Abstract
Excess nutrient uptake leads to obesity, insulin resistance, and type 2 diabetes. Mammalian target of the rapamycin (mTOR), a major component of the nutrient-sensing pathway also regulates mitochondrial oxidative function. Rapamycin, a pharmacological inhibitor of mTOR, causes glucose intolerance and inhibits mitochondrial oxidative function. While a number of studies have focused on the effect of rapamycin on control wild-type mice, ours is the first to study the effect of rapamycin on mitochondrial gene expression and insulin sensitivity in the db/db mouse, a model of diabetic dyslipidemia. Female db/+ and db/db mice were fed ad libitum a rapamycin-containing diet or a control diet for 6 months, starting at two months of age. Body weight, fat mass, lean mass and food intake were measured monthly. Effect of rapamycin or control diet on markers of adipogenesis, fatty acid oxidation and mitochondrial biogenesis in the gonadal white adipose tissue (WAT) as well as different serum parameters were assessed. Whole body insulin sensitivity was measured by insulin tolerance test. Rapamycin feeding to db/db mice decreased body weight (58%) and fat mass (33%), elevated markers of fatty acid oxidation and mitochondrial biogenesis in WAT, reduced circulating non-esterified free fatty acids (NEFA), elevated circulating adiponectin and improved insulin sensitivity, compared to control diet fed db/db mice. These data demonstrate that rapamycin exhibits an anti-obesity effect and improves whole body insulin sensitivity in db/db mice and suggest an unexpected effect of simultaneous inhibition mTOR and leptin signaling in mice.
Keywords: mitochondria, insulin sensitivity, fat oxidation, db/db mouse, obesity
1. Introduction
Mammalian target of rapamycin (mTOR) is a serine/threonine kinase that regulates cell growth, proliferation and survival in response to energy, nutrient levels and redox status [1]. mTOR functions as the catalytic subunit of two distinct multi-protein functional complexes, mTORC1 and mTORC2. Chronic activation of mTOR by growth factors and amino acids leads to excess fat accumulation in peripheral tissues as well as causes insulin resistance [2-6]. Thus, it is not surprising that dysregulation of the mTOR pathway has been associated with a number of pathological conditions, such as obesity, diabetes and cancer [7].
Rapamycin, an immunosuppresant drug that prevents organ transplant rejection, is a potent inhibitor of mTOR pathway [8, 9]. Rapamycin has been demonstrated to cause both beneficial and adverse effect on metabolism. Rapamycin inhibits differentiation of primary human adipocytes and prevent nutrient-mediated insulin resistance in skeletal muscle and adipocytes, in vitro. Similarly, rapamycin, in vivo, protects against high fat diet induced obesity in C57BL/6J mice and ameliorate age-dependent obesity in mice [5, 10-12]. In contrast, there is also evidence suggesting that rapamycin may have detrimental effects on insulin metabolism. For example, rapamycin feeding causes glucose intolerance and hyperlipidemia in rodents as well as human transplant patients [13-16].
In addition to its role in metabolism, mTOR also plays a role in the maintenance of energy balance, since mTOR is essential for maintaining mitochondrial oxidative function. In mammalian cells, mTOR controls mitochondrial transcriptional regulator peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α to maintain energy levels and inhibition of mTOR activity by rapamycin has been shown to decrease mitochondrial gene expression and oxygen consumption, both in vitro and in vivo [17, 18].
Many studies have demonstrated the effect of rapamycin on metabolism in control wild-type rodents as well as in db/db or ob/ob mouse models. Similarly, effect of rapamycin on mitochondrial gene transcription and oxidative function is known in cultured cells as well as in mouse skeletal muscle. In contrast, no study had addressed the effect of rapamycin on mitochondrial gene expression and insulin sensitivity in db/db mice that are deficient in leptin receptor and is a model of diabetic dyslipidemia. We found that administration of rapamycin to female db/db mice for six months, starting at 2 months of age, resulted in a reduction in body weight, decreased fat mass, up-regulated markers of mitochondrial biogenesis and fatty acid oxidation in WAT and improved whole body insulin sensitivity, compared to db/db mice fed the control diet. These data suggests that rapamycin may exhibit beneficial effects under conditions of insulin resistance and diabetes caused by deficiency in leptin receptor.
2. Materials and methods
2.1. Animals and diet
Female mice homozygous (db/db) or heterozygous (db/+) for a point mutation in the leptin receptor in the C57BL/KsJ background were used for all experiments (Jackson Laboratory, ME) [19]. Mice were housed in specific pathogen free facilities and given free access to water. Mice were fed ad libitum a rapamycin-containing diet or a control diet containing the microencapsulating agent Eudragit S100 for 6 months, starting at two months of age [20]. Microencapsulated rapamycin was incorporated into Purina 5LG6 chow at 14ppm (Southwest Research Institute, San Antonio, TX). Food consumption was monitored throughout the experiment. All animal testing was approved by the University of Texas Health Science Center at San Antonio and the Audie L. Murphy VA Hospital Institutional Animal Care and Use Committees.
2.2. Quantitative magnetic resonance (QMR) imaging
Fat and lean mass were quantified using quantitative magnetic resonance imaging [EchoMRI (Echo Medical Systems, Houston, TX)] with live mice.
2.3. Antibodies and ELISA kits
Antibodies to phosphorylated ribosomal protein S6 (RPS6, Ser240/244), ribosomal protein S6, peroxisome proliferator-activated receptor gamma (PPARγ) and actin were purchased from Cell signaling technology (Danvers, MA). The antibody for PGC-1α was from Abcam (Cambridge, MA, USA).
