Neuronal injury induces JNK phosphorylation of DLK, which reduces DLK ubiquitination and creates a positive feedback loop to enhance JNK signaling and increase apoptosis.
Abstract
Neurons are highly polarized cells that often project axons a considerable distance. To respond to axonal damage, neurons must transmit a retrograde signal to the nucleus to enable a transcriptional stress response. Here we describe a mechanism by which this signal is propagated through injury-induced stabilization of dual leucine zipper-bearing kinase (DLK/MAP3K12). After neuronal insult, specific sites throughout the length of DLK underwent phosphorylation by c-Jun N-terminal kinases (JNKs), which have been shown to be downstream targets of DLK pathway activity. These phosphorylation events resulted in increased DLK abundance via reduction of DLK ubiquitination, which was mediated by the E3 ubiquitin ligase PHR1 and the de-ubiquitinating enzyme USP9X. Abundance of DLK in turn controlled the levels of downstream JNK signaling and apoptosis. Through this feedback mechanism, the ubiquitin–proteasome system is able to provide an additional layer of regulation of retrograde stress signaling to generate a global cellular response to localized external insults.
Introduction
Axon degeneration and neuronal cell death occur during development to refine neuronal connections (Hamburger and Levi-Montalcini, 1949; Luo and O’Leary, 2005), after injury in the clearance of damaged cells (Quigley et al., 1995), and in neurodegenerative diseases such as Parkinson’s disease, amyotrophic lateral sclerosis (ALS), and Alzheimer’s disease (Vila and Przedborski, 2003). Although the factors that trigger neurodegeneration in these settings vary widely, many appear to converge on common conserved signaling events. Of particular interest are the Jun N-terminal kinases (JNKs), which act upstream of the BCL2 family member BAX and the transcription factor c-Jun in activating axon degeneration and neuronal apoptosis in development (White et al., 1998; Kuan et al., 1999; Eilers et al., 2001; Southwell et al., 2012) and as part of neurodegenerative disease pathology (Martin, 1999; Vila et al., 2001; Hunot et al., 2004; Yao et al., 2005). In each of these settings, BAX-dependent caspase activation appears necessary to carry out programmed cell death and axon degeneration downstream of JNK activation (Gagliardini et al., 1994; Pettmann and Henderson, 1998; Yuan and Yankner, 2000; Simon et al., 2012).
Dual leucine zipper-bearing kinase (DLK) is an evolutionarily conserved member of the mixed lineage kinase (MLK) family that is required for stress-induced JNK activity in neurons (Hirai et al., 2005; Chen et al., 2008; Ghosh et al., 2011; Shin et al., 2012; Watkins et al., 2013). Loss of DLK in mammals attenuates apoptosis and axon degeneration in development and after axon injury in the central nervous system (Chen et al., 2008; Miller et al., 2009; Ghosh et al., 2011; Watkins et al., 2013; Welsbie et al., 2013). Additional studies revealed that DLK functions as a damage sensor within the axon to initiate retrograde JNK signaling and generate a transcriptional stress response to localized insults (Shin et al., 2012; Watkins et al., 2013).
In invertebrates, a distinct function for DLK was identified through successive genetic screens that demonstrated that the PHR family of E3 ubiquitin ligases (PAM/Highwire/RPM-1) reduces DLK abundance to control synapse development (Nakata et al., 2005; Collins et al., 2006). A similar mechanism appears to regulate DLK after nerve injury in Drosophila, where DLK levels rapidly rise concomitantly with loss of Highwire from the axon (Xiong et al., 2010). In Caenorhabditis elegans, DLK activity after injury is also regulated via heterodimerization with a shorter DLK isoform that restricts DLK activation to damaged regions of the neuron (Yan and Jin, 2012).
Despite the mechanistic knowledge gained through studies in invertebrate systems, little is known about the factors that control DLK activity in mammalian neurons. In this study, we demonstrate that DLK protein is stabilized after axonal insult, and this stabilization results in amplification and propagation of the neuronal injury response. A balance of ubiquitination by PHR1 and de-ubiquitination via USP9X tightly regulate DLK protein levels, and specific JNK-dependent phosphorylation of DLK at sites distinct from those regulating kinase activity offset this balance to result in an increased abundance of DLK protein. Thus, DLK-dependent activation of JNK generates a feedback mechanism that amplifies neuronal stress signaling to convert graded or local DLK activation into a decisive cellular outcome within individual neurons.
Results
Neuronal stress increases DLK protein abundance and apparent molecular weight
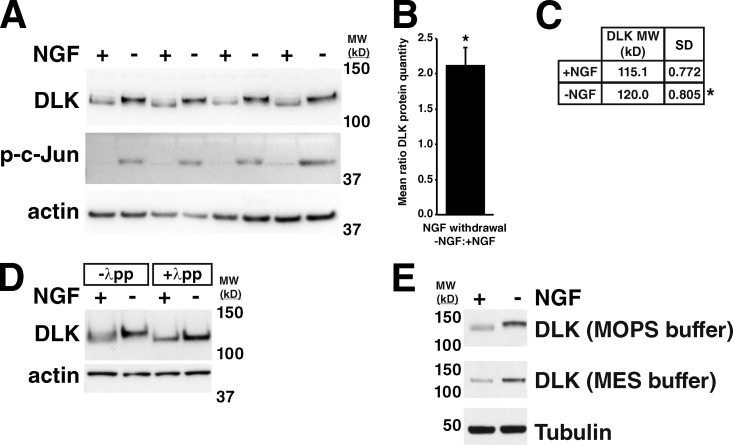
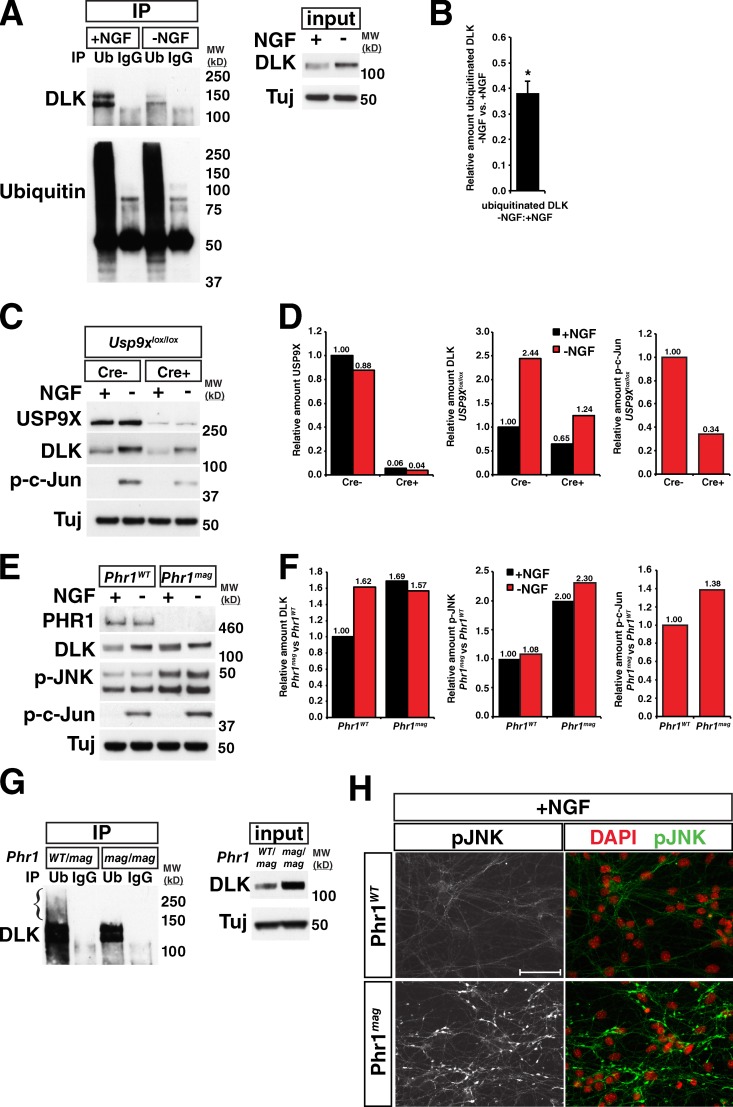
To confirm that DLK protein levels increase in response to neuronal stress, as has been observed in multiple systems (Xu et al., 2001; Xiong et al., 2010; Watkins et al., 2013; Welsbie et al., 2013), we first examined cultured embryonic sensory neurons after nerve growth factor (NGF) withdrawal. Global withdrawal of NGF from embryonic dorsal root ganglion (DRG) cells results in DLK-dependent phosphorylation of the transcription factor c-Jun, a readout of stress-induced JNK activity in neurons, by 3 h (Fig. 1 A), and neuronal degeneration at 18–24 h (Ghosh et al., 2011). At the 3-h time point, DLK protein levels increased in response to NGF withdrawal by approximately twofold compared with unstressed neurons cultured in the presence of NGF (Fig. 1, A and B). Interestingly, the increase in DLK protein quantity was accompanied by an upward shift in molecular weight of DLK by ∼5 kD (Fig. 1, A and C). Treatment of DRG lysates with lambda protein phosphatase to cleave phosphate groups equalized the molecular weights of DLK in +NGF and −NGF conditions (Fig. 1 D), demonstrating that this mobility shift was the result of phosphorylation. A similar DLK mobility shift has been observed in some instances (Mata et al., 1996; Xu et al., 2001), but not in others (Xiong et al., 2010; Watkins et al., 2013), and the basis of this phenomenon is not well understood. These conflicting results may be in part due to differences in the SDS-PAGE buffer conditions used (Fig. 1 E).
Figure 1.
