Abstract
Intraflagellar transport (IFT) is a microtubule based system that supports the assembly and maintenance of cilia. Genetic and biochemical studies have identified two distinct complexes containing multiple proteins that are part of the IFT machinery. In this study we prepared mouse pituitary cells that expressed an epitope-tagged IFT protein and immuno-purified the IFT B complex from these cells. Mass spectrometry analysis of the isolated complex led to identification of a number of well known components of the IFT B complex. In addition, peptides corresponding to mouse tetratricopeptide repeat proteins, TTC30A1, TTC30A2 and TTC30B were identified. The mouse Ttc30A1, Ttc30A2, Ttc30B genes are orthologs of Caenorhabditis elegans dyf-1, which is required for assembly of the distal segment of the cilia. We used co-immunoprecipitation studies to provide evidence that, TTC30A1, TTC30A2 or TTC30B can be incorporated into a complex with a known IFT B protein, IFT52. We also found that TTC30B can interact with mouse KIF17, a kinesin which participates in IFT. In vitro expression in a cell-free system followed by co-immunoprecipitation also provided evidence that TTC30B can directly interact with several different IFT B complex proteins. The findings support the view that mouse TTC30A1, TTC30A2 and TTC30B can contribute to the IFT B complex, likely through interactions with multiple IFT proteins and also suggest a possible link to the molecular motor, KIF17 to support transport of cargo during IFT.
Keywords: Intraflagellar transport, cilia, kinesin, tetratricopeptide repeat
Introducton
Because of the important role of cilia and flagella in a number of developmental and physiological processes, the assembly and maintenance of cilia by intraflagellar transport (IFT) has been the subject of intensive study. IFT is a microtubule-based process which moves particles along the length of a cilium. Two large protein complexes designated IFT A and IFT B have been extensively characterized [1, 2]. Movement of these IFT complexes and cargo towards the cilia tip is mediated by kinesin II motors and retrograde movement towards the basal body is mediated by dynein motor proteins. It is clear that the IFT machinery is highly conserved from unicellular eucaryotes through most metazoans with the exception of terrestrial plants. Defects in IFT have been found to lead to severe phenotypes and human disease [3, 4].
Several different experimental systems have been very useful for studying IFT. The process of IFT was first identified in the green alga, Chlamydomonas reinhardtii, [5]. Mass spectrometry studies of purified Chlamydomonas flagella have allowed determination of the flagellar proteome [6]. Genetic studies of mutations that alter flagella in Chlamydomonas or that affect sensory cilia in Caenorhabditis elegans have provided initial evidence that IFT proteins are conserved between a metazoan nematode and a unicellular alga [7]. Studies of mammalian cells and organs have shown that IFT is required for forming the primary cilia of kidney cells and that IFT mutations lead to polycystic kidney disease [8]. As variations in IFT are probably required to support the generation of structurally and functionally diverse cilia [9] it is likely that the IFT cargo and perhaps even the IFT machinery varies in different cells and tissues. To further explore the possibility of tissue- or cell-specific variations in IFT it would be useful to further characterize the IFT machinery in a variety of cells and tissues.
In the present study we have characterized proteins of the IFT complex from the αT3-1 pituitary gonadotrope cell line by preparing cells expressing a tagged component of the IFT machinery as a tool to immunochemical purification of an IFT complex.
Methods
Reagents and cell culture
AU1 monoclonal antibody was purchased from Covance and FLAG monoclonal from Sigma. The S219V mutant of TEV protease which has increased stability and catalytic activity was expressed in E. coli and purified as described [10]. The αT3-1 gonadotrope pituitary cell line (American Type Culture Collection) was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 2.5% fetal bovine serum and 15% equine serum. GP2-293 cells (Clontech) were cultured in DMEM containing 10% fetal bovine serum.
A number of different expression vectors were constructed for the studies. An expression vector for mouse IFT57 tagged with two different epitopes (FLAG and AU1) was used to prepare cells stably expressing the tagged IFT57. This vector was prepared by PCR amplification of the IFT57 coding sequence using single-stranded cDNA that was prepared from RNA from the αT3-1 pituitary cell line. The IFT57 coding sequence was fused to synthetic DNA containing the coding sequence for the calmodulin binding domain from muscle myosin light-chain kinase [11], an epitope recognized by the FLAG antibody, two copies of the recognition/cleavage sequence for the TEV protease and two copies of the epitope recognized by the AU-1 antibody (Fig. 1A, B). The tagged-IFT57 coding sequence was confirmed as correct by sequence analysis and then inserted into XhoI and BamHI sites of the pLXIN retoviral vector (Clontech). For transient transfection and co-immunoprecipation studies, expression vectors for proteins with a single epitope tag were prepared. The coding sequence for mouse mouse TTC30A1, TTC30A2, TTC30B were prepared by PCR amplication from single-stranded cDNA from αT3-1 cells and fused to synthetic DNA encoding an epitope tag (DTYRYI) recognized by the AU1 monoclonal antibody and inserted into the pcDNA3 mammalian expression vector. Similarly the coding sequence for mouse IFT52 and IFT57 were also prepared by PCR, fused to the FLAG epitope and inserted into pcDNA3. An expression vector for a short form of mouse KIF17, GenBank BAE43328, was prepared by PCR amplification of the coding sequence which was fused to the HA epitope (YPYDVPDYA) recognized by the 12CA5 monoclonal antibody and then inserted into pcDNA3. All coding sequences used for preparing expression vectors were confirmed by automated DNA sequencing.