2.4. Measurement of serum parameters
Mice were euthanized by CO2 inhalation and blood was collected by cardiac puncture. Blood was kept at room temperature for 30 minutes and serum was separated by centrifuging at 3000g for 10 minutes. Serum parameters were measured using the following kits as per manufacturer’s instructions: adiponectin and insulin using ELISA kits (Millipore, Billerica, MA, USA), triglyceride and non-esterified fatty acid (NEFA) using kits from Cayman Chemical (Ann Arbor, MI, USA) and Wako USA (Richmond, VA, USA), respectively.
2.5. Western blotting
Gonadal WAT from female mice was collected at the time of sacrifice and was immediately frozen in liquid nitrogen and stored at −80°C until use. Homogenisation of WAT and western blotting was performed as described before [21]. The bands were quantified using Image J software (NIH Image).
2.6. Quantitative real-time PCR
Total RNA was extracted from 50 mg of frozen gonadal WAT using RNeasy kit (QIAGEN, Valencia, CA). First strand cDNA synthesis was performed with SuperScript II reverse transcriptase and random hexamer primers as per manufacturer’s instructions (Life Technologies, Grand Island, NY). Quantitative real-time PCR was performed with an ABI Prism using Power SYBR Green PCR Master Mix with the primer listed in Table 1 (Applied Biosystems, Foster City, CA). Calculations were performed by a comparative method (2−ΔΔCT) using microglobin as control.
Table 1.
Primer sequences for quantitative RT-PCR
| Gene | Primer |
|---|---|
| PPARG2 | Forward 5′-CTCCTGTTGACCCAGAGCAT-3′ Reverse 5′-AATGCGAGTGGTCTTCCATC-3′ |
| SREBP1 | Forward 5′-GTAGGTCACCGTTTCTTTGTGGAC-3′ Reverse 5′-TGGGCTGAGCAATACAGTTCAAC-3′ |
| CD36 | Forward 5′-TGGAGCTGTTATTGGTGCAG-3′ Reverse 5′-TGGGTTTTGCACATCAAAGA-3′ |
| FATP1 | Forward 5′-CGCTTTCTGCGTATCGTCTGCAAG-3′ Reverse 5′-AAGATGCACGGGATCGTGTCT-3′ |
| LCAD | Forward 5′-CTTGCTTGGCATCAACATCGCAGA-3′ Reverse 5′-ATTGGAGTACGCTTGCTCTTCCCA-3′ |
| MCAD | Forward 5′-CTAACCCAGATCCTAAAGTACCCG-3′ Reverse 5′-GGTGTCGGCTTCCAAATGA-3′ |
| PGC1A | Forward 5′-CGGAAATCATATCCAACCAG-3′ Reverse 5′-TGAGGACCGCTAGCAAGTTTG-3′ |
| CRTC3 | Forward 5′-TGACTCACCTGGGGATAAGAAC-3′ Reverse 5′-GTGGCACTTGAGGGACGAG-3′ |
| CPT1-M | Forward 5′-AAGGGTAGAGTGGGCAGAGG-3′ Reverse 5′-GCAGGAGATAAGGGTGAAAGA-3′ |
| NRF1 | Forward 5′-CCACATTACAGGGCGGTGAA-3′ Reverse 5′-AGTGGCTCCCTGTTGCATCT-3′ |
| ERRalpha | Forward 5′-TTCGGCGACTGCAAGCTC-3′ Reverse 5′-CACAGCCTCAGCATCTTCAATG-3′ |
| β-ACTIN | Forward 5′-AATCGTGCGTGACATCAAAGAG-3′ Reverse 5′-GCCATCTCCTGCTCGAAGTC-3′ |
| β2-micro- globulin |
Forward 5′-CACTGACCGGCCTGTATGC-3′ Reverse 5′-GGGTGGCGTGAGTATACTTGAAT-3′ |
2.7. Insulin tolerance test
Mice were fasted for 6 hours before an intraperitoneal administration of 2 units/kg of body weight of human recombinant insulin in saline (Humalin; Eli Lilly, Indianapolis, IN, USA) to db/+ mice or db/db mice. A One-Touch Ultra glucometer was used to monitor blood glucose levels before and after the injection of insulin at the indicated time points.
2.8. Data analysis
All the data were analyzed by one-way ANOVA. Differences were regarded as significant at the P <0.05 level. All data are expressed as mean± standard error of the mean.
