Abstract
Many neurons resemble other cells in developing embryos in migrating long distances before they differentiate. However, despite shared basic machinery, neurons differ from other migrating cells. Most dramatically, migrating neurons have a long and dynamic leading process, and may extend an axon from the rear while they migrate. Neurons must coordinate the extension and branching of their leading processes, cell movement with axon specification and extension, switching between actin and microtubule motors, and attachment and recycling of diverse adhesion proteins. New research is needed to fully understand how migration of such morphologically complicated cells is coordinated over space and time.
Nervous systems are the organs through which animals perceive, interpret, and respond to the world around them. They consist of specialized, electrically active cells connected together in networks. Essentially, all nervous systems develop by four main stages: the proliferation of progenitors in an epithelium, the specification of neurons and glia, the growth and guidance of axons and dendrites, and the development and refinement of electrical and chemical synapses. However, some more complex nervous systems, including those of vertebrates, have another stage in which newly specified neurons migrate before they differentiate and form synapses. Some migrations cover long distances—up to thousands of cell diameters—and follow complex routes, changing direction at landmarks along the way (a key to the major migratory routes, terminology, and abbreviations is provided in Box 1 and Fig. 1). Because they migrate, neurons from different proliferative zones, and correspondingly distinct lineages and genetic programs, are able to position close to each other and communicate, potentially increasing efficiency. In addition, different types of neurons arrive at a particular location at different times during development, so circuits are established in a specific order. For these reasons, it is generally thought that neuron migrations facilitate circuit formation and improve nervous system function, although this hypothesis has not been critically tested by the appropriate mutation studies.
Box 1.
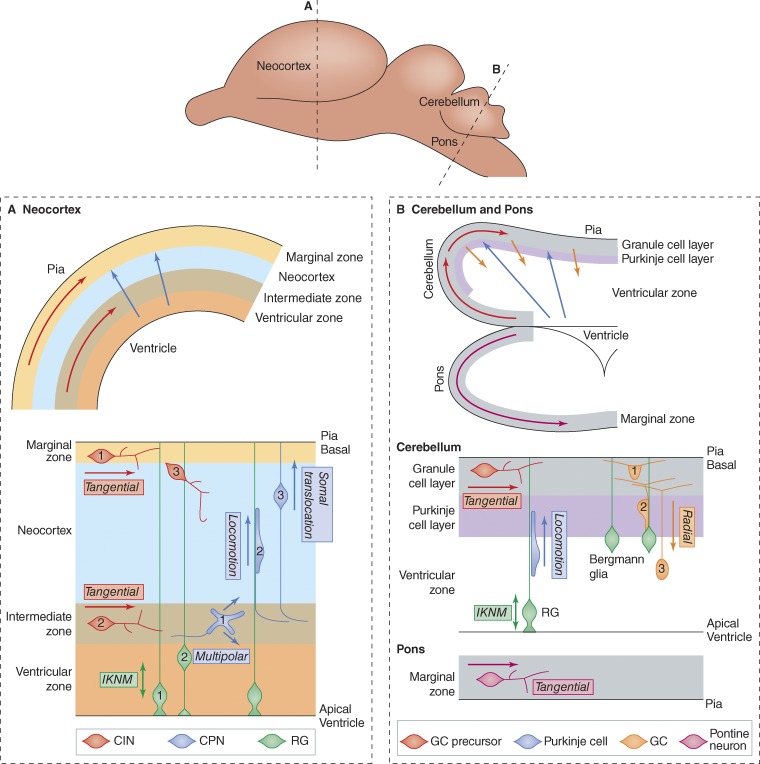
Neuron migration routes. Neuron migration routes in the developing mammalian brain are complex (Marín and Rubenstein, 2003; Ayala et al., 2007). The brain develops from the anterior end of the neural tube: a pseudo-stratified epithelial tube with its apical surface inside and basal surface outside. Neuroepithelial cells, known for much of development as radial glia (RG), span the wall of the neural tube with their apical junctional complexes at the ventricle—the fluid-filled center of the neural tube—and long processes extending to the pia—a basal lamina surrounding the tube. RG cell bodies lie close to the ventricle and undergo inter-kinetic nuclear movement (IKNM) linked to the cell cycle, moving basally (toward the pia) during G1 and apically (toward the ventricle) during G2 (Fig. 1 A, green cells). Asymmetric divisions of RG give rise to post-mitotic neurons or intermediate progenitors at the ventricular surface. In some brain regions, such as the basal forebrain, neurons coalesce into “nuclei” that may be passively displaced away from the ventricle as neurogenesis continues. However, in many other regions, newborn neurons actively migrate away from the ventricle.
Neuron migrations are broadly classified as radial (parallel to RG) or tangential (orthogonal to RG, either around the circumference of the neural tube or along its length). Radial migrations include the glial-guided locomotion phase and glia-independent somal translocation phase of cortical projection neurons (CPNs; Fig. 1 A, blue arrows and cells, steps 2 and 3), locomotion of cerebellar Purkinje cells (Fig. 1 B, blue), and locomotion of post-mitotic granule cells (GCs; Fig. 1 B, orange: note these cells are moving toward the ventricle). Tangential migrations include cortical interneurons in the marginal zone and intermediate zone (CINs; Fig. 1 A, red, steps 1 and 2), cerebellar granule cell precursors (Fig. 1 B, red), and pontine neurons (Fig. 1 B, purple). Some migrations are hard to classify: late-stage CINs switch from tangential to radial migration into the neocortex (Fig. 1 A, red, step 3), and CPNs migrate in random directions in the intermediate zone during their multipolar phase (Fig. 1 A, blue, step 1).
Migrations are termed neurophilic if they follow axons of other neurons, or gliophilic if they follow glia fibers. Gliophilic migrations include the locomotion phase of CPN migration (Fig. 1 A, blue, step 2), Purkinje cell migration (Fig. 1 B, blue, step 1), and part of the granule cell migration (Fig. 1 B, orange, step 2). Some stages of pontine neuron migration may be neurophilic. Some tangential migrations occur in direct contact with the extracellular matrix of the pia.
Figure 1.
Major neuron migrations in the developing rodent brain. Transverse sections (A and B) through the developing rodent brain (top). Top panels show the migration routes and bottom panels show the types of migration. The colors of arrows in the top panels correspond to the colors of cells in the bottom panels. (A) The neocortex. Cortical interneurons (CINs, red) migrate tangentially along the marginal zone (1) and intermediate zone (2) from their origins in the basal forebrain. Later they migrate into the cortical plate (3). Radial glia (RG, green) undergo interkinetic nuclear movement (IKNM), with mitosis apical (1) and S phase basal (2). Cortical projection neurons (CPN, blue) migrate through three phases: multipolar (1), locomotion (2), and somal translocation (3). (B) The cerebellum and pons. Granule cell precursors (red) migrate in the marginal zone, forming a granule cell layer. Purkinje cells (blue) undergo radial locomotion along RG, which also undergo IKNM. Post-mitotic granule cells (GCs, orange) migrate radially inward along Bergmann glia (steps 1, 2, and 3), leaving a bifurcated axon behind. Pontine and other precerebellar neurons (purple) migrate tangentially in the marginal zone of the pons. Note that these migrations do not all occur at the same time.
This short review presents an overview of neuron migration mechanisms for the cell biologist. For brevity, the short-range and long-range extracellular cues that guide neurons are not discussed; many of the same cues that guide neurons also guide axon growth cones and have been reviewed in depth recently (Kolodkin and Tessier-Lavigne, 2011; Vitriol and Zheng, 2012). Collective cell migrations and migrations in the peripheral nervous system are also ignored. Instead, we discuss the cellular machinery used by neurons migrating in the developing central nervous system. We focus on aspects that are peculiar or exaggerated in neurons compared with other cells: the long leading process, the linkage between the centrosome and nucleus, the use of microtubule motors and actomyosin to move the nucleus, and the variety of adhesion proteins and attachment points for traction. Not all neurons are alike, however, and exhibit almost as much variation in their migration as slime molds, keratinocytes, fibroblasts, and other cells commonly used as model systems. In addition to describing these differences, where possible we provide somewhat speculative unifying hypotheses to bring out common themes.
