Abstract
The Bardet-Biedl Syndrome (BBS) is a human developmental disorder that has been associated with fourteen BBS genes affecting the development of cilia. Three BBS genes are distant relatives of chaperonin proteins, a family of chaperones well known for the protein-folding role of their double-ringed complexes. Chaperonin-like BBS genes were originally thought to be vertebrate-specific, but related genes from different metazoan species have been identified as chaperonin-like BBS genes based on sequence similarity. Our phylogenetic analyses confirmed the classification of these genes in the chaperonin-like BBS gene family, and set the origin of the gene family earlier than the time of separation of Bilateria, Cnidaria, and Placozoa. By extensive searches of chaperonin-like genes in complete genomes representing several eukaryotic lineages, we discovered the presence of chaperonin-like BBS genes also in the genomes of Phytophthora and Pythium, belonging to the group of Oomycetes. This finding suggests that the chaperonin-like BBS gene family had already evolved before the origin of Metazoa, as early in eukaryote evolution as before separation of the lineages of Unikonts and Chromalveolates. The analysis of coding sequences indicated that chaperonin-like BBS proteins have evolved in all lineages under constraining selection. Furthermore, analysis of the predicted structural features suggested that, despite their high rate of divergence, chaperonin-like BBS proteins mostly conserve a typical chaperonin-like three-dimensional structure, but question their ability to assemble and function as chaperonin-like double-ringed complexes.
Introduction
The Bardet-Biedl Syndrome (BBS) is a human developmental disorder affecting a variety of tissues, which has been linked to a group of fourteen genes (named BBS1 to BBS14) essential for the correct development of cilia [1–3]. The recent identification of BBS genes has stimulated new interest in the genetic control of the development and functionality of the eukaryotic cilium. The products of seven of these genes, namely BBS1, BBS2, BBS4, BBS5, BBS7, BBS8 and BBS9, assemble in a newly-discovered protein complex called BBSome, which localizes to the basal body and to the axoneme of cilia [3]. Three other BBS proteins, BBS6 (also known as MKKS), BBS10 and BBS12, are related to class 2 eukaryotic chaperonin proteins, including T-complex protein 1 (TCP1), also named chaperonin containing TCP1 subunit 1 (CCT1), and CCT2 to CCT8, best known for assembling in a double hetero-8-meric ringed complex (TRiC/CCT) essential for folding nascent actin, tubulin, and other proteins (see, e.g., [4–6] for reviews). The three chaperonin-like BBS proteins (CL-BBS) are mostly found associated with the centrosome and basal body of the cilium [7,8], where they associate with selected CCT chaperonin monomers and with BBS7 to form the “BBS/CCT complex”, required for BBSome assembly [8,9].
Chaperonin-like BBS (CL-BBS) genes originated from a duplication of a progenitor of the CCT8 gene [10]. While they were originally described as vertebrate-specific [8,11–13], sequences with highest similarity to CL-BBS genes have been also reported in non-vertebrate Metazoa, including the Urochordate Ciona intestinalis [7], Lophotrochozoa, Cnidaria and Placozoa [14], suggesting that the CL-BBS gene family originated early in Metazoan evolution. In this work we supported with phylogenetic analyses the phenetic classification of these chaperonin-like genes in the CL-BBS gene family, confirming the orthology of vertebrate and non-vertebrate genes. Furthermore, we performed extensive searches and phylogenetic analyses of chaperonin-like genes found in completely sequenced genome sequences of several species belonging to a variety of anciently diverged eukaryotic lineages, and newly identified the presence of CL-BBS genes in the genomes of water molds (Oomycetes). All alternative evolutionary scenarios interpreting our findings and phylogenetic reconstructions imply that the CL-BBS gene family originated and twice duplicated at earlier stages of eukaryote evolution than previously thought.
Results
Chaperonin-like sequences in eukaryotic genomes
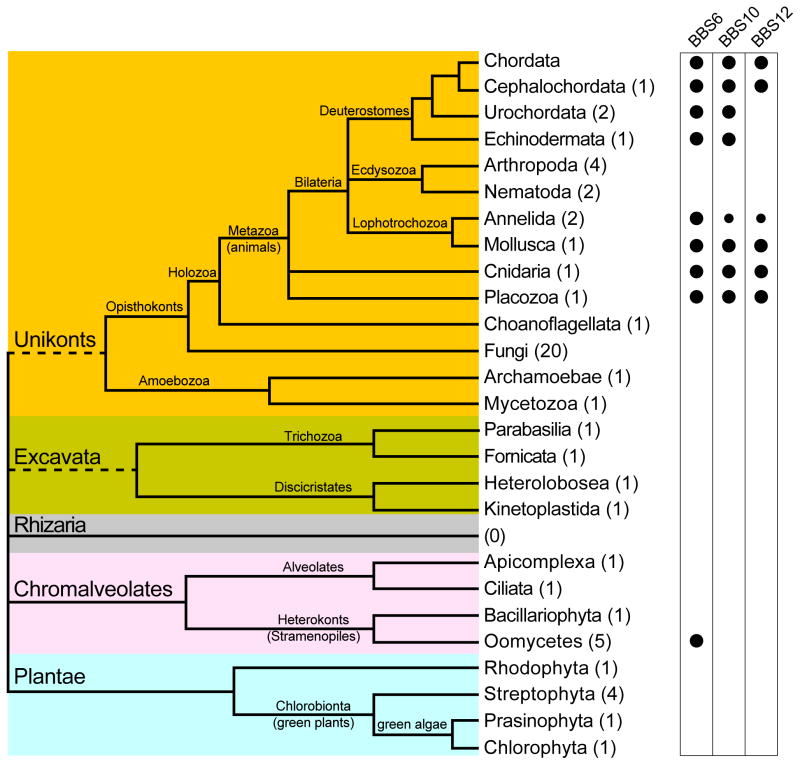
