl aliquots were extracted similarly to plasma samples. The final concentration of IS in all samples was 400 ng/ml.
Calibration curves
Fixed volumes (100 µl) of plasma samples were spiked with 10 µl of the appropriate standard solution of 13–197 to construct a calibration curve in the range of 0.1–500 ng/ml. The concentrations of QC samples were 0.1, 0.5, 10, 100, and 500 ng/ml concentration. Efavirenz (EFV) was used as an internal standard (IS) at final concentration of 400 ng/ml. Liver, kidney, lung, heart, brain, and spleen tissues were homogenized in deionized water at 1:2 w/v, whereas feces were homogenized in deionized water at 1:8 w/v ratio. The dynamic range of the calibration curve in tissues, urine, and feces was 0.2–500 ng/ml. 100 µl aliquots of the resulting homogenates were spiked with 10 µl of the appropriate standard solution containing 4.0 µg/ml IS (EFV) to yield samples with 400 ng/ml final concentration of IS. Similarly, QC samples of concentrations 0.2, 0.5, 10, 100, and 500 ng/ml were prepared in tissues, urine, and feces. Plasma, tissues, urine, and feces samples were then extracted as described in the “sample preparation” section. Calibration curves (area ratio of analyte:IS vs. nominal concentration) were fitted by least-squares linear regression using 1/x2 weighting scheme. Extraction recoveries were determined as the ratio of the analyte peak area in samples spiked before extraction compared to the corresponding peak area in untreated samples prepared in neat solution at 0.2, 0.5, 10, 100, and 500 ng/ml.
Analytical method validation
The method was validated using 5 QC points for each calibration curve and the concentrations of the QC points were 0.1, 0.5, 10, 100, and 500 ng/ml for plasma, whereas the concentrations of the QC points were 0.2, 0.5, 10, 100, and 500 ng/ml for liver, kidney, lung, heart, spleen, brain, urine, and feces. Accuracy and precision of the method were determined by assaying five replicates of each QC point using freshly prepared calibration curves in three separate runs. Intra-day accuracy and precision were calculated from the % bias [%(measured− theoretical)/measured concentration] and relative standard deviation [%RSD = %standard deviation/mean], respectively, for the 5 replicates of each QC point. Inter-day precision was calculated similarly using the 15 replicates of each QC point from the three validation runs. Accuracy and precision were considered to be acceptable when they were found to be less than 15%, except for the LLOQ, where 20% deviation was allowed. The limit of detection (LOD) was defined as the concentration that produced a signal three times above the noise level.
Animals
Eight-week-old male Sprague-Dawley rats (250±25 g) and eight-week-old male BALB/c mice were purchased from Charles River Laboratories (Wilmington, MA). Sterilized 7012 Teklad and LM-485 diets (Harlan, Madison, WI) were used for mice and rats, respectively, and water was provided ad libitum. Rat and mice were housed in the UNMC laboratory animal facility according to the American Animal Association Laboratory Animal Care guidance. All protocols and procedures were approved by the Institutional Animal Care and Use Committee at the University of Nebraska Medical Center.
Drug formulation for oral and intravenous dose administration
The oral dose of 13–197 was 50 mg/kg and the intravenous dose was 5 mg/kg in both mice and rats. In mice, the 50 mg/kg oral dose was administered as 250 µl of a 5 mg/ml 13–197 solution in a mixture of DMSO-PEG400-PG-EtOH-Cremophore-PBS (2/20/10/10/5/53 % v/v). The 5 mg/kg intravenous dose was administered as a 100 µl of 1.25 mg/ml 13–197 solution in the same DMSO-PEG400-PG-EtOH-Cremophore-PBS mixture. For rats, the 50 mg/kg oral dose was administered as 1 ml of 12.5 mg/ml 13–197 solution in a mixture of DMSO-PG-PEG400 (20/30/50 v/v). The 5 mg/kg intravenous dose was administered as 250 µl of 5 mg/ml 13–197 solution in DMSO. Oral doses were administered via oral gavage, whereas intravenous doses were administered via the tail vein for both mice and rats. After dosing, mice and rats were returned to their home cage, and cage-side observation was performed on the day of dosing and at least daily for the remainder of the study. Blood samples were collected at 0, 5, 15, 30 min, and 1, 2, 4, 8, 24, 48, and 72 h post-dose. Blood samples (100 µl) were collected from the facial vein by puncturing the skin using sterile 0.5 mm goldenrod animal lancets (MEDIpoint, Inc., Mineola, NY). Blood drops flowing out of the puncture site were collected in heparinized tubes. In rats, blood was collected via retro-ocular bleeding under isoflurane anesthesia. Plasma was separated by centrifugation of blood samples at 1500 × g for 10 min at 4°C within 1 h of sample collection and stored at –80°C until analysis. Mouse and rat liver, kidney, lung, spleen, heart, and brain, were harvested from the same animals. In mice, tissues were harvested at 8h and 72 h, whereas the same tissues were harvested from rats at 72 h only. Tissue samples were stored at −80 °C freezer until LC-MS/MS analysis.
The excretion study of 13–197 was carried out in both mice and rats after intravenous administration at 5 mg/kg dose. Mice and rats were housed in well ventilated metabolic cages in standard laboratory conditions with regular 12 h light-dark cycles. The metabolic cages are designed to collect urine and feces samples separately. Standard pellet laboratory chow and water were allowed ad libitum. The animals were acclimatized for a minimum period of 3 days prior to the experiment. Urine and feces samples were collected at 0–24, 24–48, 48–72, and 72–120 h post-dose.