DLK protein levels and molecular weight increase in response to NGF withdrawal from embryonic sensory neurons. (A) Increase in DLK abundance and molecular weight in response to NGF withdrawal from cultured embryonic DRGs. c-Jun phosphorylation (p-c-Jun) occurs as a downstream consequence of DLK and JNK activation. (B) Ratio of DLK protein quantity in −NGF vs. +NGF samples shown in A. Ratio = 2.12 ± 0.127. *, P = 0.02 by Mann-Whitney U test comparing the mean ratio to 1. Error bar represents SD. (C) Table showing molecular weight of DLK in kilodaltons (mean and SD) in +NGF and –NGF samples shown in A. *, P = 0.03 by Mann-Whitney U test comparing the molecular weights of DLK in the two conditions. (D) Lambda protein phosphatase (λpp) treatment of DRG lysates equalizes DLK molecular weight in −NGF and +NGF conditions. (E) Difference in mobility of DLK in SDS-PAGE with two different running buffers. MOPS buffer: SDS-PAGE running solution containing MOPS as a buffer. MES buffer: SDS-PAGE running solution containing MES as a buffer. MW (kD): molecular weight in kilodaltons.
DLK protein is stabilized in response to trophic factor withdrawal
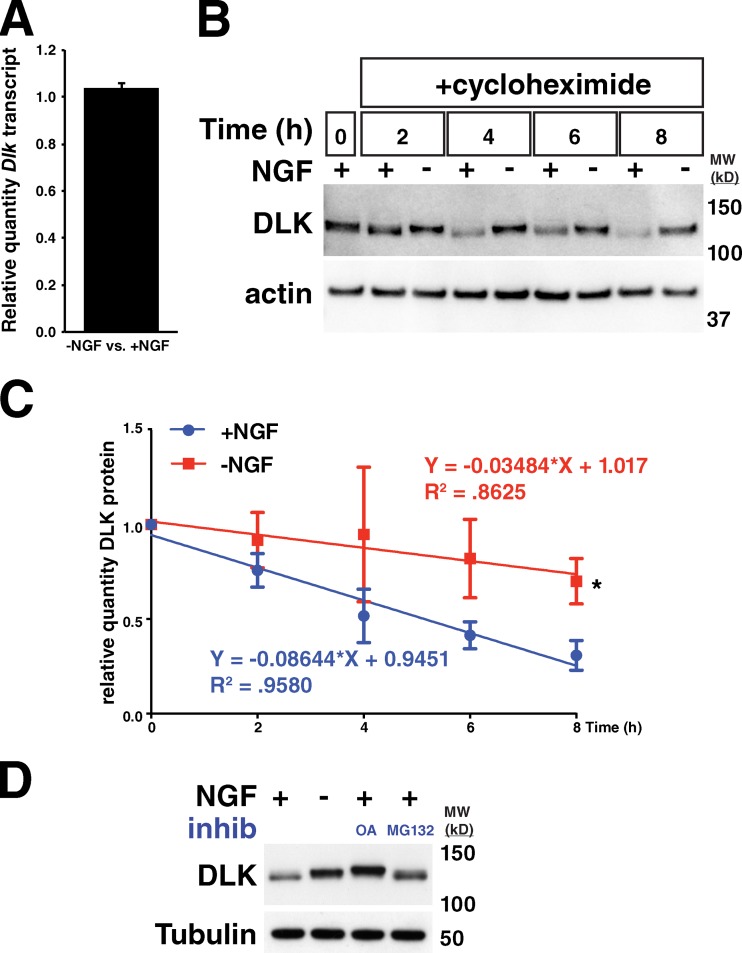
Real-time qRT-PCR to quantify the amount of Dlk transcript in DRGs undergoing trophic factor withdrawal did not reveal a detectable change in Dlk levels compared with control cells (Fig. 2 A), demonstrating that the rise in DLK protein levels is due to a post-transcriptional mechanism consistent with previous reports (Xiong et al., 2010; Welsbie et al., 2013). To determine whether the increase in DLK abundance is a result of alterations in protein stability, we assessed DLK turnover in DRGs treated with cycloheximide to inhibit protein translation in the presence or absence of NGF for 8 h. An ∼2.5-fold increase in the half-life of DLK protein was observed with NGF deprivation (Fig. 2, B and C), indicating that this mechanism is the predominant contributor to the elevation of DLK levels after neuronal stress.
Figure 2.
DLK protein is stabilized in response to trophic factor withdrawal in embryonic sensory neurons. (A) Measurement of relative amount of Dlk transcript by real-time qRT-PCR in −NGF vs. +NGF culture conditions. n = 3 technical replicates. Error bar represents SD. −NGF vs. +NGF: 1.036 ± 0.0234. (B) 8-h time course of trophic factor withdrawal in −NGF-treated DRGs and +NGF controls in the presence of 5 µM cycloheximide. (C) Quantification of three repeated trials of experiment performed in B. Plotted is the relative amount of DLK remaining at a given time compared with the amount at time 0. Error bars are SDs. Each time course was fit to a line in GraphPad Prism software. *, P = 0.0109 when comparing the slopes of the two lines with ANCOVA. (D) DRGs were treated with the given conditions for 3 h and lysed. OA, 200 nM okadaic acid. MG132 was used at 30 µM. Both OA and MG132 treatment resulted in an increase in DLK protein levels. MW (kD), molecular weight in kilodaltons.
To ascertain whether phosphorylation of DLK after trophic factor withdrawal influences the observed changes in DLK stability, unstressed DRGs cultured in the presence of NGF were treated with okadaic acid, a broad phosphatase inhibitor, to enhance DLK phosphorylation. This treatment increased both the apparent molecular weight and total amount of DLK, arguing that phosphorylation is sufficient to impart increased DLK stability (Fig. 2 D). Treatment with the proteasome inhibitor MG132 similarly increased DLK abundance without increasing DLK phosphorylation, suggesting that non-phosphorylated DLK is normally degraded by the proteasome (Fig. 2 D).
DLK stabilization amplifies JNK activation after axonal injury
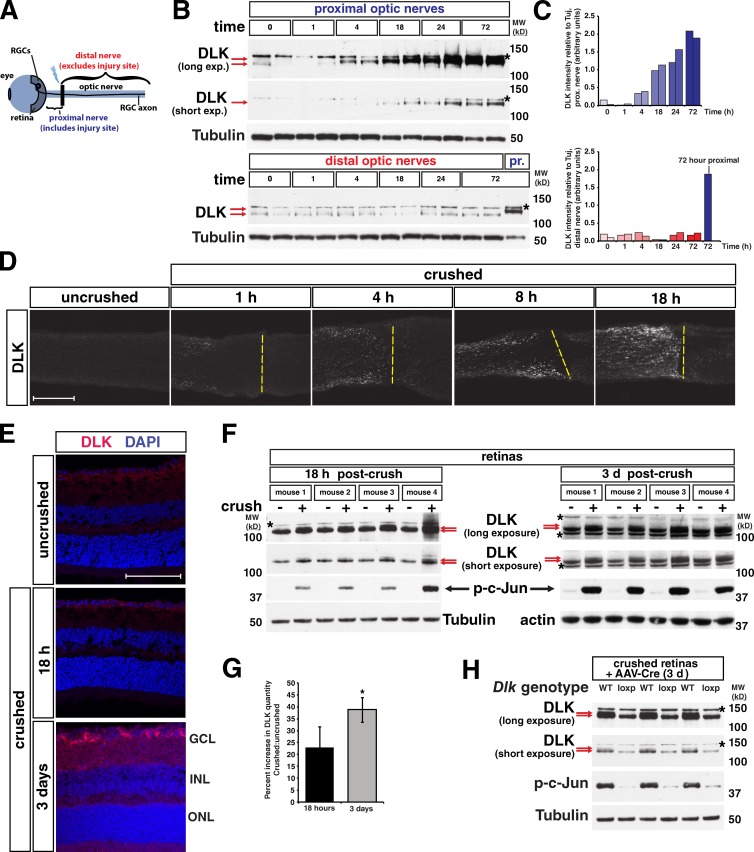
We next sought to determine whether the increase in DLK after axonal insult is a localized response or whether DLK levels increase throughout the neuron. To assess this, we used optic nerve crush, an in vivo model that severs the axons of retinal ganglion cells (RGCs), which are projection neurons that transmit visual information from the retina to the brain (Fig. 3 A). We first examined the optic nerve, where an increase in DLK abundance and molecular weight occurred as early as 4 h after injury and continued until 72 h in the proximal but not distal axons, with a the majority of the increase in protein quantity occurring by 18 h (Fig. 3, B and C; Fig. S1). This increase coincided with robust DLK staining specifically in the region of the proximal optic nerve adjacent to the injury site (Fig. 3 D). As DLK expression in the optic nerve is limited to RGC axons (Watkins et al., 2013), this elevation in DLK protein represents activation of the injury response within neurons rather than glial DLK expression. Interestingly, at the 18-h time point when a robust increase in DLK levels has occurred in the optic nerve, a similar increase in intensity of DLK staining was not observed in RGC cell bodies within the retina, and only a small amount of higher molecular weight DLK could be observed by Western blots on retina lysates (Fig. 3, E and F). By 3 d after injury, however, DLK staining in the retina was clearly increased and a more prominent higher molecular weight band could be observed in retinal lysates (Fig. 3, E–G). The incomplete molecular weight shift observed in the retina after nerve crush, as compared with that in the optic nerve, appeared to be the result of DLK phosphorylation being limited to RGCs, the cell type injured in nerve crush, and not other retinal cell types (Fig. 3 H).
Figure 3.