Figure 1. Expression of tagged mouse IFT57 to isolate an IFT complex from mouse pituitary cells.
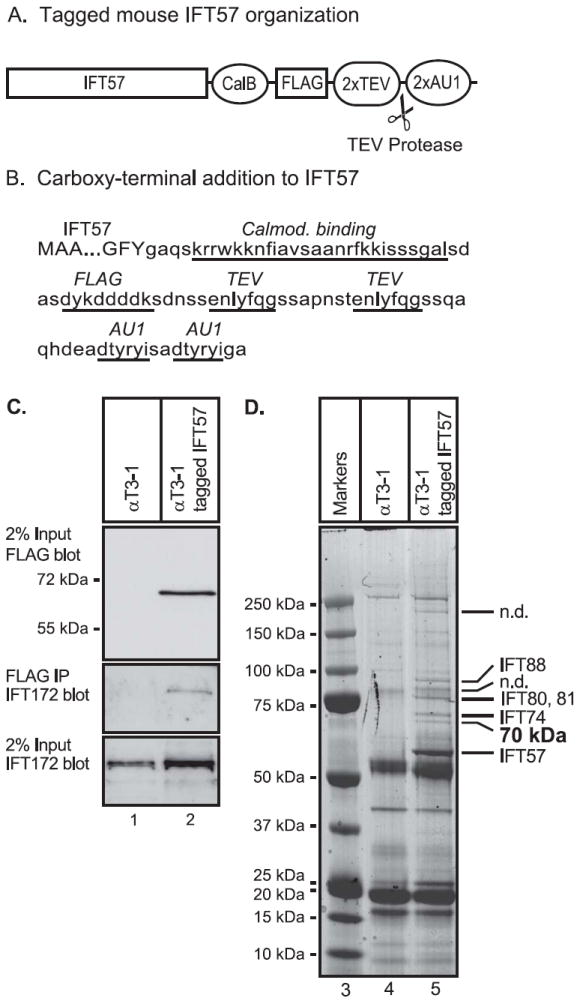
(A) Organization of the coding sequence of tagged mouse IFT57. The mouse IFT57 coding sequence was modified so that a calmodulin binding domain (CalB), an epitope recognized by the FLAG antibody (FLAG), 2 recognition/cleavage sites for the TEV protease (2xTEV) and 2 epitopes recognized by the AU1 antibody were added to the caroboxy-terminus of the protein. (B) Coding sequence for tagged IFT57. The first 3 amino acids and the last three amino acids of IFT57 are shown (upper case), and the sequence added to the carboxy-terminus of the protein is shown (lower case). The calmodulin binding domain, FLAG epitope, TEV recognition/cleavage sites and AU1 epitopes of the carboxy-terminal tag are indicated by underlines. (C) Expression of tagged IFT57 in the αT3-1 pituitary cell line. Cell extracts were prepared from either control αT3-1 cell or αT3-1 cells stably expressing tagged IFT57. Cell extracts were resolved by denaturing polyacrylamide gel electrophoresis, transferred to a membrane and then incubated with FLAG antiserum to detect input-tagged IFT57 or with antiserum to IFT172 to detect endogenous IFT172. To assay for formation of an IFT complex, the tagged-IFT57 was isolated by immunoprecipitation with FLAG monoclonal antibody and the immunoprecipitate was analyzed by gel electrophoresis and immunoblotting with antibody to IFT172 to detect co-immunoprecipitated IFT172. (D) Identification of proteins interacting with tagged IFT57 in pituitary cells. Whole cell extracts were prepared from control αT3-1 cells or αT3-1 cells expressing tagged IFT57. The cell extracts were incubated with AUI antibody immobilized on agarose beads. After washing of the beads, the bound proteins were eluted by digestion with TEV protease and the eluate incubated with FLAG antibodies immobilized on agarose beads. After washing, bound proteins were eluted with FLAG peptide and resolved by denaturing polyacrylamide gel electrophoresis followed by staining with Coomassie brilliant blue R250. Stained bands that appeared to be substantially more intense in the sample from the tagged IFT57 cells were cut from the gel and subject to trypsin digestion and mass spectrometry. Several known IFT proteins were identified (labeled to the right of the gel lanes) and one 70 kDa protein. Some bands were not determined (n.d.).