Pharmacokinetic data analysis
The pharmacokinetic parameters were determined using the extra-vascular input noncompartmental analysis module of WinNonlin (version 1.5, Pharsight, Mountain
View, CA). The elimination t0.5 was obtained from the formula of 0.693/K. The area under the curve (AUC0-∞) was estimated using the linear trapezoidal method from 0-tlast and extrapolation from tlast to infinity based on the observed concentration at the last time point divided by the terminal elimination rate constant (λ). For intravenous administration, Clearance (CL) and the apparent volume of distribution of the elimination phase (Vβ) were calculated as Dose/AUC0-∞ and Dose/K.AUC0-∞, respectively. CL, Vβ for oral data, were calculated as F.Dose/AUC0-∞ and F.Dose/Kel.AUC0-∞, respectively. Volume of distribution at steady state (Vss) was calculated as CL × MRTIV. Oral and IV mean residence time (MRT) was calculated as AUMC0-∞/AUC0-∞. The absolute bioavailability (F) was calculated as the ratio between the AUC0-∞ from oral and intravenous routes, after dose normalization using the following equation:
The percentage of the dose excreted unchanged in urine and feces was determined from the cumulative amount of the unchanged 13–197 excreted in urine and feces, which was calculated by adding the amount excreted from all collection time-intervals.
Protein binding study
The determination of the free fraction (fu) of a drug via distribution between erythrocytes and plasma was performed as previously described (Schuhmacher et al., 2000). Briefly, erythrocytes were obtained by the centrifugation of fresh, heparinized mice and rat blood, which were then washed three times in isotonic potassium phosphate buffer, pH 7.4. Isolated erythrocytes were resuspended in phosphate buffer, in10X–diluted plasma, or in undiluted plasma to yield a hematocrit (Hk) of 0.4. 13–197 was added to erythrocyte suspension to obtain a final concentration of 30 ng/ml. Suspensions were incubated at room temperature for 30 min on a laboratory shaker. After incubation, aliquots of 100 µl for determining the total concentration in the erythrocyte suspension were collected. Then, erythrocytes were separated by centrifugation at 1800g for 10 min, and 100 µl aliquots were aspirated from the resulting supernatents to determine the concentration in buffer or plasma supernatants. Concentrations in the erythrocyte suspension, buffer, and plasma supernatants were determined by LC-MS/MS.
Drug concentration in erythrocytes (CE) was calculated according to:
CB: total concentration in the blood cell suspension, Cp: concentration in plasma, and HK: volume ratio of blood cells in the suspension,
The partition coefficient between erythrocytes and plasma, PE/P, was calculated as:
Similarly the partition coefficient between buffer and erythrocytes was obtained as, PE/buffer = CE/Cbuffer. The free fraction of drug in plasma (fu) was calculated as:
The free fraction obtained in diluted plasma was corrected for dilution factor as follows;
where fu’ is the free fraction in diluted plasma and α is the dilution factor (i.e. 0.1 in the case of a 10-fold dilution of the plasma).
Results
Method development
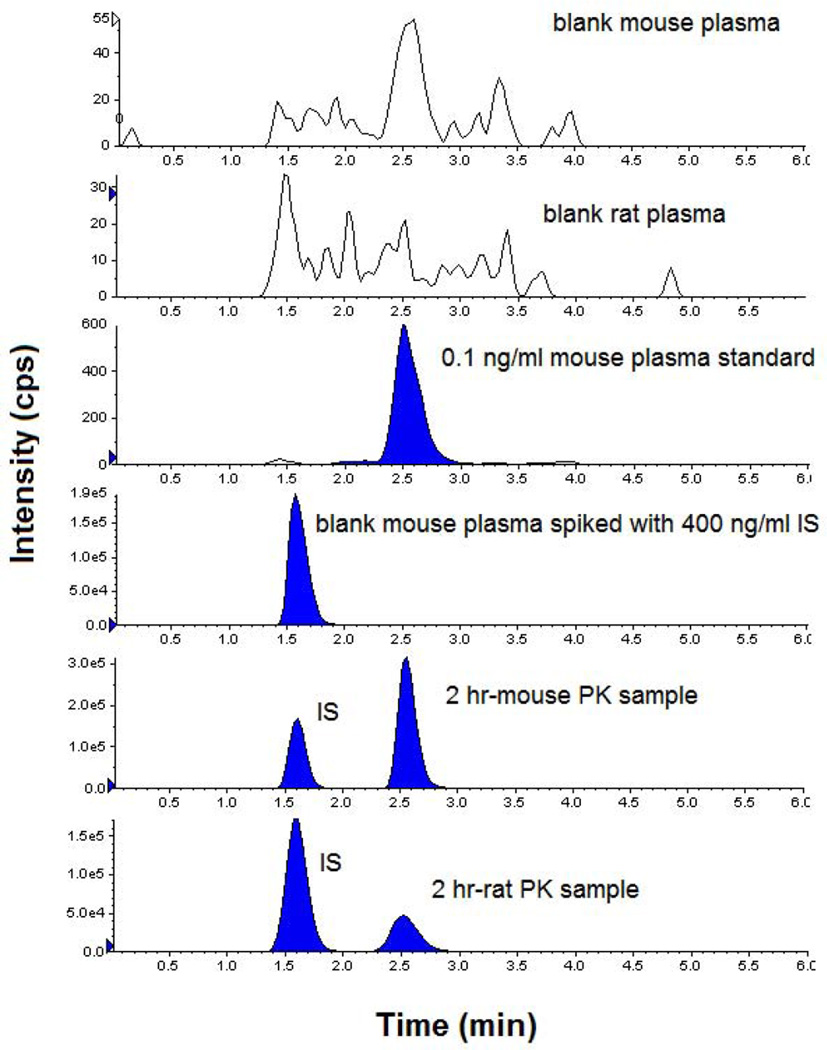
Representative chromatograms of blank mouse and rat plasma, 0.1 ng/ml 13–197 and 400 ng/ml IS standard in mouse plasma, and representative study samples in mice and rats are shown in Figure 1. The retention times of 13–197 and IS at the final chromatography conditions were 2.5 and 1.6 min, respectively. The Lower limit of detection (LOD) was 0.03 ng/ml. ACN-protein precipitation was used for the extraction of tissue and plasma samples. The extraction recovery of 13–197 in mouse and rat plasma was >90%, whereas extraction recoveries from the various tissues varied between 30 to 80%. The extraction recovery was consistent throughout the dynamic range in all matrices. This method was validated in the range of 0.1–500 ng/ml in mouse plasma and 0.2–500 ng/ml in mouse liver, kidney, lung, heart, spleen, brain, urine, and feces. Calibration curves of all analytes in all matrices were linear in this range with correlation coefficients > 0.99 (Table 1). Inter-day and intra-day accuracy and precision data in plasma and liver, are summarized in Table 2. Similarly, the inter-day and intra-day accuracy and precision obtained in other tissues (Kidney, lung, spleen, heart and brain), urine, and feces were also less than 15% (data not shown). The selectivity of the method was verified by the lack of interfering peaks that co-elute with the analyte or IS, in blank matrices. The carry-over effect was also examined and found to be < 20% of the lower limit of quantification (LLOQ). 13–197 was stable for at least 3 months in the −20 °C freezer, for 72 h in the 4 °C autosampler, and for 8 h on the laboratory bench at room temperature (data not shown).
