Abstract
Gloverins are basic, glycine-rich and heat-stable antibacterial proteins (~14-kDa) in lepidopteran insects with activity against Escherichia coli, Gram-positive bacteria, fungi and a virus. Hyalophora gloveri gloverin adopts a random coil structure in aqueous solution but has α-helical structure in membrane-like environment, and it may interact with the lipid A moiety of lipopolysaccharide (LPS). Manduca sexta gloverin binds to the O-specific antigen and outer core carbohydrate of LPS. In the silkworm Bombyx mori, there are four gloverins with slightly acidic to neutral isoelectric points. In this study, we investigate structural and binding properties and activities of B. mori gloverins (BmGlvs), as well as correlations between structure, binding property and activity. Recombinant BmGlv1–4 were expressed in bacteria and purified. Circular dichroism (CD) spectra showed that all four BmGlvs mainly adopted random coli structure (>50%) in aqueous solution in regardless of pH, but contained α-helical structure in the presence of 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP), smooth and rough mutants (Ra, Rc and Re) of LPS and lipid A. Plate ELISA assay showed that BmGlvs at pH 5.0 bound to rough mutants of LPS and lipid A but not to smooth LPS. Antibacterial activity assay showed that positively charged BmGlvs (at pH 5.0) were active against E. coli mutant strains containing rough LPS but inactive against E. coli with smooth LPS. Our results suggest that binding to rough LPS is the prerequisite for the activity of BmGlvs against E. coli.
Keywords: Gloverin, Lipopolysaccharide, Circular dichroism, Antibacterial, Random coil, α-helix, Bombyx mori
1. Introduction
Insects have the largest numbers and species on earth, and they combat a variety of pathogens mainly relying on sophisticated innate immune system. Insect defense system consists of three major parts: structural barriers, cellular and humoral immune responses (Lemaitre and Hoffmann, 2007). Structural barriers, the first protective lines, refer to cuticle, midgut epithelium and trachea. Cellular immune responses, including phagocytosis, nodulation and encapsulation, are mediated by several types of hemocytes (Lavine and Strand, 2002; Sideri et al., 2007). Humoral immune responses include melanization of hemolymph and secretion of antimicrobial peptides (AMPs) (Hoffmann, 1995). AMPs, the major and best known immune effectors induced by infection, are synthesized by fat body, hemocytes and other tissues, and regulated by the Toll and immune deficiency (IMD) pathways (Bulet et al., 1999; Lemaitre and Hoffmann, 2007).
At least 150 insect AMPs have been purified or identified. Most insect AMPs are small and cationic, and they show activities against bacteria and/or fungi (Hoffmann, 1995; Bulet and Stocklin, 2005). Based on the sequences, structures and activities, insect AMPs can be classified into four families, the α-helical peptides (e.g., cecropin, moricin and sarcotoxin), cysteine-rich peptides (e.g., insect defensin, drosomycin and heliomicin), proline-rich peptides (e.g., apidaecin, drosocin and lebocin), and glycine-rich peptides (e.g., attacin and gloverin) (Laszlo-Otvos, 2000; Bulet and Stocklin, 2005). Glycine-rich peptides with molecular masses of more than 10 kDa have become a large family of insect AMPs, including attacin, sarcotoxin II, gloverin, hymenoptaecin, coleoptericin, hemiptericin and tenecin 4 (Hultmark et al.,1983; Ando et al.,1987; Axen et al., 1997; Casteels et al., 1993; Bulet et al., 1991; Cociancich et al., 1994; Chae et al., 2012).
So far, small cationic AMPs (~4 kDa) have been the focus of structural and activity study. Cysteine-rich peptides, which are cyclic by formation of disulfide bonds, can form stable structures in aqueous solution. However, linear AMPs do not form stable structures in aqueous solution but can change to stable structures in membrane mimic environment (Nguyen et al., 2008; Haney and Vogel, 2009). Cysteine-rich AMPs, such as insect defensin A, drosomycin, termicin and heliomicin, contain “cysteine stabilized αβ motif” (CSαβ) structure with antiparallel β-sheet connected to a single α-helix by two disulfide bridges (Cornet et al., 1995; Landon et al., 1997; Silva et al., 2003; Lamberty et al., 2001). But nearly all the structures of α-helical linear AMPs are obtained by using micelle suspensions or in the presence of organic solvents. For example, Hyalophora cecropia cecropin A exists as random coli in aqueous solution but forms an amphipathic helical structure in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP)/water solution (Holak et al., 1988). The α-helical structure of moricin is also obtained in methanol and in solution containing 2,2,2-trifluoroethanol (TFE) or sodium dodecylsulphate (SDS) (Dai et al., 2008).
Since the first purification and characterization of gloverin from Hyalophora gloveri pupal hemolymph (Axen et al., 1997), a gloverin has been isolated from Helicoverpa armigera (Mackintosh et al., 1998), and two gloverins have been detected in hemolymph of septic injured Diatraea saccharialis larvae (Silva et al., 2010). Gloverin genes have also been identified in Antheraea mylitta (Gandhe-Archana et al., 2006), Galleria mellonella (Seitz et al., 2003; Brown et al., 2009), Manduca sexta (Abdel-Latief and Hilker, 2008; Xu et al., 2012; Zhu et al., 2003), Plutella xylostella (Eum et al., 2007; Etebari et al., 2011), Spodoptera exigua (Hwang and Kim, 2011), and Trichoplusia ni (Lundstrom et al., 2002). In the silkworm Bombyx mori, four gloverin genes (Bmglv1–4) have been identified, and Bmglv2–4 genes are derived from duplication of Bmglv1 (Cheng et al., 2006; Kaneko et al., 2007; Kawaoka et al., 2008; Mrinal and Nagaraju, 2008). Among the gloverins with known activities, H. gloveri gloverin is active against Escherichia coli D21f2 and D21 mutant strains with rough LPS (Axen et al., 1997), H. armigera gloverin is active against E. coli strains with smooth LPS and D22 strain that is defective in lipid A (Mackintosh et al., 1998), T. ni gloverin 1 and 2 have activity against E. coli D21f2 and D22 strains and a virus (Lundstrom et al., 2002; Moreno-Habel et al., 2012). However, S. exigua gloverin is active against a Gram-positive bacterium (Flavobacterium sp.) but inactive against E. coli strain with smooth LPS (Hwang and Kim, 2011), M. sexta gloverin shows activity against a Gram-positive Bacillus cereus and two fungi (Saccharomyces cerevisiae and Cryptococcus neoformans) but inactive against E. coli strain with smooth LPS (Xu et al., 2012).
The majority of gloverins is basic or highly basic (pI ~8.3 for H. gloveri gloverin, T. ni gloverin 1 and S. exigua gloverin, pI > 9.0 for most other gloverins) and heat-stable with high content (>18%) of glycine residues (Xu et al., 2012). H. gloveri gloverin (HgGlv, pI ~8.3) can inhibit the growth of E. coli by inhibiting synthesis of bacterial outer membrane proteins and increasing permeability of the membrane (Axen et al., 1997). Basic gloverins may interact with lipopolysaccharide (LPS) via charg–charge interaction with negatively charged lipid A (Axen et al., 1997). But direct binding of gloverin to microbial components including LPS has only been reported for M. sexta gloverin (MsGlv, pI ~9.3) (Xu et al., 2012). Recombinant MsGlv can bind to the O-specific antigen and outer core carbohydrate moieties of LPS, Gram-positive lipoteichoic acid (LTA) and peptidoglycan (PG), and laminarin, but does not bind to lipid A (Xu et al., 2012). Known gloverins with acidic or neutral pI include Heliothis virescens gloverin (pI ~7.2) (Genbank accession number: ACR78446), A. mylitta gloverin 2 (pI ~6.8) (Genbank accession number: ABG72700), and four B. mori gloverins (BmGlvs) (pI ~ 5.5, 7.0, 6.3 and 7.0 for BmGlv1–4, respectively) (Kawaoka et al., 2008). Recombinant BmGlvs show activity against E. coli strains with smooth LPS (Kawaoka et al., 2008; Mrinal and Nagaraju, 2008). However, it is not clear whether BmGlvs can interact with LPS and whether they also adopt random coil structures in aqueous solution and undergo conformational transitions in the hydrophobic environment. In this study, we investigate structural transitions of BmGlvs in the hydrophobic environment (organic solvent, detergent micelles and LPS), binding properties of BmGlvs to LPS and other microbial cell wall components, and antibacterial activities of BmGlvs against E. coli strains with smooth and rough mutant forms of LPS.
2. Materials and methods
2.1. Microorganisms and microbial components
E. coli DH5α (TIANGEN, China) and E. coli Rosetta™ (DE3) (Transgen, China) strains were used to clone and express recombinant BmGlvs. E. coli (ATCC 25922), Pichia pastoris, Serratia marcescens and Bacillus thuringiensis were from American Type Culture Collection (ATCC). Staphylococcus aureus and B. cereus were kindly provided by Professor Brian Geisbrecht, S. cerevisiae (BY4741) and C. neoformans (alpha) were provided by Professor Alexander Idnurm, Bacillus subtilis was provided by Professor Michael O’Connor, School of Biological Sciences at University of Missouri – Kansas City. E. coli D21, D21e7, D21f1 and D21f2 strains with rough mutants of LPS were purchased from E. coli Genetic Resources at Yale CGSC, The coli Genetic stock center (USA).