Five to seven distinct major clades of eukaryotic organisms are recognized in recent phylogenetic analyses [15], including Opisthokonts and Amoebozoa (clustered by some authors into the group of Unikonts), Trichozoa and Discicristates (sometimes clustered as Excavata), Rhizaria, Chromalveolates, and Plantae (Figure 1). With the exception of the clade of Rhizaria, multiple complete genome sequences from species representative of each clade have become available. We analyzed complete genome sequences from thirty-seven non-vertebrate species representative of the major eukaryotic clades, and subsequently augmented our set with two additional Oomycete genomes and 18 additional fungal genomes (Table 1, supplementary table S1, and figure 1). To identify as many chaperonin-like sequences as possible, we first searched the genomes with human and other chaperonin-like BBS proteins using a permissive E-value threshold (<1.0) (see Methods). Applying reciprocal BLAST analysis [16], we identified twenty-five sequences that were reciprocal nearest-neighbors of CL-BBS sequences, fourteen using BBS6 queries, six using BBS10 queries, and five using BBS12 queries (Figure 1 and supplementary tables S2-S4). Twenty sequences were found in Metazoan species, including Brachiostoma, Ciona and sea urchin (deuterostomes), round worm and gastropods (Lophotrochozoa), and the anciently diverged metazoan groups of Cnidaria and Placozoa, confirming previous reports of sequences similar to CL-BBS proteins encoded in non-vertebrate animal genomes [7,14]. However, we also identified five closely-related chaperonin-like sequences in genomes of the water mold genera Phytophthora and Pythium, belonging to the Oomycetes (Chromalveolates, Heterokonts), a group separate from Metazoa (Unikonts, Opisthokonts) (Figure 1 and Table 1). No reciprocal nearest-neighbors of chaperonin-like BBS sequences were identified in genomes of insects or of nematodes (Ecdysozoa), nor from genomes of non-metazoan Opisthokonts (including 20 Fungi genomes), Amoebozoa, Plants, Alveolates, Bacillariophyta, or Kinetoplastida (Figure 1).
Figure 1.
Schematic representation of the phylogenetic relations of major eukaryotic groups and presence of CL-BBS genes in the indicated groups. For each phylogenetic group the number of completely sequenced genomes analyzed in this study is shown in parentheses. Smaller bullets indicate that BBS10 and BBS12 were found in one of the two analyzed genomes of Annelida.
Table 1. Genome sequence dataset.
Dataset of complete genome sequences utilized in this study. Presence (+) or absence (−) of cilia / flagella in the corresponding organism is indicated next to the species name..
| Taxonomy | Species (abbreviation) | Data source | ||
|---|---|---|---|---|
| Opisthokonts (Unikonts) | Deuterostomes | Cephalochordata | Branchiostoma floridae (amphioxus) (Bf) + | JGI |
| Urochordata | Ciona intestinalis(Ci) + | ENSEMBL | ||
| Ciona savignyi (Cs) + | ENSEMBL | |||
| Echinodermata | Strongylocentrotus purpuratus (sea urchin) (Sp) + | HGSC | ||
| Ecdysozoa | Arthropoda | Drosophila melanogaster (fruit fly) (Dm) (+) | ENSEMBL | |
| Anopheles gambiae (Ag) (+) | FlyBase | |||
| Apis mellifera (Am) (+) | FlyBase | |||
| Tribolium casteanum (red flour beetle) (Tc) (+) | BeetleBase | |||
| Nematoda | Caenorhabditis elegans (worm) (Ce) (+) | Worm Base | ||
| Lophotrochozoa | Mollusca | Lottia gigantea+ (snail) (Lg) | JGI | |
| Annelida | Helobdella robusta + (leech) (Hr) | JGI | ||
| Capitella capitata+ (briste worm) (Cas) | JGI | |||
| Cnidaria | Nematostella vectensis+ (sea anemone) (Nv) | JGI | ||
| Placozoa | Trichoplax adhaerens (Ta) + | JGI | ||
| Choanoflagellata | Monosiga brevicollis (Mb) + | JGI | ||
| Fungi* | Ascomycota | Mycosphaerella graminicola (Mg) − | JGI | |
| Nectria haematococca (Nh) − | JGI | |||
| Amoebozoa (Unikonts) | Mycetozoa | Dictyostelium discoideum−(slime mold) (Dd) | DictyBase | |
| Archamoebae | Entamoeba histolytica− | TIGR | ||
| (Excavata) | Trichozoa | Parabasilia | Trichomonas vaginalis (Tv) + | TIGR |
| Fornicata | Giardia lamblia (Gl) + | GiardiaDB | ||
| Discicristata | Kinetoplastida | Trypanosoma cruzi (Tcr) + | TIGR | |
| Heterolobosea | Naegleria gruberi +(amoeboflagellate) (Ng) | JGI | ||
| Chromalveolates | Alveolata | Apicomplexa | Plasmodium falciparum + | PlasmoDB |
| Ciliophora | Tetrahymena thermophila (Tt) + | TGD | ||
| Paramecium tetraurelia + | ParameciumDB | |||
| Heterokonts | Oomycetes | Phytophthora sojae(Ps) + | JGI | |
| Phytophthorara morum (Pr) + | JGI | |||
| Phytophthora capsici (Pca) + | JGI | |||
| Phytophthora infestans (Pi) + | BROAD** | |||
| Pythium ultimum (Pu) + | JGI** | |||
| Bacillariophyta | Phaeodactylum tricornutum − | JGI | ||
| Plantae | Prasinophyta | Ostreococcus lucimarinus − (green algae) | JGI | |
| Chlorophyta | Chlamydomonas reinhardtii (unicellular green algae) + | JGI | ||
| Rhodophyta | Cyanidioschyzon merolae (red algae) − | Genome Project | ||
| Land Plants | Dicot | Arabidopsis thaliana (At) − | TAIR | |
| Sorghum bicolor − | PlantGDB | |||
| Monocot | Oryza sativa − | PlantGDB | ||
| Zea mays − | PlantGDB |
See Supplementary Table S1 for an expanded set of 20 genomes from Fungi included based on results.