Figure 1.
Representative chromatograms of blank mouse plasma, blank rat plasma, 13–197 (0.1 ng/ml), IS (400 ng/ml) in mouse plasma, and plasma samples at 2 h after oral administration to mice and rats.
Table 1.
Standard curves and correlation coefficients of 13–197 in plasma and tissue matrices.
| Biological samples |
Standard curve and correlation coefficient |
|---|---|
| Plasma | y = 0.00342 X + 0.00028 (R = 0.998) |
| Liver | y = 0.00272 X + 0.00062 (R = 0.996) |
| Kidney | y = 0.00448 X + 0.00024 (R = 0.997) |
| Spleen | y = 0.00482 X + 0.00056 (R = 0.994) |
| Lung | y = 0.00497 X + 0.00076 (R = 0.993) |
| Heart | y = 0.00468 X + 0.00042 (R = 0.997) |
| Brain | y = 0.00424 X + 0.00186 (R = 0.994) |
Table 2.
Summary of the inter-day and intra-day accuracy and precision of 13–197 assay in mouse plasma and liver (inter-day n=15, intra-day n=5).
| Quality controls | LLOQ | LQC | MQC-1 | MQC-2 | HQC | |
|---|---|---|---|---|---|---|
| Plasma | 0.1 ng/ml |
0.2 ng/ml |
10 ng/ml |
100 ng/ml |
500 ng/ml |
|
| Day1 |
a) Mean conc. ± %RSD |
0.107 ± 10.9 |
0.202 ± 3.3 |
10.38 ± 9.6 |
107.5 ± 4.6 |
562.3 ± 10.6 |
| Day2 | Mean conc.± %RSD |
0.105 ± 10.0 |
0.216 ± 1.3 |
10.07 ± 5.7 |
106.6 ± 2.8 |
525.0 ± 10.7 |
| Day3 | Mean conc.± %RSD |
0.098 ± 2.8 |
0.227 ± 13.4 |
10.67 ± 3.0 |
109.5 ± 3.2 |
515.0 ± 4.4 |
| Inter-day | Mean conc.± %RSD |
0.103 ± 4.6 |
0.215 ± 5.6 |
10.37 ± 2.9 |
107.9 ± 1.3 |
534.1 ± 4.7 |
| Liver | 0.2 ng/ml |
0.5 ng/ml |
10 ng/ml |
100 ng/ml |
500 ng/ml |
|
| Day1 | Mean conc.± %RSD |
0.181 ± 4.0 |
0.484 ± 12.8 |
10.8 ± 3.0 |
102.3 ± 9.7 |
542.3 ± 2.3 |
| Day2 | Mean conc.± %RSD |
0.210 ± 11.4 |
0.560 ± 3.1 |
10.2 ± 5.3 |
96.1 ± 11.9 |
527.7 ± 11.9 |
| Day3 | Mean conc.± %RSD |
0.212 ± 5.9 |
0.530 ± 5.3 |
9.7 ± 2.8 |
102.5 ± 4.3 |
548.7 ± 2.3 |
| Inter-day | Mean conc.± %RSD |
0.201 ± 8.8 |
0.525 ± 7.3 |
10.2 ± 5.1 |
100.3 ± 3.6 |
539.6 ± 2.0 |
Mean conc. in ng/ml of QC (n=5)
Pharmacokinetics in mice and rats
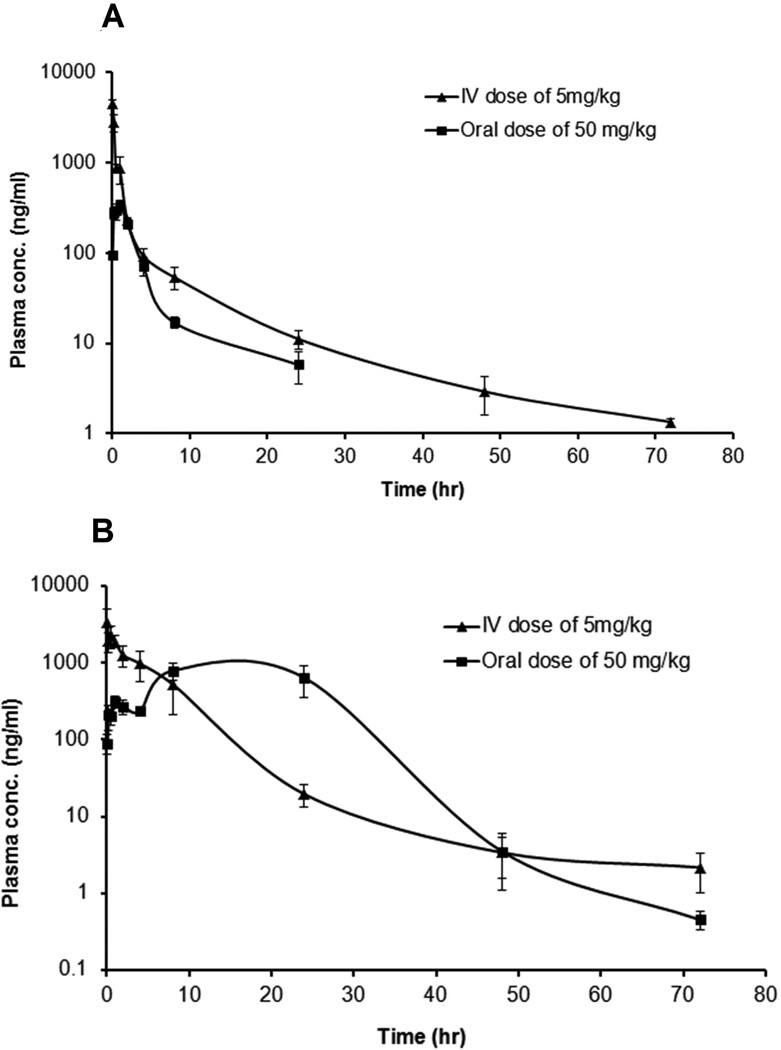
