Abstract
Background
Individual skeletal elements of the vertebrate limbs arise through a segmentation process introducing joints in specific locations. However, the molecular pathways controlling joint formation and subsequent joint maintenance are largely unknown. In this study, we focused on SOX11, and its contribution to the regulation of GDF5, a secreted signal necessary for proper joint formation and postnatal joint homeostasis.
Results
Sox11 is initially expressed broadly in the murine cartilage condensations at early stages of skeletal development, but its expression is specifically increased in the forming joint interzone as is forms. SOX11 overexpression can directly activate GDF5 expression both in vitro and in micromass cell cultures prepared from chick limb buds. Conserved SOX family binding sites are present in the 5’ UTR region of the GDF5 gene and we show SOX11 can specifically bind to one of them. While misexpression of Sox11 in developing chick limbs through RCAS virus infection does not induce Gdf5 expression in ectopic locations, it does enhance its expression. To explore the roles of Sox11 in joint homeostasis, we analyzed adult knee joints in an osteoarthritis mouse model where the medial meniscus and the medial collateral ligament were removed. We also analyzed knee joints from human subjects who underwent total knee replacement surgery. We find that SOX11 is mainly expressed in the weight-bearing areas of knee joints, and its expression is decreased in degraded cartilage during progression of knee osteoarthritis in both mice and humans.
Conclusions
This work implicates SOX11 as a potential regulator of GDF5 expression in joint maintenance and suggests a possible role in the pathogenesis of osteoarthritis.
Keywords: SOX11, GDF5, Joint maintenance, Articular cartilage
Background
Within the embryonic limb, skeletal development is initiated by aggregation of mesenchymal cells. These early condensations can subsequently adopt either of two distinct fates. They can give rise to cartilage and then bone through a process known as an endochondral ossification [1] or alternatively they can initiate a program leading to joint formation. In this latter process, a localized dense region known as an interzone is formed in the mesenchymal condensations, ultimately yielding a synovium, a capsule, ligaments and articular cartilage [2]. Although the molecular mechanisms regulating endochondral ossification have been fairly well-investigated [3,4], those controlling joint formation largely remain to be clarified. The subsequent mechanisms of joint maintenance are equally poorly understood.
Osteoarthritis (OA) is one of the most common diseases of the joint, which is characterized by cartilage destruction, synovial inflammation, and bone remodeling. Since articular cartilage has very poor regenerative capability, once the joint surface becomes degraded, it cannot be recovered. OA is a multifactorial disease which is influenced by mechanical, environmental and genetic factors [5,6].
GDF5 is one of the genes that has been implicated in joint maintenance. Gdf5 is expressed in the early developing joint interzone [7], and loss-of-function mutations in this gene cause abnormalities of a number of joints in mice [7,8]. However, misexpression of Gdf5 does not induce ectopic joints, but rather causes an increase in chondrocyte differentiation and proliferation [9-11]. Interestingly, GDF5 has also been reported to be required for joint integrity and homeostasis in humans. A single nucleotide polymorphism in the human GDF5 promoter that reduces its transcriptional activity is associated with susceptibility to osteoarthritis [5]. Moreover, human loss-of-function mutations in the GDF5 gene result in congenital skeletal disorders such as Hunter-Thompson and Grebe chondrodysplasia [12].
SOX genes are involved in cell type specification in a variety of tissues, including sex determination, neurogenesis and skeletal formation [13]. Of particular relevance, the chondrocyte lineage is established through the activity of SOX9, SOX5 and SOX6[4,14]. SOX9 binds to a regulatory element of the cartilage-specific type II collagen (COL2A1) gene [15], and its mutation in humans is known to cause a skeletal malformation called campomelic dysplasia [16,17]. Interestingly, the chondrogenic actions of SOX genes are also required for synovial joint morphogenesis [18], raising the possibility that SOX genes might similarly play integral roles in joint maintenance.
Based on analysis of their HMG domains, SOX genes can be separated into subgroups A-J [19]. Here, we examined the group-C SOX gene SOX11. We find that Sox11 is expressed in the cartilage condensations in early stages of skeletal development in mice, but at later stages Sox11 expression is notably increased in the joint interzone. We show that SOX11 can activate GDF5 expression and demonstrate that there is a direct binding site for SOX11 in the 5’UTR of the GDF5 gene in vitro. Sox11 misexpression in chicks does not induce ectopic joint formation in vivo, but slightly enhances endogenous Gdf5 expression. Finally, we find that SOX11 protein levels are decreased in degraded cartilage during osteoarthritic progression in humans and mice, indicating a possible role of SOX11 in pathogenesis of osteoarthritis.
Results
The Sox11 expression in developing limbs
To identify candidate genes that might play a role in regulating GDF5 expression in developing joints, we conducted a preliminary screen of all the known SOX transcription factors. We examined the transcriptional activities of a reporter dependent upon the human GDF5 promoter (-448/+319) following overexpression of each of the known human SOX genes in Hela and ATDC5 cells (Additional file 1). The genes that gave the most prominent results in this assay were the three group-C SOX genes (SOX4, SOX11, and SOX12) and of these, SOX11 produced the strongest enhancement in both cell lines. The increased luciferase activitities induced by co-transfection of SOX11 and the reporter construct containing the GDF5 promoter were about 18 and 3.5 times higher (in the two cell lines respectively) than that of SOX11 and the control reporter vector lacking the GDF5 promoter but containing the luciferase reporter (Additional file 1). We noted that SOX11 activated transcription in the absence of the GDF5 promoter, albeit to a lesser degree, presumably due to non-specific interactions with the backbone of the reporter plasmid. This effect, which was not specific to SOX11, might be caused by the more stable α-helical structure of its transactivation domain than other SOX genes [20]. Nonetheless, SOX11 had a clear and significant effect on the GDF5 promoter above that seen with the vector alone. On the other hand, the overexpression of SOX6 and 9, which are known to be involved in cartilage differentiation in developing limb, had less effect on the activity of the GDF5 promoter than that of SOX11. Based on this preliminary screen, we chose SOX11 for further study.
We first compared the expression pattern of Sox11 and Gdf5 by double fluorescence section in situ hybridization during an embryonic limb development of mice. At embryonic stage 13.5 (E13.5), when joint interzones are just starting to form between the metatarsals and tarsal elements, Sox11 is expressed in the cartilage condensations, showing a distinct expression pattern from the joint-specific Gdf5 (Figure 1). However, at E14.5, the level of Sox11 transcription is specifically increased at the interzone in the prospective joint region and co-localized with Gdf5 (Figure 1).
Figure 1.