The leading process
Migrating neurons generally have a long leading process, tipped by dynamic filopodia and lamellipodia, which resembles a growth cone on a dendrite or axon. In some neurons, the leading process is branched and dynamic, with different branches growing and collapsing as migration proceeds, whereas in others there is a single, stable leading process that moves forward continuously at the tip. Diverse mechanisms regulate stabilization and guidance of the leading process.
Multiple, branched leading processes as guidance sensors.
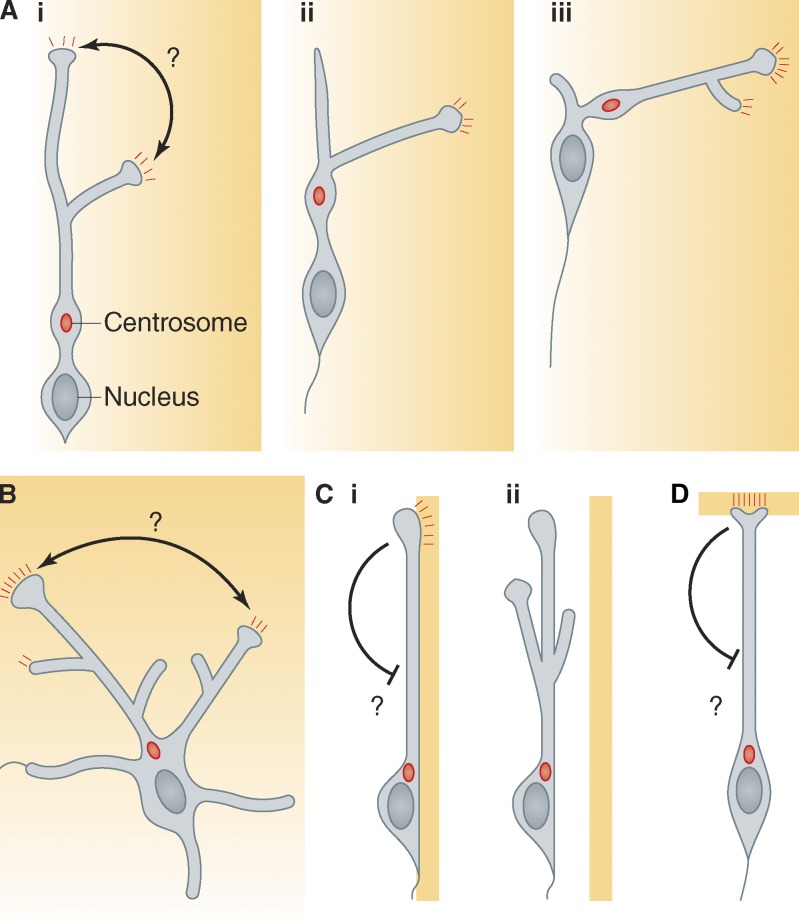
Highly branched, dynamic leading processes are characteristic of several types of neurons that migrate tangentially, including cortical interneurons (CINs), pontine neurons, and neuroblasts in the rostral migratory stream (Bellion et al., 2005; Schaar and McConnell, 2005; Ward et al., 2005; Martini et al., 2009; Watanabe and Murakami, 2009). The branching may facilitate route finding by measuring the concentrations of chemoattractants or repellents across a broad area (Valiente and Marín, 2010). Whereas a single growth cone can only compare the concentrations of attractant or repellant across its width (J.Q. Zheng et al., 1996), a cell may be able to compare attractant and repellant concentrations at widely different locations using multiple growth cones (Fig. 2 A, i). Indeed, the leading process does not turn when the source of attractant changes (Ward et al., 2005; Martini et al., 2009). Rather, branches are selectively stabilized based on proximity to the source of attractant: the branch whose growth cone is nearer the attractant is stabilized while others retract (Fig. 2 A, i). A dilation in front of the nucleus, containing the centrosome and Golgi, translocates to the branch point and then into the dominant process, with the nucleus following behind (Fig. 2 A, ii–iii). Competition between different branches also steers pontine neurons from tangential to radial paths (Watanabe and Murakami, 2009). Thus, selective stabilization of different growth cones determines the direction for moving the centrosome and nucleus. We speculate that increased growth cone stability may lead to more cytoskeletal tension in that process and more “pull” on the centrosome, steering the centrosome into the dominant process.
Figure 2.
Morphology and functions of the leading process. (A) The elaborate leading processes of tangentially migrating neurons are continuously remodeled as migration progresses, with individual branches growing and collapsing. (i) A chemoattractant (concentration gradient indicated by orange shading) stimulates signaling at growth cones (red dashes). A growth cone in a higher concentration of chemoattractant generates a stronger signal and becomes dominant. Presumably, the cell has a mechanism to compare the signals from different growth cones and determine which is stronger, but the mechanism is unknown (black arrow). In this illustration, the right-hand process has more signal and becomes dominant. (ii) A dilation in front of the nucleus, containing the centrosome and Golgi apparatus (red dot), translocates to the branch point. (iii) The nucleus moves to the branch point and the centrosome relocates into the dominant process. The cycle continues in a new direction. (B) Multipolar neurons extend and retract processes in various directions from the cell soma. At any given time one process is dominant, recruits the centrosome, and directs movement. Other processes can take over, presumably based on relative stabilization of growth cones by short-range and long-range signals. It is unclear how the signal strengths at different growth cones are compared (black arrow). (C, i) Radially migrating neurons undergoing locomotion along radial glia have a simple, relatively unbranched leading process. Branching may be suppressed by long-range signaling from receptors at the tip of the leading process (black arrow). (ii) Mutations that interfere with adhesion also destabilize the leading process and induce branching. (D) The leading process in neurons undergoing somal translocation is anchored to the cells or extracellular matrix at the pia. Branching is also suppressed, perhaps by unidentified long-range inhibitory signals (black arrow). Question marks represent unknown mechanisms.
Similar principles may apply to cortical projection neurons (CPNs) in the intermediate zone of the developing neocortex. These cells are multipolar, extending and retracting unstable processes as they thread their way between tangentially aligned axons and radially aligned glial fibers. The cells change direction frequently, one process then another taking the leading role (Nadarajah et al., 2003; Tabata and Nakajima, 2003; Kriegstein and Noctor, 2004; Noctor et al., 2004; Sakakibara et al., 2013). The growth cones on individual processes may detect short-range or long-range signals and be differentially stabilized, like the branched processes of CINs (Fig. 2 B). Cytoskeletal forces may then pull the centrosome to the base of the dominant process, thereby steering the nucleus and selecting the direction for migration.
Single, undivided processes lead radially migrating cells.
When a multipolar CPN nears the top of the intermediate zone, a radially oriented process becomes dominant, and the CPN migrates radially (Sakakibara et al., 2013). The CPN then migrates by locomotion, with the leading process entwined around radial glia (Rakic, 1972). The cells are called bipolar, although there is only one leading process and the trailing process is actually the axon, growing from the rear. The leading process is relatively unbranched and its tip moves forward continuously, without collapsing (Nadarajah et al., 2001). The leading process may help create a passage between the radial glia fiber and surrounding differentiating neurons. The base of the leading process, near the cell body, provides adhesion sites for moving the nucleus. Adhesion is discussed at the end of the review.