Newly sequenced genomes included based on maximum-likelihood and Bayesian tree results.
Evolutionary analysis
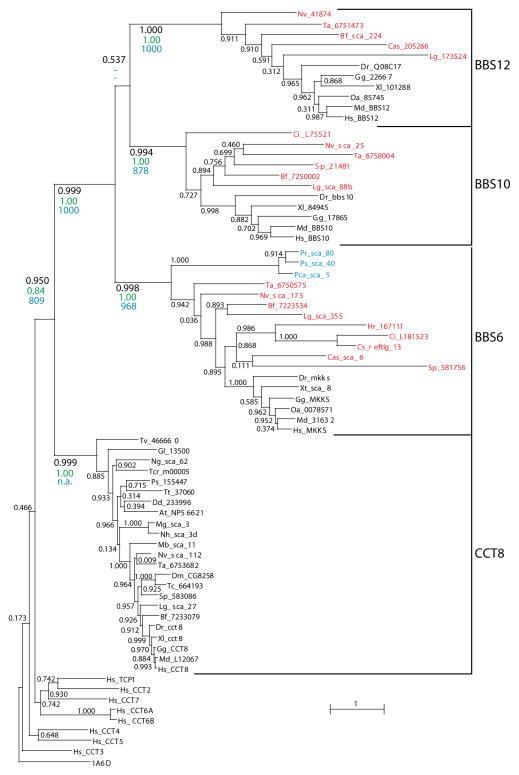
The evolutionary relations of the newly identified sequences with CL-BBS proteins were reconstructed in phylogenetic trees obtained with Maximum-likelihood (ML), Bayesian, or distance methods, based on the multiple protein alignment of the 26 newly-identified sequences with human CCT proteins, vertebrate CL-BBS proteins, and one representative of archaeal chaperonin class 2 proteins as out-group (see Methods for details, and Supplementary Material for Alignment). All methods resulted in tree topologies (Figure 2 and supplementary figures S1 and S2) implying that all chaperonin-like genes identified by our searches originated monophyletically within the CL-BBS gene family by duplication of the CCT8 chaperonin gene. They also identified the newly found sequences from Oomycetes as belonging to the BBS6 subfamily. The substantial concordance between the ML and Bayesian trees emphasized the robustness of the results on substitution model and on possible long-branch attraction effects [17,18] (see Methods). We also tested robustness of the clusters over a wide range of shapes of gamma-distributed position-specific substitution rates, used to estimate pairwise evolutionary distances for neighbor-joining distance-based tree reconstructions. Association of the twenty-six newly-identified sequences within one of the three CL-BBS clusters was robustly reproduced over values of the parameter a of the gamma distribution in the interval 1.0 to 3.0, with a value a=2.212 estimated by the ML procedure (Supplementary figure S2). All relevant clusters, including the sequences identified in Oomycetes, were supported by very high values of aLRT (approximate likelihood ratio, for the ML tree), posterior probability (for the Bayesian tree), or bootstrap (for the neighbor-joining trees). An exception was the order with which the three BBS6, BBS10 and BBS12 families were clustered, with the sister group (BBS10, BBS12) identified by the ML tree, and the sister group (BBS6, BBS12) resulting from Bayesian and neighbor-joining trees. The branching position of the Oomycete sequences outside of the cluster of metazoan BBS6 sequences reflected the phylogenetic relations of the respective species, thus suggesting that the Oomycete and Metazoan genes originated from a common ancestor predating radiation of Metazoan groups, including the anciently diverged lineages of Cnidaria and Placozoa. Thus, the topology of the tree excluded that sequences from Oomycetes have been laterally transferred from any of the Metazoan lineages represented in the trees. Furthermore, the association of the Oomycete sequences with the BBS6 subfamily indicated that when the BBS6 common ancestor separated into the Oomycetes and Metazoa lineages, the gene family had already duplicated into three paralogous subfamilies. Finally, the tree topologies confirmed that the CL-BBS gene family originated from a duplication of a CCT8 gene precursor before separation of Unikonts and Chromalveolates.
Figure 2.