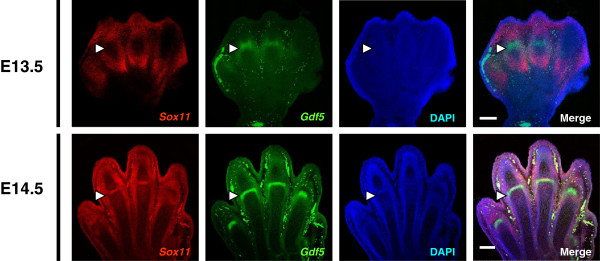
Expression patterns of Sox11 and Gdf5 during joint formation in mouse embryos. Double fluorescence section section in situ hybridization in hind limbs of mouse embryos (E13.5, 14.5). Confocal microscope images show (from left to right): Sox11 (red channel), GDF5 (green channel), DAPI staining of nuclei, and merged images. White arrowheads indicate the location of the interzone at prospective joint sites. Scale bars, 200 μm.
Increase of endogenous GDF5 expression by SOX11 overexpression
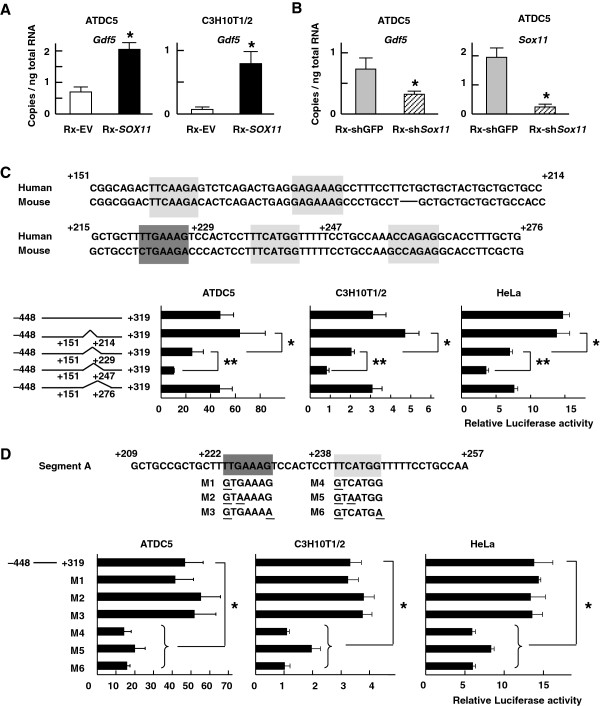
To examine the capacity of SOX11 to regulate cellular GDF5 expression, we created variants of the murine chondrogenic ATDC5 and fibroblastic C3H10T1/2 cell lines in which we stably introduced human SOX11 through retroviral infection. The resultant SOX11 overexpression increased endogenous mRNA levels of Gdf5 in both cell lines (Figure 2A). Conversely, the Gdf5 mRNA level was suppressed (to a level 44% of that seen in controls) by the retrovirally mediated shRNA knock-down of endogenous Sox11 activity (to a level 12% of that seen in controls) in ATDC5 cells (Figure 2B).
Figure 2.
Determination of a core responsive element of SOX11 in the GDF5 promoter. (A) Levels of Gdf5 mRNA in ATDC5 and C3H10T1/2 cells retrovirally transfected with the control vector (Rx-EV) or the vector expressing human SOX11 (Rx-SOX11). mRNA levels were determined by quantitative RT-PCR, normalized to Gapdh, and expressed as means (bars) ± SDs (error bars) for 4 wells/construct. *P < 0.05 vs. Rx-EV. (B) Gdf5 and Sox11 mRNA levels in ATDC5 cells retrovirally transfected with the shRNA vector targeting GFP (Rx-shGFP) or targeting mouse Sox11 (Rx-shSox11). *P < 0.05 vs. Rx-shGFP. (C) Deletion analysis using constructs lacking putative SOX-family-binding motifs in the proximal human GDF5 promoter (+151/+276) in ATDC5, C3H10T1/2, and HeLa cells. Five putative SOX family binding sites are shown (+159 to +165, +179 to +185, +222 to +228, +238 to +244, +258 to +264). The site with 1 base difference from the ideal binding site is shaded in dark gray, and those with 2 bases difference are indicated in light gray. Luciferase assays were carried out as above, in ATDC5, C3H10T1/2 and HeLa cells. *P < 0.05 vs. deletion of +151/+214 **P < 0.05 vs. deletion of +151/+229. (D) Identification of a core responsive element of SOX11 in the GDF5 promoter (segment A: +209/+257). Site-directed mutagenesis analysis with 6 mutated constructs (M1–M6). Base changes for each mutation as shown. Luciferase assays were carried out as above, in ATDC5, C3H10T1/2 and HeLa cells. The data for the luciferase assay are expressed as means (bars) ± SDs (error bars) of Relative luciferase activity for 4 assays/construct. *P < 0.05 vs. wild type promoter, –448/+319.
SOX11 has three conserved domains: an N-terminus SRY-related HMG domain (amino acids 1–125), an acid-rich domain (amino acids 218–250) and a C-terminus domain (amino acids 386–441) (Additional file 2), which contains potential transcriptional regulatory regions [21]. To determine the domains that are required for GDF5 expression, we created constructs lacking each domain, called SOX11ΔN (lacks amino acid 1–134), SOX11ΔAR (lacks amino acid 200–309) and SOX11ΔC (lacks amino acid 297–441), respectively, and examined their ability to transcriptionally regulate the GDF5 promoter. The lack of the N-terminus HMG domain or C-terminus domain abrogated the transcriptional response induced by full-length SOX11 overexpression (Additional file 3A). Moreover, the endogenous Gdf5 mRNA level was significantly suppressed by constructs carrying deletions in either of these domains in ATDC5 cells by retroviral infection (Additional file 3B). In contrast, the construct lacking the acid-rich domain did not abrogate Gdf5 expression (Additional file 3B), indicating that the N-terminus and C-terminus domains, but not the acid-rich domain, play important roles in mediating SOX11 function with respect to GDF5 expression.
Identification of a core SOX11-responsive element in the GDF5 promoter
We found five clustered sequences similar to the putative SOX family consensus binding site (A/T)(A/T)CAA(A/T)G in this region (Figure 2C), that were highly conserved between humans and mice [22]. One of them was one-base different from the published consensus binding site (shaded in dark gray), and the others were two-bases different (shaded in light gray) (Figure 2C). We created four reporter constructs deleting these motifs: deletion of first two binding sites (+151 bp and +214 bp), first three binding sites (+151 bp and +229bp), first four binding sites (+151 bp and +247 bp), or all five binding sites (+151 bp and +276 bp); and compared their transcriptional activities in a dual-luciferase assay. We observed a stepdown in the luciferase activity induced when a deletion is introduced between +215 bp and +247 bp, which contains two adjacent SOX family binding motifs. These were thus candidates for SOX11 binding sites in the GDF5 promoter. A 49 bp element (+209 bp/+257 bp), containing the identified putative binding sites, was defined as segment A (Figure 2D).