The plasma concentration vs. time profile after intravenous and oral administration of 13–197 in mice and rats are shown in Figure 2 A and Figure 2 B, respectively. The mean plasma concentration along with their standard error mean (SEM) are shown. After intravenous administration of a single 5 mg/kg dose, 13–197 was detectable up to 72 h, with an elimination t0.5 of 15.7 h in mice and 9.7 h in rats. After oral administration of a 50 mg/kg dose, 13–197 was detectable up to 24 h in mice, whereas in rats, it was detectable up to 72 h. The PK parameters of 13–197 in mice and rats are summarized in Table 3. The oral bioavailability of 13–197 was 3.14 % and 16.03 % in mice and rats, respectively. 13–197 was detected in the liver, kidney, lung, spleen, heart, and brain tissues up to 72h in both mice and rats after oral and intravenous dose administration. The percentage extrapolation of AUC from the last measured time point to infinity, which is required to calculate AUC0-∞, was less than 5 %. This indicates that AUC0-∞ and the derived PK parameters were well estimated. The concentrations of 13–197 at 8h, 48h, and 72 h in mouse tissues and at 72h in rat tissues are shown in Table 4. 13–197 demonstrated high accumulation in tissues, where higher tissues concentrations compared to the corresponding plasma concentrations were observed in both mice and rats. The percentage of the 13–197 doses excreted unchanged over a 120 h was <0.1% in urine and <1.2 % in feces for both mice and rats.
Figure 2.
Plasma concentration vs. time profile of 13–197 after 5 mg/kg intravenous and 50 mg/kg oral dose administration in (A) mice, and (B) rats (N=5, Mean ± SEM).
Table 3.
The pharmacokinetic parameters of 13–197 in mice and in rats after 5 mg/kg intravenous and 50 mg/kg oral doses (N=5).
| PK Parameters | Mice |
Rat |
||
|---|---|---|---|---|
| IV dose- 5mg/kg |
Oral Dose- 50mg/kg |
IV dose- 5mg/kg (Mean ± SEM |
Oral dose- 50mg/kg (Mean ± SEM) |
|
| Tmax (h) | * a) | 1.0 | * | 12.0 ± 4.0 |
| Cmax(ng/ml) | * | 340.2 | * | 1049.5 ± 195.1 |
| AUClast (ng.h/ml) | 3848 | 1184 | 13819 ± 5693 | 22168 ± 5455 |
| AUC0-∞ (ng.h/ml) | 3879 | 1219 | 13848 ± 5683 | 22170 ± 5454 |
| λz (1/h) | 0.044 | 0.1623 | 0.087 ± 0.020 | 0.138 ± 0.005 |
| t0.5(h) | 15.7 | 4.3 | 9.4 ± 2.1 | 5.0 ± 0.18 |
| Vβ (l/kg) | 29.26 | 252.5 | 7.5 ± 2.5 | 21.2 ± 3.4 |
| CL (l/hr/kg) | 1.28 | 40.99 | 0.50 ± 0.11 | 2.96 ± 0.53 |
| MRT0-∞ (h) | 5.14 | 4.4 | 5.7 ± 1.0 | 14.1 ± 1.7 |
| Vss (l/kg) | 6.6 | * | 2.99 ± 1.06 | * |
| % bioavailability | * | 3.14 | * | 16.03 |
not applicable
Table 4.