Elevation of DLK protein levels occurs first in the axon after optic nerve crush and then progresses to cell bodies. (A) Diagram of the retina nerve crush model showing location of the retina, crush site (black bar), proximal nerve, and distal nerve. RGCs, retinal ganglion cells, located in the ganglion cell layer (GCL) of the retina. (B) Within the crushed nerve, DLK undergoes a stress-dependent increase in molecular weight and abundance by 4 h post-crush and is notably increased by 18 h. The shift in DLK mobility (red arrows) and increase in DLK quantity can be observed after injury only in the proximal (top) but not the distal nerve (bottom). Asterisk: a background band observed in nerve lysates blotted for DLK (see Fig. S1). A proximal nerve sample collected 72 h after injury is included in the distal nerve blot for comparison. (C) Quantification of the DLK intensity relative to tubulin intensity for each well shown in B. n = 2 animals/time point. (D) Time course showing the increase in DLK quantity in the optic nerve after injury by immunostaining for DLK. Yellow dashed lines: approximate location of crush site. The increase in DLK is limited to the proximal nerve that lies to the left of the line. (E) In the retina, no increase in total DLK is observed by staining at 18 h post-crush, but by three days an increase in DLK is visible. Magenta, DLK staining; blue, DAPI nuclear stain; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. (F) Blots for DLK in crushed retinas and contralateral uncrushed controls 18 h and 3 d after nerve crush. Red arrows denote the lower molecular weight DLK and the appearance of a small amount of the higher molecular weight band at 18 h and a more prominent higher molecular weight band at 3 d. Asterisk denotes background bands. (G) Quantification of the percent increase in DLK in crushed retina vs. uncrushed retina at the given times in the blots shown in F. 18 h = 23.02 ± 8.39. 3 d = 38.68 ± 5.27. *, P = 0.028 by t test. n = 4 animals for each time point. Error bars are SD. (H) Blots of retina lysates 3 d after nerve crush from wild-type (WT) and Dlklox/lox (loxp) mice injected with AAV-Cre virus. The DLK mobility shift is seen in WT retinas in the form of a doublet (red arrows), but not in loxp mice. Because only RGCs were transduced with AAV-Cre virus in loxp mice, the top band (red arrow) that appears with retina nerve crush represents phosphorylation of DLK specifically in RGCs and not other cell types of the retina. Bars, 100 µm. MW (kD), molecular weight in kilodaltons.
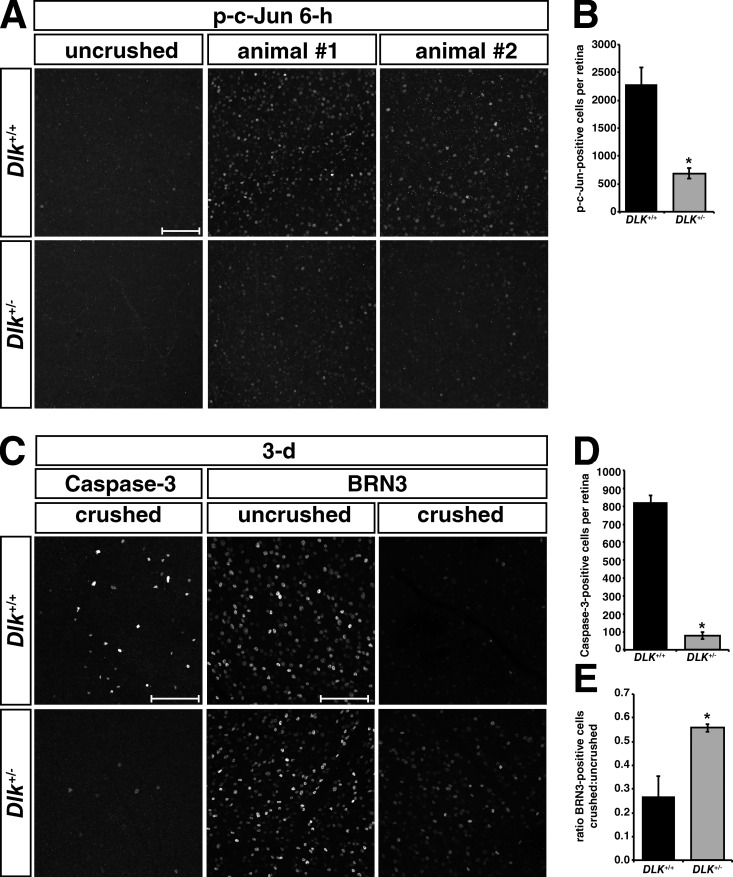
Our observation that the accumulation of DLK protein and shift in apparent molecular weight occurs first in the axon proximal to the injury site and then spreads to the retina led us to speculate that DLK protein abundance may directly affect the propagation of downstream JNK signaling after localized axonal injury. To test this hypothesis, we used Dlk knockout (KO) heterozygotes that express roughly 50% of the amount of DLK present in wild-type (WT) littermates (Fig. S2 A). After optic nerve crush, the number of strongly p-c-Jun–positive nuclei in heterozygous crushed retinas was reduced by ∼70% in Dlk heterozygous mice compared with WT retinas at 6 h, the earliest time point at which this marker could be observed (Fig. 4, A and B). This observation suggests that the increase in DLK levels serves to amplify and/or propagate DLK signaling to generate a more decisive injury response within individual neurons, a process that is delayed in heterozygous animals with reduced DLK protein. Similar observations were made in Dlk heterozygous DRGs after NGF withdrawal (Fig. S2 B). The delay in c-Jun phosphorylation after nerve crush is sufficient to generate a significant reduction in the number of RGCs undergoing apoptosis at 3 d after crush by >85% as measured by staining for active caspase 3, and a greater than twofold increase in the total number of BRN3-positive nuclei, a marker of healthy RGCs (Fig. 4, C–E; Erkman et al., 1996; Gan et al., 1996). By 14 d post-crush, however, the amount of degeneration is nearly indistinguishable in WT and Dlk heterozygous retinas (Fig. S2 C), unlike the long-lasting neuroprotection observed in Dlk knockouts (Watkins et al., 2013). Taken together, these data suggest that modulation of DLK levels spatially and temporally regulates the progression of downstream JNK signaling and induction of neuronal apoptosis after axonal injury.
Figure 4.
DLK modulates the propagation of downstream pro-apoptotic signaling in a dose-dependent manner. (A) p-c-Jun staining of retinas from Dlk+/+ and Dlk+/− mice 6 h post-crush. (B) Quantification of the mean number of p-c-Jun–positive cells per retina 6 h after crush shows significantly fewer p-c-Jun–positive cells in Dlk+/− mice. *, P = 0.0014 by t test. Dlk+/+ = 2291 ± 299. Dlk+/− = 689 ± 97.6. n = 7 Dlk+/+ and 5 Dlk+/− animals. (C) Active caspase-3 and BRN3 staining of retinas from Dlk+/+ and Dlk+/− mice 3 d post-crush. (D) Quantification of mean number of caspase-3–positive cells per retina shows a reduction in Dlk+/− mice. *, P = 0.0001 by t test. Dlk+/+ = 823 ± 36. Dlk+/− = 79 ± 20. n = 4 Dlk+/+ and 3 Dlk+/− animals. (E) Quantification of ratio of BRN3-positive cells per retina in crushed vs. uncrushed retinas from Dlk+/+ and Dlk+/− mice. *, P = 0.0253 by student’s t test. Dlk+/+ = 0.27 ± 0.09. Dlk+/− = 0.56 ± 0.02. n = 5 Dlk+/+ and 4 Dlk+/− animals. Bars, 100 µm. Error bars are SEM.
Neuronal stress regulates DLK ubiquitination
To better understand the mechanisms underlying the observed increase in DLK abundance, we next asked whether DLK stability is modulated by the ubiquitin proteasome system as suggested by the observed increase in DLK abundance after MG132 treatment. Primary DRG neurons were used for this analysis, as stress-induced changes in the abundance of DLK could be easily measured. Immunoprecipitation of ubiquitinated proteins from +NGF and −NGF DRGs showed that DLK ubiquitination was markedly reduced in the −NGF condition (Fig. 5, A and B; Fig. S3 A). In invertebrate systems, the PHR family of E3 ubiquitin ligases (PAM/Highwire/RPM-1) and the DUB Fat Facets are thought to regulate DLK protein levels (Nakata et al., 2005; Collins et al., 2006; Xiong et al., 2010). To directly investigate the role of the ubiquitin–proteasome system in regulating DLK levels in mammalian neurons, we used loss-of-function alleles of the mouse homologues of these two genes.
Figure 5.
The ubiquitin–proteasome system regulates DLK levels in a stress-dependent manner. (A) DLK ubiquitination is reduced after trophic factor withdrawal. NGF-deprived and control DRGs were collected after 3 h of treatment. Ubiquitinated proteins were immunoprecipitated from the lysates and blotted for DLK and ubiquitin. Mouse IgG controls were used as negative controls to demonstrate antibody specificity. (B) Ratio of amount of ubiquitinated DLK in −NGF vs. +NGF conditions. Mean = 0.381 ± 0.046. *, P = 0.0018 by one-sample t test comparing the mean to 1. n = 3 independent experiments. Error bar represents SD. (C) Western blots for DLK, USP9X, p-c-Jun, and tubulin (Tuj) after NGF withdrawal in Usp9xlox/lox; Rosa26-Cre/ERT2− (Cre−) and Rosa26-Cre/ERT2+ (Cre+) embryonic DRGs that had been treated with 10 µM 4-hydroxytamoxifen to induce Cre recombination at the lox sites. Blots are representative of three independent experiments. (D) Quantifications of blots for USP9X, DLK, and p-c-Jun shown in C. Loss of ∼95% of USP9X (left) results in an overall decrease in DLK levels (middle) but does not alter the relative levels of DLK in the + and −NGF conditions (compare fold change in Cre+ to that in Cre−). The reduction in DLK with loss of USP9X results in a decrease in downstream p-c-Jun (right). (E) Western blots for DLK, PHR1, p-JNK, p-c-Jun, and tubulin after NGF withdrawal in Phr1mag/mag and control embryonic DRGs. Blots shown are representative of three independent experiments. (F) Quantification of blots shown in E. DLK (left), p-JNK (middle), and p-c-Jun after NGF withdrawal (right) are elevated in Phr1mag loss-of-function mutants compared with Phr1WT. (G) Immunoprecipitation of ubiquitinated proteins shows that less ubiquitinated DLK is present in Phr1mag/mag homozygotes than in Phr1WT/mag heterozygote controls. Left: immunoprecipitation with anti-ubiquitin (Ub) followed by blotting for DLK. Immunoprecipitation with mouse IgG was used as a control for antibody specificity. Right: blots of the input lysates for DLK and tubulin. Despite the elevated level of DLK in the input for Phr1mag compared with Phr1WT/mag neurons, less ubiquitinated DLK was pulled down and no polyubiquitinated DLK could be detected at higher molecular weights (bracket). (H) Imaging of p-JNK in Phr1mag and control DRGs cultured with NGF. p-JNK is elevated in axons in the +NGF condition in Phr1mag (left), but does not coincide with DAPI-stained nuclei (right). DAPI channel has been false-colored as red for ease of visualization. Bar, 50 µm. MW (kD), molecular weight in kilodaltons.