To further determine the core responsive sequence in segment A, we created mutations in the identified motifs (mutation 1 at +222 bp, mutation 2 at both +222 bp and +224 bp, mutation 3 at both +222 bp and +228 bp, mutation 4 at +238 bp, mutation 5 at both +238 bp and +240 bp, and mutation 6 at both +238 bp and +244 bp). Transactivation by SOX11 was decreased only by mutations 4,5 and 6, not by mutations 1,2 or 3, and the inhibitory effect among mutations 4, 5 and 6 were comparable, indicating that the thymine at the +238 bp nucleotide is crucial for the binding between GDF5 promoter and SOX11 (Figure 2D).
Previous reports suggest that a single nucleotide polymorphism (SNP) rs143383 (+45 bp) in the 5’UTR region is related to GDF5 expression and its postnatal joint maintenance [5]. We found a putative SOX family binding motif at (+57/+63) just downstream of the SNP. We examined the potential ability of SOX11 to activate gene transcription through this motif in a luciferase assay. Deletion of the SOX motif near the SNP did decrease transcriptional activity induced by SOX11 overexpression. However, the one nucleotide mutation at the +238 bp (identified above) suppressed it more extensively, indicating the binding motif at +238 has a stronger affinity for the GDF5 promoter (Additional file 4).
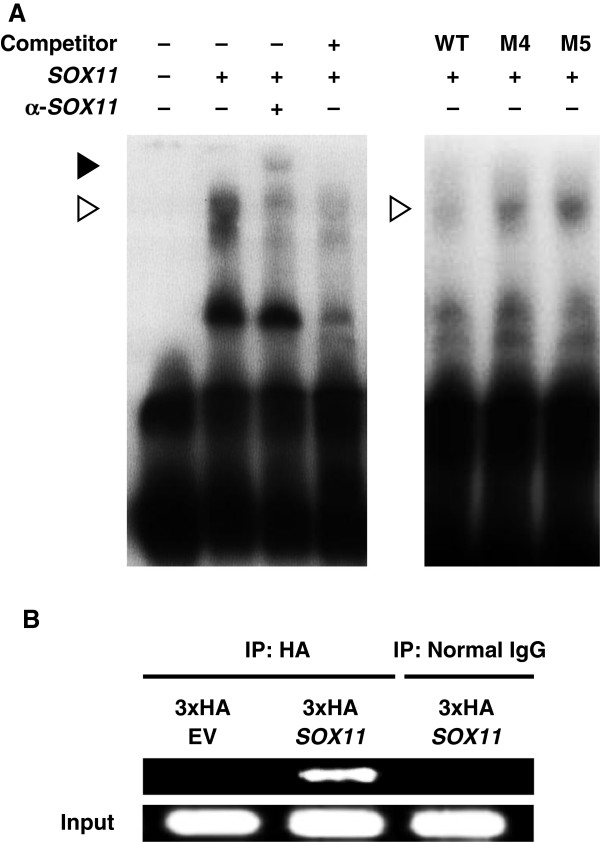
We further examined the binding of SOX11 to segment A in electromobility shift (EMSA) and chromatin immunoprecipitation (ChIP) assays. In EMSA, an oligonucleotide probe detected a complex formation between a segment A and in-vitro translated SOX11 protein, which was supershifted by an antibody to SOX11, indicating the direct binding of SOX11 to the segment A (Figure 3A). Moreover, cold competition with an excess amount of the unlabeled wild-type probe reduced the complex formation. Specificity of SOX11 antibody was confirmed by the absence of a supershift band with a non-immune normal IgG (Additional file 5). A chromatin immunoprecipitation (ChIP) assay was performed using HeLa cells transfected with SOX11 linked to three tandem repeats of the HA tag. Pull down with an antibody to the HA tag showed in vivo binding of SOX11 to the GDF5 regulatory region including segment A (Figure 3B). Specificity was confirmed, as SOX11 was not immunoprecipitated by the overexpression of the empty vector or by a control non-immune IgG antibody.
Figure 3.
Direct binding between SOX11 protein and GDF5 promoter. (A) Electro mobility shift assay (EMSA) for binding of the wild-type (WT) and mutated probes with human SOX11 protein. An open arrowhead indicates the shifted bands of the SOX11-DNA probe complex, and a solid arrowhead indicates the band super shifted by an antibody to SOX11. Cold competitions with a 50-fold excess of the unlabeled WT probe or the mutated probes are shown. Detailed information about each mutated nucleotide is indicated in Figure 2D. (B) Chromatin immunoprecipitation (ChIP) assay for in vivo binding of SOX11 and the segment A. Cell lysates of HeLa cells transfected with empty vector (3xHA EV) or SOX11 (3xHA SOX11) were amplified with a primer set (+98/+364) spanning segment A.
Involvement of SOX11 in cartilage formation in vitro
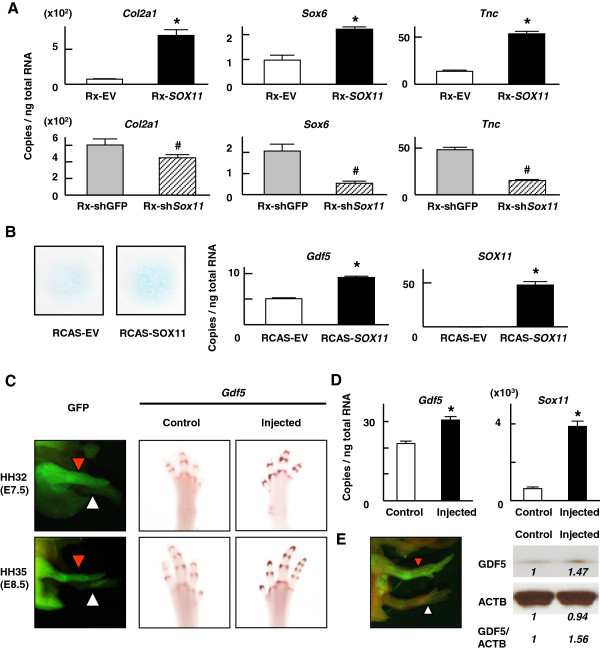
GDF5 is known to have chondrogenic ability in vitro and in vivo[9-11]. Therefore, we next examined the potential ability of SOX11 to stimulate chondrogenic differentiation in vitro. The endogenous mRNA levels of chondrogenic marker genes, Col2a1, Sox6 and TenascinC[23] were all increased by the retroviral introduction of human SOX11 into ATDC5 cells (Figure 4A). Moreover, a short hairpin RNA (shRNA) directed against the endogenous mouse Sox11 decreased the expression of the three chondrogenic marker genes (Figure 4A). A similar chondrogenic effect of SOX11 was observed in micromass cell cultures of chick limb bud cells after infection of an RCAS virus encoding human SOX11, as indicated by an increase in Alcian blue staining (Figure 4B). The endogenous Gdf5 mRNA level was concomitantly increased in the micromass cultures by the infection of RCAS virus encoding SOX11 (Figure 4B).