Smooth LPS from Salmonella enterica, S. marcescens, E. coli 055:B5, E. coli 026:B6 and E. coli 0111:B4, rough mutants of LPS from E. coli EH100 (Ra mutant), E. coli J5 (Rc mutant), E. coli F583 (Rd mutant) and S. enterica serotype minnesota Re 595 (Re mutant), as well as lipid A monophosphoryl from E. coli F583 (Rd mutant), laminarin, mannan, and zymosan were from Sigma–Aldrich (MO, USA) and used for binding assay. TLRgrade LPS and PG from E. coli K12 (LPS-K12 and PG-K12), TLRgrade peptidoglycan (PG) and lipoteichoic acid (LTA) from B. subtilis (LTA-BS and PG-BS) and S. aureus (LTA-SA and PG-SA) were from Invivogen (CA, USA) and also used for binding assay. LPS from E. coli serotype 055:B5 (smooth LPS), E. coli serotype EH100 (Ra), E. coli serotype J5 (Rc) and E. coli serotype R515 (Re), and monophosphoryl lipid A from E. coli serotype R515 were purchased from Alexis (Alexis, Switzerland) and used for circular dichroism (CD) experiments.
2.2. Construction of expression vectors for recombinant B. mori gloverins
Total RNAs were isolated from the fat body of day-3 fifth instar B. mori larvae collected at 24 h after injection of E. coli (ATCC 25922) (1 × 105 cells per larva) using TRIzol Reagent (Invitrogen), and the first strand cDNA was synthesized using M-MLV Reverse Transcriptase (TOYOBO, Japan). RT-PCR was performed to obtain cDNA sequences encoding B. mori mature gloverins using the following primer pairs: 5′-CAT GCC ATG GAT ATT CAC GAC TTT GTC AC-3′ and 5′-CGC CTC GAG CCA CTC GTG AGT AAT CTG-3′ (for mature BmGlv1, residues 44–178), 5′-CAT GCC ATG GAC GTC ACT TGG GAC AAA CAA-3′ and 5′-CAG CTC GAG CCA ATC ATG GCG GAT CTC TG-3′ (for mature BmGlv2, residues 43–173), 5′-CGA TCC ATG GAC GTC ACG TGG GAC ACG-3′ and 5′-CCG CTC GAG CCA CTC ATG CCG GAT CTC-3′ (for mature BmGlv3, residues 43–173), 5′-CAT GCC ATG GAC GTC ACC TGG GAC AAA CAA G-3′ and 5′-CCG CTC GAG CCA ATC ATG GCG GAA CTC T-3′ (for mature BmGlv4, residues 41–171). PCR products were purified using the EZNA cycle-pure kit (Omega, USA) and digested with Nco I and Xho I enzymes. After purification by agarose gel electrophoresis, these DNA fragments were ligated into the Nco I/Xho I-digested expression vector pET-21d (+) (Novagen), and the ligated products were then transformed into competent E. coli DH5 α cells. The recombinant expression vectors containing target genes from positive bacterial colonies were extracted and confirmed by restriction enzyme digestion and DNA sequencing.
2.3. Expression and purification of recombinant B. mori gloverins and production of polyclonal antibody against BmGlv2
To express soluble recombinant BmGlvs, recombinant plasmids were transformed into E. coli Rosetta™ (DE3) cells. Positive bacterial colonies were inoculated into LB medium containing ampicillin (100 µg/ml), chloramphenicol (34 µg/ml) and incubated overnight at 37 °C. The overnight cultures were diluted 1:100 into fresh LB medium containing ampicillin (100 µg/ml), chloramphenicol (34 µg/ml) and incubated at 37 °C to OD600 = 0.6–0.8, then isopropyl-D-thiogalactoside (IPTG) (1 mM final concentration) was added to induce protein expression. After incubation for another 4 h at 37 °C, bacterial cells were harvested by centrifugation at 8,000g for 10 min at 4 °C.
The bacterial pellets were re-suspended in 10 mM phosphate, 200 mM NaCl, pH 8.0 (10 ml/g) and sonicated for 3 min. After centrifugation at 16,000g for 30 min at 4 °C, the supernatant was collected and applied to Ni2+-NTA column (Amersham Bio-sciences). The column was washed sequentially with 10, 25 and 50 mM imidazole in 10 mM phosphate, 200 mM NaCl, pH 8.0. Recombinant proteins were eluted with 500 mM imidazole in 10 mM phosphate, 200 mM NaCl, pH 8.0, and then loaded to a Superdex 75 column (16/60 mm) pre-equilibrated with 10 mM phosphate, 200 mM NaCl, pH 8.0. Fractions containing recombinant BmGlvs were pooled, desalted, and concentrated to 1 mg/ml in 10 mM phosphate (pH 5.0 or 8.0) or 10 mM phosphate, 100 mM NaCl (pH 5.0 or 8.0). Protein concentrations were determined by absorbance at 280 nm with the theoretical molar extinction coefficients (http://www.expasy.org).
To produce polyclonal antibody against BmGlv2 in a rabbit, purified recombinant BmGlv2 (600 µg) was applied to a preparative SDS-PAGE, and the gel slice containing recombinant BmGlv2 was cut out and used as an antigen to inject a rabbit for polyclonal antibody production (Cocalico Biologicals, Inc., Reamstown, PA, USA).
2.4. SDS-PAGE and western blot analyses
Day 3 fifth instar B. mori Dazao larvae were injected with saline, E. coli (ATCC 25922) (1 × 105 cells/larva), S. aureus (ATCC 27217) (1 × 105 cells/larva), or yeast (Pichia pastoris) (1 × 105 cells/larva), or without treatment (naïve) and hemolymph was collected at 24 h post-injection. Hemolymph from at least four larvae of each group was combined, hemocytes were removed by centrifugation and cell-free hemolymph samples were used for Western blot analysis. Recombinant BmGlv1–4 purified from E. coli (1 µg each for SDS-PAGE analysis, 0.1 µg each for Western blot) and cell-free hemolymph samples (2 µl each) were separated on 15% SDS-PAGE and proteins were stained with Coomassie Brilliant Blue or transferred to nitrocellulose membranes (162–0097, Bio-Rad). The membrane was blocked with 5% BSA inTris-buffered saline (100 mM Tris–HCl, pH 7.6,150 mM NaCl) containing 0.1% Tween-20 (TBS-T) at room temperature for at least 3 h and then incubated overnight with rabbit anti-BmGlv2 antibody (1:2000) at 4 °C in TBS-T containing 5% BSA with gentle rocking. Then, the membrane was washed four times with TBS-T and incubated with goat anti-rabbit IgG conjugated to alkaline phosphatase (1:10,000) in TBS-T containing 5% BSA for 2 h at room temperature. After washing four times with TBS-T (10 min each time), the signal was developed by using alkaline phosphatase (AP) conjugate color development Kit (170– 6432, Bio-Rad).
2.5. CD spectroscopy
CD experiments were performed on a Jasco-810 spectropolarimeter (Jasco, Tokyo, Japan) at 25 °C using a quartz cell with a path length of 0.1 cm. Spectra were recorded over a wavelength of 190–260 nm. Each spectrum was obtained after subtracting the signal from protein-free solution. Proteins were dissolved to 0.15 mg/ml in 10 mM phosphate, pH 5.0 or 8.0, in the presence or absence of 40% HFIP, 10 mM SDS, 10 mM dodecylphosphocholine (DPC), smooth LPS, Ra-LPS, Rc-LPS, Re-LPS, or lipid A (w/w = 1/1), or in the presence of increasing concentrations of HFIP (10–40%), SDS (0.5–100 mM) or DPC (1–20 mM). Percentages of secondary structures were estimated using the Jasco protein secondary structure estimation program by the method of Yang et al. (1986).
2.6. Binding of B. mori gloverins to LPS
To test binding of BmGlvs to LPS and other microbial components, plate ELISA assays were performed. Briefly, wells of a flat bottom 96-well plate (Polysorp, Nunc) were coated with different forms of LPS, LTA, PG, laminarin, mannan, zymosan, or lipid A (2 µg/ well) as described previously (Yu and Kanost, 2000; Yu et al., 2005). The plates were placed overnight at room temperature until the water evaporated completely, heated to 60 °C for 30 min, and then blocked with 1 mg/ml BSA in Tris buffer (TB) (50 mM Tris–HCl, 50 mM NaCl, pH 8.0) for 2 h at 37 °C. Then, plates were rinsed four times with TB, purified BmGlv1, BmGlv2, BmGlv3, BmGlv4, or CP36 (a recombinant cuticle protein from M. sexta as a control protein) was diluted to 1 µg/ml in 10 mM phosphate, 100 mM NaCl, pH 5.0 or 8.0, containing 0.1 mg/ml BSA and added to the coated plates (50 µl/ well). Binding was allowed to occur for 3 h at room temperature, and the plates were rinsed four times with TB. Then monoclonal anti-polyhistidine antibody (Sigma–Aldrich, USA) (1:2000 in TB containing 0.1 mg/ml BSA) was added (100 µl/well) and incubated overnight at 4 °C. The plates were rinsed four times with TB, and alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma– Aldrich, USA) (1:3000 in TB containing 0.1 mg/ml BSA) was added (100 ml/well) and incubated for 2 h at 37 °C. The plates were rinsed, p-nitro-phenyl phosphate (1 mg/ml in 10 mM diethanolamine, 0.5 mM MgCl2) was added (50 µl/well), and absorbance at 405 nm of each well was determined every minute for 30 min period using a microtiter plate reader (Bio-Tek Instrument, Inc.). Specific binding of BmGlvs to each microbial component was obtained by subtracting the total binding of the control CP36 protein from the total binding of each BmGlv. These experiments were repeated at least three times. Figures were made with the GraphPad Prism software (GraphPad, CA, USA) with one representative set of data. Significance of difference was determined by an unpaired t-test or by one way ANOVA followed by a Tukey’s multiple comparison test using the same software (GraphPad, CA, USA).