Preparation of αT3-1 cells stably expressing tagged-IFT57
High-titer pantropic retrovirus for the FLAG-AU1-tagged-IFT57 in pLXIN was prepared using the GP2-293 packing cell line and VSV-G pseudotyped using recommended protocols (Clontech). αT3-1 cells were infected with the retroviral preparation and stably transduced cells selected using G418 (Invitrogen).
Transient transfections
Human embryonic kidney 293 cells (HEK293) were maintained in DMEM containing 10% fetal bovine serum. Cells were transfected with a total of 2 μg DNA and 5 μl of Lipofectamine 2000 (Invitrogen, Inc.) in 35 mm well plates, or 0.8 μg DNA and 2 μl Lipofectamine 2000 in 22 mm well plates using a protocol provided by the supplier. Total amount of DNA for each transfection was maintained constant by adding empty expression vector as needed.
Immunoprecipitations, and immunoblotting
Cells were scraped from the culture dishes in phosphate buffered saline and pelleted in a microfuge. The cells were resuspended in 100 mM sodium phosphate, pH 7.8, with 0.1% NP-40 and disrupted by 4 freeze-thaw cycles. After centrifugation at 10,000 × g for 5 min at 4° C, the supernatant was saved as a whole cell extract. Cell extracts were adjusted to contain 0.1% Tween-20. For small scale, analytical studies, cell extracts containing equal amounts of total protein were combined with 15 μl of a 50% slurry of anti-FLAG agarose (Sigma) or rabbit anti-AU1 serum and protein A/G agarose (Santa Cruz). The immunoprecipitation mixtures were rotated for 2 h at 4° C and the agarose bound antibodies were collected by centrifugation. The agarose beads were then washed 3 times with 1 ml each of 10 mM Tris, pH 7.4, 150 mM NaCl, 0.1 % Tween-20, 0.1% Triton X-100. Proteins bound to the agarose beads were then analyzed by electrophoresis on a denaturing, polyacrylamide gel. For immunoblotting, proteins were transferred to polyvinylidene difluoride membranes (Millipore). Blocking reactions, incubation with a 1:5,000 dilution of antiserum to FLAG, incubation with a 1:10,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse antibody (Santa Cruz) and incubation with chemiluminescent reagent (Perkin Elmer Western Lightening Plus-ECL) were all performed as suggested by the suppliers.
For preparative scale immunoprecipitation of the IFT complex to be used for mass spectrometry studies, cell extracts were prepared from 2 each 15 cm culture dishes and the cell extracts were first absorbed to a resin containing immobilized AU1 monoclonal antibody (Bethyl Laboratories) and then eluted by incubating with TEV protease using digestion conditions as described [10]. The eluate was then further purified by absorption to a resin containing immobilized FLAG monoclonal antibody.
Mass spectrometry of immunoprecipitated proteins
Immunoprecipitated proteins were resolved by one dimensional, denaturing polyacrylamide gel electrophoresis. The gel was stained with Coomassie Brilliant Blue R250 and individual bands were cut from the gel using a 2DiDx sample preparation robot (Leap Technologies). Gel slices were destained in two changes of 100 mM ammonium bicarbonate in 30% methanol, dried with neat acetonitrile, reduced with 10 mM DTT, and alkylated with 50 mM iodoacetamide. After washing and drying, approximately 100 ng of sequencing grade trypsin in 20 mM ammonium bicarbonate was added to each sample followed by incubation overnight at 37°C. Peptides were precipitated by addition of 10 μl of 1% formic acid and collected by centrifugation. Gel slices were further extracted with two additions each of 50% and 70% acetonitrile. Peptide extracts were brought to near dryness before being resuspended in 10 μl of 0.1% formic acid in preparation for MS/MS. Tandem mass spectrometry data were collected using a Qstar XL hybrid time-of-flight mass spectrometer (Applied Biosystems) under the following conditions: spray voltage 1800-1900 V; TOF-MS scan m/z 400-1600, 0.5 s; TOF-MS/MS scan m/z 50-2000, 2.0 and 90 s exclusion; data-dependent product ion acquisition of the three most abundant +2 and +3 ions from the TOF-MS scan. Monoisotopic masses for data base searching were generated using Distiller (Matrix Science) and submitted to X!Tandem for protein identification. Masses were searched against the mouse International Protein Index.