The locomotion phase ends when the leading process nears the pia and the cell body reaches a dense layer of immature neurons called the primitive cortical zone (Nadarajah et al., 2001; Sekine et al., 2011). The tip seems to anchor to the pia and the nucleus migrates smoothly up the leading process, penetrating the primitive cortical zone. The velocity is ∼60 µm/h, similar to the speed of individual forward “jumps” made during locomotion. In this form of migration, known as somal translocation, the leading process appears to be a “grappling hook” for hauling in the cell body. Somal translocation is also the main mechanism of CPN movement early in cortical development, when the intermediate zone has not developed, and there are no multipolar or locomotion phases. Instead, the new neuron inherits a long basal process from its radial glia progenitor (Miyata et al., 2001). The pial process is under tension (Miyata and Ogawa, 2007). The newborn neuron down-regulates apical junctions in the ventricular zone and the cell smoothly moves upward by somal translocation.
The role of the leading process therefore changes several times as CPNs journey from the ventricle to the marginal zone. In the intermediate zone there are multiple unstable processes, with the cell following whichever process is dominant at a particular time. Then, a single, stable leading process leads the way up the radial glia, and subsequently provides an attachment site during somal translocation.
Signals that regulate process stability.
The contrast between the multiple, branched, unstable processes of multipolar neurons and the single, relatively undivided, stable process during locomotion suggests that growth tip stability and side branching are coordinately regulated (Valiente and Marín, 2010). Single, unbranched processes are presumably stabilized by adhesion or secreted factors that stimulate actin dynamics and delivery of new membrane, as described for growth cones (Vitriol and Zheng, 2012). These inputs may also suppress branching (Fig. 2 C, i). Indeed, branching is stimulated by mutations that reduce adhesion (Fig. 2 C, ii; Elias et al., 2007). This suggests that adhesive signals from the process tip may inhibit branching along the shaft (Fig. 2 C, i). Similarly, anchoring of the tip of the leading process to the pia during somal translocation may suppress side branches (Fig. 2 D). The suppression mechanism is unknown, but one possibility, as yet untested, is that a local excitation–global inhibition process might coordinate tip stability and side branch suppression in a similar way that migrating amoebae polarize toward a chemoattractant (Xiong et al., 2010).
Chemotactic signals may also stabilize one process at the expense of others. Semaphorin 3A (Sema3A) is important to stabilize the radial process of multipolar CPNs and induce locomotion (Chen et al., 2008). Sema3A is secreted near the top of the cortical plate and forms a gradient. Multipolar CPNs express receptors for Sema3A. Either ablating Sema3A receptors or flooding the system with Sema3A inhibits radial migration, suggesting that Sema3A is a chemoattractant. Another factor is Reelin, which is synthesized near the pia (Tissir and Goffinet, 2003). Processed fragments of Reelin diffuse down to the intermediate zone (Jossin et al., 2007). Blockade of Reelin receptors or of a Reelin-activated GTPase, Rap1A, inhibits radial migration out of the intermediate zone (Jossin and Cooper, 2011). Reelin and Rap1A normally ensure the cell surface expression of N-cadherin, which is needed for radial migration. Quite how N-cadherin stimulates radial migration is not clear, however. Rap1A and N-cadherin are not needed for locomotion along radial glia, suggesting that N-cadherin–dependent adhesion is not required (Franco et al., 2011; Jossin and Cooper, 2011). Exit from the multipolar stage also requires membrane traffic, with roles for dynamin, clathrin, exocyst component Sec8, and early endosome regulator Rab5 (Letinic et al., 2009; Kawauchi et al., 2010; Shieh et al., 2011). Inhibiting Rab5 increases surface N-cadherin, which increases neuron–glia adhesion in vitro but inhibits radial migration in vivo (Kawauchi et al., 2010). On the other hand, strong knockdown of N-cadherin also inhibits radial migration (Kawauchi et al., 2010; Jossin and Cooper, 2011). It appears that too much or too little surface N-cadherin is detrimental.
Cerebellar granule neurons (CGNs) have a relatively unbranched leading process during their radial migration along Bergmann glia in vivo. The migration requires a chemoattractant, brain-derived neurotrophic factor (BDNF; Zhou et al., 2007). The BDNF gradient in vivo is very shallow: the concentration of BDNF at the front of a migrating CGN is only 6% more than at the back. Remarkably, CGNs cultured in vitro in a similarly shallow BDNF gradient show remarkable directional persistence, even though Bergmann glia are absent. The mechanisms are complex and involve positive feedback loops.
The first positive feedback loop is that external BDNF stimulates BDNF exocytosis by the migrating cells (Zhou et al., 2007). Mutant CGNs that cannot make BDNF cannot orient in an external BDNF gradient. Presumably, more BDNF is secreted on the side of the cell nearer the external BDNF source, sharpening the gradient. Another positive feedback loop is provided by the internalization of active BDNF receptor (TrkB) into signaling endosomes in the leading process. Internalization requires the endocytic adaptor Numb, which binds to and colocalizes with active TrkB (Zhou et al., 2011). Active TrkB stimulates atypical PKC (aPKC) to autophosphorylate and phosphorylate Numb. Numb phosphorylation further increases Numb binding to TrkB and sharpens the gradient of endosomal TrkB and Numb across the length of the leading process. Through these concatenated positive feedback loops, a shallow gradient of BDNF across the length of the cell can be converted to a robust difference in TrkB–Numb–aPKC activity and provide a compass for the cell to follow. However, positive feedback loops are unstable. Presumably slow negative feedback loops are required for directional persistence, but these negative loops have not been identified.
Intracellular regulation of process stability and branching.
Extracellular signals that regulate process stability are interpreted by intracellular machinery. Cell autonomous regulators of leading process stability and branching include the proneural basic helix-loop-helix gene, neurogenin2 (Ngn2; Hand et al., 2005). Ngn2 is induced soon after the last progenitor division, and stimulates a cascade of transcription events that lead ultimately to neuronal differentiation and the induction and repression of various migration genes (Ge et al., 2006). Mutation of a phosphorylation site in Ngn2, Y241, prevents migration but not differentiation (Hand et al., 2005). A key Ngn2 target gene encodes a small GTPase, Rnd2, which opposes RhoA (Heng et al., 2008). If Rnd2 is not induced, all phases of cell migration from the ventricle to the intermediate zone (multipolar and radial) are delayed. Cells lacking Rnd2 remain multipolar for an increased time, and when they start migrating radially they have a branched leading process. Somewhat paradoxically, a transcription inhibitor, RP58, is needed for the multipolar-to-radial transition, apparently by inhibiting Ngn2 and down-regulating Rnd2 (Ohtaka-Maruyama et al., 2013). Therefore, the correct ratio of Rnd2 and RhoA may promote the stabilization of a single leading process.
Cytoskeletal and membrane proteins also regulate branching and migration. Slit-Robo GAP2 (srGAP2) decreases tip stability, stimulates branching, and slows migration (Guerrier et al., 2009). Although srGAP2 regulates Rac1, it stimulates branching through its F-BAR domain, which interacts with membranes. srGAP2 expression is dynamically regulated during neuron migration, and srGAP2 is one of a small number of genes that has undergone duplication after humans diverged from other primates (Charrier et al., 2012). The human-specific paralogues act as partial dominant-negatives, stabilizing the leading process and sustaining radial migration. Regulation of srGAP2 expression or activity during migration may be important to adapt neuron migrations to increasing cortical thickness during evolution.