2.7. Antimicrobial activity assays
Antimicrobial activity of purified BmGlvs was tested against two Gram-negative and four Gram-positive bacteria (E. coli DH5a, S. marcescens, S. aureus, B. subtilis, B. cereus, and B. thuringiensis), two fungal strains (S. cerevisiae (BY4741) and C. neoformans (alpha)), and four isogenic E. coli K-12 strains with different rough mutants of LPS: E. coli D21 (with Ra-LPS), E. coli D21e7 (with Rc-LPS), E. coli D21f1 (with Rd-LPS), and E. coli D21f2 (with Re-LPS). The activities were determined using bacterial clearance assays as described by Mrinal and Nagaraju (2008) with slight modifications. Briefly, overnight bacterial or fungal cultures were subcultured in LB or YPD medium (1% yeast extract, 2% peptone and 2% dextrose) until mid-log phase. The bacterial and fungal cultures were centrifuged at 1000 g for 10 min at 4 °C and washed once with 10 mM phosphate, 100 mM NaCl, pH 5.0 or 8.0. The bacterial and fungal cells were adjusted to OD600 = 0.4 and 0.2, respectively, in 10 mM phosphate, 100 mM NaCl, pH 5.0 or 8.0. Then the prepared cell cultures (85 µl each) were mixed with purified BmGlvs (15 µl of 1 mg/ml in 10 mM phosphate, 100 mM NaCl, pH 5.0 or 8.0) (final concentrations of BmGlvs were 150 µg/ml or ~ 10 µM) or buffer alone (10 mM phosphate, 100 mM NaCl, pH 5.0 or 8.0) as controls in 96-well plates. Bacterial cells were incubated at 37 °C with 220 rpm shaking and fungal cells were cultured at 30 °C with 220 rpm shaking. OD600 was measured every hour by Powerwave XS plate reader (BioTek, VT, US). The activity assay was also performed with increasing concentrations of BmGlvs (0.4, 2 and 10 µM) against E. coli D21 and D21f2 strains. In addition, antibacterial activity of BmGlvs against E. coli D21 and D21f2 strains was verified by bacterial viability assay. At 2 h after incubation with BmGlvs (10 µM each), aliquots ofE. coli D21 and E. coli D21f2 cells were serially diluted with 10 mM phosphate, 100 mM NaCl, pH 5.0 and plated on LB-agar plates containing streptomycin sulfate (50 µg/ml) and ampicilin (10 µg/ml). The plates were incubated at 37 °C overnight and the numbers of viable bacterial cells were recorded. Microbial growth curves were generated using the Graphpad Prism version 4.0 for Windows (GraphPad Software, CA, USA).
3. Results
3.1. Expression and purification of recombinant B. mori gloverins
Based on the amino acid sequences, all reported gloverins have three forms: prepro-, pro-, and mature gloverins. The theoretical molecular masses and isoelectric points (pI) of mature BmGlv1–4 are 14.4 kDa and 5.49, 14.1 kDa and 7.03, 14.1 kDa and 6.32, and 14.1 kDa and 6.94, respectively (Kawaoka et al., 2008). In order to obtain gloverins for structural and functional studies, mature BmGlv1 (residues 44–178), BmGlv2 (residues 43—173), BmGlv3 (residues 43—173), and BmGlv4 (residues 41—171) with a C-termi-nal six-histidine tag were expressed as soluble proteins in E. coli Rosetta™ (DE3) cells. The recombinant proteins were purified by Ni-NTA affinity column followed by a Superdex 75 column using phosphate buffer (pH 5.0 or 8.0) (Fig. S1A), and all four BmGlvs were recognized by rabbit polyclonal antibody against recombinant BmGlv2 (Fig. S1B). The calculated masses and pI of recombinant BmGlv1–4 are 15.6 kDa and 6.02,15.3 kDa and 6.75, 15.3 kDa and 6.48, and 15.3 kDa and 6.7, respectively. Western blot analysis of cell-free hemolymph samples from B. mori larvae using anti-BmGlv2 antibody showed that BmGlvs were induced by E. coli but not by injury (saline-injection), S. aureus or yeast (Fig. S1C). The estimated total concentration of BmGlvs in E. coli-induced hemolymph was about 0.1–0.12 mg/ml (7–9 µM) (Fig. S1C).
3.2. CD spectroscopy and secondary structural prediction of BmGlvs in aqueous solution
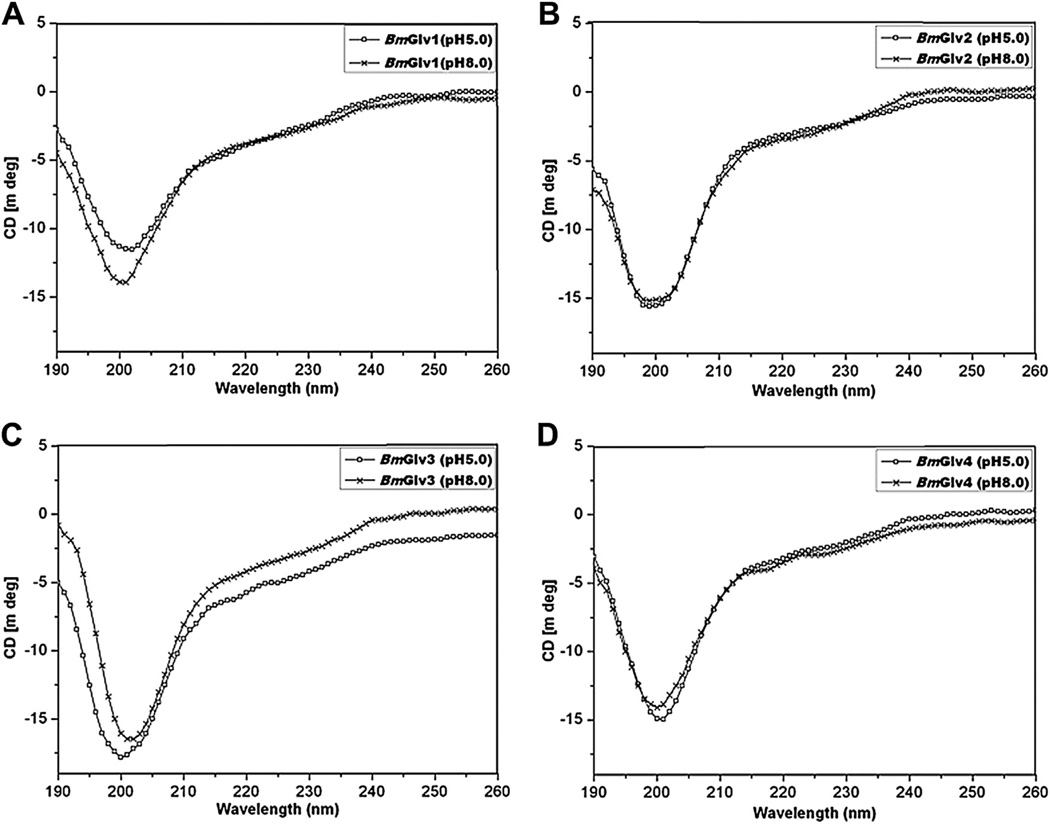
CD spectroscopy is an important tool to determine secondary structures of proteins in solutions. We performed CD experiments for BmGlv1–4 in phosphate buffer from pH 3–8, and all four BmGlvs adopted random coil conformation in aqueous solution in regardless of pH values (Fig. S2), indicating that pH alone cannot induce conformational transitions. Fig. 1 shows CD spectra of BmGlv1–4 in phosphate buffer at pH 5.0 and 8.0, in which BmGlv1–4 are positively and negatively charged, respectively, based on their theoretical pI. The following CD experiments, ELISA binding assays and antibacterial activity assays were all performed at pH 5.0 and 8.0. CD spectra of BmGlv1–4 at both pH 5.0 and 8.0 displayed negative ellipticity around 200 nm (Fig. 1), which is typical for random coil conformation (Tiffany and Krimm, 1972; Yang et al., 1986), and all four BmGlvs contained over 50% random coil as well as certain contents of β-strands (25–40%) and turns (9–19%), but did not contain α-helix conformation (Table 1).
Fig. 1.
CD spectra of BmGlvs in aqueous solutions at pH 5.0 and 8.0. Purified BmGlv1–4 were diluted to 0.15 mg/ml in 10 mM phosphate buffer (pH 5.0 or 8.0) and CD spectra were recorded on a Jasco-810 spectropolarimeter at 25 °C using a quartz cell with a path length of 0.1 cm. Each CD spectrum was obtained after subtracting the signal from protein-free solution.
Table 1.
Predicted secondary structures of BmGlv1–4 in phosphate buffer at pH 5.0 and 8.0
|
BmGlv1 |
BmGlv2 |
BmGlv3 |
BmGlv4 |
|||||
|---|---|---|---|---|---|---|---|---|
| pH 5.0 | pH 8.0 | pH 5.0 | pH 8.0 | pH 5.0 | pH 8.0 | pH 5.0 | pH 8.0 | |
| Helix (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Beta (%) | 32.6 | 35.4 | 33.0 | 39.7 | 27.7 | 25.4 | 30.1 | 25.0 |
| Turn (%) | 12.6 | 12.0 | 12.4 | 9.1 | 17.6 | 19.2 | 15.8 | 17.7 |
| Random (%) | 54.8 | 52.6 | 54.6 | 51.2 | 54.7 | 55.4 | 54.1 | 57.3 |
3.3. CD spectroscopy and secondary structural prediction of BmGlvs in membrane-like environment
Most linear antimicrobial peptides adopt random coil structures in aqueous solution, but change to more defined structures when encountering bacterial membrane. Organic solvents such as trifluoroethanol (TFE) and HFIP, and detergent micelles like SDS and DPC, are commonly used to mimic a membrane environment (Haney and Vogel, 2009). HgGlv undergoes conformational transition to α-helix in hydrophobic environment (HFIP solution) (Axen et al., 1997). To test whether BmGlvs also undergo conformational transitions in a membrane-like environment, CD experiments were performed for BmGlv1–4 in the presence of HFIP, SDS and DPC.