3. Results
3.1. Rapamycin decreases fat accumulation in female db/db mice
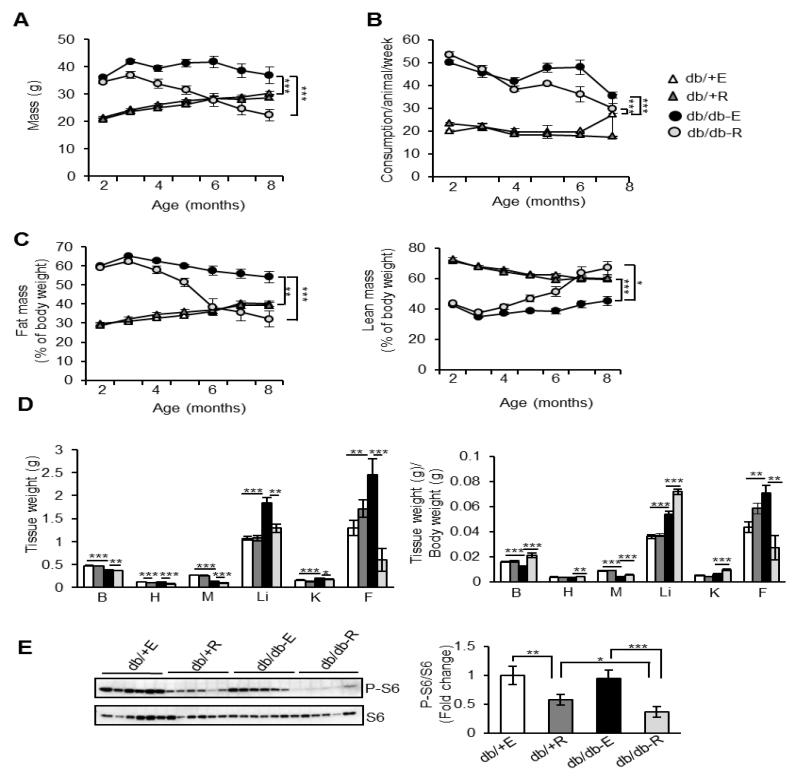
Mice were divided into four groups (n=9/group) for the study: (1) non-diabetic control mice fed eudragit diet (db/+-E), (2) non-diabetic control mice fed rapamycin diet (db/+R), (3) db/db mice fed eudragit diet (db/db-E) and (4) db/db mice fed rapamycin diet (db/db-R). Total body mass, food consumption and body composition were monitored monthly for six months starting at two months of age. Hyperphagia in the db/db model resulted in a 60% increase in body mass relative to non-diabetic control mice by two months of age and increased food consumption was observed in db/db mice relative to control mice throughout the study, regardless of treatment (Figures 1A and 1B). Rapamycin treatment had no effect on mass, body composition or food consumption in db/+ mice, and eudragit-fed db/db mice did not have a loss in total mass or a change in body composition over the course of the treatment paradigm. In stark contrast, db/db mice treated with rapamycin lost 58% of their body weight over the six months feeding period although food consumption did not differ statistically from control-fed db/db mice for the duration of the experiment (Figures 1A and 1B). Quantitative magnetic resonance imaging revealed that the loss of body mass in the rapamycin-treated db/db mice was due to a 32.7% reduction in total fat mass with a corresponding increase in percent lean mass (Figure 1C). By the end of the six month period, body composition of the db/db mice treated with rapamycin was similar to wild type animals. Tissue weights of rapamycin-fed db/+ mice were similar to eudragit-fed db/+ mice, except for heart. Tissue weights of rapamycin-fed db/db mice were significantly decreased compared to eudragit-fed db/db mice with the strongest decline of 71% in gonadal WAT (Figure 1D). Rapamycin inhibited the phosphorylation of RPS6 by 42% in WAT of db/+ mice in comparison to eudragit-fed db/+ mice, whereas in db/db mice rapamycin inhibited phosphorylation of RSP6 by 61% compared to eudragit-fed db/db mice (Figure 1E).
Fig. 1.
Rapamycin administration decreases fat accumulation in db/db mice. A) Body weights of non-diabetic control mice fed eudragit diet (db/+ E), non-diabetic control mice fed rapamycin diet (db/+ R), db/db mice fed eudragit diet (db/db-E) and db/db mice fed rapamycin diet (db/db-R) (n=9/group). B) Food consumption of db/+ E, db/+ R, db/db-E and db/db-R mice (n=9/group). C) Percentage lean mass and fat mass of db/+ E, db/+ R, db/db-E and db/db-R mice by QMR imaging. D) Tissue weights and weights of tissue normalized to body weight of db/+ E, db/+ R, db/db-E and db/db-R mice. B-brain, H-heart, M-skeletal muscle (gastroc), Li-liver, K-kidneys, F-gonadal fat. E) Left panel: Immunoblots of WAT extract from db/+ and db/db mice fed with eudragit or rapamycin with phospho-S6 (Ser 240/244; top panel) and total S6 protein (bottom panel). Right panel: quantification of phospho-protein to total protein. ***P < 0.001, **P < 0.01, *P < 0.05.
3.2. Markers of adipogenesis or fatty acid transport are unaffected in rapamycin-fed db/db mice
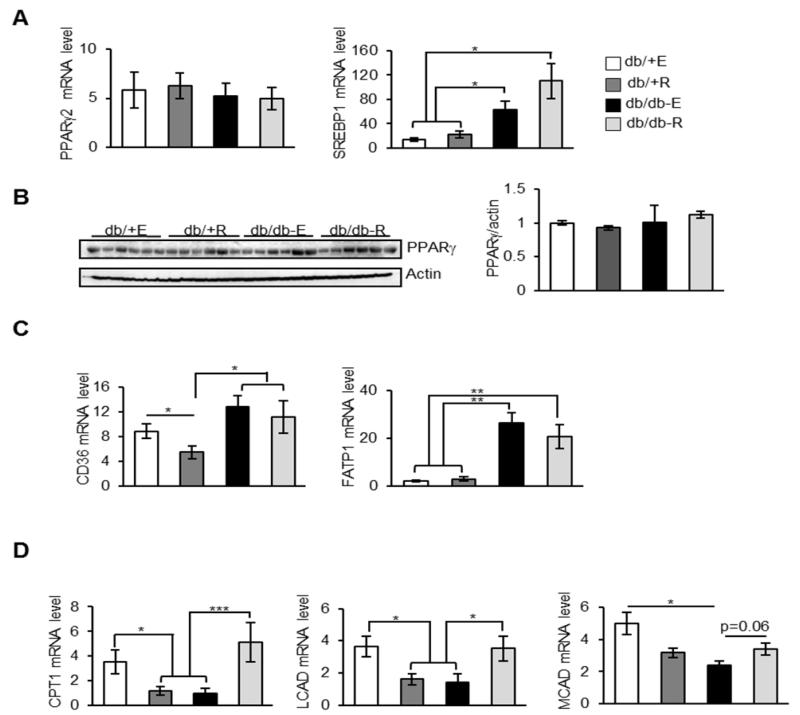