Concentrations of 13–197 in mouse and rat tissues after 5 mg/kg intravenous dose and 50 mg/kg oral dose (N=5, Mean ± SEM).
| Tissue | Time | IV-5mg/kg ng/g tissue |
Oral-50 mg/kg ng/g tissue |
|---|---|---|---|
| Mice | |||
| Liver | 8 h | 12262 ± 2601 | 209 ± 21 |
| 72 h | 1548 ± 143 | 3.0 ± 0.7 | |
| Lung | 8 h | 48217 ± 15521 | 61 ± 4.7 |
| 72 h | 4901 ± 1116 | 1.3 ± 0.9 | |
| Kidney | 8 h | 613 ± 45 | 113 ± 20 |
| 72 h | 346 ± 50 | 2.4 ± 0.8 | |
| Spleen | 8 h | 8859 ± 2718 | 30 ± 6.4 |
| 72 h | 1538 ± 96 | 1.9 ± 0.8 | |
| Heart | 8 h | 3205 ± 1033 | 69 ± 10 |
| 72 h | 404 ± 32 | 6.7 ± 2.6 | |
| Brain | 8 h | 67 ± 21 | 1.13 ± 0.16 |
| 72 h | 5.6 ± 1.1 | <LLOQ | |
| Rat | |||
| Liver | 72 h | 43 ± 25 | 7.0 ± 2.5 |
| Kidney | 72 h | 196 ± 108 | 20 ± 7.7 |
| Lung | 72 h | 52 ± 24 | 7.0 ± 2.0 |
| Spleen | 72 h | 39 ± 18 | 9.4 ± 3.7 |
| Heart | 72 h | 23 ± 11 | 4.4 ± 1.9 |
| Brain | 72 h | 93 ± 53 | 0.8 ± 0.2 |
Protein binding in mice and rats
The plasma protein binding for 13–197 was investigated using several methods including ultrafiltration, ultracentrifugation, charcoal adsorption, and blood partitioning. The blood partitioning method was selected because it is the most reliable method for the determination of unbound fraction (fu) for highly lipophilic compounds such as 13–197. Using this method, the fu of 13–197 was found to be 0.6 % and 0.4 % in mouse and rat plasma, respectively (fractions bound to plasma proteins in mice and rats were 99.4% and 99.6, respectively).
Metabolite profiling in mice and rats
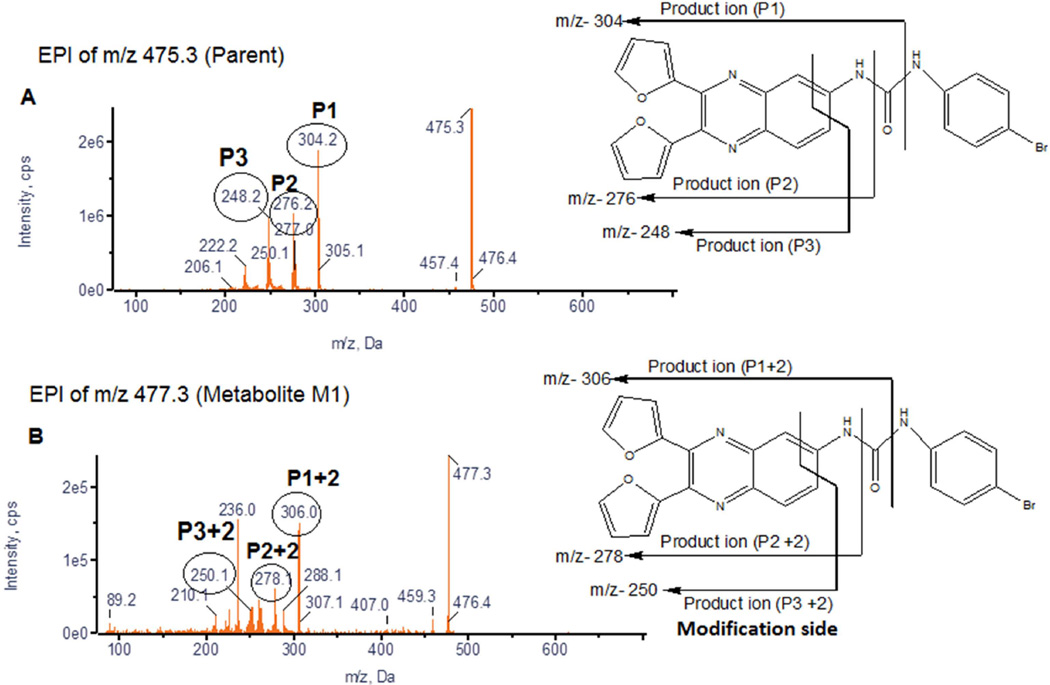
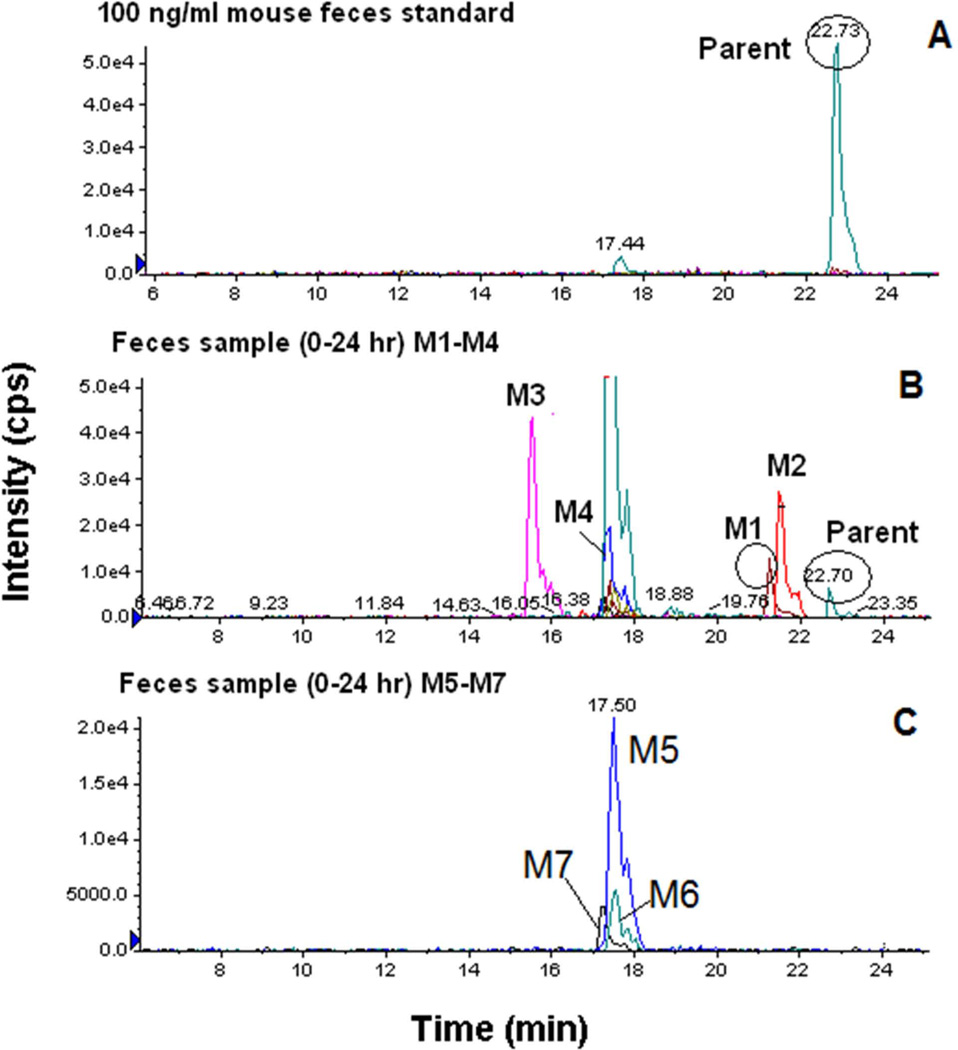
The MRM-IDA-EPI MS/MS analysis resulted in the detection of at least ten metabolites in mouse plasma, urine, and feces (Table 5). Similarly at least five metabolites were detected in the same matrices in rats (data not shown). Detected ions were labeled as metabolites based on their mass shifts as well as the similarity of their fragmentation pattern compared to the parent compound. Representative EPI spectra demonstrating the identification of metabolite (M1) with a mass shift of 2 (hydrogenation) are shown in Figure 3. Representative extracted ion chromatograms obtained from mouse feces sample (0–24 h) demonstrating the detection of seven metabolites (M1-M7) are shown in Figure 4. Figure 4 also shows that the peak area of the parent ion is markedly lower than those for the detected metabolites. Similar data were observed in plasma and urine (data not shown)