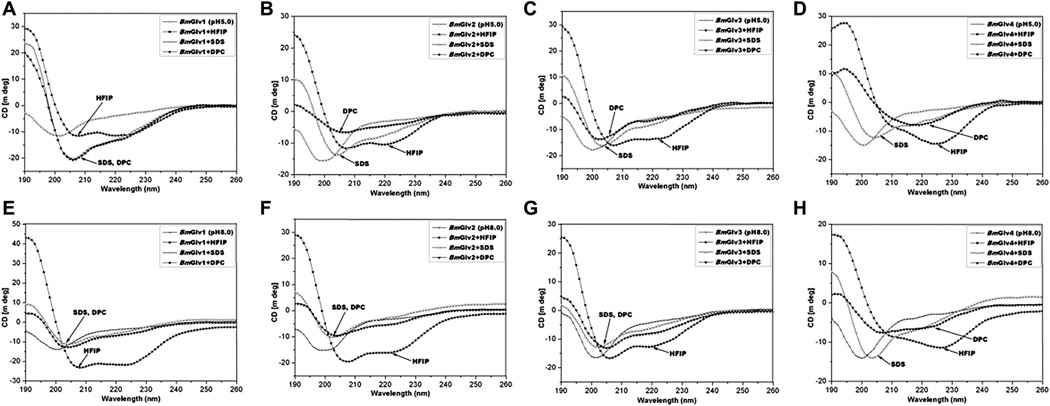
We first collected CD spectra of BmGlvs at pH 5.0 and 8.0 in the presence of increasing concentrations of HFIP (10–40%), SDS (0.5– 100 mM) or DPC (1–20 mM). HFIP at 10% already caused conformational transitions of BmGlvs, but DPC and SDS even at high concentrations did not cause conformational transitions of BmGlvs (Fig. S3) with the exception of BmGlv4, which underwent conformational transitions at pH 5.0 even with 1 mM DPC (Fig. S3F). Fig. 2 shows the CD spectra of BmGlvs in40% HFIP,10mMSDS and 10 mM DPC. CD spectra of BmGlv1–3 inphosphate buffer at both pH 5.0 and 8.0 containing 40% HFIP showed two minima at 208 and 222 nm (Fig. 2A–C, E–G) and CD spectra of BmGlv4 in the presence of 40% HFIP also showed a major negative peak around 222 nm (Fig. 2D and H), which are characteristics of α-helix (Holzwarth and Doty, 1965; Yang et al., 1986), indicating that BmGlv1–4 adopted α-helix conformation in the presence of HFIP in a pH-independent manner. The predicted secondary structures showed that BmGlv1–4 contained 57.6, 57.6, 32.7 and 59.5% α-helix at pH 5.0 (Table 2), and 44.9, 25.6, 26.5 and 46.6% α-helix at pH 8.0 (Table 3), respectively. These results suggest that more BmGlvs converted to α-helical structure at pH 5.0 when proteins were positively charged than at pH 8.0 when proteins were negatively charged in the presence of HFIP.
Fig. 2.
CD spectra of BmGlvs in the presence of HFIP, SDS and DPC at pH 5.0 and 8.0. Purified BmGlv1–4 were diluted to 0.15 mg/ml in 10 mM phosphate buffer (pH 5.0 or 8.0) in the presence or absence of 40%HFIP, 10 Mm SDS or 10 mM DPC and CD spectra were recorded on a Jasco-810 spectropolarimeter at 25 °C. Each CD spectrum was obtained after subtracting the signal from protein-free solution.
Table 2.
Predicted secondary structures of BmGlv1–4 in the presence of HFIP and Ra-LPS at pH 5.0
| BmGlv1 |
BmGlv2 |
BmGlv3 |
BmGlv4 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH 5.0 | +HFIP | +Ra-LPS | pH 5.0 | +HFIP | +Ra-LPS | pH 5.0 | +HFIP | +Ra-LPS | pH 5.0 | +HFIP | +Ra-LPS | |
| Helix (%) | 0.0 | 57.6 | 54.8 | 0.0 | 57.6 | 58.4 | 0.0 | 32.7 | 67.0 | 0.0 | 59.5 | 62.1 |
| Beta (%) | 32.6 | 0.0 | 0.0 | 33.0 | 0.0 | 0.0 | 27.7 | 25.9 | 0.0 | 30.1 | 0.0 | 0.0 |
| Turn (%) | 12.6 | 0.0 | 45.2 | 12.4 | 11.0 | 41.6 | 17.6 | 0.0 | 33.0 | 15.8 | 24.5 | 37.9 |
| Random (%) | 54.8 | 42.4 | 0.0 | 54.6 | 31.4 | 0.0 | 54.7 | 41.4 | 0.0 | 54.1 | 16.0 | 0.0 |
Table 3.
Predicted secondary structures of BmGlv1–4 in the presence of HFIP and Ra-LPS at pH 8.0
|
BmGlv1 |
BmGlv2 |
BmGlv3 |
BmGlv4 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH 8.0 | +HFIP | +Ra-LPS | pH 8.0 | +HFIP | +Ra-LPS | pH 8.0 | +HFIP | +Ra-LPS | pH 8.0 | +HFIP | +Ra-LPS | |
| Helix (%) | 0.0 | 44.9 | 14.0 | 0.0 | 25.6 | 63.0 | 0.0 | 26.5 | 65.4 | 0.0 | 46.6 | 63.4 |
| Beta (%) | 35.4 | 2.1 | 52.1 | 39.7 | 34.7 | 0.0 | 25.4 | 30.7 | 0.0 | 25.0 | 0.0 | 0.0 |
| Turn (%) | 12.0 | 10.9 | 0.7 | 9.1 | 0.9 | 37.0 | 19.2 | 0.0 | 34.6 | 17.7 | 19.5 | 36.6 |
| Random (%) | 52.6 | 42.0 | 33.2 | 51.2 | 38.8 | 0.0 | 55.4 | 42.8 | 0.0 | 57.3 | 34.0 | 0.0 |
In the presence of 10 mM SDS, CD spectra of all four BmGlvs at both pH 5.0 and pH 8.0 did not display negative peaks typical for α-helix conformation (Fig. 2), and the predicted secondary structures showed that BmGlv1–4 at both pH 5.0 and 8.0 contained mainly random coils (38–45%) and β-strands (43–56%) with low contents of α-helix (2–18%) but no turns (Tables S1 and S2). In the presence of 10 mM DPC, CD spectra of BmGlv1–3 at both pH 5.0 and 8.0 and CD spectrum of BmGlv4 at pH 8.0 were similar to CD spectra of BmGlv1–4 with SDS (Fig. 2A–C, E–H), but the spectrum of BmGlv4 at pH 5.0 showed a distinct minimum around 220 nm (Fig. 2D). Predicted secondary structures showed that BmGlv1–3 at pH 5.0 and 8.0 and BmGlv4 at pH 8.0 in the presence of 10 mM DPC mainly cotained random coils (38–44%) and β-strands (34–57%) with low contents of α-helix (2–16%) and turns (0–13%) (Tables S1 and S2). But BmGlv4 at pH 5.0 (with positive net charge) contained 47% α-helical conformation and 17.5% random coil, indicating that most random coil in BmGlv4 was converted to a-helix conformation in the presence of DPC similar to that in the presence of HFIP (Table S1).
3.4. Binding of BmGlvs to LPS
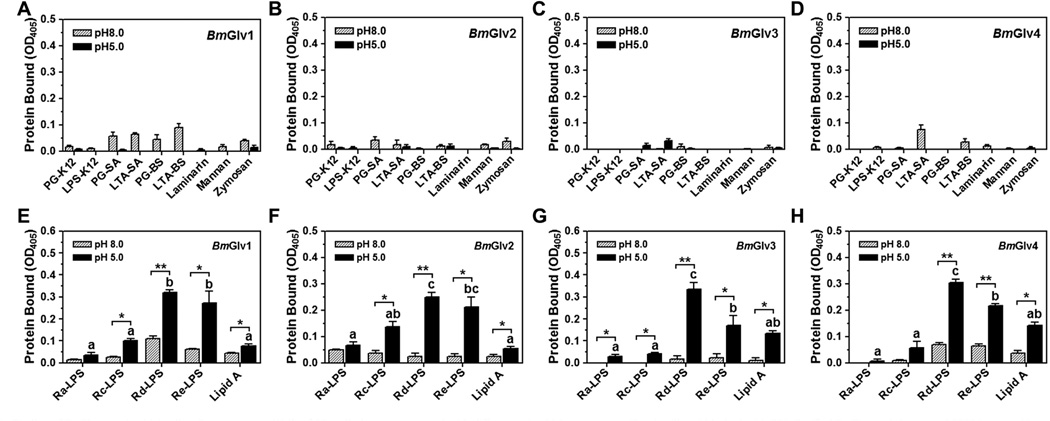
LPS is a major component of Gram-negative bacteria cell walls, and it is composed of three moieties: the O-specific antigen, the core (outer and inner core) carbohydrate, and the lipid A (Raetz, 1990; Yu and Kanost, 2002). HgGlv may interact with the lipid A moiety of LPS to exert its activity (Axen et al., 1997), and MsGlv can bind to the O-specific antigen and outer core carbohydrate moieties of LPS as well as Gram-positive LTA and PG, but does not bind to lipid A (Xu et al., 2012). To test binding of BmGlvs to different forms of LPS and other microbial components, plate ELISA assays were performed using purified recombinant BmGlvs in phosphate buffer at pH 5.0 and 8.0. Almost no or very little BmGlv1–4 bound to E. coli LPS-K12 and PG-K12, S. aureus LTA-SA and PG-SA, B. subtilis LTA-BS and PG-BS, laminarin, mannan, or zymosan at both pH 5.0 and 8.0 (Fig. 3A – D), and no or very little BmGlv1–4 bound to smooth LPS from several E. coli strains and Gram-negative bacteria (Fig. S4). Also, almost no or very little BmGlv1–4 bound to Ra-, Rc-, Rd- and Re-LPS and lipid A at pH 8.0; however, all four BmGlvs bound to rough mutants of LPS and lipid A at pH 5.0, with more proteins bound to Rd-LPS and Re-LPS than to Ra-LPS, Rc-LPS and lipid A (Fig. 3E – H). Together, these results suggest that positively charged (at pH 5.0) BmGlv1–4 may bind to the inner core carbohydrate of LPS and lipid A.