Even though rapamycin could affect the whole body physiology, since gonadal WAT tissue exhibited the strongest reduction in terms of tissue weight, we focused our studies on gonadal WAT. We measured markers of adipogenesis and fatty acid transporters in gonadal WAT to probe the underlying mechanisms for decreased fat accumulation in rapamycin-fed db/db mice. mTORC1 regulates the activity or translation of PPARγ and sterol regulatory element-binding transcription factor 1 (SREBP1), which are two transcription factors that regulate adipogenesis and fat accumulation [22, 23]. Previous studies have shown that expression of PPARγ remain unaltered in the WAT of genetic or acquired models of obesity [24]. Consistent with this, mRNA or protein levels of PPARγ were similar in eudragit-fed db/+ and db/db mice and rapamycin feeding had no effect on PPARγ levels in db/+ and db/db mice (Figures 2A and 2B). On the contrary, mRNA level of SREBP1 was elevated ~1.9-fold in eudragit-fed db/db mice compared to eudragit-fed db/+ mice and is consistent with the elevated SREBP1 levels in the liver tissue of db/db mice [25]. Interestingly, rapamycin feeding did not alter the expression levels of SREBP1 in db/db mice (Figure 2A). We also found no changes in the mRNA or protein levels of PPARγ or the mRNA level of SREBP1 in the db/+ mice fed rapamycin (Figures 2A and 2B). Collectively these results suggest that adipogenesis is not altered by rapamycin in the control or db/db mice. Consistent with this observation, mRNA levels of fatty acid transporters cluster of differentiation 36 (CD36) and fatty acid transport protein 1 (FATP1) were elevated in eudragit-fed db/db mice and their expression levels were suppressed by rapamycin diet (Figure 2C).
Fig. 2.
Rapamycin feeding does not affect adipogenesis in the WAT of db/db mice. A) mRNA levels of PPARγ2 and SREBP1 in the WAT of db/+ E, db/+ R, db/db-E and db/db-R mice (n=8/group) measured by quantitative RT-PCR, normalized to macroglobulin. B) Left panel: Immunoblots of WAT extract from db/+ and db/db mice fed with eudragit or rapamycin with PPARγ (top panel) and loading control (bottom panel). Right panel: quantification of PPARγ (top panel) to actin. C) mRNA levels of CD36 and FATP1 in the WAT of db/+ E, db/+ R, db/db-E and db/db-R mice (n=8/group) measured by quantitative RT-PCR, normalized to macroglobulin. D) mRNA levels of CPT1, LCAD and MCAD in the WAT of db/+ E, db/+ R, db/db-E and db/db-R mice (n=8/group) measured by quantitative RT-PCR, normalized to macroglobulin.***P < 0.001, **P < 0.01, *P < 0.05.
3.3. Markers of mitochondrial fatty acid oxidation are up-regulated in WAT in rapamycin-fed db/db mice
A reduction in the transcript level of enzymes regulating mitochondrial fatty acid oxidation has been reported in the WAT of db/db mice, in comparison to db/+ mice [26]. To test whether alteration in fat oxidation is a casual factor for decreased fat accumulation in rapamycin-fed db/db mice, we measured the mRNA levels of enzymes involved in mitochondrial fatty acid transport and oxidation such as, carnitine acyltransferase I (CPT1), long-chain acyl-CoA dehydrogenase (LCAD) and medium-chain acyl-CoA dehydrogenase (MCAD). Surprisingly, rapamycin exhibited opposite effects on mitochondrial fatty acid oxidation in db/+ mice and db/db mice (Figure 2E). Levels of CPT1, LCAD and MCAD were decreased by ~3-, 2- and 1.5-fold, respectively, in rapamycin-fed db/+ mice compared to eudragit-fed db/+ mice. On the contrary, levels of CPT1, LCAD and MCAD were increased by ~5-, 2.5-, and 1.4-fold, respectively, in rapamycin-fed db/db mice compared to eudragit-fed db/db mice (Figure 2E). Collectively, this suggests that increased mitochondrial fatty acid oxidation could be a potential mechanism for decreased fat accumulation in rapamycin-fed db/db mice.