Table 5.
List of ions detected in plasma, urine, and feces after 5 mg/kg dose in mice using MRM-IDA-EPI scans, which represent potential metabolites of 13–197.
| Matrix | Mass Shift |
m/z | Biotransformation | Peak Area |
% Area Compared to parent |
|---|---|---|---|---|---|
| Plasma | −ve ion mode | ||||
| 0 | 473 | Parent | 9.26E+06 | 100 | |
| 46 | 519 | Keto (Ox-2H) + Di-Oxidation | 1.68E+06 | 18 | |
| 4 | 477 | Di-Demethylation + Di-Oxidation | 1.39E+06 | 15 | |
| 48 | 521 | Tri-Oxidation | 1.29E+06 | 14 | |
| 323 | 796 | Oxidation + Glutathione | 3.97E+05 | 4.3 | |
| 28 | 501 | CO | 3.41E+05 | 3.7 | |
| 107 | 580 | Taurine | 2.34E+05 | 2.5 | |
| 30 | 503 | Keto (Ox-2H) + Oxidation | 1.72E+05 | 1.9 | |
| Urine | +ve ion mode | ||||
| 0 | 475 | Parent | 0.00E+00 | * a) | |
| 30 | 505 | Keto (Ox-2H) + Oxidation | 1.83E+06 | * | |
| −14 | 461 | Demethylation | 6.94E+05 | * | |
| 107 | 582 | Taurine | 1.61E+05 | * | |
| 46 | 521 | Keto (Ox-2H) + Di-Oxidation | 1.31E+05 | * | |
| 32 | 507 | Di-Oxidation | 1.12E+05 | * | |
| 2 | 477 | Hydrogenation | 1.04E+05 | * | |
| −ve ion mode | |||||
| 0 | 473 | Parent | 0.00E+00 | * | |
| −46 | 427 | Debromination plus H + Di-Oxidation | 3.58E+05 | * | |
| −2 | 471 | Dehydrogenation | 1.60E+05 | * | |
| Feces | +ve ion mode | ||||
| 0 | 475 | Parent | 7.40E+04 | 100 | |
| 107 | 582 | Taurine | 7.20E+05 | 973 | |
| 14 | 489 | Methylation | 4.43E+05 | 599 | |
| 34 | 509 | Oxidation + OH+H | 4.31E+05 | 582 | |
| −30 | 445 | Loss of NO | 3.94E+05 | 532 | |
| 2 | 477 | Hydrogenation | 1.62E+05 | 219 | |
| 46 | 521 | Keto (Ox-2H) + Di-Oxidation | 7.56E+04 | 102 | |
| −42 | 433 | Tri-Demethylation | 6.41E+04 | 87 | |
| −ve ion mode | |||||
| 0 | 473 | Parent | 1.51E+05 | 100 | |
| −46 | 427 | Debromination plus H + Di-Oxidation | 9.04E+04 | 60 | |
| 42 | 515 | Acetylation | 7.84E+04 | 52 | |
| 323 | 796 | Oxidation + Glutathione | 6.42E+04 | 43 | |
| 321 | 794 | Oxidation + Glutathione - 2H | 3.88E+04 | 26 | |
Parent not detectable
Figure 3.
Representative EPI spectra of 13–197 and its metabolite (M1).
Figure 4.