DRGs lacking USP9X, the closest mouse homologue to Fat Facets, were derived from mice that expressed the tamoxifen-inducible recombinase Cre/ERT2 ubiquitously from the Rosa26 locus and had Usp9x exon 31, encoding catalytic Cys 1560, flanked by lox sites (see Materials and methods). As would be predicted for knockout of a DUB that controls the turnover of DLK, a reduction in DLK abundance was observed in DRGs lacking USP9X (Cre+) versus Cre− DRGs in both the +NGF and −NGF conditions (Fig. 5, C and D). Importantly, this decrease in DLK resulted in reduced c-Jun phosphorylation in the −NGF condition in Cre+ DRGs, consistent with the hypothesis that alterations in DLK protein levels through ubiquitination are sufficient to modulate downstream signaling events. However, while the knockout of Usp9x had an effect on the overall levels of DLK and p-c-Jun, the −NGF/+NGF ratio of DLK protein quantity in Cre+ neurons was nearly identical to that in Cre− neurons (Fig. 5 D). As would be predicted based on this observation, cross-linking with ubiquitin vinyl sulfone revealed that USP9X activity is unaltered by NGF withdrawal (Fig. S3 B). Therefore, we conclude that USP9X regulates DLK abundance, but that this function is not altered by neuronal stress.
In contrast, DRGs homozygous for the loss-of-function Phr1mag allele (Lewcock et al., 2007) contained more DLK in the presence of NGF than wild-type controls without showing an NGF deprivation–dependent increase in DLK, resulting in roughly equivalent DLK abundance in stressed and nonstressed conditions (Fig. 5, E and F). In addition, Phr1mag/mag homozygous neurons contained less ubiquitinated DLK, and lacked higher molecular weight polyubiquitinated forms of DLK (Fig. 5 G). These observations, together with the decrease in DLK ubiquitination with neuronal stress (Fig. 5 A), suggest that the change in ubiquitination of DLK is at least in part due to modulation of PHR1 activity or its interaction with DLK. The increased DLK abundance observed in Phr1mag/mag neurons was sufficient to result in elevated JNK phosphorylation (p-JNK) even in the presence of NGF (Fig. 5, E and F). Interestingly, this increase did not coincide with c-Jun phosphorylation (Fig. 5 E) or cause axon degeneration in DRGs in the presence of NGF (Fig. S3 C). Further examination of Phr1mag/mag neurons by immunostaining revealed that the p-JNK in neurons is largely localized to axons (Fig. 5 H) and is not retrogradely transported to the nucleus as occurs after NGF withdrawal (Ghosh et al., 2011). This relocalization of p-JNK is required for phosphorylation of c-Jun and these data suggest that additional inputs must be required to trigger DLK-dependent degeneration. However, the elevated p-JNK did result in a modest increase in p-c-Jun 3 h after NGF withdrawal (Fig. 5, E and F). Despite these observations, no noticeable difference in degeneration of NGF-deprived DRGs was apparent at the end point of 18 h (Fig. S3 C). This could be a result of the acute nature of this model or a potential role of additional PHR1 substrates such as NMNAT2 that also influence neuronal degeneration (Murthy et al., 2004; Xiong et al., 2012; Babetto et al., 2013).
DLK activity and JNK activity are required for stabilization of DLK
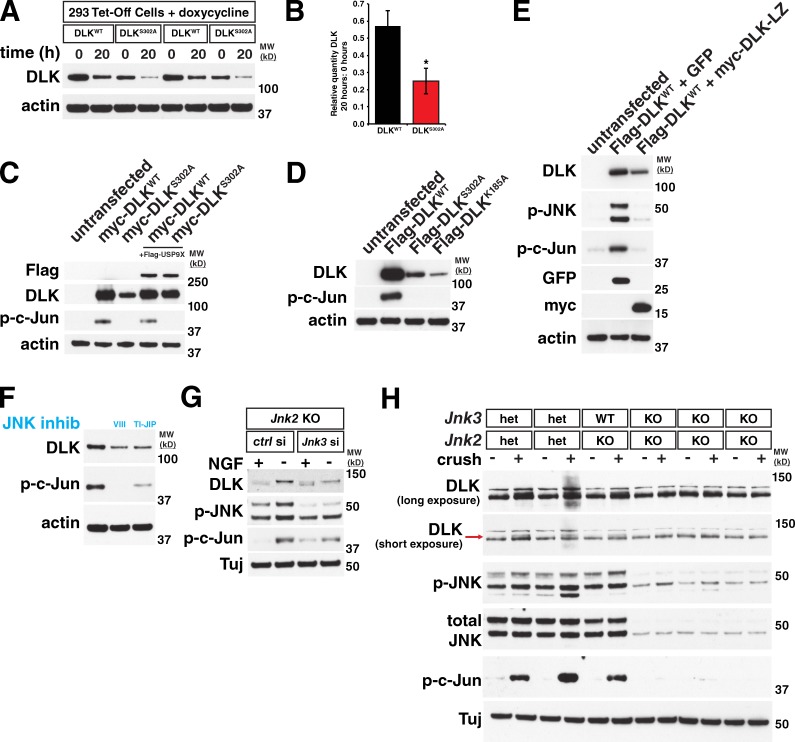
The correlation between DLK phosphorylation and stabilization after neuronal stress suggests a possible link between these events. To investigate this hypothesis, we overexpressed Flag-tagged mouse DLK in human embryonic kidney 293T (293T) cells, which is sufficient for DLK activation and phosphorylation of c-Jun (Mata et al., 1996). To mimic, to some extent, the unstressed versus stressed conditions in neurons, we generated a kinase-dead version of DLK by mutating phosphorylation sites in the putative activation loop that we identified by homology with MLK3 (Leung and Lassam, 2001). One such point mutant, DLKS302A, did not cause phosphorylation of c-Jun and other downstream targets in 293T cells, confirming that it lacked kinase activity. Interestingly, DLKS302A reproducibly expressed at lower levels than wild-type DLK in 293T cells. Using a doxycycline-dependent repression system in HEK293 cells to turn off expression of transiently transfected DLK constructs, we found that 20 h after doxycycline addition, ∼2.7-fold more wild-type DLK than DLK302A was remaining, demonstrating that DLKS302A has a greater degradation rate than wild-type DLK (Fig. 6, A and B). Co-expression with USP9X increased expression of DLKS302A to wild-type levels, suggesting that the lower protein levels and greater degradation rate observed are due to increased ubiquitination of the inactive DLK (Fig. 6 C). To confirm that the reduction in protein levels observed with DLKS302A was a result of a loss in kinase activity rather than an effect of this specific mutation, we expressed another kinase-dead point mutant, K185A, and found that it too expresses at lower levels than wild-type DLK (Fig. 6 D; Fan et al., 1996; Mata et al., 1996). We also coexpressed wild-type DLK with a truncated construct containing only the DLK leucine zipper domain (DLK-LZ), which acts as a dominant-negative by preventing full-length DLK dimerization (Nihalani et al., 2000). Similar to what was observed with DLKS302A, decreased DLK activity in the presence of DLK-LZ resulted in lowered expression of wild-type DLK when compared with DLK coexpressed with GFP (Fig. 6 E).
Figure 6.
DLK stabilization depends on DLK activity and on downstream targets of DLK. (A) Two replicates of wild-type DLK (DLKWT) and a kinase-dead point mutant (DLKS302A) transiently expressed in 293 Tet-Off cells followed by treatment with doxycycline for the given times to prevent expression of new DLK protein. (B) Kinase-dead DLKS302A turns over more rapidly than WT DLK. The mean amount of DLK remaining after 20 h doxycycline treatment relative to the amount at 0 h for each DLK construct was calculated. DLKWT = 0.571 ± 0.092. DLKS302A = 0.251 ± 0.075. *, P < 0.001 by t test. n = 6 independent replicates. Error bars represent SD. (C) Transient transfection of HEK 293T cells shows that a kinase-dead version of DLK (DLKS302A) is expressed at lower levels than wild-type DLK. Co-expression with USP9X rescues this effect. (D) Similar to the S302A point mutant, the kinase-dead K185A point mutant also shows reduced expression compared with wild-type DLK when transiently transfected. (E) Co-expression of DLK with a dominant-negative DLK leucine zipper domain N-terminally tagged with myc epitope (myc-DLK-LZ) decreases DLK expression compared with coexpression with GFP. (F) Inhibition of JNK activity with two structurally distinct JNK inhibitors (VIII and TI-JIP, see Materials and methods) in a stable cell line after doxycycline-induced DLK expression reduces levels of DLK protein. (G) Knockdown of Jnk3 in a Jnk2 KO background blocks the increase in DLK quantity observed with NGF withdrawal in embryonic DRGs. (H) At 18 h after nerve crush, Jnk2/3 double-knockout retinas do not have activated DLK. The long exposure DLK blot shows the overall change in DLK levels at this time point. The short exposure shows the appearance of the phosphorylated, higher molecular weight DLK band (red arrow) in control animals that is missing in JNK2/3 double knockouts. MW (kD), molecular weight in kilodaltons.
Given that DLK kinase activity appeared necessary for DLK protein stabilization, we examined whether downstream JNK signaling plays any role in DLK stability. To better normalize DLK levels, we generated 293T cells that stably express doxycycline-inducible DLK and treated these cells with two structurally distinct JNK inhibitors (Fig. 6 F). Surprisingly, both JNK inhibitors reduced the amount of DLK protein expressed. To establish the relevance of this finding in a neuronal system, we used siRNA to knock down JNK3 expression in Jnk2 knockout DRGs, removing the two JNK family members that regulate the majority of stress-induced neuronal degeneration (Coffey et al., 2002; Chang et al., 2003). Compared with control siRNA, JNK3 knockdown attenuated the increase in DLK after NGF withdrawal. In the JNK3 knockdown −NGF condition, some change in DLK molecular weight and c-Jun phosphorylation was still observed, which may be a result of residual JNK activity in these neurons (Fig. 6 G). In the optic nerve crush model, Jnk2/3 double knockouts showed no increase in DLK levels or molecular weight compared with littermate controls at 18 h post-crush (Fig. 6 H, red arrow), demonstrating that JNK-dependent phosphorylation of DLK also occurs in an adult in vivo injury paradigm. Based on these findings, we hypothesize that JNK-dependent feedback results in the phosphorylation of specific sites on DLK that are required for DLK stabilization, though additional JNK-independent phosphorylation events may also occur.