3.4. Rapamycin increases markers of mitochondrial biogenesis in WAT from db/db mice
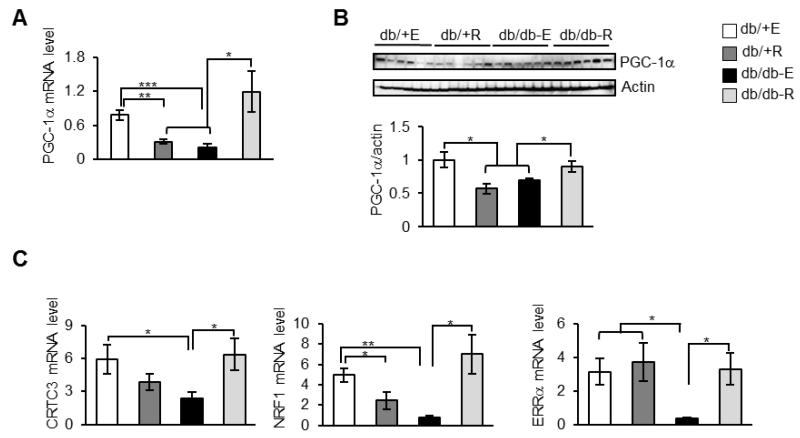
Studies have shown that WAT of db/db mice has attenuated markers of mitochondrial biogenesis compared to db/+ mice [26]. Because rapamycin diet up-regulated markers of fatty acid oxidation in db/db mice, we asked whether mitochondrial biogenesis markers were affected by rapamycin. To test this, we used PGC-1α as a marker for mitochondrial biogenesis, since expression of PGC-1α is suppressed in the WAT of db/db mice [26]. In agreement with the previous study, we also found that mRNA and protein level of PGC-1α were ~3.7- and 1.5-fold lower, respectively, in eudragit-fed db/db mice, in comparison to eudragit-fed db/+ mice (Figures 3A and 3B). While rapamycin-feeding attenuated mRNA and protein levels of PGC-1α by ~2.5- and 1.5-folds in db/+ mice, compared to eudragit-fed db/+ mice, an opposite effect was observed in db/db mice. mRNA and protein levels of PGC-1α were elevated by ~5.7- and 1.3-folds, respectively, in rapamycin-fed db/db mice compared to eudragit-fed db/db mice (Figures. 3A and 3B). Effects of rapamycin on the mRNA levels of CREB-regulated transcription coactivator 3 (CRTC3) that initiates mitochondrial biogenesis [27], nuclear respiratory factor 1 (NRF1) a direct target of PGC-1α that regulates nuclear genes required for mitochondrial respiration, and estrogen-related receptor alpha (ERRα) that regulates genes involved in mitochondrial biogenesis were also tested [28]. In eudragit-fed db/db mice, mRNA levels of CRTC3, NRF1 and ERRα were ~2.5-, 6.5- and 8.7-fold lower, respectively, when compared to eudragit-fed db/+ (Figure 3C). Rapamycin up-regulated mRNA levels of CRTC3, NRF1 and ERRα by ~2.6-, 9.2 and 9.1-fold, respectively, in db/db mice compared to eudragit-fed db/db mice (Figure 3C). While the mRNA level of NRF1 was down-regulated by ≈2-fold in rapamycin-fed db/+ compared with eudragit-fed non-diabetic controls, levels of CTRC3 and ERRα were not affected significantly (Figure 3C).
Fig. 3.
Rapamycin improves mitochondrial biogenesis in the WAT of db/db mice. A) mRNA levels of PGC-1α in the WAT of db/+ E, db/+ R, db/db-E and db/db-R mice (n=8/group) measured by quantitative RT-PCR, normalized to macroglobulin. B) Top panel: Immunoblots of WAT extract from db/+ E, db/+ R, db/db-E and db/db-R mice with PGC-1α (top panel) and β-actin (bottom panel). Bottom panel: quantification of PGC-1α to β-actin. C) mRNA levels of CRTC3, NRF1 and ERRα in the WAT of db/+ E, db/+ R, db/db-E and db/db-R mice (n=8/group) measured by quantitative RT-PCR, normalized to macroglobulin.***P < 0.001, **P < 0.01, *P < 0.05.
3.5. Circulating levels of NEFA are reduced while adiponectin levels are elevated by rapamycin feeding in db/db mice
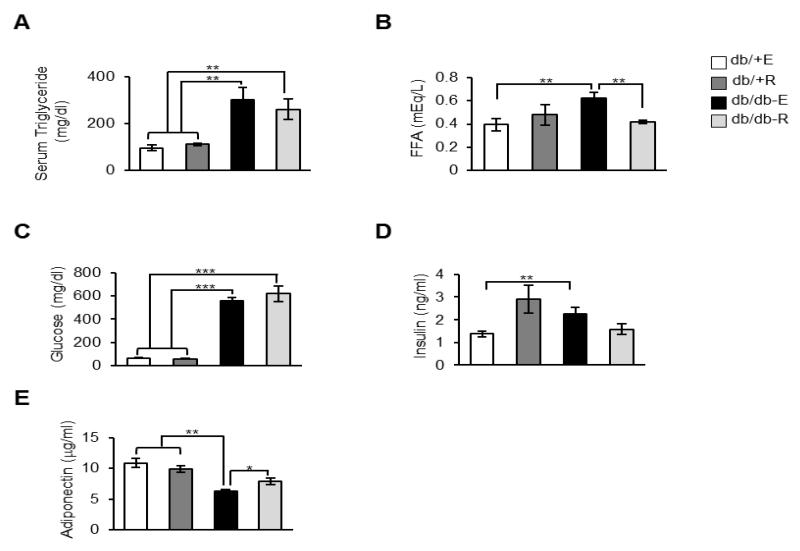
High levels of circulating triglycerides and free fatty acids were reported in db/db mice and are one of the characteristics of insulin resistance [29]. Consistent with this, we also found elevated levels of triglycerides (~3-fold), NEFA (~1.5-fold), glucose (~8.8-fold) and insulin (~1.6-fold) in eudragit-fed db/db mice, compared to eudragit-fed db/+ mice (Figures 4A-4D). While rapamycin feeding exerted no effect on the elevated triglyceride, glucose or insulin levels in db/db mice, rapamycin significantly reduced serum NEFA levels in db/db mice (~1.5-fold) when compared to eudragit-fed db/db mice Figures 4A-4D). Adiponectin, an adipokine secreted by the WAT, is an insulin sensitizer and low levels of circulating adiponectin are a causal factor in the development of insulin resistance [30]. Consistent with previous reports, we also found that eudragit-fed db/db mice have decreased levels of circulating adiponection as compared to eudragit-fed db/+ mice (~1.7-fold), and interestingly, rapamycin feeding up-regulated adiponectin levels in db/db mice (~2.6-fold) (Figure 4E).
Fig. 4.