Fig. 3.
Binding of BmGlvs to microbial cell wall components. Wells of 96-well plates were coated with different microbial components (2 µg/well) and blocked with BSA. Purified BmGlvs and the control CP36 (a recombinant cuticle protein from M. sexta) were diluted to 1 µg/ml in 10 mM phosphate, 100 mM NaCl, pH 5.0 or 8.0, and the diluted proteins were added to the coated plates. Binding of proteins to microbial components was determined by plate ELISA assays as described in the materials and methods. The figure showed specific binding of BmGlvs to each microbial component after subtracting the total binding of the control CP36 protein from the total binding of BmGlvs. Each bar represents the mean of at least three individual measurements ± SEM. Comparing binding of each BmGlv to rough LPS and lipid A at pH 5.0 (panels E–H), identical letters are not significant difference (p > 0.05), while different letters indicate significant difference ( < 0.05) determined by one way ANOVA followed by a Tukey’s multiple comparison test. Comparing binding of BmGlv to each rough LPS or lipid A between pH 5.0 and 8.0 (panels E–H), ‘*’ (p < 0.05) and ‘**’ (p < 0.01) indicate significant differences determined by an unpaired t-test
3.5. CD spectroscopy and secondary structural prediction of BmGlvs in the presence of LPS
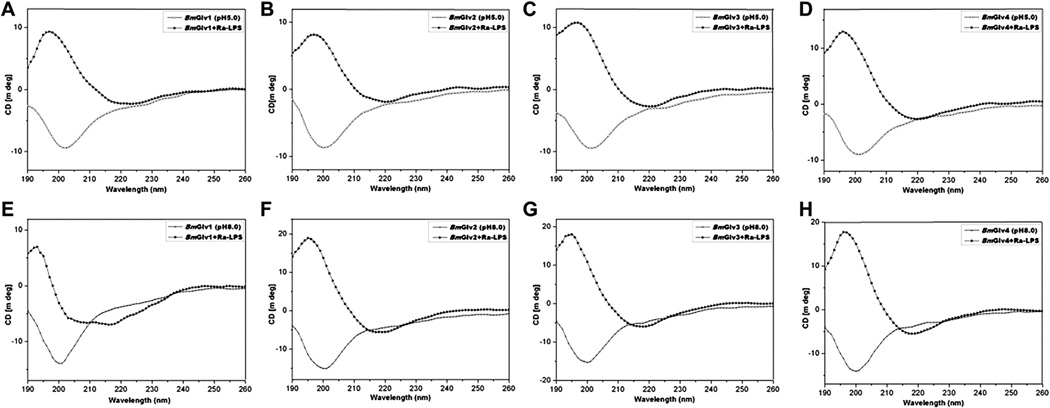
To determine whether interaction between BmGlvs and LPS can induce conformational transitions to α-helix, CD experiments were performed for BmGlv1–4 in the presence of smooth LPS, rough mutants (Ra, Rc and Re) of LPS and lipid A. In the presence of Ra-LPS at both pH 5.0 and 8.0, BmGlv1–4 all underwent conformational transitions to α-helix (Fig. 4) with predicted secondary structures of mainly α-helix (55–67%) and turns (33–45%), but no random coil or β-strand (except BmGlv1 at pH 8.0) (Tables 2 and 3). BmGlv1 at pH 8.0 in the presence of Ra-LPS contained high contents of β-strands (52%) and random coil (33%) but low content of α-helix (14%) (Table 3).
Fig. 4.
CD spectra of BmGlvs in the presence of Ra-LPS at pH 5.0 and 8.0. Purified BmGlv1–4 were diluted to 0.15 mg/ml in 10 mM phosphate buffer (pH 5.0 or 8.0) in the presence or absence of Ra-LPS (w/w= 1:1) and CD spectra were recorded on a Jasco-810 spectropolarimeter at 25 °C. Each CD spectrum was obtained after subtracting the signal from protein-free solution.
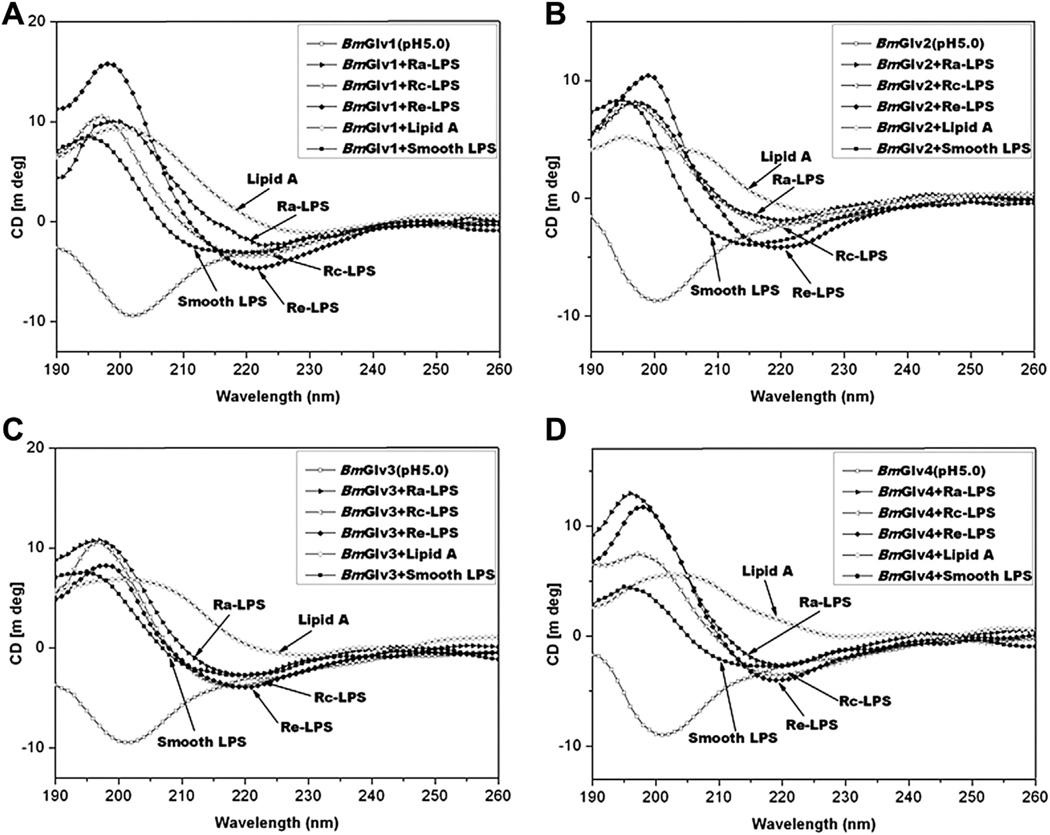
In the presence of smooth LPS, Rc- and Re-LPS, the CD spectra of BmGlv1–4 at pH 5.0 were very similar to those with Ra-LPS (Fig. 5), although BmGlv1–4 bound to rough mutants of LPS but not smooth LPS (Fig. 3). Lipid A also changed the CD spectra of BmGlv1–4 comparing to those in aqueous solutions (Fig. 5). The predicted secondary structures showed that in the presence of smooth LPS, rough LPS (Ra-, Rc- and Re-LPS) and lipid A at pH 5.0, BmGlv1–4 contained predominantly α-helix (50–72%) and turns (28–50%), with no β-strand or random coil (except for BmGlv2 and BmGlv4 with smooth LPS) (Table 4), suggesting that LPS and lipid A can induce conformational transitions from random coil to α-helix in BmGlv1–4. The concentrations of LPS and lipid A used in the CD experiment were 0.15 mg/ml (~15 µM for smooth LPS), which were higher than the critical micelle concentration (CMC) of LPS (CMC for smooth LPS from E. coli 0111:B4 is 1.3–1. 6 µM) (Yu et al., 2006). Together, these results suggest that hydrophobic property of LPS and lipid A is required for conformational transitions from random coil to α-helix in BmGlv1–4. We also tried the CD experiments with LTA and PG, but could not obtain CD spectra due to poor solubility of LTA and PG and precipitation problem after mixing LTA and PG with BmGlvs proteins.
Fig. 5.
CD spectra of BmGlvs in the presence of smooth LPS, Ra-, Rc- and Re-LPS and lipid A at pH 5.0. Purified BmGlv1–4 were diluted to 0.15 mg/ml in 10 mM phosphate buffer at pH 5.0 in the presence or absence of smooth LPS, Ra-, Rc- and Re-LPS or monophosphoryl lipid A (w/w= 1:1) and CD spectra were recorded on a Jasco-810 spectropolarimeter at 25 °C. Each CD spectrum was obtained after subtracting the signal from protein-free solution.
Table 4.