Figure 4.
Contribution of Sox11 to Gdf5 expression in developing limbs. (A) mRNA levels of Col2a1, Sox6 and TenasinC (Tnc) in ATDC5 cells that were retrovirally transfected with a control vector (Rx-EV) or human SOX11 (Rx-SOX11) and the short hairpin RNA directed against GFP (Rx-shGFP) or against mouse Sox11 (Rx-shSox11). mRNA levels were determined by quantitative RT-PCR and expressed as means (bars) ± SDs (error bars) for 4 wells/construct. *P < 0.05 vs. Rx-EV. #P < 0.05 vs. Rx-shGFP. (B) Alcian blue staining of micromass cell cultures and mRNA levels of Gdf5 and SOX11 in those cultures after infection of RCAS virus encoding human SOX11. *P < 0.05 vs. RCAS-EV. (C) Whole mount in situ hybridization for Sox11 and Gdf5 in wild type chick embryos at HH stage 32 or 35 and in embryos after injection of RCAS virus encoding GFP-fused chick Sox11 at HH stage 10. The GFP fluorescence after the injection is also shown. Red arrows indicate the injected right hind limbs, while white arrows indicate the uninjected left hind limbs for control. Scale bars, 1 mm. (D) mRNA levels of chick Sox11, and Gdf5 genes of hind limbs at HH stage 35 (E8.5) after the RCAS injection of GFP-fused chick Sox11. The uninjected left hind limb is used as control. *P < 0.05 vs. control. (E) Western blotting for Gdf5 and β-Actin (ACTB) in HH stage 35 (E8.5) chick hind limbs after the RCAS injection of GFP-fused chick Sox11. The uninjected left hind limb is used as control. The GFP fluorescence in the embryos used for Western blotting analysis is also shown. Red arrow indicates the injected hind limb and white arrow shows the control hind limb. Densitometric analysis was performed with Image J software.
Misexpression of Sox11 in chicks enhances Gdf5 expressions in vivo
To know the effect of Sox11 overexpression in vivo, we injected RCAS virus encoding GFP fused to the chick Sox11 cDNA into right chick lateral plate mesoderm at HH stage 10 (day 1.5), and harvested the limb at HH stage 32 or 35 (day 7.5 or 8.5, respectively). After confirming GFP fluorescence (Figure 4C), we performed whole mount in situ hybridization on the injected limbs with a riboprobe for chick Gdf5. We used the non-infected contralateral side of the same embryo as a control. Misexpression of Sox11 did not induce additional domains of Gdf5 expression, indicating Sox11 is not sufficient to drive its expression in ectopic locations (Figure 4C). However, when we harvested the hind limbs and examined the expression level of Gdf5, we saw a slight but significant enhancement following Sox11 overexpression (Figure 4D). The increased expression of GDF5 protein was also confirmed by Western blotting (Figure 4E). These results suggest that Sox11 can at least partially contribute to the regulation of Gdf5 expression but it is more than likely that there are other molecules involved in its expression during embryonic development.
Decrease of SOX11 expression during cartilage degradation
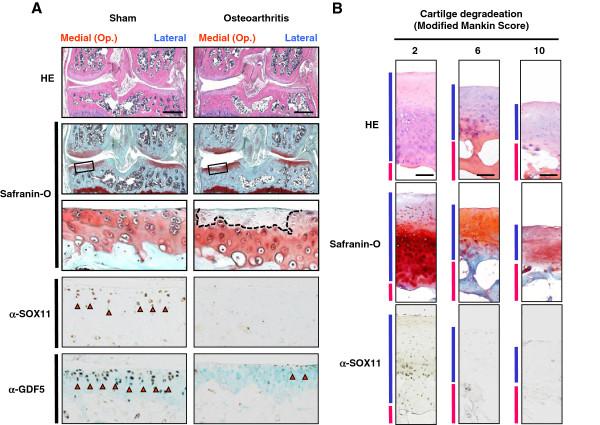
To know the role of SOX11 in cartilage homeostasis, we next created an experimental osteoarthritis (OA) model through surgical induction of instability in the knee joints of 8-week-old mice [24]. Sox11 is normally expressed at a high level in the weight-bearing regions of the mature joint, although it is less expressed in the peripheral weight bearing zone or non-weight bearing zone such as edge of femoral condyle, tibial plateau or intercondylar space (Additional file 6). Four weeks after surgery, the medial knee joints exhibited cartilage degradation, as demonstrated by a decrease in Safranin-O staining (Figure 5). Immunohistochemical analysis revealed that both Sox11 and Gdf5 expression levels were decreased in the degraded cartilage in the medial knee joint, whereas they were still strongly expressed in the intact cartilage from the sham-operated knees or from the lateral knee joint of OA-operated knees (Figure 5A) (Additional file 7). These results were consistent among multiple sections obtained from the center of the weight-bearing area in different mouse knee joints (n = 6/6 slides) (Figure 5A).
Figure 5.
Contribution of SOX11 to articular cartilage homeostasis in mice and humans. (A) Histological findings of articular cartilage after creating an experimental osteoarthritis model that induces instability in the knee joints of 8-week-old mice. HE and Safranin-O stainings, as well as immunohistochemical staining with an antibody to SOX11 (α-SOX11) or GDF5 (α-GDF5) were performed 4 weeks after surgery. A sham operation was performed using the same approach. Operations were performed in the medial side of knee joints. Dotted line highlights the region where Safranin-O staining is lost in the arthritic model. Red arrowheads indicate positive signal for SOX11 or GDF5. Scale bars, 500 μm. (B) H&E staining, Safranin-O staining and immunostaining with an antibody to human SOX11 in human knee cartilages, obtained as surgical specimens of total knee arthroplasty. Cartilage degradation was evaluated by the modified Mankin scoring system. The blue and red side bars indicate the articular cartilage layer and subchondral bone layer, respectively. Scale bars, 500μm.
Finally we analyzed 10 samples of human knee joints that had undergone total knee arthroplasty. Immunohistochemical analysis revealed that SOX11 expression was decreased from early stages of osteoarthritis progression, as assessed by decreased Safranin-O staining and modified Mankin’s cartilage degradation scores (Figure 5B) (Additional file 8). Specificity of SOX11 antibody was confirmed by using the antibody of non-immune IgG (Additional file 9).