Effect of rapamycin on serum parameters in db/+ and db/db mice. A) Circulating levels of triglycerides in db/+ E, db/+ R, db/db-E and db/db-R mice (n=9/group) under fed conditions. B) Free fatty acid levels in db/+ E, db/+ R, db/db-E and db/db-R mice (n=9/group) under fed conditions. C) Glucose levels in db/+ E, db/+ R, db/db-E and db/db-R mice (n=9/group) serum after 16 h without feeding. D) Insulin levels in db/+ E, db/+ R, db/db-E and db/db-R mice (n=9/group) serum under fed condition E) Serum adiponectin levels db/+ E, db/+ R, db/db-E and db/db-R mice (n=9/group) under fed condition. ***P < 0.001, **P < 0.01, *P < 0.05.
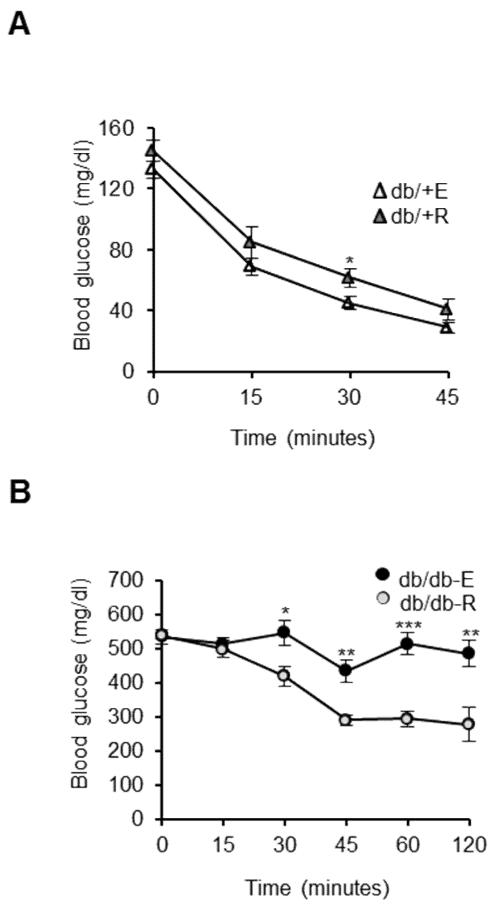
3.6. Rapamycin administration improves insulin sensitivity in db/db mice
Because we found that rapamycin could significantly decrease fat mass, lower circulating levels of NEFA and up-regulate circulating adiponectin levels, we hypothesized that rapamycin-fed db/db mice would exhibit improved insulin sensitivity compared to eudragit-fed db/db mice, despite high levels of circulating glucose. We measured insulin sensitivity using the insulin tolerance test. While the insulin tolerance test showed mild insulin intolerance in rapamycin-fed non-diabetic controls compared to eudragit-fed db/+ mice, rapamycin feeding significantly improved insulin sensitivity in db/db mice in comparison to eudragit-fed db/db mice (Figures 5A and 5B).
Fig. 5.
Rapamycin administration improves insulin sensitivity in db/db mice. A, B) Insulin tolerance test in db/+ and db/db mice fed with eudragit or rapamycin. The mice were subject to ITT after 5h without feeding. Mice were injected with insulin (1U/kg for db/+ mice and 2U/kg for db/db mice), and blood glucose was measured before and 15, 30, 45, 60, and 120 min after glucose injection (n=9/group). ***P < 0.001, **P < 0.01, *P < 0.05.
4. Discussion
In the db/db mouse model, the db gene carries a point mutation of the leptin receptor thereby disrupting leptin signaling, and loss of leptin signaling in the hypothalamus causes hyperphagia and obesity in db/db mice [31]. In addition, excess nutrient-mediated activation of mTOR pathway can lead to excess fat accumulation and play an important role in the development of insulin resistance and diabetes [32, 33]. In this study, we discovered an unexpected effect of simultaneous inhibition of leptin signaling and mTOR signaling on markers of mitochondrial biogenesis and fatty acid oxidation in mice. Our results suggest that long-term administration of the mTOR inhibitor rapamycin to female db/db mice diminished obesity in these mice, without affecting food intake, and improved whole body insulin sensitivity. Importantly, this phenotype was not observed for non-diabetic control mice fed with rapamycin. While the molecular mechanism(s) for these contrary effects of rapamycin in db/+ and db/db mice is not known, it is possible that the concurrent inhibition of leptin and mTOR signaling causes beneficial effects in rapamycin-fed db/db mice in terms of fat utilization and insulin signaling.
Reduced body weight and fat mass are important phenotypic markers of rapamycin-fed db/db mice in our study. These results are consistent with a previous study in db/db mice that were fed with rapamycin for 17 weeks [34]. In the current study, we found that in db/db mice, rapamycin feeding elevated markers of mitochondrial fatty acid oxidation in the WAT that could contribute to decreased fat mass in these mice. Previous studies have reported that under normal conditions, rapamycin could inhibit fat accumulation in rodents, both in vitro and in vivo, through multiple mechanisms such as inhibition of adipocyte differentiation, induction of lipolysis, downregulation of genes involved in lipid storage or decreasing food intake [10, 12, 13, 35, 36]. Surprisingly, in our study, rapamycin-feeding did not inhibit fat accumulation in db/+ mice. One possible explanation could be the genetic background of the mice used in our study.