Most mutations that inhibit exit from the intermediate zone simply prolong multipolar migration (LoTurco and Bai, 2006); however, removing or inhibiting the actin regulatory protein lammellipodin (Lpd) causes a remarkable phenotype. The cells stabilize a single process sooner than normal (at the bottom of the intermediate zone), and the stable process is oriented perpendicular to the radial glia, parallel to the axons that are abundant in the intermediate zone (Pinheiro et al., 2011). The mutant cells bind and align with axon bundles in vitro and move at the same speed in vivo as they would normally migrate along radial glia. Absence of Lpd causes an increase in monomeric actin and thereby inhibits serum response factor (SRF), a transcription factor. Inhibition of SRF causes the same mis-oriented phenotype as absence of Lpd (Pinheiro et al., 2011). The results suggest that actin polymerization and SRF activity are required for radial orientation, and the default is to migrate along axons. Because SRF regulates the transcription of many genes and micro-RNAs, it is not clear which specific SRF target genes are involved. As in normal radial migration, the leading process may be stabilized by adhesion, but the adhesion has switched from glia to axons.
Orienting migration: The microtubule cytoskeleton and the centrosome
Many cultured cells, and most neurons, migrate with the centrosome/microtubule-organizing center, Golgi apparatus, and recycling endosomes in front of the nucleus (Tsai and Gleeson, 2005). The centrosome may be “pulled” to the front by microtubule-binding proteins such as Dcx and IQGAP at the leading edge (Schaar et al., 2004; Kholmanskikh et al., 2006). This orientation allows microtubule-directed membrane traffic to supply new membrane and protein complexes to the front of the cell for forward movement (Li and Gundersen, 2008). Indeed, the centrosome seems to regulate nuclear translocation when CGNs and CPNs migrate along glia and CINs migrate ex vivo. Nuclear movement is intermittent or “saltatory” (Nadarajah et al., 2001). The centrosome moves forward into a dilation in the proximal part of the leading process, then the nucleus catches up, in a “two-stroke” cycle (Rivas and Hatten, 1995; Solecki et al., 2004; Bellion et al., 2005; Schaar and McConnell, 2005). Microtubules extend from near the centrosome to surround the nucleus in a “cage” in CGNs (Rivas and Hatten, 1995; Solecki et al., 2004) or a “fork” in CPNs (Xie et al., 2003). Apical polarity complex proteins such as Par6 localize to the centrosome, and knockdown or overexpression of Par6 perturbs the microtubule cage and inhibits CGN migration in vitro (Solecki et al., 2004). Microtubules connect to the nucleus by SUN and Nesprin proteins in the outer nuclear envelope, which are required for interkinetic nuclear movement (IKNM) of radial glia and locomotion of CPNs (Zhang et al., 2009).
On some occasions, the nucleus moves with the centrosome behind it, for example, when CGNs reverse their usual direction of migration (Umeshima et al., 2007). CPNs also sometimes overtake their centrosomes during locomotion (Sakakibara et al., 2013). The centrosome also lags behind the nucleus when a CPN precursor first divides from its radial glia sister and moves away from the ventricle (Ochiai et al., 2007). This motion is similar to the basal movement of the nucleus of radial glia progenitors during IKNM, discussed in more detail in the next section. Overall, migration with the centrosome behind the nucleus seems to be the exception rather than the rule.
In addition to orienting migration, the centrosome also dictates the site of axon emergence (de Anda et al., 2005). However, many neurons, including CPNs and CGNs, extend their axons from the rear of the cell during migration (Zmuda and Rivas, 1998; Hatanaka and Murakami, 2002). How does the centrosome regulate axon emergence as well as migration? Careful observation shows that multipolar neurons first enter the lower intermediate zone with their centrosome toward the pia (de Anda et al., 2010). The centrosome then rotates, aligning with the dominant process as the cells move randomly in the intermediate zone (Sakakibara et al., 2013). After some time, a lateral process with the centrosome at its base extends dramatically to form an axon. Manipulations that change the centrosome position alter the axon position (de Anda et al., 2010). After the axon emerges, the centrosome reorients again to the base of a pia-directed process that becomes dominant. The neuron begins migrating radially, other side processes are lost, and the axon grows from the rear (Sakakibara et al., 2013). In vitro studies show that axons require the centrosome for specification but not for extension (Stiess et al., 2010). The centrosome thus appears to designate the dominant process, whether it is the incipient axon or the leading process for migration (Sakakibara et al., 2013).
Moving the nucleus: Microtubule and actin motors
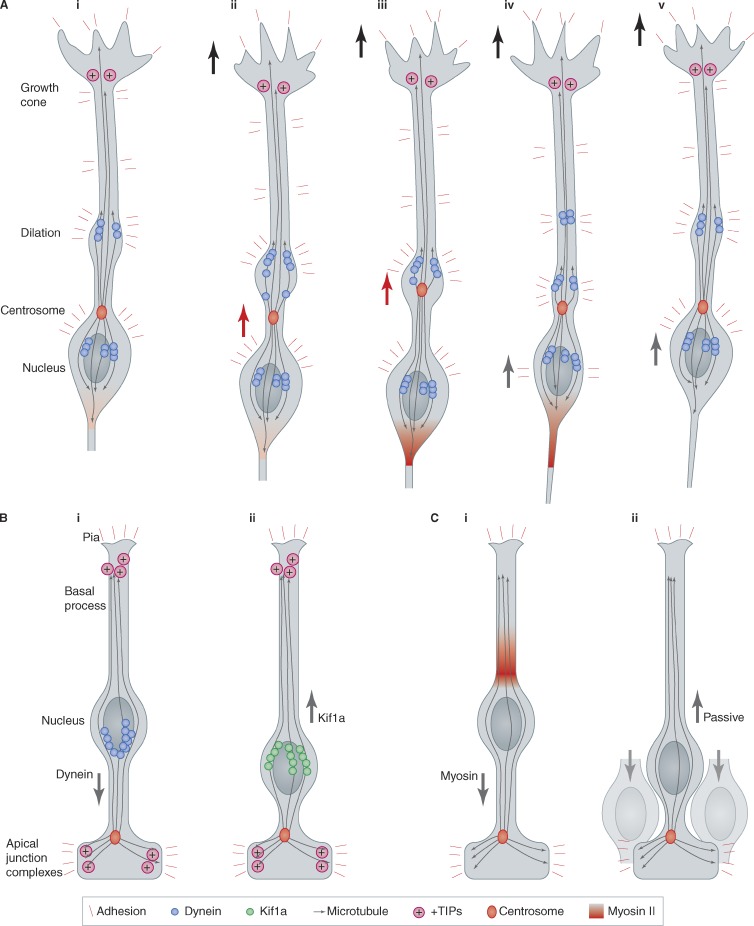
Like other cells, neurons migrate using forces generated by microtubule and actin motors, but their relative importance and sites of action are highly variable (Fig. 3). Microtubule motors are paramount during IKNM in radial glia in the rodent cortex (Fig. 3 B; Tsai et al., 2005, 2010; Xie et al., 2007). Microtubules emanate from the centrosome near the apical pole, and extend past the nucleus toward the basal end feet. Dynein and a kinesin, Kif1a, localize to the nuclear envelope. Consistent with microtubule orientation, minus end–directed dynein is required for apical movement and plus end–directed Kif1a for basal movement (Tsai et al., 2010). Presumably, in order for Kif1a to pull effectively, the microtubules must be anchored near their plus ends. The plus end anchor has not been identified, but +TIPs (plus end tubule-interacting proteins), dynein, or IQGAP in the basal process may perform such a function (Kholmanskikh et al., 2006). IKNM is coordinated with the cell cycle, so there must be a switch in motor use. In this regard, the microtubule-associated protein Tpx2 is required specifically for apical movement (Kosodo et al., 2011). Tpx2 is detected on apical microtubules during apical nuclear movement but is down-regulated during basal nuclear movement. Cell cycle regulation of Tpx2 may coordinate IKNM with the cell cycle.
Figure 3.