Results and discussion
To isolate the IFT complex from a mouse pituitary cell line, we prepared a modified coding sequence for IFT57, also known as Hippi, a component of the IFT B complex [2]. The IFT57 coding sequence was altered by addition of two AU1 epitopes, two tobacco etch virus (TEV) protease cleavage sites, a FLAG epitope and a calmodulin binding domain to the carboxy terminus of the coding sequence (Fig 1A,B). The AU1 and FLAG epitopes permit immunopurification using commercially available, immobilized antibodies. The TEV protease site allows selective and efficient elution of a protein bound to an immuno-absorbent and therefore also provides a quite useful purification step. The calmodulin binding domain could provide another purification step, but was not utilized in these studies. The tagged-IFT57 coding sequence was used to produce a retroviral vector which was used to infect the mouse αT3-1 pituitary gonadotrope cell line. Stably tranduced cells were selected with G418 and individual colonies were expanded for analysis of the expression of the tagged-IFT57. Cell extracts were prepared and expression of the tagged-IFT57 was identified by denaturing gel electrophoresis followed by immunoblotting with a monoclonal FLAG antibody. A cell line that expressed tagged-IFT57 at an intermediate level was selected for further study (data not shown). To determine if the tagged-IFT57 associates with a known member of the IFT complex B in αT3-1 pituitary cells, we tested for co-immunoprecipitation with endogenous IFT172 (Fig. 2C). IFT172 is another component of the complex B, although presumably it has lower affinity than most complex B proteins as IFT172 dissociates from the complex at modestly elevated salt concentrations [7]. As expected, FLAG antibody detected a strong band with the appropriate mobility for the tagged-IFT57 only in cell extracts from the transduced cells expressing the tagged protein and not from the control cells (Fig. 1C, lane 2 vs lane 1). Also as expected, both extracts contained IFT172 in similar amounts (Fig. 1C). Following immunoprecipitation of the tagged IFT57 with FLAG antibody, immuno-detection of IFT172 was observed only with the sample from the tagged-IFT57 cells, not the control cells. Therefore, it appeared that the tagged-IFT57 was capable of incorporation into a complex that includes IFT172, an endogenous IFT complex B protein.
Figure 2. The tetratricopeptide repeat-containing proteins TTC30A1, TTC30A2 and TTC30B interact with IFT52.
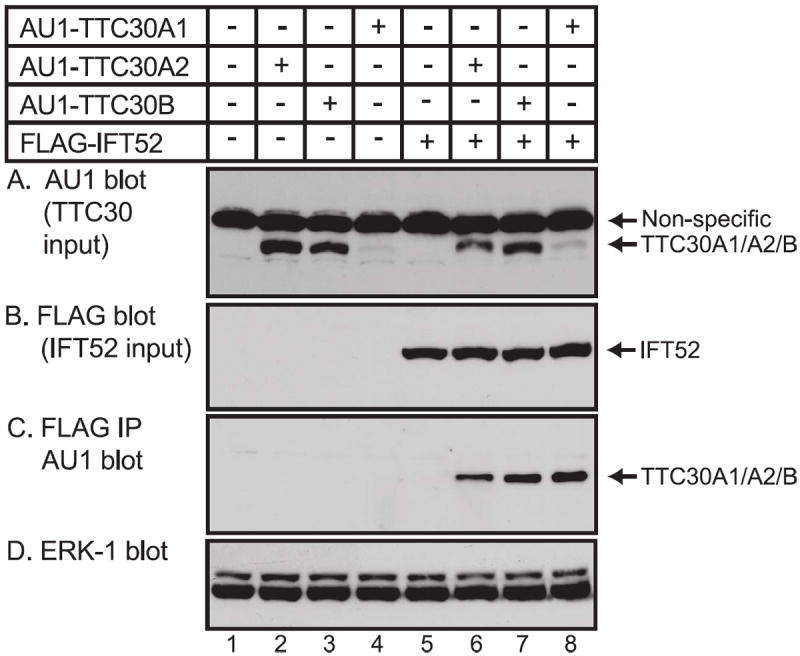
(A) HEK293 cells were transfected with expression vectors for AU1-TTC30A1, AU1-TTC30A2 or AU1-TTC30B and FLAG tagged-IFT52 as indicated. At 20 hours after transfection, whole cell extracts were prepared. (A) Samples were analyzed by denaturing gel electrophoresis and immunoblotting with antibody to AU1 to detect the input expression of the tagged-TTC30 isoforms. A non-specific band was detected in all lanes. (B) Input expression of FLAG-IFT52 was detected by immunoblotting with FLAG monoclonal antibody. (C) To detect interaction of AU-tagged TTC30 isoforms with IFT52, FLAG-tagged proteins were isolated by immunoprecipitation with FLAG antibody and the immunoprecipitate was analyzed by gel electrophoresis and immunoblotting with AU1 antibody to detect co-immunoprecipitated AU1-TTC30A1, AU1-TTC30A2 or AU1-TTC30B. (D) Imunoblotting of cell extracts with antibody to ERK-1 as a loading control.