Maximum-likelihood (ML) phylogenetic tree of chaperonin-like sequences including canonical CCT subunits and CL-BBS proteins, rooted with the thermosome alpha-subunit sequence from Thermoplasma acidophilum (PDB code 1A6D). TCP1 is synonymous with CCT1. MKKS is synonymous with BBS6. Names of CL-BBS sequences identified in this study from non-vertebrate Metazoa are indicated in red and those from Oomycetes in blue. Sequences representing all major eukaryotic groups are included for CCT8. Values associated with branches indicate levels of support measured by aLTR (black) and are compared at relevant branches with posterior probabilities (green) of a Bayesian tree (Supplementary Figure S1) and with bootstrap values (blue, 1000 replicates) of a neighbor-joining tree based on pairwise distances estimated with the JTT matrix and gamma distributed rates with parameter a = 2.212 (see Methods and Supplementary Figure S2). BBS6 and BBS12 are sister groups in the Bayesian and NJ trees (with low support). Species names are abbreviated as indicated in Table 1. Other species abbreviations are: Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Md, Monodelphis domestica; Oa, Ornithorhynchus anatinus; Xl, Xenopus laevis; Xt, Xenopus tropicalis.
Since the CL-BBS genes evolved at a much higher rate than the canonical CCT sequences, as indicated by the respective branch lengths, their clustering in the phylogenetic trees could be the artifactual result of long-branch attraction [19]. Although some of the models (the CAT-GTR model in Bayesian analysis) used in this analysis are expected to reduce or eliminate this effect [17,18], long-branch attraction remains a potential alternative explanation to the clustering of Oomycete and animal CL-BBS sequences. The long-branch attraction hypothesis, however, is contradicted by the reciprocal closest similarity of the fast evolving vertebrate and Oomycete CL-BBS sequences, which cluster together also in a phenogram (Supplementary figure S3). The highest sequence similarity of non-vertebrate metazoan and Oomycete chaperonin-like sequences to vertebrate CL-BBS sequences despite the high divergence rate of these sequences provides further support to their monophyletic origin.
Signature BBS6 sequence in Oomycetes
To further support the classification of Oomycete sequences we looked in the alignment of CL-BBS and CCT protein sequences for signature motifs unique to BBS6 proteins. We identified BBS6 sequence signatures within two regions conserved across CL-BBS and CCT proteins (Supplementary figure S4). One signature sequence, QK[IV] [IV] x16[DE]R[LIVA], was found within a conserved region of the predicted chaperonin structural Apical Domain, corresponding to the C-terminal ends of two adjacent parallel beta-strands [12,13]. Two other signature positions, not structurally connected, corresponded, respectively, to a Leu and a His amino acid residue uniquely conserved in BBS6 sequences, within a conserved region including parts of the chaperonin C-terminal Intermediate and Equatorial structural domains (Supplementary figure S4).
Functionality of non-vertebrate chaperonin-like BBS sequence
The high rate of divergence of CL-BBS proteins suggests that their evolution was either driven by positive selection, as in functional differentiation, or by neutral differentiation, as could be expected in case of loss of functionality. To establish functionality of the newly identified CL-BBS sequences we evaluated (i) presence of codon-position-specific compositional contrasts in the predicted coding regions, (ii) the ratio between non-synonymous and synonymous evolutionary rates (Ka/Ks ratio), and (iii) presence of corresponding gene transcripts.
We tested all coding sequences newly predicted in Oomycetes for the presence of significant association of nucleotide usages with codon position typical of coding regions (see Supplementary Methods). Surprisingly and despite their sequence similarity, we identified great heterogeneity of codon-position-specific nucleotide usages among Oomycete CL-BBS coding sequences, which often conformed to expectations only within non-significant sequence stretches (Supplementary figure S5). However, we also found non-significant codon-position association of nucleotide usages in human BBS6, in sharp contrast to the high significance of the associations observed instead for the canonical chaperonin gene CCT8 (Supplementary figure S5).
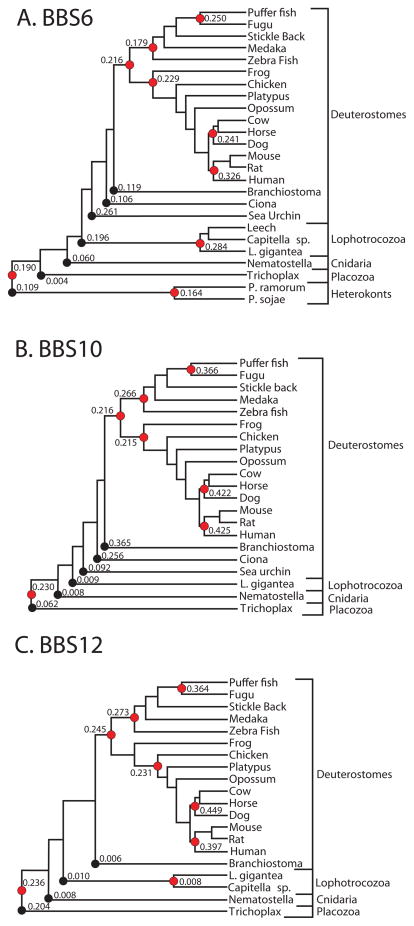
Using the PAML4 [20] software we estimated the overall ratio of non-synonymous and synonymous substitution rates (Ka/Ks ratio) during the evolution of lineage-specific CL-BBS genes within the BBS6, BBS10 and BBS12 gene families, based on the complete tree (including the root-branch) and on sub-trees of the same groups of sequences (excluding the root-branch) (Figure 3). All analyses resulted in highly significant (p≪0.001) reduction of non-synonymous compared to synonymous substitution rates (Ka/Ks≪1.0), indicating that the evolution of CL-BBS proteins within the corresponding lineages was characterized by strong constraining selection, despite their overall fast evolutionary rate.
Figure 3.
Ratios of non-synonymous and synonymous substitution rates (ω=Ka/Ks) along different lineages of chaperonin-like BBS genes, estimated with PAML4 [20]. Black circles identify rooted clusters of “foreground” branches for which ω was estimated in comparison to all other branches (“background”) of the complete tree, using PAML4 branch model 2 and one-rate model M0. Red circles identify unrooted subtrees for which ω was independently determined with model M0.