PGC1-α and NRF1 play a role in mitochondrial biogenesis and reduced transcript levels of PGC-1α and markers of mitochondrial biogenesis were previously reported in the WAT of db/db mice as well as in type 2 diabetic skeletal muscle [26, 37, 38]. Interestingly, rapamycin treatment reduced transcript levels of PGC-1α and mitochondrial biogenesis markers in skeletal muscle cells, both in vitro and in vivo [18]. Consistent with this observation, we also observed a decline in the markers of mitochondrial biogenesis in the gonadal WAT of rapamycin-fed db/+ animals. Conversely, in rapamycin-fed db/db mice transcript levels of PGC1-α and NRF1 levels are elevated in the gonadal WAT. The mTOR pathway play a critical role in the maintenance of mitochondrial oxidative function, since inhibition of mTOR with rapamycin attenuated oxygen consumption and ATP generation under normal conditions and decreased mitochondrial gene expression through down-regulation of PGC1α in skeletal muscle cells [17, 18]. The transcription factor Ying Yang 1 (YY1) is reported to be involved in rapamycin-mediated inhibition of PGC-1α transcription and related mitochondrial genes in skeletal muscle cells [18]. Whether YY1 plays such a role in the gonadal WAT of rapamycin-fed non-diabetic mice and what is the role of YY1 in rapamycin-mediated elevation of PGC-1α in db/db mice needs to be investigated. Most importantly, how concurrent inhibition of leptin and mTOR signaling mediate mitochondrial biogenesis in the WAT of db/db mice is an open question.
Even though rapamycin-feeding to rodents and humans causes insulin resistance under normal conditions, rapamycin is an effective agent to prevent diabetic nephropathy in mice [34]. An important finding of our study is the improved insulin sensitivity of db/db mice and insulin resistance in non-diabetic control mice on rapamycin diet. It is interesting to note that even though insulin sensitivity was improved in rapamycin-fed db/db mice, circulating levels of glucose were unaffected by rapamycin in db/db mice. The reasons for these contradictory effects are not clear. One potential contributor to improved insulin sensitivity in rapamycin-fed db/db mice is an increase in circulating adiponectin. Elevated levels of the insulin sensitizer adiponectin have been positively correlated with improved insulin sensitivity. Circulating adiponectin is lower in db/db mice and elevation of adiponectin levels improves insulin sensitivity in db/db mice [30, 39]. Mechanisms by which rapamycin elevates levels of adiponectin in db/db mice and contributes to improved insulin sensitivity warrants further investigation.
Continued administration of rapamycin has been demonstrated to be protective against high fat diet-induced obesity in mice, a condition that induces leptin resistance [11, 40]. Similarly, our current findings in db/db mice suggest that rapamycin protects against obesity when leptin signaling is blocked. Altogether, it suggests that rapamycin can be an attractive weight loss strategy in a leptin resistant condition. It will be of interest to test whether long-term administration of rapamycin exerts a similar effect in leptin deficient ob/ob mice, since an acute injection of rapamycin to ob/ob two hours prior to insulin injection did not improve insulin sensitivity [41]. It is also possible that the effects of rapamycin in our study are due to long-term changes, as observed by altered gene expression. Additionally, the role of peripheral tissues such as skeletal muscle and liver in exerting these beneficial effects needs to be further investigated. Identification of signaling pathways that mediate beneficial effects in response to nutrition when leptin and mTOR signaling are inhibited will help to elucidate potential mechanisms and interventions to treat obesity and associated metabolic syndrome.
Acknowledgments
S.S.D. and H.V.R. contributed to the study design. S.S.D., M.E.W., R.T.H., D.P., Y.S., S.H. and Y.L. were responsible for data collection and analysis. S.S.D. and M.E.W. wrote the manuscript and H.V.R. reviewed/edited the manuscript. The authors would like to thank Vivian Diaz (Animal Core Supervisor, Nathan Shock Center for Excellence in the Biology of Aging) for excellent care and maintenance of the mice. This research was supported by the National Institutes of Health (NIH) RC2 Grand Opportunity grant (AG036613).
Footnotes
Conflict of interest: None declared.
References
- [1].Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- [2].Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;CB19:R1046–52. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Patti ME, Kahn BB. Nutrient sensor links obesity with diabetes risk. Nat Med. 2004;10:1049–1050. doi: 10.1038/nm1004-1049. [DOI] [PubMed] [Google Scholar]
- [5].Tremblay F, Marette A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem. 2001;276:38052–38060. doi: 10.1074/jbc.M106703200. [DOI] [PubMed] [Google Scholar]
- [6].Tremblay F, Krebs M, Dombrowski L, Brehm A, Bernroider E, Roth E, et al. Overactivation of S6 kinase 1 as a cause of human insulin resistance during increased amino acid availability. Diabetes. 2005;54:2674–2684. doi: 10.2337/diabetes.54.9.2674. [DOI] [PubMed] [Google Scholar]
- [7].Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: At the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- [8].Bodziak KA, Hricik DE. New-onset diabetes mellitus after solid organ transplantation. Transpl Int. 2009;22:519–530. doi: 10.1111/j.1432-2277.2008.00800.x. [DOI] [PubMed] [Google Scholar]
- [9].Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- [10].Bell A, Grunder L, Sorisky A. Rapamycin inhibits human adipocyte differentiation in primary culture. Obes Res. 2000;8:249–254. doi: 10.1038/oby.2000.29. [DOI] [PubMed] [Google Scholar]