Discussion
In the present study, we identify SOX11 as a molecule contributing to the regulation of GDF5 expression during joint formation and maintenance. In previous studies, SOX11 was identified as an essential molecule for neuronal maturation, however its molecular function in developing limbs had not been explored [25,26]. We found that SOX11 is expressed in the skeletal elements of the developing limbs and that its expression is increased in joint tissue as it develops (Figure 1). Sox11 deficient mice show a range of skeletal morphological abnormalities [27]. However, no defects were observed in the program of joint formation per se, and the resulting limb phenotype is not severe [27]. It is more than likely that many genes are involved in joint formation and their function may be redundant.
We show that SOX11 can enhance GDF5 expression in vitro (Figure 2). SOX proteins are known to bind the 5’ UTR of target genes to modulate their transcriptional activities. [28,29] We find that, indeed, SOX11 can bind the 5’UTR of the GDF5 gene. Although SOX11 enhanced chondrogenic proliferation in vitro, its misexpression in vivo did not affect the gross morphology of limb nor the mRNA expression patterns of Gdf5 (Figure 4C). However, SOX11 overexpression increased the mRNA levels of Gdf5 both in micromass cell culture from chick limb bud cells and in misexpression experiments in vivo, albeit to a small extent (Figure 4). While SOX11 is not requisite for joint formation, our results suggest that SOX11 is likely to at least partially contribute to the regulation of GDF5 expression during that process.
Finally, to investigate the roles of SOX11 in joint homeostasis, we examined its expression pattern in an osteoarthritis experimental model of knee joints in adult mice and human samples after total knee replacement surgery (Figure 5). A single nucleotide polymorphism in the GDF5 gene has been reported to be related to susceptibility to osteoarthritis [5,30]. Although the SOX11 binding site in our study seems to be independent of this reported SNP, reverse phase protein arrays in osteoarthritic and normal cartilage tissue identified SOX11 as one of the molecules down-regulated in osteoarthritic cartilage [31]. Consistent with these results, SOX11 expression was decreased in degraded cartilage both in mouse experimental osteoarthritis model and in human samples of osteoarthritis. These data support the speculation that SOX11 could be important not only for embryonic joint formation, but also for joint homeostasis in articular cartilage in adults. This could be tested by assessing whether Sox11 deficient mice have increased incidence of developing OA relative to their wild type littermates.
Joint formation is a complex and multistep process. Another molecule essential for joint formation is Wnt9a[32]. Wnt signaling inhibits chondrogenesis and promotes joint formation [32,33]. Several joints are abnormally formed in the Wnt9a-deficient mice [34]. The process of joint formation likely includes local induction of β-catenin/Wnt9a activity, which serves to block chondrogenesis and to subsequently maintain the interzone space. In parallel, signals secreted from the interzone, potentially including Gdf5, act on neighboring cartilage elements to prevent induction of a new interzone in the vicinity. Cells taken from prospective autopod joint sites using a microsurgical procedure displayed a propensity to undergo chondrogenesis following treatment with exogenous recombinant Gdf5[35], indicating that the formation of the mature articular cartilage is induced by the stimulation of chondrogenesis by secreted proteins such as Gdf5. We suggest that SOX11 could be one of the molecules involved in the later stages of joint development, contributing to the maturation of articular cartilage, at least in part through enhancing GDF5 expression.
Conclusions
In this study, we have identified SOX11 as being capable of directly regulating GDF5 in vitro and showed its partial contribution to the in vivo regulation of GDF5 expression during joint formation and homeostasis. The further identification of molecules acting upstream of GDF5 will not only provide further understanding of joint formation, but will also potentially lead to insight into the pathogenesis of human joint diseases such as osteoarthritis.
Methods
Animals
All animal experiments were performed according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School and the Animal Care and Use Committee of the University of Tokyo.
Cell cultures
DF-1 cells were obtained from the American Type Culture Collection (ATCC), and ATDC5, C3H10T1/2, and HeLa cells were obtained from the RIKEN BioResource Center Cell Bank (Ibaraki, Japan). DF-1, C3H10T1/2 and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS (Invitrogen, Carlsbad, CA), supplemented with L-glutamine (2 mM), penicillin (100 U/mL), and streptomycin (100 mg/mL). ATDC5 cells were maintained in DMEM/Ham’s F-12 (1:1) containing 5% FBS, human transferrin (10 μg/mL), sodium selenite (30 nM) and the above supplements.
Plasmid construction
The human SOX11 gene was PCR-amplified, and cloned into pCMV-HA plasmid (Clontech, Mountain View, CA). The SOX11 cDNA was then excised from pCMV-HA and cloned into pMx-IRES-Bsr retroviral expression plasmid [36] and pCMV-3x HA tag vector, which was modified from the original pCMV-3Tag-1A vector (Stratagene, La Jolla, CA). Human GDF5 cDNA was cloned into the pMx-IRES-Bsr. Chick Sox11 cDNA was cloned into the pCAG-GFP vector [37], and chick cDNA with GFP fragment was excised and cloned into RCAS-BP(A) vector [38]. The deleted mutants of the functional domains in SOX11 were created by PCR. For gene silencing, short hairpin RNA (shRNA) sequences directed against the mouse Sox11 gene and GFP for control were designed by BLOCK-iT RNAi Designer (Invitrogen) and their expression vectors were constructed with piGENEmU6 vector (iGENE Therapeutics, Tokyo). The U6 promoter and the insert were excised from the piGENEmU6 and cloned into the pMx-IRES-Bsr. The 5’-flanking sequence (-448 bp/+319 bp) of the human GDF5 promoter was amplified by PCR and cloned into pGL3-basic plasmid (Promega, Madison, WI). Mutated constructs were created by PCR. Sequences of all the plasmids were confirmed by an ABI PRISM 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA). Primers for plasmid construction and shRNA design are shown in Additional file 10.
Luciferase assay
Transfection of ATDC5, C3H10T1/2, or HeLa cells was performed in duplicate in 24-well plates using FuGene 6 with plasmid DNA (200 ng of pGL3 reporter vector, 100 ng of effector vector and 4 ng of pRL-TK vector (Promega) for internal control per well). Cells were harvested 48 hours after transfection. Luciferase assays were performed with a dual luciferase reporter assay system (Promega) using the LMax luminometer (Molecular Devices, Sunnyvale, CA). Results were shown as the ratio of firefly activity to Renilla activity. Four wells were transfected with each construct (2 wells for Figure S1) and experiments were repeated at least twice in each cell line for each construct.