To identify other proteins that are a component of the IFT complex in mouse pituitary cells, control cells and cells expressing the tagged-IFT57 were grown in large scale. Cell pellets were extracted with a detergent-containing buffer. Extracts were absorbed to a resin containing immobilized AU1 monoclonal antibody and then eluted by incubating with TEV protease. The eluate was then further purified by absorption to a resin containing immobilized FLAG monoclonal antibody. Bound proteins were resolved on a denaturing polyacrylamide gradient gel (Fig. 1D). Stained bands that were selectively isolated from cells expressing tagged-IFT57 as compared to controls cells were analyzed by protease digestion/elution and mass spectrometry. Analysis of the mass spectrometry data indentified several known IFT complex B proteins including IFT57, IFT74, IFT80, IFT81 and IFT88. In addition, an approximately 70 kDa protein was found to contain peptides QHPELGVGMTTEGIDVR, AAIEYQLR, LNVAHVLFMQENK, EAIGFYEPIVK, GNYDFGISR and NIPAVIEQPLEEER which correspond to isoforms of murine tetratricopeptide repeat protein 30 (TTC30). Although the genomes of a number of vertebrate species have genes encoding two TTC30 isoforms, the mouse genome contains genes encoding three isoforms, TTC30A1, TTC30A2 and TTC30B. The three TTC30 encoding genes are located adjacent to each other on chromosome 2 and encode closely related proteins (each encodes 664 amino acid proteins which are 94-95% identical to each other). Of the six peptides identified for TTC30 isoforms, five of the peptides are common to all three isoforms. The other peptide, NIPAVIEQPLEEER, is found in both TTC30A2 and TTC30B, but is not contained in TTC30A1. This data suggests that the IFT B complex in mouse αT3-1 cells contains TTC30A2 and/or TTC30B and may also contain TTC30A1.
The Caenorhabditis elegans ortholog of the mouse Ttc30 genes is dyf-1 which is thought to regulate the kinesin-II motor protein, OSM-3, possibly involving an interaction with the IFT complex B [12, 13]. Subsequent biochemical studies have provided evidence that DYF-1 is a stable component of the IFT complex B [14, 15]. Knockdown of Chlamydomonas DYF-1 was found to greatly shorten flagella. These observations provide evidence that DYF-1 is a component of the IFT complex B and it is required for normal assembly of flagella. Therefore, as a functional component of the IFT B complex, DYF-1 has been designated IFT70 [14]. Interestingly, the Zebrafish ortholog of dyf-1 is fleer [16]. Loss of function studies have shown that both Zebrafish fleer and Caenorhabditis elegans dyf-1 function as regulators of cilia tubulin polyglutamylation [16]. There is also evidence that mouse TTC30 has a role in IFT. Immunoprecipitation studies have shown that mouse TTC30A1 can contribute to the IFT B complex in murine kidney cells [17]. Mouse TTC30A1 was also identified in an informatics screen of genes that are highly expressed in tissues with highly ciliated cells such as olfactory sensory neurons, trachea and lung [18]. Although there is evidence that TTC30A1 can be incorporated into the IFT B complex, a possible role for TTC30A2 and TTC30B in the complex has not been tested.
To determine if TTC30A2 or TTC30B can interact with a known component of the IFT B complex, expression vectors were prepared for AU1-tagged TTC30A1, TTC30A2 and TTC30B and used to transfect HEK293 cells in the presence or absence of an expression vector for FLAG-IFT52. All three TTC30 isoforms were detected in the input cell extract, although interestingly TTC30A2 and TTC30B were expressed at considerably higher levels than TTC30A1 (Fig. 2A). All three TTC30 isoforms were also detected in co-immunoprecipitates with IFT52 (Fig 2C). Although TTC30A1 was expressed at much lower levels than TTC30A2 and TTC30B, it was quite efficiently enriched by immunoprecipitation of IFT52 (Fig 2D). The poor expression and efficient co-immunoprecipitation of TTC30A1 with IFT52 were confirmed in several experiments (data not shown) suggesting that TTC30A1 may have a higher affinity for components of the IFT B complex than TTC30A2 and TTC30B. Alternatively, the subcellular location of TTC30A1 may facilitate incorporation into the IFT B complex. In any case, these findings provide evidence that TTC30A1, TTC30A2 and TTC30B can participate in a complex containing IFT52 and are consistent with a possible role for all three mouse TTC30 isoforms as components of the IFT complex B.