We identified in public databases ESTs corresponding to many of the non-vertebrate CL-BBS genes here described (Supplementary table S5) suggesting, by expression, functionality of the corresponding proteins. Most relevantly, we found among these ESTs corresponding to the BBS6 genes newly identified in the genomes of Phytophthora and Pythium species.
Structural features of non-vertebrate chaperonin-like BBS proteins
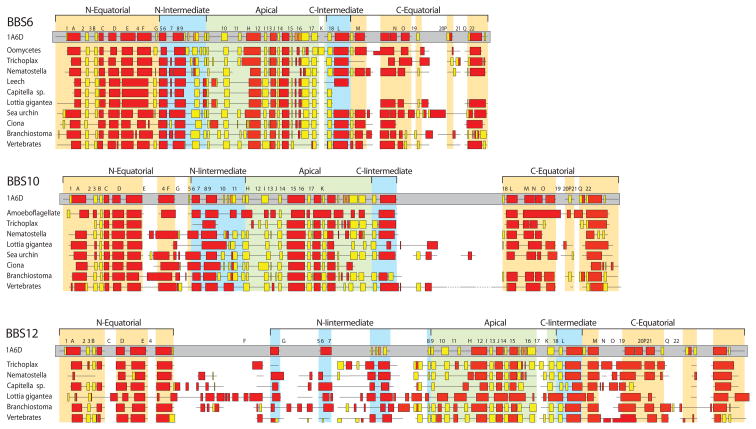
Homology modeling of CL-BBS structures based on the available chaperonin structures is biased by the implicit assumption that chaperonin and CL-BBS proteins share similar core structures. The considerable sequence divergence of CL-BBS proteins from canonical chaperonin proteins makes the assumption of homology modeling problematic. We chose instead to assess the structural features of CL-BBS proteins by predicting their secondary structure elements, reasoning that conservation of secondary structure elements typical of chaperonin structures would strongly indicate that CL-BBS proteins also conserve the tertiary structure of chaperonin proteins. Having previously shown that current prediction methods can successfully identify the secondary structure elements of chaperonin proteins [10], we independently predicted secondary structure elements from CL-BBS sequences of different taxonomic groups, excluding information from known structures and from alignments with chaperonin or with CL-BBS proteins from other groups. We found that, with the exception of the BBS12 sequence from Lottia gigantea, predicted secondary structure elements of non-vertebrate (as well as vertebrate) CL-BBS sequences corresponded in many instances to those of chaperonin proteins (Figure 4). However, we also identified significant differences. In the case of BBS6, most structural elements appeared to be conserved in correspondence to the N-terminal part of the chaperonin Equatorial and Intermediate domains, and in correspondence to the Apical domain. The C-terminal part of the Intermediate and Equatorial domain regions was instead either not recognizable (Leech, Capitella) or perturbed by a variety of insertions and deletions (indels) specific to the different taxonomic groups. A similar pattern was observed in BBS10 proteins, with the addition of non-conserved indels also in the N-terminal Equatorial or Intermediate domains, and of deletions in the Apical domain of the sequence from Ciona. BBS12 sequences were characterized by the greatest occurrence of non-conserved indels affecting the Intermediate and Equatorial domains, and, as previously mentioned, by failed recognition of typical chaperonin structural elements in the sequence from Lottia gigantea. Thus, with few exceptions, structural elements of the chaperonin Apical domain appeared to be remarkably conserved across most CL-BBS sequences, whereas other structural domains, particularly the C-terminal part of the Equatorial domain, showed the greatest amount of perturbations, in the form of missing sequences and of indels of different size, generally not conserved across phyla.
Figure 4.
Secondary structure prediction representations of BBS6, BBS10 and BBS12 proteins from different phylogenetic groups, compared to the secondary structure description and to the typical structural-domain architecture (Equatorial, Intermediate and Apical domains) from the crystal structure of the archaeal chaperonin alpha subunit from Thermoplasma acidophilum (PDB code 1A6D). Alpha-helices are represented as red boxes, beta-strands as yellow boxes, and loops as black lines. Gaps are identified by line breaks. Horizontal half-boxes indicate that the corresponding structure has been predicted only in a subset of the sequences within the group.
Chaperonin proteins are ATPases with well-characterized ADP/ ATP-binding and ATP-hydrolysis motifs, well conserved across eukaryotic CCT protein sequences. In contrast, the ADP/ATP-binding and, in particular, the ATP-hydrolysis motifs, are not as conserved among vertebrate CL-BBS proteins [7,10,12,13]. We compared profiles of amino acid usage (logos) in the ATP binding and hydrolysis motifs of a large collection of CCT proteins to those of non-vertebrate CL-BBS proteins (Supplementary figure S6). In BBS6 and BBS10 proteins we observed within the ADP/ATP-binding motif –[LYFMI]GPx[GAS] xxK[ILM] – substantial conservation of the GP dipeptide, which is shown in chaperonin structures to be in direct contact with ATP and to entail an unusual conformation of the protein backbone (phi/psi angles). In BBS12, the ADP/ATP-binding motif was less conserved, including substantial variability of the crucial GP dipeptide. The ATP-hydrolysis motif – GDGT[TN][TSG] – was less conserved than the ADP/ATP-binding motif. In BBS6 proteins aspartate (D) was conserved, but not glycine (G), which in canonical chaperonin structures also corresponds to unusual protein backbone conformation. In BBS10 proteins, although the ATP-hydrolysis motif was conserved as a consensus, in individual sequences substantial variability was observed at most positions. The ATP-hydrolysis motif was not conserved in BBS12 proteins.