Identification of phosphorylation sites required for DLK stabilization
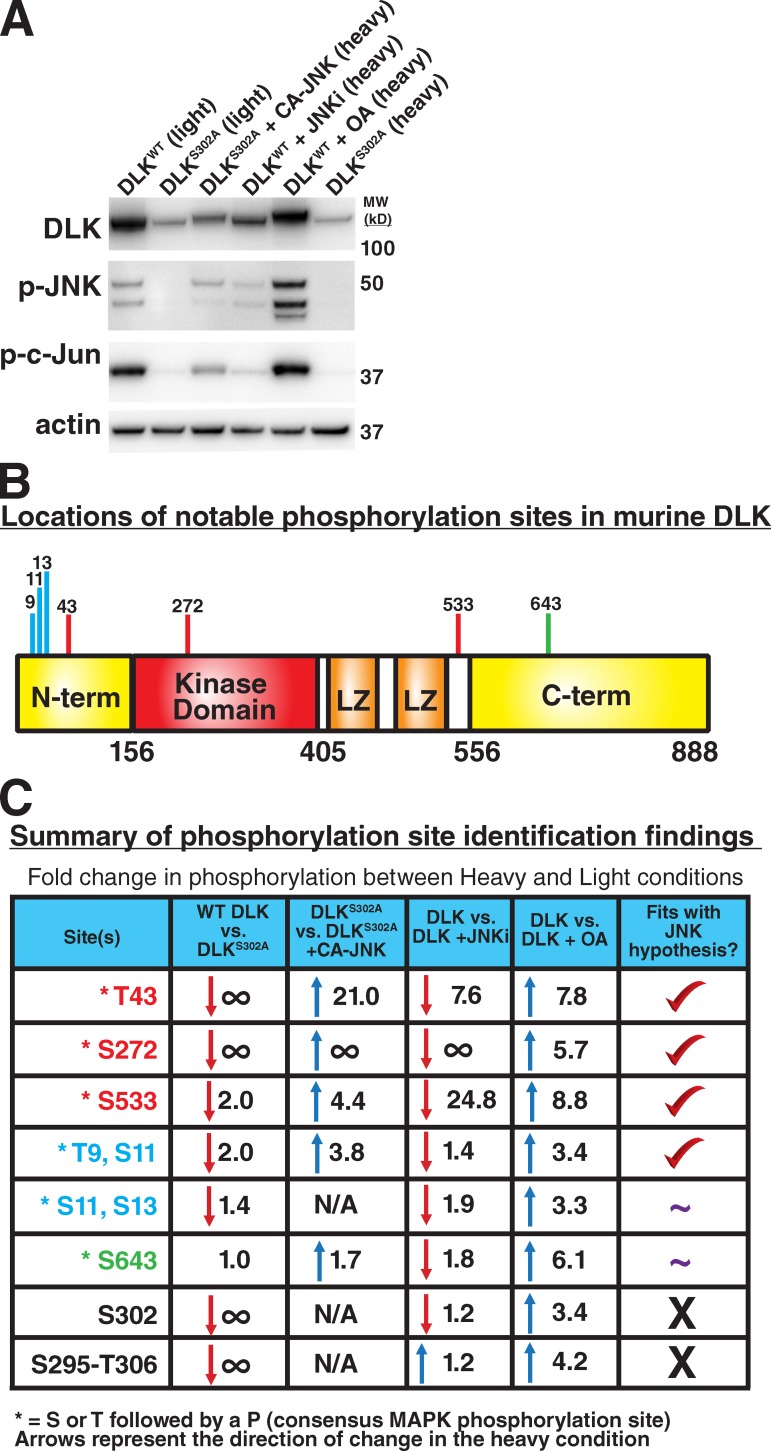
To identify functionally relevant phosphorylation sites on DLK, we used mass spectrometry in conjunction with stable isotope labeling by amino acids in cell culture (SILAC). For these studies, 293T cells expressing FLAG-tagged DLK were cultured in SILAC media containing isotopically enriched (heavy) versions of lysine (13C615N2) and arginine (13C615N4) or their unlabeled counterparts (light). Four paired conditions (light vs. heavy) were analyzed: (1) WT DLK vs. DLKS302A to identify DLK-dependent phosphorylation sites, (2) DLKS302A vs. DLKS302A coexpressed with constitutively active JNK (Lei et al., 2002) to identify JNK-dependent phosphorylation sites on DLK, (3) WT DLK vs. WT DLK with JNK inhibitor to identify JNK-dependent phosphorylation sites on active DLK, and (4) WT DLK vs. WT DLK with okadaic acid to enrich for stabilizing phosphorylation sites on DLK (Fig. 7 A).
Figure 7.
Identification of phosphorylation sites in mouse DLK that are modulated by DLK or JNK activity. (A) Expression of DLK in 293T cells in the heavy and light SILAC conditions used for mass spectrometry. After expression, Flag-tagged DLK was immunoprecipitated (IP) and IPs of the given conditions were combined for ratiometric analysis of phosphorylation sites in DLK. WT DLK, wild-type DLK; DLKS302A, kinase-dead point mutant; CA-JNK, constitutively active JNK (see Materials and methods); JNKi, JNK inhibitor VIII, 10 µM; OA, okadaic acid, 200 nM; MW (kD), molecular weight in kilodaltons. (B) Schematic of DLK domains and locations of noted phosphorylation sites. N-term, N-terminal domain of DLK; kinase domain, catalytic DLK domain; LZ, leucine zipper motifs; C-term, C-terminal domain. Domains and numbering are based on mouse DLK according to Holzman et al. (1994). (C) Summary of identified phosphorylation site changes. Sites listed show the largest effects between conditions, or in the case of S295-T306, are known sites within the kinase activation loop. ∞, phosphorylation of this site was observed in condition A but not in condition B; N/A, phosphorylation of this site was not observed in either condition. Numbers given are the fold changes in phosphorylation of the site in condition B vs. condition A, and up or down arrows denote the direction of change (e.g., for top-right box, there is 7.8-fold more phosphorylation of T43 in okadaic acid–treated cells expressing DLK than in cells expressing DLK with no okadaic acid). Asterisk denotes that this site contains a threonine (T) or serine (S) followed by a proline. A flanking proline is found in the majority of MAPK phosphorylation sites. The column entitled “Fits with JNK hypothesis?” summarizes whether the pattern of phosphorylation across the four conditions fits with the hypothesis that phosphorylation of this site by JNK occurs and is responsible for stabilization of DLK. Check mark, pattern of phosphorylation is consistent with this hypothesis; ∼, pattern of phosphorylation is partially consistent with this hypothesis; X, pattern of phosphorylation is inconsistent with this hypothesis. See Materials and methods for details on how individual sites were identified.
A series of phosphopeptides on DLK were identified whose abundance changed in response to the conditions tested. After correction for differences in overall DLK abundance among conditions (see Materials and methods), we identified multiple sites whose phosphorylation state changed in a manner consistent with DLK and JNK-dependent phosphorylation (Fig. 7, B and C; Fig. S4). The top three sites in terms of the magnitude with which they changed between conditions were T43 in the N-terminal domain, S272 in the kinase domain, and S533 immediately C-terminal to the leucine zipper domains. JNK-dependent changes were also observed for a peptide containing multiple phosphorylation sites (T9, S11). Interestingly, all of these sites contain a flanking proline consistent with a MAPK substrate motif (Songyang et al., 1996). Phosphorylation sites within the kinase activation loop (S295–T306) were found to be dependent on the kinase activity of DLK but independent of JNK. This observation fits with a model in which JNK does not directly modulate the activity of DLK, but rather controls factors that affect DLK stability.
Identified sites are phosphorylated in stabilized DLK
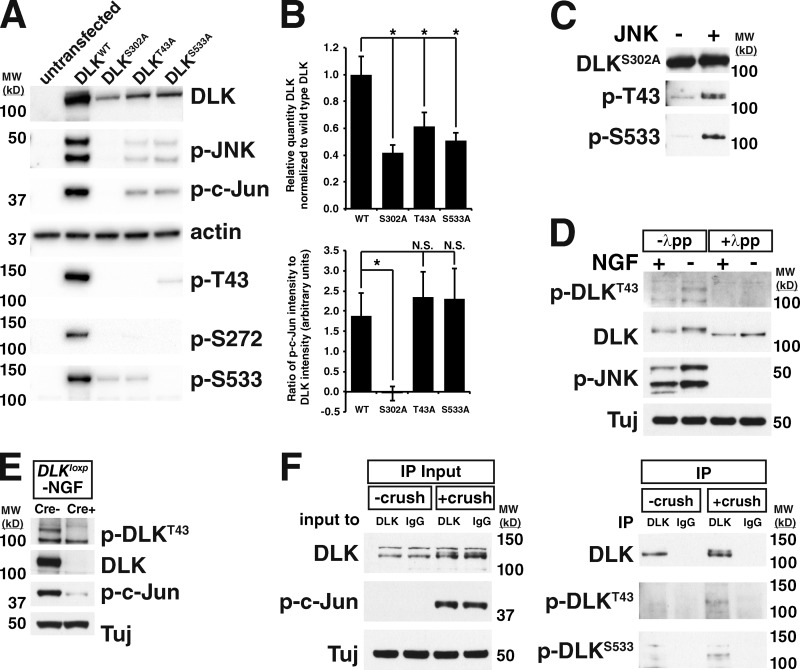
To determine the effect of phosphorylation of each of the top three identified sites on the stability of DLK, we expressed alanine point mutants of each in 293T cells. DLKS272A was inactive as measured by c-Jun phosphorylation (data not shown), so this point mutation was not pursued further. In contrast, DLKT43A and DLKS533A retained normal levels of kinase activity, but were expressed at lower levels than wild-type DLK (Fig. 8, A and B), consistent with a decrease in protein stability. Phospho-specific antibodies raised against T43, S272, and S533 failed to detect DLKT43A, DLKS272A, and DLKS533A, respectively, demonstrating specificity of the antibodies. In addition, these antibodies showed reduced detection of their phosphorylated antigens in DLKS302A, providing independent confirmation of our mass spectrometry results (Fig. 8 A). An in vitro kinase assay using purified JNK and DLK showed that both T43 and S533 could be phosphorylated directly by JNK (Fig. 8 C). Together, these data suggest that direct phosphorylation of these sites by JNKs contributes to enhanced DLK protein stability, although we cannot exclude the possibility of indirect phosphorylation mechanisms.