- [11].Chang GR, Chiu YS, Wu YY, Chen WY, Liao JW, Chao TH, et al. Rapamycin protects against high fat diet-induced obesity in C57BL/6J mice. J Pharmacol Sci. 2009;109:496–503. doi: 10.1254/jphs.08215fp. [DOI] [PubMed] [Google Scholar]
- [12].Yang SB, Tien AC, Boddupalli G, Xu AW, Jan YN, Jan LY. Rapamycin ameliorates age-dependent obesity associated with increased mTOR signaling in hypothalamic POMC neurons. Neuron. 2012;75:425–436. doi: 10.1016/j.neuron.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Houde VP, Brule S, Festuccia WT, Blanchard PG, Bellmann K, Deshaies Y, et al. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes. 2010;59:1338–1348. doi: 10.2337/db09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Deblon N, Bourgoin L, Veyrat-Durebex C, Peyrou M, Vinciguerra M, Caillon A, et al. Chronic mTOR inhibition by rapamycin induces muscle insulin resistance despite weight loss in rats. Br J Pharmacol. 2010;165:2325–2340. doi: 10.1111/j.1476-5381.2011.01716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Morrisett JD, Abdel-Fattah G, Hoogeveen R, Mitchell E, Ballantyne CM, Pownall HJ, et al. Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J Lipid Res. 2002;43:1170–1180. [PubMed] [Google Scholar]
- [16].Teutonico A, Schena PF, Di Paolo S. Glucose metabolism in renal transplant recipients: Effect of calcineurin inhibitor withdrawal and conversion to sirolimus. J Am Soc Nephrol. 2005;16:3128–3135. doi: 10.1681/ASN.2005050487. [DOI] [PubMed] [Google Scholar]
- [17].Schieke SM, Phillips D, McCoy JP, Aponte AM, Shen RF, Balaban RS, et al. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- [18].Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- [19].Chen H, Charlat O, Tartagli LA, Woolf EA, Weng X, Ellis SJ, et al. Evidence that the diabetes gene encodes the leptin receptor: Identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- [20].Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deepa SS, Pulliam D, Hill S, Shi Y, Walsh ME, Salmon A, et al. Improved insulin sensitivity associated with reduced mitochondrial complex IV assembly and activity. FASEB J. 2013;27:1371–1380. doi: 10.1096/fj.12-221879. [DOI] [PubMed] [Google Scholar]
- [22].Kim JE, Chen J. Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53:2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- [23].Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- [24].Vidal-Puig A, Jimenez-Linan M, Lowell BB, Hamann A, Hu E, Spiegelman B, et al. Regulation of PPAR gamma gene expression by nutrition and obesity in rodents. J Clin Invest. 1996;97:2553–2561. doi: 10.1172/JCI118703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li S, Ogawa W, Emi A, Hayashi K, Senga Y, Nomura K, et al. Role of S6K1 in regulation of SREBP1c expression in the liver. Biochem Biophys Res Commun. 2011;412:197–202. doi: 10.1016/j.bbrc.2011.07.038. [DOI] [PubMed] [Google Scholar]
- [26].Rong JX, Qiu Y, Hansen MK, Zhu L, Zhang V, Xie M, et al. Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes. 2007;56:1751–1760. doi: 10.2337/db06-1135. [DOI] [PubMed] [Google Scholar]
- [27].Than TA, Lou H, Ji C, Win S, Kaplowitz N. Role of cAMP-responsive element-binding protein (CREB)-regulated transcription coactivator 3 (CRTC3) in the initiation of mitochondrial biogenesis and stress response in liver cells. J Biol Chem. 2011;286:22047–22054. doi: 10.1074/jbc.M111.240481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- [29].Tabatabai-Mir H, Sataranatarajan K, Lee HJ, Bokov AF, Fernandez E, Diaz V, et al. Rapamycin selectively alters serum chemistry in diabetic mice. Pathobiol Aging Age Relat Dis. 2012;2:10. doi: 10.3402/pba.v2i0.15896. 3402/pba.v2i0.15896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sharma K, McCue P, Dunn SR. Diabetic kidney disease in the db/db mouse. Am J Physiol Renal Physiol. 2003;284:F1138–1144. doi: 10.1152/ajprenal.00315.2002. [DOI] [PubMed] [Google Scholar]
- [32].Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Um SH, D’Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- [34].Mori H, Inoki K, Masutani K, Wakabayashi Y, Komai K, Nakagawa R, et al. The mTOR pathway is highly activated in diabetic nephropathy and rapamycin has a strong therapeutic potential. Biochem Biophys Res Commun. 2009;384:471–475. doi: 10.1016/j.bbrc.2009.04.136. [DOI] [PubMed] [Google Scholar]
- [35].Yeh WC, Bierer BE, McKnight SL. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc Natl Acad Sci U S A. 1995;92:11086–11090. doi: 10.1073/pnas.92.24.11086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Soliman GA, Acosta-Jaquez HA, Fingar DC. mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids. 2010;45:1089–1100. doi: 10.1007/s11745-010-3488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- [38].Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Awazawa M, Ueki K, Inabe K, Yamauchi T, Kubota N, Kaneko K, et al. Adiponectin enhances insulin sensitivity by increasing hepatic IRS-2 expression via a macrophage-derived IL-6-dependent pathway. Cell Metab. 2011;13:401–412. doi: 10.1016/j.cmet.2011.02.010. [DOI] [PubMed] [Google Scholar]
- [40].Chang GR, Chiu YS, Wu YY, Chen WY, Liao JW, Chao TH, et al. Rapamycin protects against high fat diet-induced obesity in C57BL/6J mice. J Pharmacol Sci. 2009;109:496–503. doi: 10.1254/jphs.08215fp. [DOI] [PubMed] [Google Scholar]
- [41].Miller AM, Brestoff JR, Phelps CB, Berk EZ, Reynolds TH. Rapamycin does not improve insulin sensitivity despite elevated mammalian target of rapamycin complex 1 activity in muscles of ob/ob mice. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1431–1438. doi: 10.1152/ajpregu.90428.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]