To test the possible interaction of TTC30B with individual members of the IFT B complex, epitope-tagged proteins were expressed using an in vitro, cell-free, mRNA translation system from wheat germ. We used the wheat germ system for protein expression as flowering plants, angiosperms, do not have cilia [19, 20]. Our initial assumption was that wheat germ extracts would not contain IFT proteins and therefore we could test for binary interactions of the expressed proteins without the complication of possible interactions with an endogenous IFT complex in the extract. However, there is evidence that at least some ciliary proteins are conserved in plants that have no cilia [19]. Therefore, we cannot totally exclude possible involvement of a conserved wheat germ ciliary protein in any complexes that are formed in the wheat germ extract. We observed co-immunoprecipitation of TTC30B with each of the tested IFT proteins, except IFT74. These findings are consistent with previous studies providing evidence that Chlamydomonas DYF-1 can bind directly to IFT46 [14] and IFT52 [15]. Based on the present immunoprecipitation studies, it is possible that TTC30B may also directly interact with IFT27, IFT57, IFT80, IFT81 and IFT88. However, as indicated above, it remains possible that some of the interactions may be mediated by conserved ciliary proteins or other proteins in wheat germ extracts. While it is surprising to find that TTC30B appears to directly interact with multiple proteins in the IFT B complex, analysis of the domain composition of IFT proteins has indicated that many IFT proteins contain multiple protein-protein interaction domains [21]. In view of this, it is conceivable that a single component of the IFT complex could interact with multiple proteins. Indeed, biochemical studies have provided evidence for multiple, direct interactions between several IFT proteins [15, 22, 23]. Further studies using purified proteins as well as mutation of individual protein interaction domains will be required to further assess the interaction of TTC30B with individual proteins of the IFT B complex.
We also explored possible interaction of murine TTC30B with the kinesin, KIF17, a kinesin that plays a role in IFT. In Caenorhabditis elegans, two different kinesin motors contribute to anterograde intraflagellar transport in sensory cilia [24]. Mutation of one of these kinesins, OSM-3, has the same phenotype as mutation of DYF-1 [12]. Although DYF-1 does not appear to be required for the localization of OSM-3 to sensory cilia, it seems to be required for the function of OSM-3. DYF-1 may function as an adapter protein to link to cargo to be delivered to the distal regions of cilia. The present and previous findings [17]provide evidence that mouse TTC30 isoforms have properties similar to DYF-1. As KIF17, the vertebrate homologue of OSM-3, has been shown to play a role in intraflagellar transport [25, 26], we sought to test for a possible formation of a complex containing both KIF17 and TTC30B. In mice, alternative splicing generates several different isoforms of KIF17. Mouse KIF17 isoform 1 contains 1038 amino acids while a shorter isoform contains 511 amino acids. The first 462 amino acids of these two murine KIF17 isoforms are identical and contain the complete motor domain of the protein. Based on studies of KIF17 truncations [27], the short form of mouse KIF17 is predicted to form dimers and to bind microtubules. In preliminary studies the short form of KIF17 was well expressed, but difficulty was encountered in obtaining adequate expression of the long form of KIF17. Therefore we focused attention on characterizing the interaction of IFT proteins with the short form of KIF-17. HEK 293 cells were transfected with expression vectors for AU1-TTC30B, HA-KIF17, and either FLAG-IFT52 or FLAG-IFT57. Use of monoclonal anti-FLAG antibody to isolate FLAG-IFT52 or FLAG-IFT57 led to co-immunoprecipitation of AU1-TTC30B (Fig. 4B) and HA-KIF7 (Fig 4,D). The co-expression of TTC30B with KIF17 does not significantly reduce the amount of either protein co-immunoprecipitating with IFT52 or IFT57 (Fig. 4B & 4D). The observation that TTC30B does not compete with KIF17 for formation of a complex with IFT52 or IFT57 is consistent with the possible presence of all of the proteins in the same complex, likely the IFT B complex.
Figure 4. Interaction of TTC30B and KIF17 with IFT52 and IFT57 in cells.