Figure 8.
Sites identified in DLK are phosphorylated after neuronal stress. (A) Western blots on lysates from HEK 293T cells in which DLKWT and the given phospho-incompetent alanine point mutants were transiently expressed. p-T43, p-S272, p-S533: blots with phospho-specific antibodies for each of these sites. (B) The expression shown in A was repeated three times and for each point mutant the amount of DLK expressed relative to DLKWT (top) or the intensity of p-c-Jun relative to the intensity of DLK (bottom) was quantified. WT: DLK = 1.00 ± 0.135. p-c-Jun = 1.89 ± 0.552. S302A: DLK = 0.417 ± 0.060. *, P = 0.0024. p-c-Jun = -0.046 ± 0.173. *, P = 0.0044. T43A: DLK = 0.614 ± 0.104. *, P = 0.017. p-c-Jun = 2.33 ± 0.629. S533A: DLK = 0.508 ± 0.057. *, P = 0.0044. p-c-Jun = 2.29 ± 0.773. n = 3 independent replicates. Error bars represent SD. (C) T43 and S533 of DLK can be directly phosphorylated by JNK. Purified DLKS302A was incubated with (right lane) or without (left lane) purified JNK3 and blotted with the shown phospho-specific antibodies. (D) Western blots on lysates from trophic factor–deprived sensory neurons demonstrate anti–phospho-T43 immunoreactivity specifically in the −NGF condition. Treatment of lysates with lambda protein phosphatase demonstrates the specificity of the antibody for phosphorylated protein. (E) Blotting trophic factor–deprived DRGs from Dlkloxp/loxp; Cre- and Cre+ embryos shows that the anti–p-T43 antibody is specific for DLK. (F) Immunoprecipitation of DLK from crushed retina lysates and uncrushed controls followed by blotting with phospho-T43– and phospho-S533–specific antibodies shows that these sites are phosphorylated after optic nerve crush. Left: input to immunoprecipitations showing the increase in DLK levels and molecular weight with optic nerve crush. Right: immunoprecipitation from crushed lysates or uncrushed controls with anti-DLK or a control rabbit IgG followed by blotting with the given phospho-DLK antibodies. MW (kD), molecular weight in kilodaltons.
We next asked whether phosphorylation of T43 and S533 occurs in a stress-dependent manner in neurons. To answer this question, lysates from DRGs cultured in the presence and absence of NGF were immunoblotted with the phospho-specific antibodies for T43 and S533 (Fig. 8 D). The antibody targeting phospho-T43 showed immunoreactivity specifically in the −NGF condition that was eliminated by lambda phosphatase treatment, demonstrating that DLK is indeed phosphorylated at this site after neuronal stress. Blotting trophic factor–deprived Dlklox/lox; Cre− and Cre+ lysates demonstrated the specificity of this antibody for DLK (Fig. 8 E). In addition, phosphorylation of both T43 and S533 was detected in DLK immunoprecipitated from retinas after optic nerve crush (Fig. 8 F). Thus, T43 and S533 are phosphorylated after neuronal stress in vivo, consistent with the hypothesis that phosphorylation of these sites contributes to DLK stability in neurons.
Discussion
DLK is a MAP3K that senses neuronal damage and triggers both degenerative and regenerative signaling (Chen et al., 2008; Hammarlund et al., 2009; Miller et al., 2009; Xiong et al., 2010, 2012; Ghosh et al., 2011; Shin et al., 2012; Watkins et al., 2013; Welsbie et al., 2013). Loss of DLK suppresses JNK activation and downstream responses in a strikingly wide variety of neuronal stress paradigms (Miller et al., 2009; Ghosh et al., 2011; Shin et al., 2012; Watkins et al., 2013; Welsbie et al., 2013), but it has remained unclear how DLK is itself regulated by neuronal stress in mammalian neurons. In this study, we demonstrate that JNK-dependent phosphorylation of DLK results in rapid stabilization and accumulation of DLK proximal to the site of axonal injury, and DLK abundance in turn directly controls the propagation of the retrograde stress response. These results suggest that the stabilization of DLK is an essential step in allowing neurons to mount a rapid and decisive response to localized insults.
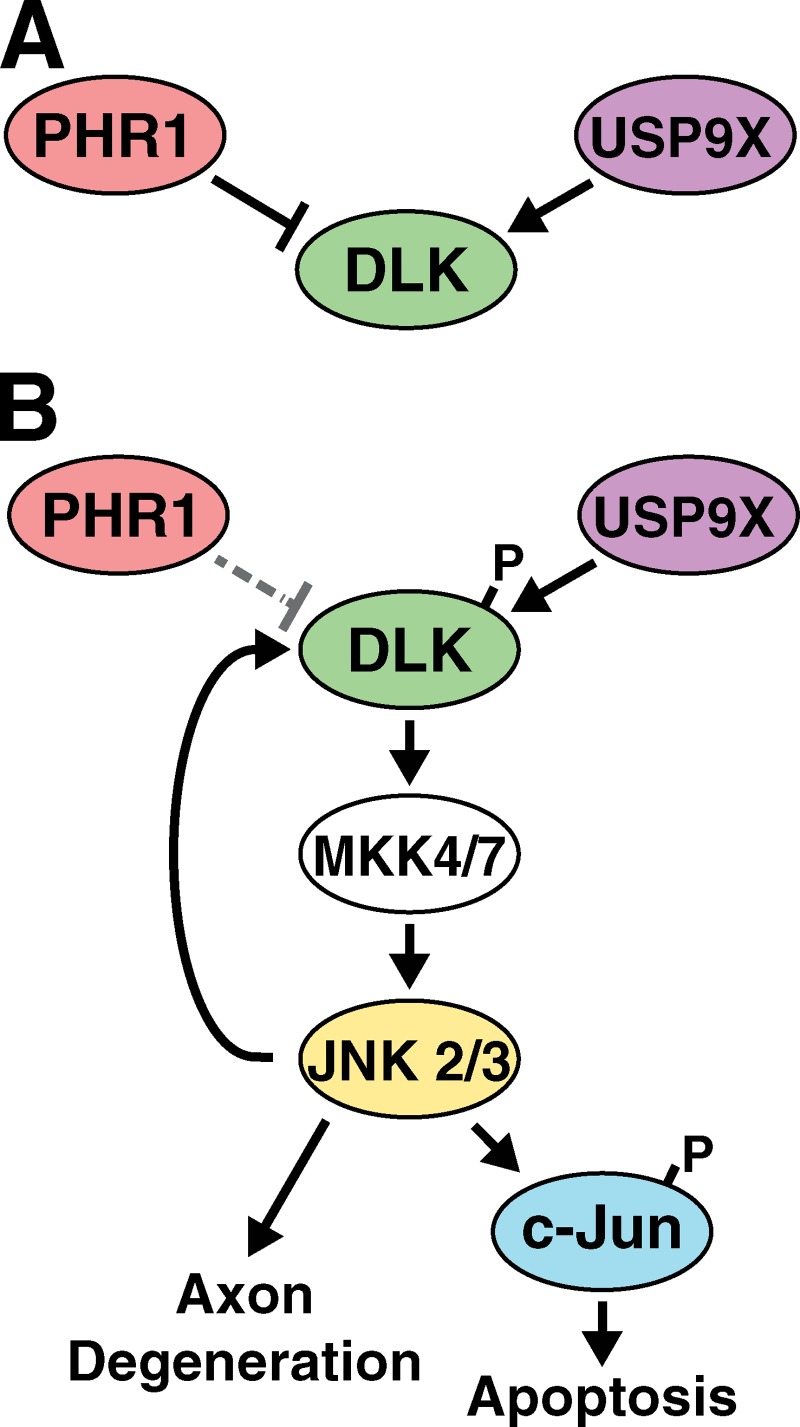
We propose the following model for regulation of DLK protein abundance and propagation of DLK/JNK signaling after axonal injury: DLK protein levels within the axon are tightly regulated under normal conditions via the opposing actions of PHR1 and USP9X (Fig. 9 A). Neuronal stresses (e.g., NGF deprivation or injury) lead to local activation of DLK and phosphorylation of the downstream targets MKK4/7 and JNK. A JNK-dependent feedback mechanism then results in phosphorylation of DLK on specific residues, causing reduced ubiquitination of DLK and enhanced DLK stability (Fig. 9 B). Unlike studies in invertebrates, which have focused on PHR1-dependent regulation of DLK function (Nakata et al., 2005; Collins et al., 2006; Xiong et al., 2010), this study demonstrates a role for DLK activity in regulating its own stability, generating a positive feedback loop. The change in DLK ubiquitination we observe after phosphorylation likely occurs through alteration of PHR1 activity or substrate availability of DLK for PHR1, although the presence of some ubiquitinated species of DLK in Phr1 mutants (Fig. 5 G) implies that additional E3 ubiquitin ligases may participate in this process. Our data suggest that USP9X continues de-ubiquitinating DLK at the same rate regardless of the conditions (Fig. 5, C and D) and when PHR1-dependent ubiquitination of DLK is reduced, de-ubiquitination by USP9X causes a rapid rise in DLK levels.
Figure 9.
Proposed model for a feedback loop mechanism for amplification and propagation of DLK stabilization and downstream JNK activation. (A) Before the onset of stress, DLK levels are kept at a constant low level by a “tug of war” between PHR1 and USP9X, and DLK is inactive. (B) After neuronal stress, DLK is activated, which drives downstream activation of JNKs. JNKs then phosphorylate DLK on sites that regulate stability. This feedback mechanism causes a rapid up-regulation of DLK levels.