Discussion
The identification of fourteen genes associated with the multisystemic developmental disorder Bardet-Biedl Syndrome (BBS) highlights the broad role of ciliary and other microtubule-based processes in cellular homeostasis and in organism development [21]. The identification of these genes prompted the discovery of an essential ciliary complex, the BBSome [3], and of chaperonin-gene paralogs mostly localized to the basal body and to the centrosome [7,12,13]. Phylogenetic studies indicated that the chaperonin-like BBS (CL-BBS) gene family originated from duplication of a progenitor of the CCT8 chaperonin gene [10], and its identification among vertebrates [7,8,12,13] and other metazoan species [7,14] suggested a metazoan origin. Our discovery of chaperonin-like BBS6 sequences also in Oomycetes and their position within the phylogenetic tree suggests instead that chaperonin-like BBS genes originated and triplicated before separation of the lineages of Opisthokonts (or Unikonts) and Chromalveolates (>2300 Ma ago [22]), hence much earlier than the time of origin of vertebrates (~500 Ma ago [21]) or of Metazoa (~1450 Ma ago [22]). The possibility that the BBS6 gene has been acquired by Oomycetes by Lateral Gene Transfer (LGT) from a different organism cannot however be excluded. LGT events between eukaryotic species are not common and most of the times they involve either transfer to a protist phagotrophic recipient species, or transfer between plant species [23]. However, events of LGT have been recognized to play a significant role in the evolution of plant-parasitism in Oomycetes. These involved transfer of genetic material from fungi (Ascomycetes) [24,25] and through an ancestral photosynthetic plastid derived from an endosymbiont red alga [25,26]. The phylogenetic tree of BBS6 genes (Figure 2) would be consistent with both the hypothesis that the oomycete BBS6 genes originated from a lineage of fungi, and the hypothesis that they originated from a red alga (Rhodophyta). These hypotheses could be tested and verified by identifying the BBS6 donor gene from fungi or red algae and demonstrating that the Oomycete gene cluster with one or the other in the phylogenetic tree. Cilia were present in the common progenitor of Archaeplastida (plants) and are commonly found in lower plants, but they have been secondarily lost in red algae and in many land plants, where genes for proteins with an ancestral ciliary function are still found [27]. Cilia must also have been present in the common ancestor of Fungi, and they are still found in Chytridiomycota, the only group of real fungi known to develop flagellated zoospores. However, although we searched for CL-BBS genes in the available genome of the red alga Cyanidio schyzonmerolae as well as in the genomes of land plants and green algae, including the flagellated unicellular green alga Chlamydomonas reinhardtii (Table 1), we could not identify any CL-BBS gene in these genomes. We also searched for CL-BBS genes in twenty available genomes from Fungi (mostly Ascomycota) (Supplementary table S1), including the genome of Batrachochytrium dendrobatidis, a chytridiomycete with flagellated zoospores, and again we could not identify any CL-BBS gene in any of these genomes. Although lack of a corresponding gene prevents positive verification of the LGT hypothesis, it cannot be excluded that a BBS6 gene was present in the ancestral red alga endosymbiont or in an ancestral fungus, and that it was transferred to the water mold genome before being secondarily lost. The scenario of LGT from a red alga would still imply an ancient origin and very early triplication of the CL-BBS gene family, which would have occurred before separation of the lineages of Archaeplastida (leading to red algae) and Opisthokonts (leading to Metazoa). The hypothesis of LGT from an ancestral fungal species would set a somewhat later origin and triplication of the CL-BBS genes, but pre-dating the time of separation of the lineages of Fungi and Holozoa (including Metazoa) possibly in the Opisthokont lineage. Thus, any of the three hypotheses explaining the origin of the BBS6 gene in Oomycetes (vertical descent or LGT from red algae or from Fungi) imply that the CL-BBS gene family origin and triplication predated the origin of Metazoa, and depict scenarios of gene losses that are consistent with the more recent history of the gene, including loss of all three CL-BBS paralogs in Ecdysozoa, and of BBS12, independently in Echinodermata and in Urochordata (Figure 1).
Conservation of secondary structure elements (Figure 4) indicated that, despite their sequence divergence, CL-BBS proteins from different phylogenetic groups conserve a typical chaperonin “Apical Domain”. The isolated apical domain is sufficient in canonical chaperonin proteins for retaining substrate-binding properties [28], and in BBS proteins for conferring centrosomal localization [7]. The conservation of a chaperonin-like structural apical domain in CL-BBS proteins suggests that CL-BBS proteins bind to their substrates in a similar way than canonical chaperonin proteins. In contrast, the putative ATP-binding “Equatorial Domain” of non-vertebrate CL-BBS proteins is disrupted by proliferation of non-conserved deletions and insertions, and by divergence of the ADP/ATP binding site of BBS12 and of the ATP-hydrolysis sites of BBS6, BBS10 and BBS12, as also previously noted for their vertebrate orthologs [7,10,12,13]. Since most intra-ring and all inter-ring interactions in the canonical chaperonin complex involve the Equatorial Domain [29] and the ATP binding and hydrolysis sites are necessary for the folding activity of the chaperonin complex [30–32], divergence of the Equatorial domain and of the ATP binding/hydrolysis sites suggest that CL-BBS proteins do not assemble in a functional chaperonin-like complex. This conclusion is supported by early reports that CL-BBS proteins are not found associated in a complex [7]. However, more recently it has been reported that CL-BBS proteins associate with selected CCT monomers and with the BBSome component BBS7 in a “BBS/CCT complex” [8,9]. To reconcile the strong experimental evidence of formation of a protein complex with the apparent loss of sequence and structure integrity of the Equatorial domain of CL-BBS proteins, we suggest that CL-BBS and CCT proteins may aggregate in a non-chaperonin complex through their interaction with BBS7 by means of their relatively conserved substrate-binding apical domains, rather than in a hybrid BBS/CCT chaperonin-like conformation. This hypothesis would also be consistent with the observation that CL-BBS and CCT proteins aggregate only in the presence of BBS7 [8,9], suggesting that they are unable to assemble into a multimeric complex stabilized by monomer-monomer interactions as in chaperonins.