Adhesion points, the centrosome, microtubules, and motors. (A) Composite of events described for somal movement during migration of various neuron types. (i–v) Sequential stages in migration. The growth cone moves forward, stimulated by short-range adhesive interactions or long-range chemotactic signals. The centrosome and nucleus move forward in a “two stroke” cycle, with dynein pulling the centrosome into a dilated region where the leading process is attached to the substrate, while myosin squeezes the nucleus forward from behind. Note the need to recycle adhesion proteins from the rear of the soma into the leading process. (B) Interkinetic nuclear movement (IKNM) in rodent cortex. (i) Apical movement during G2 is driven by dynein. (ii) Basal movement during G1 is driven by Kif1a. (C) IKNM in zebrafish neuroepithelia. (i) Apical movement is driven by myosin. (ii) Passive basal movement results from apical movement of surrounding cells. +TIPs, plus-end tubule-binding proteins. See text for references and details.
Microtubule motors are also important for linking the centrosome and nucleus during the “two-stroke” saltatory movement of CGNs, CPNs, and CINs (Fig. 3 A). Microtubules in the leading process and trailing axon are oriented with their minus ends toward the centrosome (Tsai et al., 2007). Dynein, a minus end–directed motor, is concentrated at the nucleus as well as in the dilation near the base of the leading process. The dilation contains mitochondria, rough ER, and part or all of the Golgi and may be a major point of attachment to generate traction forces (Bellion et al., 2005). Dynein in the dilation may “pull” the centrosome forward, while dynein at the nucleus pulls the nucleus toward the centrosome (Tsai et al., 2007). Indeed, knockdown of dynein or associated proteins Lis1 and Ndel1 loosens the linkage between the centrosome and nucleus and inhibits radial migration of CPNs (Shu et al., 2004; Tanaka et al., 2004).
Although dynein and kinesin drive IKNM in rodent cortex, actin and myosin are needed for IKNM in zebrafish retina or hindbrain (Fig. 3 C; Norden et al., 2009). Apical movement is continuous and rapid (∼40 µm/h), consistent with an active process. Actin and myosin are recruited to the basal side of the nucleus a few minutes before it starts moving apically, as though actomyosin is pushing from behind (Leung et al., 2011). In contrast, movement in the basal direction is slow, discontinuous, and interrupted by reversals of direction (Leung et al., 2011), and actin and myosin are on both sides of the nucleus (Norden et al., 2009). It appears that basal movement is stochastic and passive, and is caused by pressure from the surrounding cells as they move apically (Leung et al., 2011). The same passive mechanism may explain the slow rate of basal IKNM in mouse cortex (Kosodo et al., 2011), although an alternative explanation is that Kif1a is a slow motor compared with dynein (Tsai et al., 2010). The relative importance of microtubule motors for IKNM in rodents and of actomyosin in zebrafish may be related to the respective cell shapes (Lee and Norden, 2013). Actomyosin might be effective for moving the nucleus in the relatively squat cells of zebrafish but not in the skinny cells of mammals, where microtubule motors may be more effective.
Actin and myosin are also needed for nuclear movement in migrating CGNs, CINs, and CPNs in vivo and in vitro (Vallee et al., 2009). However, a major unresolved question is whether myosin pushes the nucleus from behind or pulls it from in front, and the answer may vary according the neuron type and the conditions of the experiment. For CPNs migrating in neocortical slices and CINs migrating in slices or in vitro, myosin seems to act at the rear. Cells in the act of nuclear translocation have condensed actin, phosphorylated myosin light chains, and membrane blebs at their rear (Bellion et al., 2005; Schaar and McConnell, 2005; Tsai et al., 2007; Martini and Valdeolmillos, 2010). This suggests that myosin squeezes the sides of the cell behind the nucleus, pushing the nucleus forward like toothpaste from a tube (Fig. 3 A). In contrast, CGNs migrate using actomyosin contraction in front of the nucleus. The pulling point is not clear: when CGNs migrate on glia fibers in vitro, the proximal part of the leading process, ahead of the nucleus, contains active myosin, and actin flows forward toward the dilation (Solecki et al., 2009). This is consistent with myosin pulling actin filaments toward the dilation. However, when CGNs migrate on polylysine/laminin, pulling forces come from the process tip (He et al., 2010). Severing or damaging the leading tip, or inhibiting myosin, actin, or Rho kinase anywhere along the leading process, stops nuclear movement. Actin flows toward the tip, which is where most phosphorylated myosin light chains are localized. This suggests that adhesions at or near the tip of the leading process provide attachments for actin filaments that create tension on the cell body, as in fibroblasts (Munevar et al., 2001) or axon growth cones (Bray, 1979; Lamoureux et al., 1989). It is not clear whether the differences in pulling points are due to the different geometry—a “one-dimensional” glia fiber or a two-dimensional coverslip—or different substrates (glia versus polylysine/laminin), but the evidence favors a pulling model for CGNs, versus a pushing model for CPNs and CINs.
Adhesion sites and membrane traffic
Neurons migrate along a variety of substrates, including radial glia, other neurons, axons, and ECM. In many cases, the sites of attachment and adhesion molecules are still poorly understood despite years of effort (Solecki, 2012). Electron microscopy has revealed apparent junctional complexes, resembling the apical junctional complexes in an epithelium, between migrating Purkinje cells and radial glia in vivo (Yuasa et al., 1996) and between CGNs and Bergmann glia ex vivo (Gregory et al., 1988). Unlike the classical focal adhesions of cultured fibroblasts, however, it is difficult to discern cytoskeletal connections, so it is not clear how traction forces are generated.
Two adhesion molecules, astrotactin and JAM-C, are required for CGN migration along Bergmann glia in vitro and in vivo (Fishell and Hatten, 1991; C. Zheng et al., 1996; Adams et al., 2002; Famulski et al., 2010). Both proteins are regulated so they are available when CGNs reach the Bergmann glia. Astrotactin is induced transcriptionally (C. Zheng et al., 1996), while JAM-C is up-regulated on the cell surface by Pard3-dependent exocytosis (Famulski et al., 2010). Before migration, Pard3 levels are low due to ubiquitylation by the Siah E3 ubiquitin ligase and proteasomal degradation. During migration, Siah activity decreases, Pard3 levels increase, and JAM-C traffics to the surface. Because Pard3 forms an apical polarity complex with Par6 in polarized epithelial cells, and Par6 regulates polarity of migrating granule cells (Solecki et al., 2004), Pard3 might also be important for orienting granule cell migration.
Further evidence that membrane traffic is important for neuron migration comes from electron and fluorescence microscopy. Endocytic structures have been detected near cell–cell contacts between CGNs and Bergmann glia, Purkinje cells, and radial glia, and between chain-migrating adult neural progenitors (Yuasa et al., 1996; Doetsch et al., 1997; Gregory et al., 1988). Integrins, endocytic adaptor Dab2, clathrin, and endocytic cargo transferrin colocalize at potential endocytic sites near the leading tip and proximal dilation when CINs migrate in vitro (Shieh et al., 2011). CPN radial migration requires a type I plasma membrane protein KIAA0319, which associates with endocytic adaptor AP2 (Paracchini et al., 2006). Dominant-negative dynamin and clathrin mutants inhibit radial migration in vivo (Shieh et al., 2011). Dynamin is also needed for somal translocation of serotonergic neurons in the ventral brain stem to form the Raphe nuclei (Hawthorne et al., 2010). Live imaging of CGNs expressing fluorescent astrotactin shows adhesion structures forming in the proximal part of the leading process and disappearing under the soma (Wilson et al., 2010). JAM-C also localizes to punctate structures in the proximal leading process and larger junctions under the soma (Famulski et al., 2010). F-actin is enriched near the forming junctions, and also at the rear of the soma, where disassembly occurs. The results support the idea that adhesion proteins actively recycle and attach to actin to provide traction forces for cell movement.