Representative extracted ion chromatograms (EICs) obtained from feces sample (0–24 h) for 13–197 using the MRM-IDA-EPI method: (A) parent spiked 100 ng/ml (475 m/z); (B) metabolites M1(477.1 m/z), M2 (509.1 m/z), M3 (582 m/z), M4 (445 m/z); (C) metabolite M5 (489.1 m/z), M6 (521.1 m/z) and M7 (433.1 m/z).
Discussion
High sensitivity is required for the bioanalytical support of the various preclinical and clinical drug development studies. Our goal was to maximize the assay sensitivity through optimizing MS, chromatographic, and sample preparation conditions without compromising the assay accuracy and precision. The pH and the composition of the mobile phase were investigated to maximize column retention, improve peak shape, and enhance signal intensity of 13–197. Based on the peak area, peak shape, and retention time of the selected ions, pH 3–7.5 mM ammonium formate buffer and 5% ACN in MeOH were selected for the mobile phase (Figure 1). This validated LC-MS/MS method was utilized for the quantification of 13–197 in plasma and tissues to support the preclinical PK studies of 13–197. This method will also be further utilized to support all preclinical development studies of this compound.
After oral administration of 13–197, Tmax was 1 h in mice and 12 h in rats indicating that absorption was faster in mice compared to rats. In addition, the difference between MRT oral and MRT iv was about 10 times higher in rats than in mice, which also indicates slower absorption in rats compared to mice. In rats, two peaks were observed in the plasma concentration vs. time profile after oral administration (Figure 2 B). One explanation to this phenomenon may be related to the slow absorption and low aqueous solubility of 13–197, which typically results in the precipitation of the dose in the intestine (Wang et al., 1999). Other explanations for the 2nd peak in the concentration vs. time profile may be related to the redistribution of drug from deep depot sites, entero-hepatic recycling, and/or variable absorption in the different areas of the gastrointestinal tract (Lin, 1995). Despite its faster absorption in mice, oral bioavailability of 13–197 in mice was 5.1-fold lower than that in rats. This may be due to the 2-fold higher clearance in mice compared to rats (Cl in mouse and rats were 1.28 and 0.5 L/h/kg, respectively). Vβ and Vss after intravenous administration were 2–4 fold higher in mice than in rats, which indicates that 13–197 was more distributed outside the plasma compartment in mice. This conclusion was supported by tissue concentrations, which show that liver, lung, kidney, spleen, and heart have 1.3–69.2 fold higher concentrations in mice compared to the same tissues in rats, 72h after intravenous administration. In general, 13–197 was well distributed to the peripheral tissues and had relatively high tissue: plasma concentration ratios, ranging from 1.8 to 3634, in both mice and rats, possibly due to the hydrophobic nature of this compound. t0.5 is determined by both Cl and Vd. Therefore, despite the higher Cl in mice, the elimination t0.5 was longer in mice compared to rats, due to the higher volume of distribution in mice (Vss = 6.6 l/kg in mice vs. 2.99 l/kg in rats). Over the 72 h period we did not observe any visible / obvious signs of toxicity in mice and rats treated with 13–197. There was no death or weight loss observed in either mice or rats during the study. These preliminary data from two rodent species indicate that 13–197 has favorable toxicity profile.
Only fractions unbound to plasma proteins are available for elimination and disposition as well as for their target receptors to exert pharmacological and/or toxiciological effects. Therefore, the protein binding profiles of drug candidates are determined early on in discovery. Commonly used methods for the determination of protein binding include ultrafiltration, equilibrium dialysis, ultracentrifugation, and charcoal adsorption. However, all these methods have major limitation in the case of highly lipophilic drugs, due to solubility issues, non-specific binding to vial walls, filters, and/or membranes, or binding of drug to low density lipoproteins (Yuan et al., 1995). An alternative technique to determine protein binding is via measuring the partitioning of the drug between plasma and blood cells (Roos and Hinderling, 1981; Schuhmacher et al., 2000; Schuhmacher et al., 2004). 13–197 demonstrated high nonspecific binding to filters and vial walls in the absence of plasma proteins and therefore we used the blood partitioning method for the determination of protein binding. fu of 13–197 in mouse and rat plasma was found to be 0.6% and 0.4%, respectively. Despite the extensive protein binding of 13–197, it was highly distributed to peripheral tissues indicating that 13–197 affinity to these tissues was even higher than binding to plasma proteins.
To characterize the routes of 13–197 elimination, excretion studies were performed, where urine and feces were collected in both mice and rats. Less than 1% of the dose was excreted unchanged in urine and feces in both mice and rats. In addition, metabolite profiling analyses were performed in plasma, urine, and feces. The hybrid Q-trap mass spectrometer combines the scanning capabilities of triple quadrupole and linear ion trap instruments into a single platform capable of performing several types of mass spectrometric analyses, including EMS (enhanced MS), EPI (enhanced product ion), PI (precursor ion), NL (neutral loss), and MRM (multiple reaction monitoring) scans. In IDA, one of these scans can be used as a survey scan, which triggers the acquisition of EPI spectra if the signal intensity of the survey scan exceeds a predefined threshold value (Huang et al., 2010). Due to the superior sensitivity and selectivity of MRM scans, the highest number of metabolites for 13–197 was detected using MRM-based survey methods. Total of 26 metabolites in mouse and 19 in rats plasma, urine, and feces were detected for 13–197. Further investigations using high resolution MS and NMR are underway for more definitive structural elucidation of the detected metabolites. In addition the peak are of the parent compound was markedly lower than those of the detected metabolites in all three matrices in both mice and rats (Figure 4), which supports the urinary and fecal excretion data that clearly indicates 13–197 is extensively metabolized and primarily eliminated by metabolism rather than by excretion.