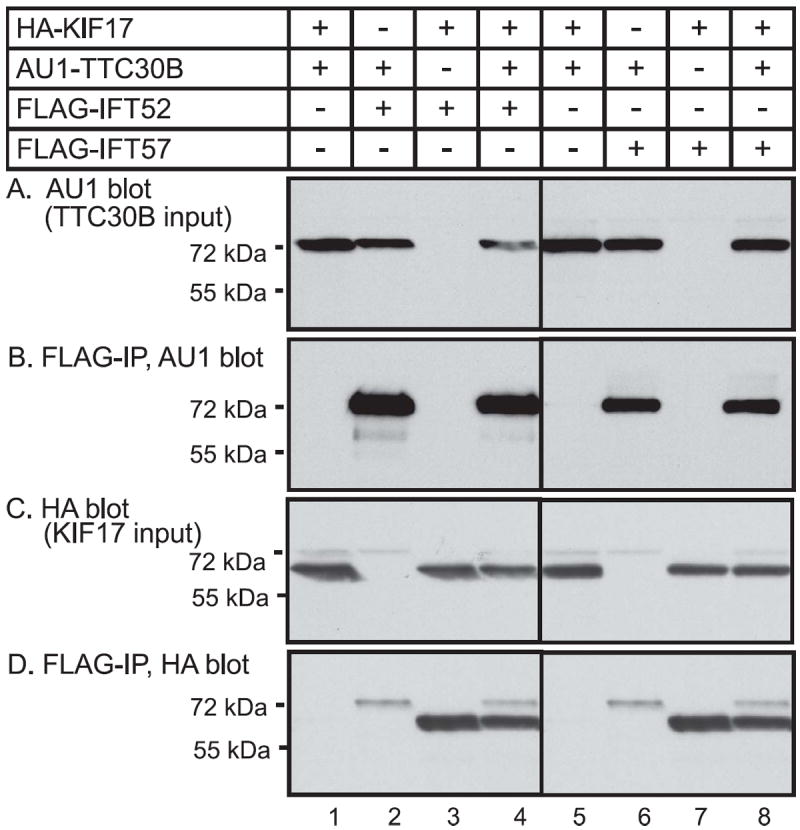
(A) HEK293 cells were transfected with expression vectors for AU1-tagged TTC30B or HA-tagged KIF17 and FLAG tagged-IFT52 or FLAG-tagged IFT57 as indicated. At 20 hours after transfection, whole cell extracts were prepared. (A) Samples were analyzed by denaturing gel electrophoresis and immunoblotting with antibody to AU1 to detect the input expression of TTC30B. (B) To detect interaction of TTC30B with IFT52 or IFT57, FLAG-tagged proteins were isolated by immunoprecipitation with FLAG antibody and the immunoprecipitate was analyzed by gel electrophoresis and immunoblotting with AU1 antibody to detect co-immunoprecipitated AU1-TTC30B. (C) Input expression of HA-KIF17 was detected by gel electrophoresis and immunoblotting with HA antibody. (D) Interaction of KIF17 with the IFT complex was analyzed by immunoprecipitation with FLAG antibody and the immunoprecipitate was analyzed by gel electrophoresis and immunoblotting with HA antibody to detect co-immunoprecipitated HA-KIF17.
Conclusions
In summary, we used immunopurification to isolate the IFT B complex from mouse pituitary cells. Using mass spectrometry, we identified peptides corresponding to the tetratricopeptide repeat-containing proteins, TTCA30A1, TTC30A2 and TTC30B. A previous study found that TTC30A1 can interact with a known IFT protein [17]. The present findings provide evidence that TTC30A2 and TTC30B can also be incorporated into a complex with known IFT B complex proteins. Immunoprecipitation of proteins produced in a cell-free system also showed that TTC30B appears to be able to interact with IFT27, IFT46, IFT52, IFT57, IFT80, IFT81 and IFT88. Interestingly, the present studies also demonstrate that mouse TTC30B can be incorporated in a complex with the kinesin motor, KIF17, consistent with a possible functional interaction in cargo transport as has been suggested by studies in C. elegans [24]. Thus the studies contribute to further understanding of the protein complexes that support the essential process of intraflagellar transport.
Figure 3. Interaction of TTC30B with multiple IFT proteins in a cell-free, in vitro system.
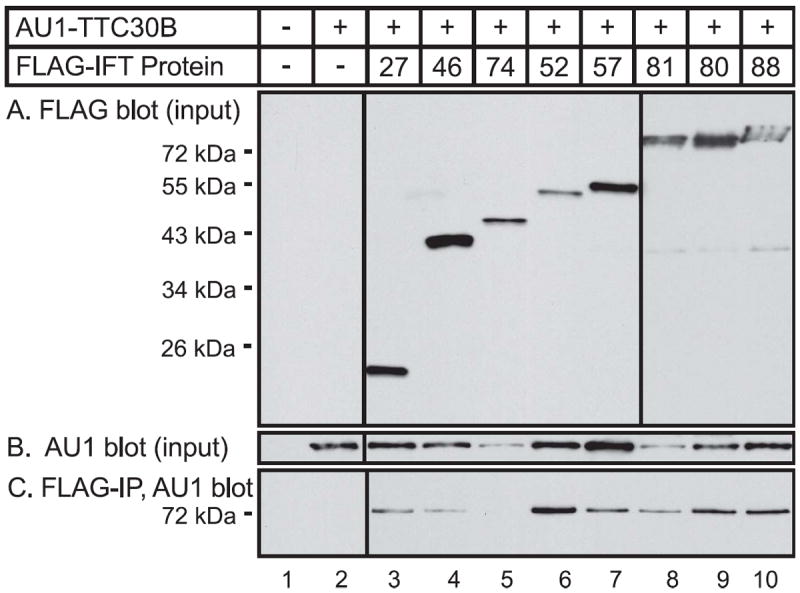
Coding sequences for tagged proteins were prepared by PCR and fused to a promoter for bacteriophage SP6 polymerase. The promoter-tagged protein DNA was used to direct protein synthesis in a cell-free coupled transcription-translation system from wheat germ. The in vitro reactions contained either coding sequences for AU1-tagged TTC30B or FLAG-tagged IFT protein as indicated. (A) To analyze protein expression, aliquots of the in vitro reactions were resolved by denaturing gel electrophoresis and immunoblotting with antibody to FLAG to detect the input expression of IFT proteins or with AU1 antibody to detect input expression of TTC30B. non-specific band was detected in all lanes and serves as a loading control. (B) To detect interaction of TTC30B with IFT proteins, FLAG-tagged IFT proteins were isolated by immunoprecipitation with FLAG antibody and the immunoprecipitate was analyzed by gel electrophoresis and immunoblotting with AU1 antibody to detect co-immunoprecipitate, AU1-tagged-TTC30B.