CL-BBS proteins are required for BBSome assembly [8,9] and localize to various tubulin-dense structures, including, besides the pericentriolar material of centrosomes and basal bodies [7,33], also the intercellular bridge at mitosis [7] and dendrites of mature neurons [34]. Intriguingly, it has been observed that CCT proteins, besides being essential for the folding of several proteins in their TRiC/CCT complex conformation [6], also bind as individual monomers to microtubule filaments [35] or to the growing ends of actin polymerizing filaments [36]. These observations suggest that CCT monomers and chaperonin-like BBS proteins are also capable of association with microtubules and other filamentous structures in a yet-to-be-characterized manner.
CL-BBS genes have been so far identified only in organisms developing cilia or flagella at some stage of their development. For example, Phytophthora and Pythium develop motile flagellated zoospores from sporangia. However, CL-BBS genes are not found in all organisms that develop cilia or flagella. For example, they are not found in species from Ciliates, Choanoflagellates, or in the flagellated green alga Chlamydomonas or in the flagellated fungus Batrachochytrium. In the case of Ciliates, it is known that specific chaperonin monomers are essential for cilium development [37], suggesting that in this group certain CCT monomers may be the functional equivalent to CL-BBS proteins. If chaperonin-like BBS genes emerged early in eukaryote evolution from a pre-adapted CCT gene, the poor correlation of their distribution with the distribution of ciliary structures in different lineages, might reflect some functional overlap with CCT monomers in affecting cilium development and functionality.
Material and Methods
We searched chaperonin-like BBS gene orthologs in 37 completely sequenced eukaryotic genomes. To these we added at a later stage of the analyses two Oomycete genomes that became available (for a total of five Oomycete genomes, Table 1), and 18 genomes from Fungi, based on results (for a total of 20 Fungus genomes, Supplementary table S1). Query targets were identified using TBLASTN [38] with the method of reciprocal best hit [16], according to the following procedure. Human chaperonin-like BBS (CL-BBS) proteins were used as queries and BLAST hits were collected with a liberal cut-off value (E-value<1.0). Whenever candidate CL-BBS gene homologs were not identified using human CL-BBS proteins as queries, we mined the genomes with CL-BBS proteins from other vertebrate species or, when available, CL-BBS proteins identified with previous searches in non-vertebrate genomes most closely related to the target genome. An extended region around each hit (up to ± 5000 bp) was excised from the genome and the corresponding query protein was used to guide the prediction of the complete structure of the newly-identified gene, based on homology and on intron-exon junction signals, using the gene-prediction software FGENESH+ [39] at the Softberry web-site (linux1.softberry.com). Reverse BLAST analyses were performed using the extended predicted protein sequence as queries against the NCBI non redundant (nr) database.
Multiple sequence alignments were obtained using MUSCLE [40]. Pairwise similarity of CL-BBS and CCT proteins was calculated from the alignment and the corresponding pairwise dissimilarity (1.0–similarity) matrix was used to produce a phenogram using the UPGMA method [41].
Phylogenetic trees were obtained using Maximum-likelihood (ML) and Bayesian probabilistic methods, and by the neighbor-joining distance method [42]. Maximum-likelihood evolutionary trees were produced with PHYML 3.0 [43] with the LG substitution matrix [44], simultaneously estimating tree topology and branch lengths, amino acid equilibrium frequencies, fraction of invariable sites and discrete-gamma distributed substitution rates (8 states). Support for tree branches of the ML tree was obtained with the approximate Likelihood-Ratio Test (aLRT) [45]. The Bayesian tree was generated using PHYLOBAYES 3.2 [46] based on the CAT-GTR model, inferring from sequence data amino acid substitutability matrix coefficients (GTR model) and position-specific equilibrium frequencies of amino acids (CAT model). Support values for the Bayesian tree topology were obtained as branch marginal posterior probabilities calculated from the distribution sampled from two converged MCMC chains of 20,000 cycles sampled every 10 steps after a burn in of 4,000 cycles. Thus, while for the ML method we used a model with generalized amino acid equilibrium frequencies, the Bayesian method was instead based on a highly-parameterized profile mixture-model of position-specific amino acid equilibrium frequencies, expected to be more resistant to long-branch attraction effects [17,18]. Neighbor-joining trees were obtained using MEGA5.1 [47] with a distance matrix based on the JTT substitution model and gamma distributed rates with parameter a=0.5, 1.0, 1.5, 2.0, 2.212 (the maximum likelihood estimate), 2.5, or 3.0, with bootstrap branch supports from 1000 sampling replicates.