Adhesion of migrating projection neurons to radial glia requires connexins, which are better known for forming gap junctions (Dere and Zlomuzica, 2012). Connexins assemble into rings in the membrane called hemi-channels, and complete gap junctions form when hemi-channels on different cells adhere to each other. Cx43 and Cx26 are required for radial migration of CPNs, and rescue experiments show that Cx43 adhesive but not channel functions are required (Elias et al., 2007). Cx43 expression allows homotypic adhesion of neurons to glia in vitro. Cx43 localizes to puncta between the radial glia and tip of the leading process in vivo, while Cx26 localizes to puncta between the radial glia and proximal leading process (Elias et al., 2007). Focal adhesion kinase (FAK) regulates traffic of Cx26 hemi-channels to the cell surface, and FAK inhibition causes similar phenotypes to Cx26 inhibition (Valiente et al., 2011). FAK binds Cx26 and the actin-regulatory protein paxillin. Mutation of the paxillin-binding sites on FAK prevents it from recruiting Cx26 to the membrane, reducing adhesion (Valiente et al., 2011). It is not clear whether Cx26 also interacts with FAK and paxillin to exert traction forces on the actin cytoskeleton. Cx43 may also bind the cytoskeleton, but it would presumably bind via its C-terminal cytoplasmic tail and there is disagreement whether the cytoplasmic tail is required for migration (Elias et al., 2007; Cina et al., 2009). Cx43 is also needed for IKNM, multipolar migration, and neuron layering in the cerebellum, but it is unknown whether adhesion or channel functions are involved (Wiencken-Barger et al., 2007; Liu et al., 2010, 2012).
When CPNs undergo somal translocation, the tip appears to be stuck near the pia. This adhesion has been variously reported to require N-cadherin or α5β1 integrin (Franco et al., 2011; Sekine et al., 2012). Both groups report that stabilization of the leading process in the marginal zone requires a previously identified signaling mechanism involving Reelin and Rap1A (Ballif et al., 2004). However, Franco et al. (2011) report that N-cadherin is also required and point out that adherens junction–like structures (which typically contain cadherins) are detected by electron microscopy, apparently linking radial glia end feet to neuronal processes. In contrast, Sekine et al. (2012) detect activated integrin α5β1 and its ligand, fibronectin, in the marginal zone. They provide evidence that Reelin-dependent Rap1A signaling activates integrin α5β1, suggesting an inside-out mechanism to secure the tip of the leading process and provide anchorage for somal translocation.
Moving forward
Despite the varied morphologies and dynamics of different types of migrating neurons, the underlying mechanisms are likely very similar. While the leading process can be dynamic or stable, branched or simple, myosin or microtubule motors may be dominant, and the migration substrate may be extracellular matrix, neurons, or glia, the intracellular machinery for migration is apparently much the same in neurons as in most other cells. However, additional research will be required to understand how a particular process becomes dominant, how process branching is regulated, how the growth tip communicates with the centrosome, how motors are switched at different stages of migration cycles, and how adhesions are regulated and recycled.
Acknowledgments
I thank Sergi Simó, Anjali Teckchandani, and Yves Jossin for comments on the manuscript.
The author is supported by grant no. NS080194 from the National Institutes of Health. Illustrations were provided by Neil Smith, www.neilsmithillustration.co.uk.
Footnotes
Abbreviations used in this paper:
- BDNF
- brain-derived neurotrophic factor
- CGN
- cerebellar granule neuron
- CIN
- cortical interneuron
- CPN
- cortical projection neuron
- IKNM
- interkinetic nuclear movement
References
- Adams N.C., Tomoda T., Cooper M., Dietz G., Hatten M.E. 2002. Mice that lack astrotactin have slowed neuronal migration. Development. 129:965–972 [DOI] [PubMed] [Google Scholar]
- Ayala R., Shu T., Tsai L.H. 2007. Trekking across the brain: the journey of neuronal migration. Cell. 128:29–43 10.1016/j.cell.2006.12.021 [DOI] [PubMed] [Google Scholar]
- Ballif B.A., Arnaud L., Arthur W.T., Guris D., Imamoto A., Cooper J.A. 2004. Activation of a Dab1/CrkL/C3G/Rap1 pathway in Reelin-stimulated neurons. Curr. Biol. 14:606–610 10.1016/j.cub.2004.03.038 [DOI] [PubMed] [Google Scholar]
- Bellion A., Baudoin J.P., Alvarez C., Bornens M., Métin C. 2005. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 25:5691–5699 10.1523/JNEUROSCI.1030-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D. 1979. Mechanical tension produced by nerve cells in tissue culture. J. Cell Sci. 37:391–410 [DOI] [PubMed] [Google Scholar]
- Charrier C., Joshi K., Coutinho-Budd J., Kim J.E., Lambert N., de Marchena J., Jin W.L., Vanderhaeghen P., Ghosh A., Sassa T., Polleux F. 2012. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 149:923–935 10.1016/j.cell.2012.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Sima J., Jin M., Wang K.Y., Xue X.J., Zheng W., Ding Y.Q., Yuan X.B. 2008. Semaphorin-3A guides radial migration of cortical neurons during development. Nat. Neurosci. 11:36–44 10.1038/nn2018 [DOI] [PubMed] [Google Scholar]
- Cina C., Maass K., Theis M., Willecke K., Bechberger J.F., Naus C.C. 2009. Involvement of the cytoplasmic C-terminal domain of connexin43 in neuronal migration. J. Neurosci. 29:2009–2021 10.1523/JNEUROSCI.5025-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda F.C., Pollarolo G., Da Silva J.S., Camoletto P.G., Feiguin F., Dotti C.G. 2005. Centrosome localization determines neuronal polarity. Nature. 436:704–708 10.1038/nature03811 [DOI] [PubMed] [Google Scholar]
- de Anda F.C., Meletis K., Ge X., Rei D., Tsai L.H. 2010. Centrosome motility is essential for initial axon formation in the neocortex. J. Neurosci. 30:10391–10406 10.1523/JNEUROSCI.0381-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Zlomuzica A. 2012. The role of gap junctions in the brain in health and disease. Neurosci. Biobehav. Rev. 36:206–217 10.1016/j.neubiorev.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Doetsch F., García-Verdugo J.M., Alvarez-Buylla A. 1997. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J. Neurosci. 17:5046–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias L.A., Wang D.D., Kriegstein A.R. 2007. Gap junction adhesion is necessary for radial migration in the neocortex. Nature. 448:901–907 10.1038/nature06063 [DOI] [PubMed] [Google Scholar]
- Famulski J.K., Trivedi N., Howell D., Yang Y., Tong Y., Gilbertson R., Solecki D.J. 2010. Siah regulation of Pard3A controls neuronal cell adhesion during germinal zone exit. Science. 330:1834–1838 10.1126/science.1198480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G., Hatten M.E. 1991. Astrotactin provides a receptor system for CNS neuronal migration. Development. 113:755–765 [DOI] [PubMed] [Google Scholar]
- Franco S.J., Martinez-Garay I., Gil-Sanz C., Harkins-Perry S.R., Müller U. 2011. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 69:482–497 10.1016/j.neuron.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W., He F., Kim K.J., Blanchi B., Coskun V., Nguyen L., Wu X., Zhao J., Heng J.I., Martinowich K., et al. 2006. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. USA. 103:1319–1324 10.1073/pnas.0510419103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory W.A., Edmondson J.C., Hatten M.E., Mason C.A. 1988. Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro. J. Neurosci. 8:1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N.V., Chen K., Jin W.L., Frost A., Polleux F. 2009. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 138:990–1004 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R., Bortone D., Mattar P., Nguyen L., Heng J.I., Guerrier S., Boutt E., Peters E., Barnes A.P., Parras C., et al. 2005. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 48:45–62 10.1016/j.neuron.2005.08.032 [DOI] [PubMed] [Google Scholar]