Retroviral gene transfer
Two x 106 Plat-E cells were plated in 60-mm dishes, and were transfected with 3 μg pMx vector using Fugene (Roche) on the following day. Forty-eight hours after transfection, the medium was collected and used as the retrovirus supernatant, which was applied to the ATDC5 or C3H10T1/2 cells. Selection of the retrovirally introduced ATDC5 or C3H10T1/2 cells was started after 2 days in the medium containing 10 μg/ml of blasticidin. After 2 weeks, the surviving cells were collected and assayed.
Quantitative RT-PCR analysis
Total RNAs from cells were isolated with an RNeasy Mini Kit (Qiagen) and each sample was treated by DNase, reverse-transcribed with a QuantiTect reverse transcription kit (Qiagen), and used as a template for the 2nd step PCR. Full-velocity SYBR Green PCR Master Mix (Stratagene, La Jolla, CA) was used for the 2nd step PCR and fluorescence detection was performed with MX3000P (Stratagene). Copy numbers of target genes’ mRNA were quantified by a dilution series of standard templates and adjusted with human, rodent or chick Gapdh as an internal control. Four pipetting replicates were performed for each reaction. Primers for quatitative RT-PCR are shown in Additional file 10.
Electrophoretic mobility shift assay (EMSA)
SOX11 protein was in-vitro translated using the TNT Quick coupled transcription/translation system (Promega). EMSA was carried out using a DIG Gel Shift kit (Roche), according to manufacturer’s instructions. Binding reactions were incubated for 30 minutes at room temperature. For competition analyses, a 50-fold excess of unlabeled competitor probe was included in the binding reaction. For the super shift experiments, 200ng of anti-SOX11 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was added after 30 minutes of binding reaction, and the reaction was incubated for an additional 60 minutes at room temperature. Samples were loaded onto Novex 6% Tris–borate–EDTA gels (Invitrogen) and electrophoresed at 120V for 60 minutes.
Chromatin immunoprecipitation (ChIP) assay
A ChIP assay was performed with an OneDay ChIP kit, according to manufacturer’s instructions (Diagenode, Liege, Belgium). HeLa cells were transfected with 3x HA tagged empty vector or the SOX11 vector using FuGene 6 as described above. In vivo crosslinking was performed 72 hours after transfection. To shear genomic DNA, the lysates were then sonicated on ice ten times for 30 seconds each. For immunoprecipitation, 1μg of anti-HA antibody (Abcam, Cambridge, MA) or non-immune normal rabbit IgG were used.
Histological analyses of mouse embryos
For section in situ hybridization, partial coding sequences of mouse Sox11 and Gdf5 gene with the T7 promoter sequence were obtained by PCR amplification. A labeled antisense riboprobe for Sox11 was prepared with T7 RNA polymerase using a DIG RNA labeling mix (Roche), and that for Gdf5 was prepared using a Fluorescein RNA labeling mix (Roche). The C57BL/6 mouse embryos (E13.5-E14.5) were prepared as previously described [39]. The Sox11 probe was detected using an anti-DIG antibody-coupled POD and TSA plus cyanine 3 system (PerkinElmer, Waltham, MA). The Gdf5 probe was detected using an anti-Fluorescein antibody-coupled POD and TSA plus Fluorescein system (PerkinElmer). Images were obtained using an SP2 inverted confocal microscope (Leica, Wetzlar, Germany). For immunohistochemistry, tissues were fixed in 4% paraformaldehyde, embedded in paraffin. The sections were incubated with primary antibodies to SOX11 (Sigma; 1:200) or GDF5 (Abcam; 1:100). Immunodetection was performed using CSA II (DakoCytomation, Carpinteria, CA).
Misexpression experiment in chick embryos
A chicken fibroblast cell line DF-1 was plated in a 60 mm dish the day before transfection. Three μg of RCAS BP (A) plasmids were introduced with Fugene6 transfection regent (Roche). The cells were cultured for 3 weeks, and the supernatant was collected and used as a virus stock. Preparation of micromass cell culture and the infection of RCAS virus were performed as previously described [32]. The RCAS viruses were injected into the lateral plate mesoderm of Hamburger Hamilton (HH) stage 10 chick embryos (day 1.5) to target gene misexpression to developing limbs as previously reported [38,40]. Embryos were harvested at HH stage 32 or 35 (or day 7.5, 8.5, respectively). Tissue preparations for whole mount in situ hybridization were performed as described above. Digoxigenin (DIG)-labeled antisense riboprobes were detected using anti-DIG antibody-coupled AP and NBT/BCIP solution (Roche). For quatitative RT-PCR analysis, total RNAs from chick limbs at HH stage 35 were isolated with an RNeasy Mini Kit. For Western blotting, hind limbs of HH stage 35 were homogenized with TissueLyser (Qiagen). Anti-GDF5 antibody (Abcam; 1:1000) and Anti-β-Actin antibody (Abcam; 1:1000) were used for primary antibodies and Goat anti-Rabbit antibody (Jackson ImmunoResearch; 1:10000) was used for a secondary antibody. Densitometry analysis was performed with ImageJ software.
Osteoarthritis experiment
The surgical procedure to create an osteoarthritis experimental model of knee joints was carried out as previously reported [24], transecting the medial collateral ligament and medial meniscus of 8-week-old mice. A sham operation was performed on the contralateral knee. Four weeks after surgery, the entire knee joints were dissected, decalcified, and underwent hematoxylin-eosin (HE) and Safranin-O-fast green stainings, and/or immunohistochemical staining with the antibody to SOX11 and GDF5.
Human samples
We obtained human samples from individuals undergoing total knee arthroplasty after obtaining written informed consent as approved by the Ethics Committee of the University of Tokyo. We histologically assessed cartilage degradation according to the modified Mankin scoring system. [41,42].
Statistical analysis
A Mann-Whitney test was performed for the statistical analyses of luciferase assays and quantitative RT-PCR. p < 0.05 was deemed significant.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AK carried out the experiments described in this report. TI, KN and UC participated in the design of the study. AF created the experimental model of osteoarthritis in mice and TN collected human samples. AK, CT and HK analyzed the data. AK and CT wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Screening for candidate SOX genes activating GDF5 promoter. Luciferase assays for the transcriptional activity of the GDF5 promoter by the overexpression of different SOX family molecules in ATDC5 and HeLa cells transfected with GDF5 promoter region (-448/+319) ligated to the luciferase-reporter gene. Data are expressed as means (bars) ± SDs (error bars) of Relative luciferase activity for two assays/construct. EV: empty vector, the vector containing the luciferase transgene but lacking the GDF5 promoter.
Conserved domains in the SOX11 protein sequence among humans, mice and chicks. Amino acid sequences coding SOX11 protein is shown and the conserved domains among species are indicated by black boxes.