Conclusions
The LC-MS method we developed provided the required sensitivity and was suitable for the support of the preclinical discovery and development of 13–197. 13–197 has low bioavailability in both mice and rats, which needs to be addressed by further structural modifications if intended for oral administration. 13–197 is more than 99% bound to plasma proteins and yet is well distributed to peripheral tissues. 13–197 was absorbed more slowly in rats compared to mice. 13–197 is primarily eliminated by metabolism and less than 1% is excreted unchanged in urine and feces.
Acknowledgments
The authors gratefully acknowledge to Animal Facility of University of Nebraska medical center. This work was supported in part by NIH R01 CA127239.
Abbreviations used
- DMSO
Dimethyl sulfoxide
- EtOH
Ethyl alcohol
- PBS
phosphate buffered saline
- PEG400
Polyethylene glycol 400
- PG
Propylene glycol
Footnotes
Financial support This work was supported in part by NIH R01CA127239
References
- Aggarwal BB, Takada Y, Shishodia S, Gutierrez AM, Oommen OV, Ichikawa H, Baba Y, Kumar A. Nuclear transcription factor NF-kappa B: role in biology and medicine. Indian J Exp Biol. 2004;42:341–353. [PubMed] [Google Scholar]
- Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Cheng A, Diller DJ, Dixon SL, Egan WJ, Lauri G, Merz KM., Jr Computation of the physio-chemical properties and data mining of large molecular collections. J Comput Chem. 2002;23:172–183. doi: 10.1002/jcc.1164. [DOI] [PubMed] [Google Scholar]
- Clark DE, Grootenhuis PD. Progress in computational methods for the prediction of ADMET properties. Curr Opin Drug Discov Devel. 2002;5:382–390. [PubMed] [Google Scholar]
- el-Kamar FG, Grossbard ML, Kozuch PS. Metastatic pancreatic cancer: emerging strategies in chemotherapy and palliative care. Oncologist. 2003;8:18–34. doi: 10.1634/theoncologist.8-1-18. [DOI] [PubMed] [Google Scholar]
- Farrow B, Evers BM. Inflammation and the development of pancreatic cancer. Surg Oncol. 2002;10:153–169. doi: 10.1016/s0960-7404(02)00015-4. [DOI] [PubMed] [Google Scholar]
- Holcomb B, Yip-Schneider M, Schmidt CM. The role of nuclear factor kappaB in pancreatic cancer and the clinical applications of targeted therapy. Pancreas. 2008;36:225–235. doi: 10.1097/MPA.0b013e31815b3207. [DOI] [PubMed] [Google Scholar]
- Huang J, Bathena SP, Alnouti Y. Metabolite profiling of praziquantel and its analogs during the analysis of in vitro metabolic stability using information-dependent acquisition on a hybrid triple quadrupole linear ion trap mass spectrometer. Drug Metab Pharmacokinet. 2010;25:487–499. doi: 10.2133/dmpk.dmpk-10-rg-041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Hu L, Xu L, Sun M, Fan Z, Qiu J, Li G, Si L. In vitro enantioselective metabolism of TJ0711 hydrochloride by human liver microsomes using a novel chiral liquid chromatography-tandem mass spectrometry method. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2012;891–892:98–101. doi: 10.1016/j.jchromb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- Kunnumakkara AB, Guha S, Krishnan S, Diagaradjane P, Gelovani J, Aggarwal BB. Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, angiogenesis, and inhibition of nuclear factor-kappaB-regulated gene products. Cancer Res. 2007;67:3853–3861. doi: 10.1158/0008-5472.CAN-06-4257. [DOI] [PubMed] [Google Scholar]
- Kwok BH, Koh B, Ndubuisi MI, Elofsson M, Crews CM. The anti-inflammatory natural product parthenolide from the medicinal herb Feverfew directly binds to and inhibits IkappaB kinase. Chem Biol. 2001;8:759–766. doi: 10.1016/s1074-5521(01)00049-7. [DOI] [PubMed] [Google Scholar]
- Lin JH. Species similarities and differences in pharmacokinetics. Drug Metab Dispos. 1995;23:1008–1021. [PubMed] [Google Scholar]
- Roos A, Hinderling PH. Protein binding and erythrocyte partitioning of the antirheumatic proquazone. J Pharm Sci. 1981;70:252–257. doi: 10.1002/jps.2600700306. [DOI] [PubMed] [Google Scholar]
- Schuhmacher J, Buhner K, Witt-Laido A. Determination of the free fraction and relative free fraction of drugs strongly bound to plasma proteins. J Pharm Sci. 2000;89:1008–1021. doi: 10.1002/1520-6017(200008)89:8<1008::aid-jps5>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Schuhmacher J, Kohlsdorfer C, Buhner K, Brandenburger T, Kruk R. High-throughput determination of the free fraction of drugs strongly bound to plasma proteins. J Pharm Sci. 2004;93:816–830. doi: 10.1002/jps.10588. [DOI] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Roy A, Sun L, Lau CE. A double-peak phenomenon in the pharmacokinetics of alprazolam after oral administration. Drug Metab Dispos. 1999;27:855–859. [PubMed] [Google Scholar]
- Yuan J, Yang DC, Birkmeier J, Stolzenbach J. Determination of protein binding by in vitro charcoal adsorption. J Pharmacokinet Biopharm. 1995;23:41–55. doi: 10.1007/BF02353785. [DOI] [PubMed] [Google Scholar]
- Zheng YT, Zhu JH, Ma G, Zhu Q, Yang P, Tan B, Zhang JL, Shen HX, Xu JL, Zhu YZ, Cai WM. Preclinical assessment of the distribution, metabolism, and excretion of S-propargyl-cysteine, a novel H2S donor, in Sprague-Dawley rats. Acta Pharmacol Sin. 2012;33:839–844. doi: 10.1038/aps.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]