- Hatanaka Y., Murakami F. 2002. In vitro analysis of the origin, migratory behavior, and maturation of cortical pyramidal cells. J. Comp. Neurol. 454:1–14 10.1002/cne.10421 [DOI] [PubMed] [Google Scholar]
- Hawthorne A.L., Wylie C.J., Landmesser L.T., Deneris E.S., Silver J. 2010. Serotonergic neurons migrate radially through the neuroepithelium by dynamin-mediated somal translocation. J. Neurosci. 30:420–430 10.1523/JNEUROSCI.2333-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Zhang Z.H., Guan C.B., Xia D., Yuan X.B. 2010. Leading tip drives soma translocation via forward F-actin flow during neuronal migration. J. Neurosci. 30:10885–10898 10.1523/JNEUROSCI.0240-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J.I., Nguyen L., Castro D.S., Zimmer C., Wildner H., Armant O., Skowronska-Krawczyk D., Bedogni F., Matter J.M., Hevner R., Guillemot F. 2008. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 455:114–118 10.1038/nature07198 [DOI] [PubMed] [Google Scholar]
- Jossin Y., Cooper J.A. 2011. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat. Neurosci. 14:697–703 10.1038/nn.2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y., Gui L., Goffinet A.M. 2007. Processing of Reelin by embryonic neurons is important for function in tissue but not in dissociated cultured neurons. J. Neurosci. 27:4243–4252 10.1523/JNEUROSCI.0023-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T., Sekine K., Shikanai M., Chihama K., Tomita K., Kubo K., Nakajima K., Nabeshima Y., Hoshino M. 2010. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron. 67:588–602 10.1016/j.neuron.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Kholmanskikh S.S., Koeller H.B., Wynshaw-Boris A., Gomez T., Letourneau P.C., Ross M.E. 2006. Calcium-dependent interaction of Lis1 with IQGAP1 and Cdc42 promotes neuronal motility. Nat. Neurosci. 9:50–57 10.1038/nn1619 [DOI] [PubMed] [Google Scholar]
- Kolodkin A.L., Tessier-Lavigne M. 2011. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb. Perspect. Biol. 3:a001727 10.1101/cshperspect.a001727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y., Suetsugu T., Suda M., Mimori-Kiyosue Y., Toida K., Baba S.A., Kimura A., Matsuzaki F. 2011. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. EMBO J. 30:1690–1704 10.1038/emboj.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A.R., Noctor S.C. 2004. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 27:392–399 10.1016/j.tins.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Lamoureux P., Buxbaum R.E., Heidemann S.R. 1989. Direct evidence that growth cones pull. Nature. 340:159–162 10.1038/340159a0 [DOI] [PubMed] [Google Scholar]
- Lee H.O., Norden C. 2013. Mechanisms controlling arrangements and movements of nuclei in pseudostratified epithelia. Trends Cell Biol. 23:141–150 10.1016/j.tcb.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Letinic K., Sebastian R., Toomre D., Rakic P. 2009. Exocyst is involved in polarized cell migration and cerebral cortical development. Proc. Natl. Acad. Sci. USA. 106:11342–11347 10.1073/pnas.0904244106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung L., Klopper A.V., Grill S.W., Harris W.A., Norden C. 2011. Apical migration of nuclei during G2 is a prerequisite for all nuclear motion in zebrafish neuroepithelia. Development. 138:5003–5013 10.1242/dev.071522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Gundersen G.G. 2008. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 9:860–873 10.1038/nrm2522 [DOI] [PubMed] [Google Scholar]
- Liu X., Hashimoto-Torii K., Torii M., Ding C., Rakic P. 2010. Gap junctions/hemichannels modulate interkinetic nuclear migration in the forebrain precursors. J. Neurosci. 30:4197–4209 10.1523/JNEUROSCI.4187-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sun L., Torii M., Rakic P. 2012. Connexin 43 controls the multipolar phase of neuronal migration to the cerebral cortex. Proc. Natl. Acad. Sci. USA. 109:8280–8285 10.1073/pnas.1205880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoTurco J.J., Bai J. 2006. The multipolar stage and disruptions in neuronal migration. Trends Neurosci. 29:407–413 10.1016/j.tins.2006.05.006 [DOI] [PubMed] [Google Scholar]
- Marín O., Rubenstein J.L. 2003. Cell migration in the forebrain. Annu. Rev. Neurosci. 26:441–483 10.1146/annurev.neuro.26.041002.131058 [DOI] [PubMed] [Google Scholar]
- Martini F.J., Valdeolmillos M. 2010. Actomyosin contraction at the cell rear drives nuclear translocation in migrating cortical interneurons. J. Neurosci. 30:8660–8670 10.1523/JNEUROSCI.1962-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini F.J., Valiente M., López Bendito G., Szabó G., Moya F., Valdeolmillos M., Marín O. 2009. Biased selection of leading process branches mediates chemotaxis during tangential neuronal migration. Development. 136:41–50 10.1242/dev.025502 [DOI] [PubMed] [Google Scholar]
- Miyata T., Ogawa M. 2007. Twisting of neocortical progenitor cells underlies a spring-like mechanism for daughter-cell migration. Curr. Biol. 17:146–151 10.1016/j.cub.2006.11.023 [DOI] [PubMed] [Google Scholar]
- Miyata T., Kawaguchi A., Okano H., Ogawa M. 2001. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 31:727–741 10.1016/S0896-6273(01)00420-2 [DOI] [PubMed] [Google Scholar]
- Munevar S., Wang Y.L., Dembo M. 2001. Distinct roles of frontal and rear cell-substrate adhesions in fibroblast migration. Mol. Biol. Cell. 12:3947–3954 10.1091/mbc.12.12.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B., Brunstrom J.E., Grutzendler J., Wong R.O., Pearlman A.L. 2001. Two modes of radial migration in early development of the cerebral cortex. Nat. Neurosci. 4:143–150 10.1038/83967 [DOI] [PubMed] [Google Scholar]
- Nadarajah B., Alifragis P., Wong R.O., Parnavelas J.G. 2003. Neuronal migration in the developing cerebral cortex: observations based on real-time imaging. Cereb. Cortex. 13:607–611 10.1093/cercor/13.6.607 [DOI] [PubMed] [Google Scholar]
- Noctor S.C., Martínez-Cerdeño V., Ivic L., Kriegstein A.R. 2004. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7:136–144 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Norden C., Young S., Link B.A., Harris W.A. 2009. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 138:1195–1208 10.1016/j.cell.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai W., Minobe S., Ogawa M., Miyata T. 2007. Transformation of pin-like ventricular zone cells into cortical neurons. Neurosci. Res. 57:326–329 10.1016/j.neures.2006.10.015 [DOI] [PubMed] [Google Scholar]
- Ohtaka-Maruyama C., Hirai S., Miwa A., Heng J.I., Shitara H., Ishii R., Taya C., Kawano H., Kasai M., Nakajima K., Okado H. 2013. RP58 regulates the multipolar-bipolar transition of newborn neurons in the developing cerebral cortex. Cell Rep. 3:458–471 10.1016/j.celrep.2013.01.012 [DOI] [PubMed] [Google Scholar]
- Paracchini S., Thomas A., Castro S., Lai C., Paramasivam M., Wang Y., Keating B.J., Taylor J.M., Hacking D.F., Scerri T., et al. 2006. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum. Mol. Genet. 15:1659–1666 10.1093/hmg/ddl089 [DOI] [PubMed] [Google Scholar]
- Pinheiro E.M., Xie Z., Norovich A.L., Vidaki M., Tsai L.H., Gertler F.B. 2011. Lpd depletion reveals that SRF specifies radial versus tangential migration of pyramidal neurons. Nat. Cell Biol. 13:989–995 10.1038/ncb2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 1972. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 145:61–83 10.1002/cne.901450105 [DOI] [PubMed] [Google Scholar]