Identification of functional SOX11 domains which are required for GDF5 expression. (A) Luciferase assays for the transcriptional activity of the reporter driven by the GDF5 (-448/+319) promoter, in the context of co-transfection with deletion mutants that lack N-terminus domain (Rx-ΔN), Acid-rich domain (Rx-ΔAR), or C-terminus domain (Rx-ΔC) in ATDC5, C3H10T1/2 and HeLa cells. Data are expressed as means (bars) ± SDs (error bars) of Relative Luciferase activity for four assays/construct. (B) Endogenous Gdf5 mRNA expressions in ATDC5 cells retrovirally transfected with the deletion mutants of SOX11. mRNA levels were determined by quantitative RT-PCR and expressed as means (bars) ± SDs (error bars) for 4 wells/construct.
Luciferase assay for deletion of SOX family binding site near the rs143383. Data are expressed as means (bars) ± SDs (error bars) of Relative Luciferase activity for four assays/construct. *P < 0.05 vs.–448/+319 promoter. **P < 0.05 vs.–448/+319 promoter lacking the +51/+75 region.
Electromobility shift assay (EMSA) for specific binding of the GDF5 promoter with human SOX11 protein. The wild-type probe for segment A was used. (Detailed information is shown in Figure 2). An open arrowhead indicates the shifted bands of the SOX11-DNA probe complex, and a solid arrowhead indicates the band supershifted by an antibody to SOX11. The supershift band is absent by a non-immune normal IgG.
Immunostaining with an antibody to SOX11 in multiple regions of mouse knee joints. Red arrowheads indicate positive signal for SOX11, while white arrowheads showed scarcity of signal for SOX11. Immunostaining was detected by DAB, which was followed by counterstaining with methyl green. mm: medial meniscus, lm: lateral meniscus, ACL: anterior cruciate ligament.
Immunostaining in the lateral compartment of knee joints in osteoarthritis model of mice. Immunostainings for SOX11 or GDF5 were detected by DAB, which was followed by counterstaining with methyl green. Red arrowheads showed positive signal for SOX11 or GDF5.
Decrease of the SOX11 expression during osteoarthritis progression. Safranin-O staining and immunostaining, with an antibody to SOX11, of human knee cartilages. Other representative samples are shown in Figure 5B. Cartilage degradation was evaluated by the modified Mankin scoring system. Red arrowheads indicate positive signal for SOX11. Scale bars, 500μm.
Immunostaining, with an antibody to SOX11 or non-immune normal IgG, of human cartilages. Immunostaining was performed on human knee cartilage with a modified Mankin Score = 2.
Primer sequences for plasmid construction, shRNA or RT-PCR analysis used in this study.
Contributor Information
Akinori Kan, Email: akinori@genetics.med.harvard.edu.
Toshiyuki Ikeda, Email: ikedat-ko@umin.ac.jp.
Atsushi Fukai, Email: fukai-ort@umin.ac.jp.
Takumi Nakagawa, Email: takumin-tky@umin.ac.jp.
Kozo Nakamura, Email: nakamurak-ort@h.u-tokyo.ac.jp.
Ung-il Chung, Email: uichung-tky@umin.ac.jp.
Hiroshi Kawaguchi, Email: kawaguchi-ort@h.u-tokyo.ac.jp.
Clifford J Tabin, Email: tabin@receptor.med.harvard.edu.
Acknowledgments
We thank Jimmy K. Hu and Jessica A. Lehoczky for critical suggestions and technical advice. We are grateful to Dr. Veronique Lefebvre for insightful suggestions and discussion of unpublished materials. The pMx-IRES-Bsr retroviral plasmid and the Plat-E cells were generous gifts from Dr. Toshio Kitamura (The Institute of Medical Science, The University of Tokyo). This work was supported by a grant PO1DK056246 to Clifford J. Tabin and Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (#17591549 & #21591931).
Grant information
NIH: Clifford J. Tabin; #PO1DK056246.
Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; #17591549 & #21591931.
References
- Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Pacifici M, Koyama E, Iwamoto M. Mechanisms of synovial joint and articular cartilage formation: recent advances, but many lingering mysteries. Birth Defects Res C Embryo Today. 2005;75:237–248. doi: 10.1002/bdrc.20050. [DOI] [PubMed] [Google Scholar]
- De Crombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13:721–727. doi: 10.1016/S0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S, Chung UI. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, Fujioka M, Sudo A, Uchida A, Yamamoto S. A functional polymorphism in the 5’ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Shi D, Nakajima M, Ozaki K, Sudo A, Kotani A, Uchida A, Tanaka T, Fukui N, Tsunoda T. Common variants in DVWA on chromosome 3p24.3 are associated with susceptibility to knee osteoarthritis. Nat Genet. 2008;40:994–998. doi: 10.1038/ng.176. [DOI] [PubMed] [Google Scholar]
- Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee SJ. Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- Masuya H, Nishida K, Furuichi T, Toki H, Nishimura G, Kawabata H, Yokoyama H, Yoshida A, Tominaga S, Nagano J. A novel dominant-negative mutation in Gdf5 generated by ENU mutagenesis impairs joint formation and causes osteoarthritis in mice. Hum Mol Genet. 2007;16:2366–2375. doi: 10.1093/hmg/ddm195. [DOI] [PubMed] [Google Scholar]
- Tsumaki N, Tanaka K, Arikawa-Hirasawa E, Nakase T, Kimura T, Thomas JT, Ochi T, Luyten FP, Yamada Y. Role of CDMP-1 in skeletal morphogenesis: promotion of mesenchymal cell recruitment and chondrocyte differentiation. J Cell Biol. 1999;144:161–173. doi: 10.1083/jcb.144.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton P, Edwards C, Archer CW, Francis-West P. Growth/differentiation factor-5 (GDF-5) and skeletal development. J Bone Joint Surg Am. 2001;83-A(Suppl 1):S23–S30. [PubMed] [Google Scholar]
- Coleman CM, Tuan RS. Growth/differentiation factor 5 enhances chondrocyte maturation. Dev Dyn. 2003;228:208–216. doi: 10.1002/dvdy.10369. [DOI] [PubMed] [Google Scholar]
- Thomas JT, Lin K, Nandedkar M, Camargo M, Cervenka J, Luyten FP. A human chondrodysplasia due to a mutation in a TGF-beta superfamily member. Nat Genet. 1996;12:315–317. doi: 10.1038/ng0396-315. [DOI] [PubMed] [Google Scholar]
- Kiefer JC. Back to basics: SOX genes. Dev Dyn. 2007;236:2356–2366. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, De Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, De Crombrugghe B. Toward understanding SOX9 function in chondrocyte differentiation. Matrix Biol. 1998;16:529–540. doi: 10.1016/S0945-053X(98)90065-8. [DOI] [PubMed] [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Dy P, Smits P, Silvester A, Penzo-Mendez A, Dumitriu B, Han Y, de la Motte CA, Kingsley DM, Lefebvre V. Synovial joint morphogenesis requires the chondrogenic action of Sox5 and Sox6 in growth plate and articular cartilage. Dev Biol. 2010;341:346–359. doi: 10.1016/j.ydbio.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P, Goze C, Marsollier C, Taviaux S, Hardelin JP, Koopman P, Berta P. The human SOX11 gene: cloning, chromosomal assignment and tissue expression. Genomics. 1995;29:541–545. doi: 10.1006/geno.1995.9970. [DOI] [PubMed] [Google Scholar]
- Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici M. Tenascin-C and the development of articular cartilage. Matrix Biol. 1995;14:689–698. doi: 10.1016/S0945-053X(05)80011-3. [DOI] [PubMed] [Google Scholar]
- Kamekura S, Hoshi K, Shimoaka T, Chung U, Chikuda H, Yamada T, Uchida M, Ogata N, Seichi A, Nakamura K. Osteoarthritis development in novel experimental mouse models induced by knee joint instability. Osteoarthritis Cartilage. 2005;13:632–641. doi: 10.1016/j.joca.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski MP, Cornuet PK, McIlwrath S, Koerber HR, Albers KM. SRY-box containing gene 11 (Sox11) transcription factor is required for neuron survival and neurite growth. Neuroscience. 2006;143:501–514. doi: 10.1016/j.neuroscience.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Rettig SD, Enderich J, Bosl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valor LM, Castillo M, Ortiz JA, Criado M. Transcriptional regulation by activation and repression elements located at the 5’-noncoding region of the human alpha9 nicotinic receptor subunit gene. J Biol Chem. 2003;278:37249–37255. doi: 10.1074/jbc.M307043200. [DOI] [PubMed] [Google Scholar]
- Kou I, Ikegawa S. SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J Biol Chem. 2004;279:50942–50948. doi: 10.1074/jbc.M406786200. [DOI] [PubMed] [Google Scholar]
- Southam L, Rodriguez-Lopez J, Wilkins JM, Pombo-Suarez M, Snelling S, Gomez-Reino JJ, Chapman K, Gonzalez A, Loughlin J. An SNP in the 5’-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum Mol Genet. 2007;16:2226–2232. doi: 10.1093/hmg/ddm174. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3:e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/S0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, Yang Y. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spater D, Hill TP, O’Sullivan JR, Gruber M, Conner DA, Hartmann C. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development. 2006;133:3039–3049. doi: 10.1242/dev.02471. [DOI] [PubMed] [Google Scholar]
- Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree RB. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Tabin C. Targeted gene misexpression in chick limb buds using avian replication-competent retroviruses. Methods. 1998;14:407–420. doi: 10.1006/meth.1998.0595. [DOI] [PubMed] [Google Scholar]
- McGlinn E, Mansfield JH. Detection of gene expression in mouse embryos and tissue sections. Methods Mol Biol. 2011;770:259–292. doi: 10.1007/978-1-61779-210-6_10. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Mankin HJ, Dorfman H, Lippiello L, Zarins A. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. II. Correlation of morphology with biochemical and metabolic data. J Bone Joint Surg Am. 1971;53:523–537. [PubMed] [Google Scholar]
- Ostergaard K, Andersen CB, Petersen J, Bendtzen K, Salter DM. Validity of histopathological grading of articular cartilage from osteoarthritic knee joints. Ann Rheum Dis. 1999;58:208–213. doi: 10.1136/ard.58.4.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Screening for candidate SOX genes activating GDF5 promoter. Luciferase assays for the transcriptional activity of the GDF5 promoter by the overexpression of different SOX family molecules in ATDC5 and HeLa cells transfected with GDF5 promoter region (-448/+319) ligated to the luciferase-reporter gene. Data are expressed as means (bars) ± SDs (error bars) of Relative luciferase activity for two assays/construct. EV: empty vector, the vector containing the luciferase transgene but lacking the GDF5 promoter.
Conserved domains in the SOX11 protein sequence among humans, mice and chicks. Amino acid sequences coding SOX11 protein is shown and the conserved domains among species are indicated by black boxes.
Identification of functional SOX11 domains which are required for GDF5 expression. (A) Luciferase assays for the transcriptional activity of the reporter driven by the GDF5 (-448/+319) promoter, in the context of co-transfection with deletion mutants that lack N-terminus domain (Rx-ΔN), Acid-rich domain (Rx-ΔAR), or C-terminus domain (Rx-ΔC) in ATDC5, C3H10T1/2 and HeLa cells. Data are expressed as means (bars) ± SDs (error bars) of Relative Luciferase activity for four assays/construct. (B) Endogenous Gdf5 mRNA expressions in ATDC5 cells retrovirally transfected with the deletion mutants of SOX11. mRNA levels were determined by quantitative RT-PCR and expressed as means (bars) ± SDs (error bars) for 4 wells/construct.
Luciferase assay for deletion of SOX family binding site near the rs143383. Data are expressed as means (bars) ± SDs (error bars) of Relative Luciferase activity for four assays/construct. *P < 0.05 vs.–448/+319 promoter. **P < 0.05 vs.–448/+319 promoter lacking the +51/+75 region.
Electromobility shift assay (EMSA) for specific binding of the GDF5 promoter with human SOX11 protein. The wild-type probe for segment A was used. (Detailed information is shown in Figure 2). An open arrowhead indicates the shifted bands of the SOX11-DNA probe complex, and a solid arrowhead indicates the band supershifted by an antibody to SOX11. The supershift band is absent by a non-immune normal IgG.
Immunostaining with an antibody to SOX11 in multiple regions of mouse knee joints. Red arrowheads indicate positive signal for SOX11, while white arrowheads showed scarcity of signal for SOX11. Immunostaining was detected by DAB, which was followed by counterstaining with methyl green. mm: medial meniscus, lm: lateral meniscus, ACL: anterior cruciate ligament.
Immunostaining in the lateral compartment of knee joints in osteoarthritis model of mice. Immunostainings for SOX11 or GDF5 were detected by DAB, which was followed by counterstaining with methyl green. Red arrowheads showed positive signal for SOX11 or GDF5.
Decrease of the SOX11 expression during osteoarthritis progression. Safranin-O staining and immunostaining, with an antibody to SOX11, of human knee cartilages. Other representative samples are shown in Figure 5B. Cartilage degradation was evaluated by the modified Mankin scoring system. Red arrowheads indicate positive signal for SOX11. Scale bars, 500μm.
Immunostaining, with an antibody to SOX11 or non-immune normal IgG, of human cartilages. Immunostaining was performed on human knee cartilage with a modified Mankin Score = 2.
Primer sequences for plasmid construction, shRNA or RT-PCR analysis used in this study.