Highlights.
An intraflagellar transport (IFT) complex was purified from mouse pituitary cells.
The complex contained tetratricopeptide repeat protein 30 isoforms.
TTC30A1, TTC30A2 and TTC30B were found to enter into the IFT B complex.
In vitro, TTC30B was found to interact with several IFT proteins.
TTC30B was found to co-immunoprecipitate with the kinesin, KIF17.
Acknowledgments
This work was supported by National Institutes of Health Grant DK062779 (to RAM). We thank Bobbi Maurer for assistance in preparing the manuscript.
Abbreviations
- IFT
intraflagellar transport
- TEV
tobacco etch virus
- PCR
polymerase chain reaction
- MS
mass spectrometry
- TPR
tetratricopeptide repeat
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 2.Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 3.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 4.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 5.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci U S A. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman MA, Leroux MR. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 2009;19:306–316. doi: 10.1016/j.tcb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 11.Stofko-Hahn RE, Carr DW, Scott JD. A single step purification for recombinant proteins. Characterization of a microtubule associated protein (MAP 2) fragment which associates with the type II cAMP-dependent protien kinase. FEBS Lett. 1992;302:274–278. doi: 10.1016/0014-5793(92)80458-s. [DOI] [PubMed] [Google Scholar]
- 12.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 13.Ou G, Koga M, Blacque OE, Murayama T, Ohshima Y, Schafer JC, Li C, Yoder BK, Leroux MR, Scholey JM. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol Biol Cell. 2007;18:1554–1569. doi: 10.1091/mbc.E06-09-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan ZC, Behal RH, Geimer S, Wang Z, Williamson SM, Zhang H, Cole DG, Qin H. Chlamydomonas IFT70/CrDYF-1 is a core component of IFT particle complex B and is required for flagellar assembly. Mol Biol Cell. 2010;21:2696–2706. doi: 10.1091/mbc.E10-03-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taschner M, Bhogaraju S, Vetter M, Morawetz M, Lorentzen E. Biochemical mapping of interactions within the intraflagellar transport (IFT) B core complex: IFT52 binds directly to four other IFT-B subunits. J Biol Chem. 2011;286:26344–26352. doi: 10.1074/jbc.M111.254920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pathak N, Obara T, Mangos S, Liu Y, Drummond IA. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol Biol Cell. 2007;18:4353–4364. doi: 10.1091/mbc.E07-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Follit JA, Xu F, Keady BT, Pazour GJ. Characterization of mouse IFT complex B. Cell Motil Cytoskeleton. 2009;66:457–468. doi: 10.1002/cm.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClintock TS, Glasser CE, Bose SC, Bergman DA. Tissue expression patterns identify mouse cilia genes. Physiol Genomics. 2008;32:198–206. doi: 10.1152/physiolgenomics.00128.2007. [DOI] [PubMed] [Google Scholar]
- 19.Hodges ME, Wickstead B, Gull K, Langdale JA. Conservation of ciliary proteins in plants with no cilia. BMC Plant Biol. 2011;11:185. doi: 10.1186/1471-2229-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodges ME, Wickstead B, Gull K, Langdale JA. The evolution of land plant cilia. New Phytol. 2012;195:526–540. doi: 10.1111/j.1469-8137.2012.04197.x. [DOI] [PubMed] [Google Scholar]
- 21.Jekely G, Arendt D. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. Bioessays. 2006;28:191–198. doi: 10.1002/bies.20369. [DOI] [PubMed] [Google Scholar]
- 22.Lucker BF, Miller MS, Dziedzic SA, Blackmarr PT, Cole DG. Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46. J Biol Chem. 2010;285:21508–21518. doi: 10.1074/jbc.M110.106997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behal RH, Miller MS, Qin H, Lucker BF, Jones A, Cole DG. Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. J Biol Chem. 2012;287:11689–11703. doi: 10.1074/jbc.M111.287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, Brust-Mascher I, Scholey JM. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 25.Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Insinna C, Humby M, Sedmak T, Wolfrum U, Besharse JC. Different roles for KIF17 and kinesin II in photoreceptor development and maintenance. Dev Dyn. 2009;238:2211–2222. doi: 10.1002/dvdy.21956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]