To respond to axonal inputs that may originate a significant distance from the cell body, neurons use retrograde molecular motors such as dynein, which are able to rapidly transport signaling molecules from the axon (Chowdary et al., 2012). After axonal stress or injury, mediators of stress-response signaling such as p-JNK are retrogradely transported from the site of axonal injury through interaction with scaffolding proteins such as JIP3 (Cavalli et al., 2005). However, it is unclear what fraction of p-JNK originating at the injury site is able to reach the cell body. Previous studies have demonstrated that p-JNK can be observed in the optic nerve head 1 h after injury, but p-c-Jun cannot be detected in RGC nuclei until 6 h after crush (Fig. 4 A; Fernandes et al., 2012). In DLK heterozygous animals, the number of p-c-Jun–positive neurons at this time point is significantly reduced (Fig. 4, A and B), demonstrating that DLK protein quantity governs the amount of active JNK reaching the nucleus in the immediate response to injury. Thus, the feedback mechanism identified here may serve to facilitate propagation of this signaling by amplifying the levels of p-JNK as molecules are retrogradely transported to the cell body.
Similar feedback loops are used in other contexts to rapidly respond to changing conditions within cells or in the cellular environment. For example, JNK participates in a positive feedback loop in the Xenopus oocyte to trigger a bistable switching response to external stressful stimuli such as progesterone treatment or hyperosmolarity (Bagowski and Ferrell, 2001). In general, positive feedback loops contribute to bistability in cellular signaling, which can result in all-or-nothing, irreversible responses (Ferrell, 2002). Such mechanisms make quick changes in cellular state possible in environments with noisy inputs (D. Kim et al., 2007). Interestingly, in the contexts of Alzheimer’s disease and ALS, staining for c-Jun phosphorylation, which is indicative of cells that are responding to a stressful environment, displays an all-or-nothing salt-and-pepper pattern of a few highly positive cells in a largely negative background whose numbers increase as the disease progresses, as opposed to higher and higher p-c-Jun among many cells (Vlug et al., 2005; Thakur et al., 2007). These patterns of c-Jun activation are consistent with a bistable state model of progressive cellular pathology. Here we present evidence that DLK activation and subsequent downstream signaling participate in an activity-dependent feedback loop that confers bistability through amplification of neuronal stress signaling after localized insults to quickly generate a coordinated cellular response. Given the pattern of p-c-Jun activation in ALS and Alzheimer’s disease, this mechanism may also function to regulate axon degeneration and cell death in the disease setting.
Materials and methods
Mouse models
Dlk heterozygous mice contain a deletion of exons 2 through 5 that removes the DNA encoding the kinase domain and results in no protein expression (Ghosh et al., 2011). Dlk conditional knockout mice (Dlklox) contain loxp sites flanking exons 2 through 5 bred to chicken β-actin–cytomegalovirus (CMV) hybrid (CAG) driven Cre recombinase–estrogen receptor (Cre-ERT) mice obtained from The Jackson Laboratory. To induce Dlk recombination, both Dlklox: Crepos mice and their Dlklox: Creneg littermate controls were fed tamoxifen chow for 7 d (animals ingest roughly 40 mg/kg/d) and dosed intraperitoneally (i.p.) with 100 mg/kg tamoxifen in corn oil on days 1 and 3 (Watkins et al., 2013). Jnk2 knockout mice have portions of exons 1 and 2, which encode part of the kinase domain, replaced with a lacZ gene fused in-frame with exon 1 and a neo cassette driven by the Rous sarcoma virus promoter (Sabapathy et al., 1999). Phr1mag mice were a gift from the Salk Institute for Biological Studies (San Diego, CA) and contain a point mutation in the 63rd exon that results in a premature stop codon and deletion of the RING zinc finger domain that is required for PHR1 function (Lewcock et al., 2007). Jnk3 knockout mice were generated in C57BL/6 ES cells by genOway (Lyon, France) by homologous recombination with a targeting vector (Fig. S5 A). The targeting vector contained homology arms of 3.8 and 6.5 kb and replaced most of exon 11 with a neomycin resistance cassette. The deleted region encodes the T-P-Y tripeptide dual phosphorylation motif required for JNK activity. The neo cassette insertion creates a frameshift when exons 10 and 12 are spliced together, producing an early stop codon in exon 14. Neomycin-resistant ES cells clones were screened by PCR and Southern blot to validate homologous recombination of the cassette. Determination of Jnk3 genotype was performed by PCR with the following primers (diagrammed in Fig. S5 A): Primer 1: 5′-CCAGTAACATTGTAGTCAAGTCT-3′; Primer 2: 5′-TGGTCTTCCGCTTGGTAT-3′; Primer 3: 5′-CGCCTTCTATCGCCTTCT-3′.
Primers 1 and 2 produce a 249-bp fragment in the WT allele and no product in the KO allele. Primers 1 and 3 produce a 435-bp fragment in the KO allele and no product in the WT allele. Blotting for JNK2 and JNK3 in retina samples from Jnk2/3 double knockout and a littermate control shows loss of JNK2 and JNK3 protein in the knockout mice (Fig. S5 B).
Usp9x conditional knockout mice were generated from C57BL/6 ES cells by Lexicon Pharmaceuticals (Fig. S5 C). They contain a Usp9x allele with loxp sites flanking exon 31, which encodes catalytic Cys 1560. loxp sites were inserted by homologous recombination in ES cells using a FRT-flanked neomycin cassette with homology arms of 4.7 kb 5′ and 4.0 kb 3′ of exon 31. Neomycin-resistant ES cell clones were screened by Southern blot for homologous recombination of the cassette. Mice containing the floxed allele were crossed to a Flp deleter strain to remove the neomycin cassette. To achieve inducible recombination of the floxed Usp9x allele, Usp9x conditional knockout mice were crossed to a Rosa26-Cre/ERT2 line, which contains an insertion of a construct encoding a Cre–estrogen receptor fusion protein into the Rosa26 locus (Seibler et al., 2003). For DRG experiments, Usp9xloxp/loxp; Cre- and Usp9xloxp/loxp; Cre+ mice were crossed for timed pregnancies and embryos were genotyped for the presence or absence of Cre. Recombination in cultured DRGs was induced by adding 10 µM 4-hydroxytamoxifen (4-OHT, catalogue no. H7904; Sigma-Aldrich) to the cells for 48 h. 4-OHT was also added to Cre− control cells.
Western blotting
DRG cultures were lysed by incubation on ice for 30 min in buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, and 0.1% Triton X-100. HEK 293T cells were lysed by incubation on ice for 30 min in radioimmunoprecipitation assay (RIPA) buffer. Retina and nerve tissue samples were lysed in RIPA buffer using a TissueLyser (QIAGEN) with a 3-mm tungsten carbide bead (QIAGEN) for 6 min. Unless otherwise noted, all lysis solutions contained Complete protease inhibitor cocktail and PhosSTOP phosphatase inhibitor cocktail (Roche). Protein concentrations of 293T and retina lysates were determined by BCA assay (Thermo Fisher Scientific). Samples were loaded on NuPAGE 4–12% Bis-Tris gels (Invitrogen) and subjected to standard immunoblotting procedures. Except where noted, gels blotted for DLK were run in MOPS buffer (Invitrogen). Due to the large size of PHR1, samples blotted for PHR1 were run on 3–8% Tris-Acetate gels (Invitrogen). Blots were visualized with chemiluminescence and exposure to film. For relative protein expression and molecular weight quantification shown in Figs. 1 [B and C], 2 C, 3 G, 5 [D and F], 6 B, and 8 B, blots were also visualized on a ChemiDoc (Bio-Rad Laboratories) and quantified using ImageLab software (detailed below). For all other Western blots, band intensities were measured in ImageJ (National Institutes of Health) using the gel analysis plug-in.
For quantifications in Figs. 1 and 2, the quantity of DLK was measured in ImageLab software (Bio-Rad Laboratories), normalized to the actin loading control, and the ratio of DLK in −NGF and +NGF conditions was calculated. Molecular weight was calculated in ImageLab using the molecular weight analysis tool with Precision Plus Protein WesternC Ladder (Bio-Rad Laboratories) as a standard. For Fig. 3 G, the percent increase in the amount of total DLK in crushed and uncrushed samples was calculated after normalization to loading control. For Fig. 5 B, the intensity of the DLK signal (doublet in Fig. 5 A) was measured and normalized to the amount of input DLK for three separate experimental repetitions. The ratio shown is the normalized DLK intensity in the −NGF vs. +NGF conditions in the ubiquitin IP lanes. For Fig. 5, D and F, quantifications shown are those for blots shown in the figure but are representative of results from three experimental replicates.
Antibodies and inhibitors
The following antibodies were used for staining and Western blotting: anti-DLK (1:1,000 for Western and 1:500 for staining, produced at Genentech according to Hirai et al. [2002]); anti–p-JNK (1:250, #9251; Cell Signaling Technology); anti–p-c-Jun (1:250 for Western and 1:500 for staining, #9261; Cell Signaling Technology); anti–total JNK (1:500, #9252; Cell Signaling Technology); anti-JNK2 (1:500, #4672; Cell Signaling Technology); anti-JNK3 (1:500, #2305; Cell Signaling Technology); anti–β-tubulin (Tuj, 1:1,000, #MMS-435P-250; Covance); anti-actin (1:5,000, #612656; BD); anti–cleaved caspase-3 (1:500, #9664; Cell Signaling Technology); ant-BRN3 (1:100, #sc-6026; Santa Cruz Biotechnology, Inc.); anti–γ-synuclein (1:200, Abcam); anti–NF-M (1:200, #MMS-583S; Covance). Anti-USP9X (rat monoclonal 4B3) was produced at Genentech and was raised against the 198 C-terminal amino acids of human USP9X. Antibodies to PHR1 were generated by immunizing rabbits with a fragment of PHR1 comprising amino acids D3812–Q3961, which consists of the DOC domain, expressed in baculovirus. Serum was then affinity purified using a column loaded with the same peptide before use. This portion of the protein is absent in Phr1mag mutants. Antibodies to T43, S272, and S533 phosphorylation sites on DLK were generated through immunization of rabbits with the following peptides: PEKDL– pT–PTHVLQLHC, HRDLK–pS–PNMLITYDC, RNVPQKL– pS–PHSKRPC and affinity purified before use.