Ratios of non-synonymous and synonymous substitution rates (ω=Ka/Ks) were estimated using the program CODEML from the PAML 4.0 package [20,48]. Significance of the estimates was tested with the Likelihood Ratio Test (LRT) [49] comparing the one ratio model M0 (Ka/Ks=x) with the null model Ka/Ks=1.0. Ka/Ks ratios were calculated testing the evolutionary tree of each group of interest independently, and using a branch-specific model where “foreground” branches in turn represented each group within the complete tree. Consensus secondary structure predictions were independently obtained for each of the sequences identified in different taxonomic groups with the secondary structure prediction tool JPRED3 [50] excluding any supporting information from other homologous sequences, i.e., excluding aligned sequences not belonging to the group of interest, and excluding BLAST database searches. Predictions were compared with the secondary structures described for the crystal structure of the Thermoplasma acidophilum thermosome (PDB code 1a6d, chain A), a class 2 archaeal chaperonin.
Supplementary Material
Acknowledgments
This work was supported in part by NIH Research Grant 5R01GM087485.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Blacque OE, Leroux MR. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mykytyn K, Sheffield VC. Establishing a Bardet-Biedl Syndrome. Trends Mol Med. 2004;10:106–109. doi: 10.1016/j.molmed.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 4.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 5.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 6.Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JC, Ou YY, Badano JL, Esmail MA, Leitch CC, et al. MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet-Biedl syndrome, is a novel centrosomal component required for cytokinesis. J Cell Sci. 2005;118:1007–1020. doi: 10.1242/jcs.01676. [DOI] [PubMed] [Google Scholar]
- 8.Seo S, Baye LM, Schulz NP, Beck JS, Zhang Q, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci U S A. 2010;107:1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Yu D, Seo S, Stone EM. Sheffield interaction-mediated and chaperonin-assisted sequential assembly of stable bardet-biedl syndrome protein complex, the BBSome. J Biol Chem. 287:20625–20635. doi: 10.1074/jbc.M112.341487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee K, Conway de Macario E, Macario AJ, Brocchieri L. Chaperonin genes on the rise: new divergent classes and intense duplication in human and other vertebrate genomes. BMC Evol Biol. 2010;10:64. doi: 10.1186/1471-2148-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin H, Nachury MV. The BBSome. Curr Biol. 2009;19:R472–473. doi: 10.1016/j.cub.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- 13.Stoetzel C, Muller J, Laurier V, Davis EE, Zaghloul NA, et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeling PJ. Genomics. Deep questions in the tree of life. Science. 2007;317:1875–1876. doi: 10.1126/science.1149593. [DOI] [PubMed] [Google Scholar]
- 16.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- 17.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lartillot N, Philippe H. Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Philos Trans R Soc Lond B Biol Sci. 2008;363:1463–1472. doi: 10.1098/rstb.2007.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein J. Inferring Phylogenies. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- 20.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 21.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 22.Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards TA, Soanes DM, Jones MD, Vasieva O, Leonard G, et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci U S A. 2011;108:15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 26.Richards TA, Talbot NJ. Plant parasitic oomycetes such as phytophthora species contain genes derived from three eukaryotic lineages. Plant Signal Behav. 2007;2:112–114. doi: 10.4161/psb.2.2.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodges ME, Wickstead B, Gull K, Langdale JA. Conservation of ciliary proteins in plants with no cilia. BMC Plant Biol. 2011;11:185. doi: 10.1186/1471-2229-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spiess C, Miller EJ, McClellan AJ, Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol Cell. 2006;24:25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira JH, Ralston CY, Douglas NR, Meyer D, Knee KM, et al. Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J Biol Chem. 2010;285:27958–27966. doi: 10.1074/jbc.M110.125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, et al. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 32.Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, et al. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell. 2003;113:369–381. doi: 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- 33.Marion V, Stoetzel C, Schlicht D, Messaddeq N, Koch M, et al. Transient ciliogenesis involving Bardet-Biedl syndrome proteins is a fundamental characteristic of adipogenic differentiation. Proc Natl Acad Sci U S A. 2009;106:1820–1825. doi: 10.1073/pnas.0812518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.May-Simera HL, Ross A, Rix S, Forge A, Beales PL, et al. Patterns of expression of Bardet-Biedl syndrome proteins in the mammalian cochlea suggest noncentrosomal functions. J Comp Neurol. 2009;514:174–188. doi: 10.1002/cne.22001. [DOI] [PubMed] [Google Scholar]
- 35.Roobol A, Sahyoun ZP, Carden MJ. Selected subunits of the cytosolic chaperonin associate with microtubules assembled in vitro. J Biol Chem. 1999;274:2408–2415. doi: 10.1074/jbc.274.4.2408. [DOI] [PubMed] [Google Scholar]
- 36.Grantham J, Ruddock LW, Roobol A, Carden MJ. Eukaryotic chaperonin containing T-complex polypeptide 1 interacts filamentousactin and with reduces the initial rate of actin polymerization in vitro. Cell Stress Chaperones. 2002;7:235–242. doi: 10.1379/1466-1268(2002)007<0235:ecctcp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seixas C, Cruto T, Tavares A, Gaertig J, Soares H. CCTalpha and CCTdelta chaperonin subunits are essential and required for cilia assembly and maintenance in Tetrahymena. PLoS One. 2010;5:e10704. doi: 10.1371/journal.pone.0010704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sokal R, Michener C. A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull. 1958;38:1409–1438. [Google Scholar]
- 42.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 43.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 44.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 45.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 46.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 49.Huelsenbeck JP, Crandall KA. Phylogeny estimation and hypothesis testing using maximum likelihood. Annu Rev Ecol Syst. 1997;28:437–466. [Google Scholar]
- 50.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.