Predicted secondary structures of BmGlv1–4 in the presence of LPS and lipid A at pH 5.0
|
BmGlv1 |
BmGlv2 |
BmGlv3 |
BmGlv4 |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Smooth | Ra | Rc | Re | Lipid A | Smooth | Ra | Rc | Re | Lipid A | Smooth | Ra | Rc | Re | Lipid A | Smooth | Ra | Rc | Re | Lipid A | |
| Helix (%) | 68.3 | 54.8 | 61.2 | 60.5 | 55.6 | 65.8 | 58.4 | 62.4 | 57.6 | 60.5 | 71.9 | 67.0 | 59.3 | 58.5 | 62.5 | 52.6 | 62.1 | 66.0 | 57.9 | 49.9 |
| Beta (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Turn (%) | 31.7 | 45.2 | 38.8 | 39.5 | 44.4 | 24.4 | 41.6 | 37.6 | 42.4 | 39.5 | 28.1 | 33.0 | 40.7 | 41.5 | 37.5 | 31.7 | 37.9 | 34.0 | 42.1 | 50.1 |
| Random (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.7 | 0.0 | 0.0 | 0.0 | 0.0 |
3.6. Activity of BmGlvs against E. coli with rough LPS
Gloverins from different lepidopteran species show activities against E. coli, Gram-positive bacteria, fungi or a virus (Axen et al., 1997; Mackintosh et al., 1998; Lundstrom et al., 2002; Hwang and Kim, 2011; Xu et al., 2012; Moreno-Habel et al., 2012). It is not clear whether the activity of gloverin against different microbes is related to pH (net charge), conformational transition to α-helix, and/or binding to microbial surface. We showed above that pH alone cannot cause conformational transitions to α-helix in BmGlvs (Fig. 1 and Fig. S2) but pH is important for binding of BmGlvs to rough LPS (Fig. 3), and hydrophobic environment (HFIP, smooth and rough LPS, or lipid A) is required for conformational transitions to α-helix in BmGlvs (Figs. 2, 4 and 5, Tables 2–4, S1 and S2). To determine correlations between the activity of BmGlvs and structural conformation/binding ability, antimicrobial activity of BmGlv1–4 was tested against several Gram-negative and Gram-positive bacteria as well as fungi. BmGlv1–4 were inactive against Gram-negative E. coli DH5α and S. marcescens with smooth LPS at pH 5.0 and 8.0, or the fungi S. cerevisiae (at pH 5.0 and 8.0) and C. neoformans (at pH 8.0), but were active against C. neoformans at pH 5.0 (Fig. S5). BmGlv1–4 were also inactive against Gram-positive S. aureus, B. subtilis, B. cereus, and B. thuringiensis at both pH 5.0 and 8.0 (Fig. S6). These results were consistent with the binding properties of BmGlv1–4 as they did not bind to smooth LPS, LTA, PG, laminarin, mannan and zymosan (Fig. 3A – D and S4).
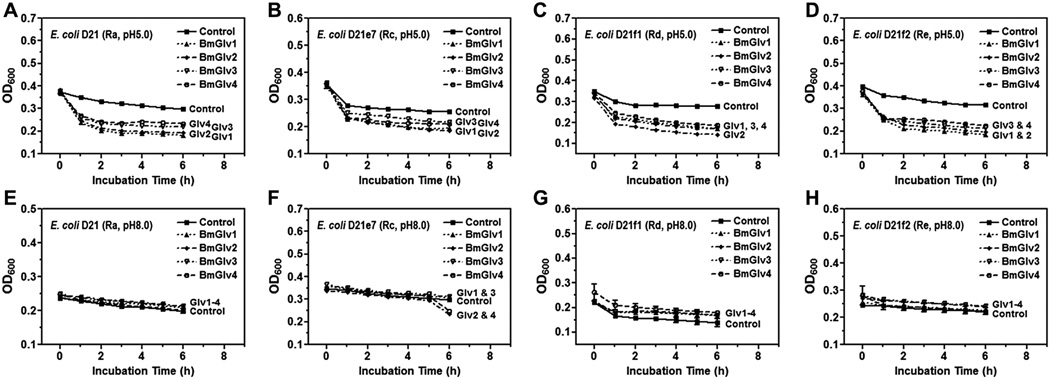
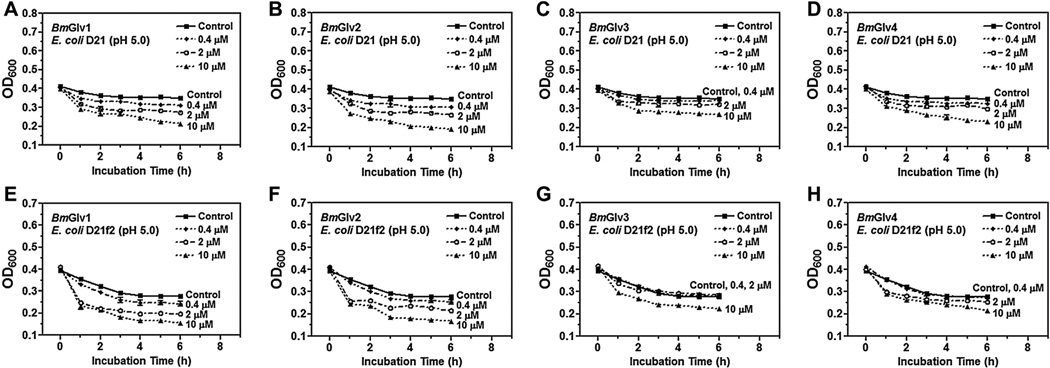
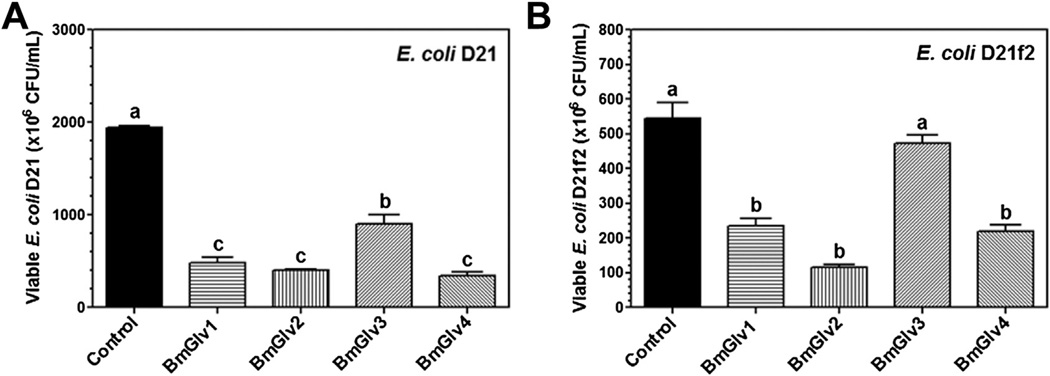
We then tested the activity of BmGlvs against E. coli mutant strains containing rough LPS since BmGlv1–4 bound to rough LPS at pH 5.0 (Fig. 3E – H). BmGlv1–4 (~10 µM each) at pH 5.0 were active against E. coli D21 (with Ra-LPS), E. coli D21e7 (with Rc-LPS), E. coli D21f1 (with Rd-LPS) and E. coli D21f2 (with Re-LPS) mutant strains, but were inactive against these E. coli mutant strains at pH 8.0 (Fig. 6). The concentration of BmGlvs used in the bacteria clearance assay was 10 µM for each protein, which was within the physiological concentration of total BmGlvs in E. coli-induced hemolymph (7–9 µM) (Fig. S1C). Using lower concentrations of recombinant BmGlvs at pH 5.0, the activity of BmGlvs against E. coli D21 and E. coli D21f2 was observed at 2 µM for BmGlv1, 2 and 4 and even at 0.4 µM for BmGlv1 and 2 (Fig. 7), indicating that the activity of BmGlvs against E. coli mutant strains is dose-dependent. We also confirmed the activity of BmGlvs using bacterial viability assay. Significantly lower numbers of viable E. coli D21 and E. coli D21f2 cells were observed when the bacterial cells were treated with BmGlvs (~10 µM) at pH 5.0 for 2 h (Fig. 8). The number of viable E. coli D21f2 cells was decreased but not significantly after treating with BmGlv3 compared to the control. In addition, the activity of BmGlv3 was significantly lower than that of BmGlv1, 2 or 4 (Fig. 8). Since BmGlv1–4 bound to rough mutants of LPS at pH 5.0 (Fig. 3E – H), together these results suggest that binding of BmGlvs to E. coli surface (via rough LPS) is required for the activity of BmGlvs against E. coli.
Fig. 6.
The activity of BmGlvs against E. coli mutant strains containing rough LPS. Mid-log phase E. coli D21, D21e7, D21f1 and D21f2 cells with different rough mutant forms of LPS were diluted to OD600 = 0.4 in 10 mM phosphate, 100 mM NaCl (pH 5.0 or 8.0), and then incubated with purified BmGlvs (final concentration of 150 µg/ml, ~10 µM) or buffer (Control) in 96-well plates with 220 rpm shaking at 37 °C OD600 was recorded every hour after incubation. The points represent the mean of four individual measurements ± SEM.
Fig. 7.
Dose-dependent activity of BmGlvs against E. coli D21 and D21f2 mutant strains. Mid-log phase E. coli D21 and D21f2 cells were diluted to OD600 = 0.4 in 10 mM phosphate, 100 mM NaCl, pH 5.0, and then incubated with increasing concentrations of purified BmGlvs (final concentration of 0.4,2 and 10 µM)or buffer (Control) in 96-well plates with 220 rpm shaking at 37 °C OD600 was recorded every hour after incubation. The points represent the mean of four individual measurements ± SEM.
Fig. 8.
Viability of E. coli D21 andD21f2 cells after treatment with BmGlvs. Mid-log phase E. coli D21 and D21f2 cells were diluted to OD600 = 0.4 in 10 mM phosphate, 100 mMNaCl, pH 5.0, and then incubated with purified recombinant BmGlvs (final concentration of ~10 mM) or buffer (Control) in 96-well plates with 220 rpm shaking at 37 °C for 2 h. The bacterial cells were serially diluted with 10 mM phosphate, 100 mM NaCl, pH 5.0, and aliquots of diluted bacterial cells were plated on LB-agar plates containing streptomycin sulfate (50 µg/ml) and ampicilin (10 µg/ml). The plates were incubated at 37 °C overnight and the numbers of viable bacterial cells were recorded. The bars represent the mean of three individual measurements ± SEM. Comparing the control group and BmGlvs treated groups, identical letters are not significant difference (p > 0.05), while different letters indicate significant difference (p < 0.05) determined by one way ANOVA followed by a Tukey’s multiple comparison test
4. Discussion
Gloverin belongs to the glycine-rich AMP family and has been identified only in lepidopteran insects so far. Gloverins from most lepidopteran species are basic (pI ~8.3) or highly basic (pI > 9.0) (Xu et al., 2012), but H. virescens gloverin, A. mylitta gloverin 2, and four B. mori gloverins are slightly acidic to neutral (pI ~ 5.5–7.2). Even basic gloverins from different lepidopteran species show activities against different microbes, including E. coli, Gram-positive bacteria, fungi, and a virus (Axen et al., 1997; Mackintosh et al., 1998; Lundstrom et al., 2002; Hwang and Kim, 2011; Xu et al., 2012; Moreno-Habel et al., 2012). B. mori gloverins are also active against E. coli (Kawaoka et al., 2008; Mrinal and Nagaraju, 2008). Thus, it is not clear whether the activity of gloverin is related to its pI or not. HgGlv (pI ~8.3) is active against E. coli, and it adopts random coil conformation in solution but has α-helical conformation in the presence of HFIP (Axen et al., 1997). MsGlv (pI ~9.3) can bind to LPS but is inactive against E. coli (Xu et al., 2012). Therefore, it is also not clear whether the activity of gloverin is related to its binding ability to microbial surface and/or conformational transition to α-helix.