- Rivas R.J., Hatten M.E. 1995. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J. Neurosci. 15:981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara A., Sato T., Ando R., Noguchi N., Masaoka M., Miyata T. 2013. Dynamics of centrosome translocation and microtubule organization in neocortical neurons during distinct modes of polarization. Cereb. Cortex. In press [DOI] [PubMed] [Google Scholar]
- Schaar B.T., McConnell S.K. 2005. Cytoskeletal coordination during neuronal migration. Proc. Natl. Acad. Sci. USA. 102:13652–13657 10.1073/pnas.0506008102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B.T., Kinoshita K., McConnell S.K. 2004. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 41:203–213 10.1016/S0896-6273(03)00843-2 [DOI] [PubMed] [Google Scholar]
- Sekine K., Honda T., Kawauchi T., Kubo K., Nakajima K. 2011. The outermost region of the developing cortical plate is crucial for both the switch of the radial migration mode and the Dab1-dependent “inside-out” lamination in the neocortex. J. Neurosci. 31:9426–9439 10.1523/JNEUROSCI.0650-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K., Kawauchi T., Kubo K., Honda T., Herz J., Hattori M., Kinashi T., Nakajima K. 2012. Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin α5β1. Neuron. 76:353–369 10.1016/j.neuron.2012.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh J.C., Schaar B.T., Srinivasan K., Brodsky F.M., McConnell S.K. 2011. Endocytosis regulates cell soma translocation and the distribution of adhesion proteins in migrating neurons. PLoS ONE. 6:e17802 10.1371/journal.pone.0017802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T., Ayala R., Nguyen M.D., Xie Z., Gleeson J.G., Tsai L.H. 2004. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 44:263–277 10.1016/j.neuron.2004.09.030 [DOI] [PubMed] [Google Scholar]
- Solecki D.J. 2012. Sticky situations: recent advances in control of cell adhesion during neuronal migration. Curr. Opin. Neurobiol. 22:791–798 10.1016/j.conb.2012.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki D.J., Model L., Gaetz J., Kapoor T.M., Hatten M.E. 2004. Par6alpha signaling controls glial-guided neuronal migration. Nat. Neurosci. 7:1195–1203 10.1038/nn1332 [DOI] [PubMed] [Google Scholar]
- Solecki D.J., Trivedi N., Govek E.E., Kerekes R.A., Gleason S.S., Hatten M.E. 2009. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 63:63–80 10.1016/j.neuron.2009.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiess M., Maghelli N., Kapitein L.C., Gomis-Rüth S., Wilsch-Bräuninger M., Hoogenraad C.C., Tolić-Nørrelykke I.M., Bradke F. 2010. Axon extension occurs independently of centrosomal microtubule nucleation. Science. 327:704–707 10.1126/science.1182179 [DOI] [PubMed] [Google Scholar]
- Tabata H., Nakajima K. 2003. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J. Neurosci. 23:9996–10001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Serneo F.F., Higgins C., Gambello M.J., Wynshaw-Boris A., Gleeson J.G. 2004. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165:709–721 10.1083/jcb.200309025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F., Goffinet A.M. 2003. Reelin and brain development. Nat. Rev. Neurosci. 4:496–505 10.1038/nrn1113 [DOI] [PubMed] [Google Scholar]
- Tsai L.H., Gleeson J.G. 2005. Nucleokinesis in neuronal migration. Neuron. 46:383–388 10.1016/j.neuron.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Tsai J.W., Chen Y., Kriegstein A.R., Vallee R.B. 2005. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 170:935–945 10.1083/jcb.200505166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.W., Bremner K.H., Vallee R.B. 2007. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10:970–979 10.1038/nn1934 [DOI] [PubMed] [Google Scholar]
- Tsai J.W., Lian W.N., Kemal S., Kriegstein A.R., Vallee R.B. 2010. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 13:1463–1471 10.1038/nn.2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeshima H., Hirano T., Kengaku M. 2007. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc. Natl. Acad. Sci. USA. 104:16182–16187 10.1073/pnas.0708047104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M., Marín O. 2010. Neuronal migration mechanisms in development and disease. Curr. Opin. Neurobiol. 20:68–78 10.1016/j.conb.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Valiente M., Ciceri G., Rico B., Marín O. 2011. Focal adhesion kinase modulates radial glia-dependent neuronal migration through connexin-26. J. Neurosci. 31:11678–11691 10.1523/JNEUROSCI.2678-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Seale G.E., Tsai J.W. 2009. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 19:347–355 10.1016/j.tcb.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol E.A., Zheng J.Q. 2012. Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 73:1068–1081 10.1016/j.neuron.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M.E., Jiang H., Rao Y. 2005. Regulated formation and selection of neuronal processes underlie directional guidance of neuronal migration. Mol. Cell. Neurosci. 30:378–387 10.1016/j.mcn.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Watanabe H., Murakami F. 2009. Real time analysis of pontine neurons during initial stages of nucleogenesis. Neurosci. Res. 64:20–29 10.1016/j.neures.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Wiencken-Barger A.E., Djukic B., Casper K.B., McCarthy K.D. 2007. A role for Connexin43 during neurodevelopment. Glia. 55:675–686 10.1002/glia.20484 [DOI] [PubMed] [Google Scholar]
- Wilson P.M., Fryer R.H., Fang Y., Hatten M.E. 2010. Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. J. Neurosci. 30:8529–8540 10.1523/JNEUROSCI.0032-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Sanada K., Samuels B.A., Shih H., Tsai L.H. 2003. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 114:469–482 10.1016/S0092-8674(03)00605-6 [DOI] [PubMed] [Google Scholar]
- Xie Z., Moy L.Y., Sanada K., Zhou Y., Buchman J.J., Tsai L.H. 2007. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 56:79–93 10.1016/j.neuron.2007.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Huang C.H., Iglesias P.A., Devreotes P.N. 2010. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc. Natl. Acad. Sci. USA. 107:17079–17086 10.1073/pnas.1011271107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa S., Kawamura K., Kuwano R., Ono K. 1996. Neuron-glia interrelations during migration of Purkinje cells in the mouse embryonic cerebellum. Int. J. Dev. Neurosci. 14:429–438 10.1016/0736-5748(96)00021-4 [DOI] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., Han M. 2009. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 64:173–187 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Heintz N., Hatten M.E. 1996. CNS gene encoding astrotactin, which supports neuronal migration along glial fibers. Science. 272:417–419 10.1126/science.272.5260.417 [DOI] [PubMed] [Google Scholar]
- Zheng J.Q., Wan J.J., Poo M.M. 1996. Essential role of filopodia in chemotropic turning of nerve growth cone induced by a glutamate gradient. J. Neurosci. 16:1140–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Porcionatto M., Pilapil M., Chen Y., Choi Y., Tolias K.F., Bikoff J.B., Hong E.J., Greenberg M.E., Segal R.A. 2007. Polarized signaling endosomes coordinate BDNF-induced chemotaxis of cerebellar precursors. Neuron. 55:53–68 10.1016/j.neuron.2007.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Alfaro J., Chang E.H., Zhao X., Porcionatto M., Segal R.A. 2011. Numb links extracellular cues to intracellular polarity machinery to promote chemotaxis. Dev. Cell. 20:610–622 10.1016/j.devcel.2011.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmuda J.F., Rivas R.J. 1998. The Golgi apparatus and the centrosome are localized to the sites of newly emerging axons in cerebellar granule neurons in vitro. Cell Motil. Cytoskeleton. 41:18–38 [DOI] [PubMed] [Google Scholar]