Most linear AMPs are unfolded in aqueous solutions and they can change to defined structures only after interacting with bacterial membrane (Nguyen et al., 2008). Organic solvents such as TFE and HFIP, and detergent micelles like SDS and DPC, are commonly used to mimic bacterial membrane. SDS is used to mimic bacterial membrane as it contains a negative head group, while DPC is used to mimic eukaryotic membrane that does not contain net negative charge (Haney and Vogel, 2009). We showed by CD experiments that BmGlv1–4 in aqueous solution adopted mainly random coil (>50%) conformation with certain contents of β-strands and turns but no α-helix in regardless of pH values (pH 3–8) (Fig. 1 and Fig. S2, Table 1), suggesting that pH alone cannot cause conformational transitions to α-helix in BmGlv1–4. BmGlvs adopted α-helical conformations in the presence of organic solvent HFIP but not detergent micelles SDS or DPC (Fig. 2), a result consistent with α-helical conformations of HgGlv and H. cecropia cecropin in solution containing HFIP (Steiner, 1982; Holak et al., 1988; Axen et al., 1997). In addition, conformations of BmGlvs in the presence of HFIP differ from those in the presence of SDS and DPC, while moricin adopts similar conformations in the presence of TFE and SDS (Dai et al., 2008).
Interestingly, in the presence of smooth LPS, rough LPS (Ra-, Rc-and Re-LPS) and lipid A, BmGlv1–4 at both pH 5.0 and 8.0 (except BmGlv1 with Ra-LPS at pH 8.0) underwent conformational transitions from random coil to α-helix (Figs. 4 and 5) with predominantly α-helix (50–72%) and turns (28–50%), but almost no random coil or β-strand (Tables 2–4). The exception is BmGlv1, which contained mainly β-strands (52%) and random coil (33%) with only 14% α-helix in the presence of Ra-LPS at pH 8.0 (Fig. 4E and Table 3). Thus, in the hydrophobic membrane-like environment (smooth LPS, rough LPS and lipid A, as well as HFIP), BmGlvs adopt mainly α-helix structure. BmGlv4 may differ from BmGlv1–3, since in the presence of HFIP and DPC, BmGlv4 at pH 5.0 (with positive net charge) contained low contents of random coil, but BmGlv1–3 at pH 5.0 contained high contents of random coil (Table S1).
The mode of action of basic AMPs may involve electrostatic interaction between the positively charged AMPs and the negatively charged head groups of bacterial membranes. For example, attacin (~20 kDa), another typical AMP of the glycine-rich family, acts on the outer membrane of E. coli by electrostatic binding to lipid A and interaction with the acyl chain of lipid A and phospholipids in the outer membrane (Carlsson et al., 1998). Our ELISA assays showed that BmGlvs did not bind to smooth LPS, LTA, PG, laminarin, mannan and zymosan at both pH 5.0 and 8.0 (Fig. 3A – D and S4), but bound to rough mutants of LPS and lipid A at pH 5.0 but not pH 8.0, with more proteins bound to Rd-LPS and Re-LPS than to Ra-LPS, Rc-LPS and lipid A (Fig. 3E – H). These results suggest that positively net charge in BmGlvs may contribute to binding of BmGlvs to rough LPS and lipid A, and BmGlvs may bind to the inner core carbohydrate of LPS. CD experiments showed that in the presence of smooth LPS, Ra-, Rc- and Re-LPS and lipid A, random coils in BmGlvs were converted to α-helix (Figs. 4 and 5), while BmGlvs did not bind to smooth LPS (Fig. 3A – D and S4). These results suggest that binding to LPS/lipid A is not required but membrane-like environment is necessary for conformational transitions of BmGlvs from random coil to α-helix.
Activity assays showed that BmGlvs were inactive against E. coli DH5α and S. marcescens with smooth LPS, and inactive against two fungal strains and several Gram-positive bacteria, but were active against C. neoformans at pH 5.0 (Figs. S5 and S6). These results are consistent with the binding properties of BmGlvs as they did not bind to smooth LPS, LTA, PG, laminarin, mannan or zymosan (Fig. 3A – D and S4). However, BmGlvs at pH 5.0 were active against E. coli mutant strains with rough LPS (Ra-, Rc-, Rd- and Re-LPS) (Fig. 6A – D), a result consistent with HgGlv (Axen et al., 1997), but inactive at pH 8.0 against these E. coli mutant strains (Fig. 6E – H). The activity of recombinant BmGlvs against E. coli mutant strains was dose-dependent (Fig. 7) and within physiological concentrations in hemolymph, as the high concentration of recombinant BmGlvs used in the assay was ~10 µM, which was closed to the estimated concentration (7–9 µM) of total native BmGlvs in the E. coli-induced hemolymph of B. mori larvae (Fig. S1C). Compared the four BmGlvs, BmGlv1, 2 and 4 had similar high activity against E. coli, while BmGlv3 had lower activity than BmGlv1, 2 and 3 (Fig. 8).
BmGlvs bound to rough mutants of LPS at pH 5.0 but not at pH 8.0 (Fig. 3E – H), but rough LPS caused conformational transition of BmGlvs to α-helix at both pH 5.0 and 8.0 (Figs. 4 and 5). These results suggest that binding of BmGlvs to rough LPS on E. coli surface is required for the activity of BmGlvs. BmGlvs at acidic pH were active against E. coli, suggesting that they may have higher activity against bacteria in the midgut. We believe that conformational transition from random coil to α-helix in BmGlvs is the key for pore formation on bacterial membrane, but binding of BmGlvs to bacterial surface via rough LPS is the prerequisite for the activity of BmGlvs. Thus, whether a gloverin is active against Gram-negative bacteria, Gram-positive bacteria, fungi or viruses may depend on its binding to microbial surface and conformational transition to α-helix. We also tried CD experiments for BmGlvs in the presence of LTA, PG, laminarin, mannan or zymosan to test interactions between BmGlvs and these microbial components and determine whether conformational transitions to α-helix occur, but failed to obtain CD spectra due to poor solubility of these microbial components. In order to better understand the mode of action of gloverins and the mechanisms of conformational transitions to α-helix, it is necessary to determine three-dimensional structure of gloverin, which will be one of our future goals.
Supplementary Material
Acknowledgments
This work was supported, in whole or in part, by National Institutes of Health Grant GM066356 (to X.Q.Y.). Support for the work was also derived from the “973” National Basic Research Program of China (No. 2012CB114600) (to Y.C., X.J.D. and W.Y.Y.) and the Project of Science and Technology New Star in Zhu Jiang Guangzhou City (2012J2200083) (to W.Y.Y.).
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ibmb.2013.03.013.
References
- Abdel-Latief M, Hilker M. Innate immunity: eggs of Manduca sexta are able to respond to parasitism by Trichogramma evanescens. Insect Biochem. Mol. Biol. 2008;38:136–145. doi: 10.1016/j.ibmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Ando K, Okada M, Natori S. Purification of sarcotoxin II, antibacterial proteins of Sarcophaga peregrina (flesh fly) larvae. Biochemistry. 1987;26:226–230. doi: 10.1021/bi00375a030. [DOI] [PubMed] [Google Scholar]
- Axen A, Carlsson A, Engstrǒm A, Bennich H. Gloverin, an antibacterial protein from the immune hemolymph of Hyalophora pupae. Eur. J. Biochem. 1997;247:614–619. doi: 10.1111/j.1432-1033.1997.00614.x. [DOI] [PubMed] [Google Scholar]
- Brown SE, Howard A, Kasprzak AB, Gordon KH, East PD. A peptidomics study reveals the impressive antimicrobial peptide arsenal of the wax moth Galleria mellonella . Insect Biochem. Mol. Biol. 2009;39:792–800. doi: 10.1016/j.ibmb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Bulet P, Stocklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept. Lett. 2005;12:3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- Bulet P, Cociancich S, Dimarcq JL, Lambert J, Reichhart JM, Hoffmann D, Hetru C, Hoffmann JA. Isolation from a coleopteran insect of a novel inducible antibacterial peptide and of new members of the insect defensin family. J. Biol. Chem. 1991;266:24520–24525. [PubMed] [Google Scholar]
- Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial peptides in insects: structure and function. Dev. Comp. Immunol. 1999;23:329–344. doi: 10.1016/s0145-305x(99)00015-4. [DOI] [PubMed] [Google Scholar]
- Carlsson A, Nystrom T, Cock H, Bennich H. Attacin, an insect immune protein-binds LPS and triggers the specific inhibition of bacterial outer membrane protein synthesis. Microbiology. 1998;144:2179–2188. doi: 10.1099/00221287-144-8-2179. [DOI] [PubMed] [Google Scholar]
- Casteels P, Ampe C, Jacobs F, Tempst P. Functional and chemical characterization of hymenoptaecin, an antibacterial polypeptide that is infectioninducible in the honeybee (Apis mellifera) J. Biol. Chem. 1993;268:7044–7054. [PubMed] [Google Scholar]
- Chae J-H, Kurokawa K, So Y-I, Hwang HO, Kim M-S, Park J-W, Jo Y-H, Lee YS, Lee BL. Purification and characterization of tenecin 4, a new anti-gram-negative bacterial peptide from the beetle Tenebrio nolitor . Dev. Comp. Immunol. 2012;36:540–546. doi: 10.1016/j.dci.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Cheng T, Zhao P, Liu C, Xu P, Gao Z, Xia Q, Xiang Z. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori . Genomics. 2006;87:356–365. doi: 10.1016/j.ygeno.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Cociancich S, Dupont A, Hegy G, Lanot R, Holder F, Hetru C, Hoffmann JA, Bulet P. Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus . Biochem. J. 1994;300:567–575. doi: 10.1042/bj3000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet B, Bonmatin JM, Hetru C, Hoffmann JA, Ptak M, Vovelle F. Refined three-dimensional solution structure of insect defensin A. Structure. 1995;3:435–448. doi: 10.1016/s0969-2126(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Dai H, Rayaprolu S, Gong Y, Huang R, Prakash O, Jiang H. Solution structure, antibacterial activity, and expression profile of Manduca sexta moricin. J. Pept. Sci. 2008;10:1002–1016. doi: 10.1002/psc.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etebari K, Palfreyman RW, Schlipalius D, Nielsen LK, Glatz RV, Asgari S. Deep sequencing-based transcriptome analysis of Plutella xylostella larvae parasitized by Diadegma semiclausum . BMC Genomics. 2011;12:446. doi: 10.1186/1471-2164-12-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eum JH, Seo YR, Yoe SM, Kang SW, Han SS. Analysis of the immune-inducible genes of Plutella xylostella using expressed sequence tags and cDNA microarray. Dev. Comp. Immunol. 2007;31:1107–1120. doi: 10.1016/j.dci.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Gandhe-Archana S, Arunkumar KP, John-Serene H, Nagaraju J. Analysis of bacteria-challenged wild silkmoth, Antheraea mylitta (Lepidoptera) transcriptome reveals potential immune genes. BMC Genomics. 2006;184:1471–2164. doi: 10.1186/1471-2164-7-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney EF, Vogel HJ. NMR of antimicrobial peptides. ISSN: 0066–4103. 2009;65 http://dx.doi.org/10.1016/S0066-4103(08)00201-9. [Google Scholar]
- Hoffmann JA. Innate immunity of insects. Immunology. 1995;7:4–10. doi: 10.1016/0952-7915(95)80022-0. [DOI] [PubMed] [Google Scholar]
- Holak TA, Engstrom A, Kraulis PJ, Lindeberg G, Bennich H, Jones A, Gronenborn AM, Clore GM. The solution conformation of the antibacterial peptide cecropin a: a nuclear magnetic resonance and dynamical simulated annealing study. Biochemistry. 1988;27:7620–7629. doi: 10.1021/bi00420a008. [DOI] [PubMed] [Google Scholar]
- Holzwarth G, Doty P. The ultraviolet circular dichroism of polypeptides. J. Am. Chem. Soc. 1965;87:218–228. doi: 10.1021/ja01080a015. [DOI] [PubMed] [Google Scholar]
- Hultmark D, Engstrom A, Andersson K, Steiner H, Bennich H, Boman HG. Insect immunity. attacins, a family of antibacterial proteins from Hyalophora cecropia . EMBO J. 1983;2:571–576. doi: 10.1002/j.1460-2075.1983.tb01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Kim Y. RNA interference of an antimicrobial peptide, gloverin, of the beet armyworm, Spodoptera exigua, enhances susceptibility to Bacillus thuringiensis . J. Invertebr Pathol. 2011;108:194–200. doi: 10.1016/j.jip.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Furukawa S, Tanaka H, Yamakawa M. Expression of antimicrobial peptide genes encoding Enbocin and Gloverin isoforms in the silkworm, Bombyx mori . Biosci. Biotechnol. Biochem. 2007;71:2233–2241. doi: 10.1271/bbb.70212. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Katsuma S, Daimon T, Isono R, Omuro N, Mita K, Shimada T. Functional analysis of four Gloverin-like genes in the silkworm, Bombyx mori . Arch. Insect Biochem. Physiol. 2008;67:87–96. doi: 10.1002/arch.20223. [DOI] [PubMed] [Google Scholar]
- Lamberty M, Caille A, Landon C, Tassin-Moindrot S, Hetru C, Bulet P, Vovelle F. Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities. Biochemistry. 2001;40:11995–12003. doi: 10.1021/bi0103563. [DOI] [PubMed] [Google Scholar]
- Landon C, Sodano P, Hetru C, Hoffmann J, Ptak M. Solution structure of drosomycin, the first inducible antifungal protein from insects. Protein Sci. 1997;6:1878–1884. doi: 10.1002/pro.5560060908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo-Otvos JR. Antibacterial peptides isolated from insects. J. Pept. Sci. 2000;6:497–511. doi: 10.1002/1099-1387(200010)6:10<497::AID-PSC277>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 2002;32:1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster . Annu. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lundstrom A, Liu G, Kang D, Berzins K, Steiner H. Trichoplusia ni gloverin, an inducible immune gene encoding an antibacterial insect protein. Insect Biochem. Mol. Biol. 2002;32:795–801. doi: 10.1016/s0965-1748(01)00162-x. [DOI] [PubMed] [Google Scholar]
- Mackintosh JA, Gooley AA, Karuso PH, Beattie AJ, Jardine DR, Veal DA. A gloverin-like antibacterial protein is synthesized in Helicoverpa armigera following bacterial challenge. Dev. Comp. Immunol. 1998;22:387–399. doi: 10.1016/s0145-305x(98)00025-1. [DOI] [PubMed] [Google Scholar]
- Moreno-Habel DA, Biglang-Awa IM, Dulce A, Luu DD, Garcia P, Weers PM, Haas-Stapleton EJ. Inactivation of the budded virus of Autographa californica M nucleopolyhedrovirus by gloverin. J. Invertebr Pathol. 2012;110:92–101. doi: 10.1016/j.jip.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrinal N, Nagaraju J. Intron loss is associated with gain of function in the evolution of the gloverin family of antibacterial genes in Bombyx mori . J. Biol. Chem. 2008;283:23376–23387. doi: 10.1074/jbc.M801080200. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Prenner EJ, Vogel HJ. In: Structural characterization of antimicrobial peptides by NMR spectroscopy. Webb Graham A., editor. Modern Magnetic Resonance, pp; 2008. pp. 1315–1323. [Google Scholar]
- Raetz CR. Biochemistry of endotoxins. Annu. Rev. Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- Seitz V, Clermont A, Wedde M, Hummel M, Vilcinskas A, Schlatterer K, Podsiadlowski L. Identification of immunorelevant genes from greater wax moth (Galleria mellonella) by a subtractive hybridization approach. Dev. Comp. Immunol. 2003;27:207–215. doi: 10.1016/s0145-305x(02)00097-6. [DOI] [PubMed] [Google Scholar]
- Sideri M, Tsakas S, Markoutsa E, Lampropoulou M, Marmaras VJ. Innate immunity in insects: surface-associated dopa decarboxylase-dependent pathways regulate phagocytosis, nodulation and melanization in medfly haemocytes. Immunology. 2007;123:528–537. doi: 10.1111/j.1365-2567.2007.02722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva PD, Jouvensal L, Lamberty M, Bulet P, Caille A, Vovelle F. Solution structure of termicin, an antimicrobial peptide from the termite Pseudacan-thotermes spiniger . Protein Sci. 2003;12:438–446. doi: 10.1110/ps.0228303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JL, Barbosa JF, Bravo JP, Souza EM, Huergo LF, Pedrosa FO, Esteves E, Daffre S, Fernandez MA. Induction of a gloverin-like antimicrobial polypeptide in the sugarcane borer Diatraea saccharalis challenged by septic injury. Braz. J. Med. Biol. Res. 2010;43:431–436. doi: 10.1590/s0100-879x2010005000010. [DOI] [PubMed] [Google Scholar]
- Steiner H. Secondary structure of the cecropins: antibacterial peptides from the moth Hyalophora cecropia . FEBS Lett. 1982;137:283–287. doi: 10.1016/0014-5793(82)80368-2. [DOI] [PubMed] [Google Scholar]
- Tiffany ML, Krimm S. Effect of temprature on the circular dichroism spectra of polypeptides in the extended state. Biopolymers. 1972;11:2309–2316. doi: 10.1002/bip.1972.360111109. [DOI] [PubMed] [Google Scholar]
- Xu XX, Zhong X, Yi HY, Yu XQ. Manduca sexta gloverin binds microbial components and is active against bacteria and fungi. Dev. Comp. Immunol. 2012;38:275–284. doi: 10.1016/j.dci.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Wu CS, Martinez HM. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Kanost MR. Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J. Biol. Chem. 2000;275:37373–37381. doi: 10.1074/jbc.M003021200. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Kanost MR. Binding of hemolin to bacterial lipopolysaccharide and lipoteichoic acid. An immunoglobulin superfamily member from insects as a pattern-recognition receptor. Eur. J. Biochem. 2002;269:1827–1834. doi: 10.1046/j.1432-1033.2002.02830.x. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Tracy ME, Ling E, Scholz FR, Trenczek T. A novel C-type immulectin- 3 from Manduca sexta is translocated from hemolymph into the cytoplasm of hemocytes. Insect Biochem. Mol. Biol. 2005;35:285–295. doi: 10.1016/j.ibmb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Yu L, Tan M, Ho B, Ding JL, Wohland T. Determination of critical micelle concentrations and aggregation numbers by fluorescence correlation spectroscopy: aggregation of a lipopolysaccharide. Anal. Chim. Acta. 2006;556:216–225. doi: 10.1016/j.aca.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem. Mol. Biol. 2003;33:541–559. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.