The following inhibitors were used at the given concentrations: cycloheximide (5 µM, #239764; EMD Millipore); okadaic acid (OA, 200 nM, #O8010; Sigma-Aldrich); MG132 (30 µM, #NC9937881; Thermo Fisher Scientific); TAT-TI-JIP153-163 (TI-JIP, 10 µM, #420134; EMD Millipore); and JNK inhibitor VIII (10 µM, #420135; EMD Millipore).
Primary neuron culture
Dorsal root ganglia were dissected from E12.5 to E13.5 mouse embryos, trypsinized (except in the case of explants), and cultured in F12 medium containing N3 supplement, 40 mM glucose, and 25 ng/ml NGF on chamber slides coated with poly-d-lysine and laminin (BioCoat; BD). The day after plating, 3 µM arabinofuranoside (AraC; Sigma-Aldrich) was added to the medium, removed two days later, and the medium was replaced with N3/F12/NGF without AraC. For NGF withdrawal experiments, after 4 to 5 d in vitro, medium was replaced with medium containing no NGF and 25 µg/ml anti-NGF antibody (Genentech) for between 1 and 3 h.
For siRNA experiments, dissociated DRGs were transfected using the Amaxa nucleofection system (Lonza). Jnk3 siRNA (sense 5′-ACATCGTAGTCAAGTCTGATTT-3′, antisense 5′-ATCAGACTTGACTACGATGTTT-3′; M.J. Kim et al., 2007) was synthesized at Genentech. Control siRNA was ON-TARGETplus Non-Targeting siRNA #1 from Thermo Fisher Scientific.
Cell culture
For stable DLK expression cells, human Dlk was cloned into pTRE2hyg vector (Takara Bio Inc.) and transfected into 293 Tet-ON cells (Takara Bio Inc.). Stable clones were selected using hygromycin resistance. Inducible cells were grown in DMEM supplemented with 10% Tet System Approved fetal bovine serum (Takara Bio Inc.), 25 µg/ml hygromycin, 100 µg/ml G418, 1× Glutamax (Gibco), and 1× penicillin/streptomycin solution (Gibco). Expression was induced by addition of 3 µM doxycycline to cells grown to 70–80% confluence, and cell were lysed 24 h later. Inhibitors were added at the time of addition of doxycycline.
Transient transfections of HEK 293T cells were performed using Fugene6 (Promega). Cells were grown to ∼70% confluency in DMEM supplemented with 1× Glutamax (Gibco), 1× penicillin/streptomycin (Gibco), and 10% fetal bovine serum. After transfection, cells were processed for Western blotting (see above) or Flag immunoprecipitation (see below). For inducible repression of DLK expression using doxycycline, 293 Tet-Off cells (Takara Bio Inc.) were transiently transfected with a vector containing Flag-tagged mouse DLK cloned into the pTRE2hyg vector (Takara Bio Inc.), which contains the Tet-responsive PhCMV-1 promoter. 20 h after transfection, 2 ug/ml doxycycline was added and cell lysates were immediately collected or collected 20 h later. Dlk expression vectors contained either N-terminal Flag-tagged or N-terminal myc-tagged mouse Dlk behind a CMV promoter in a pRK vector backbone. The Flag-USP9X expression vector was described in Schwickart et al. (2010) and contains C-terminally Flag-tagged USP9X behind a CMV promoter in a pRK vector backbone. The constitutively active JNK (CA-JNK) construct encoded a myc-tagged fusion of MKK7 and JNK2 as described in Lei et al. (2002). This fusion construct was cloned into pCAGGS, which contains a CMV promoter with an actin enhancer. The DLK leucine zipper construct (DLK-LZ) encoded residues 405–520 of mouse DLK as in Nihalani et al. (2000), plus an N-terminal myc tag, and was cloned into pCAGGS.
For stable isotope labeling by amino acids in cell culture (SILAC), cells were grown and transfected in either “heavy” medium or “light” medium, as noted in the figures. Heavy medium composition was as follows: DMEM without Lys or Arg (catalogue no. PI89985; Thermo Fisher Scientific) supplemented with 10% dialyzed FBS (Invitrogen), 2 mM l-glutamine, 50 µg/ml l-lysine-13C6, 15N2 hydrochloride (catalogue no. 608041, +8.01416 D; Sigma-Aldrich), 40 µg/ml l-arginine-13C6, 15N4 hydrochloride (catalogue no. 608033, +10.00822 D; Sigma-Aldrich), 200 mg/L proline, and 1× pen/strep (Gibco). Light medium was identical to heavy medium except supplemented with unlabeled isotopes of l-lysine and l-arginine (catalogue nos. L5501 and A5006, respectively; Sigma-Aldrich). To ensure complete labeling, cells were grown in SILAC medium long enough to allow for at least five doubling times.
Real-time qRT-PCR
RNA samples from dissociated DRGs were collected using the RNeasy Plus Mini kit (QIAGEN). Pre-designed Taqman primer sets were ordered from Applied Biosystems. Catalog numbers for primer sets were as follows: Dlk, Mm00437378_m1 (FAM labeled); Gapdh, 4352339E (VIC labeled). Comparative Ct (ΔΔCt) assays were performed using the Taqman RNA-to-Ct One-Step kit (#4392938; Applied Biosystems) on a 7500 Real-Time PCR system (Applied Biosystems) and analyzed in 7500 Software. Gapdh endogenous control and Dlk primers were multiplexed. Error bars represent the standard deviation of the relative quantities calculated from these technical replicates.
Lambda protein phosphatase assay
Lambda protein phosphatase, 10× NEBuffer for PMP, and 10 mM MnCl2 were all obtained from New England Biolabs, Inc. For DRGs, lysates were collected without phosphatase inhibitors or EDTA but otherwise under the same conditions as other DRG lysates in this manuscript. Lysates were incubated with 1× PMP buffer and 1 mM MnCl2 with either 800 U lambda protein phosphatase or the equivalent volume of 50% glycerol as a mock control at 30°C for 30 min. Reactions were stopped by heating with sample buffer and loading on a gel.
Cycloheximide time course to determine DLK stability
At time 0, DRG culture medium was replaced with medium containing no NGF, anti-NGF, and cycloheximide. Lysates were collected at the given time points and blotted for DLK. The experiment was performed three times. Bands at each time point were quantified relative to actin loading controls, and the mean band intensity at each time point was calculated and divided by the intensity at t = 0. The average quantity of DLK relative to the amount at time 0 was calculated for each time point. Linear regression and statistical analysis to compare the slopes of the two lines was performed in Prism software (GraphPad Software).
USP9X activity assay using HA-Ub-vinyl sulfone
HA-tagged ubiquitin vinyl sulfone was obtained from Enzo Life Sciences. The assay was performed as in Borodovsky et al. (2002). In brief, cultured DRGs were treated with anti-NGF or with NGF. DRGs were then resuspended in lysis buffer (50 mM Tris, pH 7.5, 5 mM MgCl2, 250 mM sucrose, 1 mM DTT, 2 mM ATP, and 100 µM PMSF) and dounced for lysis. Lysates were incubated with 6.6 µg/ml HA-ubiquitin vinyl sulfone at 25°C for 2 h. N-ethylmaleimide was added at 5 µM as a negative control. Reactions were stopped by boiling in sample buffer.
Mass spectrometry
30 h after transfection of the given vectors with Fugene 6, cells were harvested in ice-cold PBS and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, Complete protease inhibitors, PhosSTOP phosphatase inhibitors, 30 mM MG132, and 2 mM N-ethylmaleimide). They were rotated for 45 min at 4°C and centrifuged at 20,000 g for 10 min at 4°C. The protein content of the supernatant was measured by BCA assay and samples were taken from each lysate for Western blot analysis. Samples to be compared for phosphorylation of DLK were combined 1:1 based on total protein as determined by the BCA assay. 20 µl washed anti-Flag resin (catalogue no. A2220; Sigma-Aldrich) were incubated with the combined lysates for 1 h at 4°C. Beads were then washed 2× in lysis buffer and 2× in lysis buffer without Triton X-100. Supernatant was then completely removed from the beads and DLK was eluted by heating in sample buffer. Proteins in the samples were separated by SDS-PAGE and stained using Coomassie SafeStain (Invitrogen).
The gel regions containing DLK protein were excised completely, cut into 1-mm cubes, and destained at room temperature for 30 min using 50% methanol (MeOH)/50 mm ammonium bicarbonate (AMBIC). Gel pieces were dehydrated with 100% acetonitrile (ACN), reduced with 1.5 mM dithiothreitol (DTT)/50 mM AMBIC for 30 min at 60°C, rinsed, and dehydrated with 100% ACN, and then alkylated with 7.5 mM iodoacetamide (IDAC)/50 mM AMBIC for 15 min at room temperature in the dark. Gel pieces were rinsed again and dehydrated with 100% ACN before overnight digest at 37°C in 50 mM AMBIC/5% ACN/20 µg/ml modified sequencing grade trypsin (Promega). Peptides were extracted for analysis twice, first by gently shaking for 20 min in 1× gel volume of 50% ACN/5% formic acid (FA) and subsequently by dehydration using 1× gel volume of 100% ACN. Extracted peptides were combined in autosampler vials and dehydrated completely by speed vac. For mass spectrometry analysis, dried peptides were resuspended in 5% ACN/0.1% trifluoroacetic acid (TFA) and injected onto a 0.1-mm × 100-mm column packed with 1.7 mm BEH-139 C18 resin (Waters) using a NanoAcquity UPLC as described previously (Phu et al., 2011). Peptides were separated by reversed phase chromatography across a 35-min gradient and subsequently analyzed on an LTQ-Orbitrap XL (Thermo Fisher Scientific) operating in a data-dependent top 8 mode. Data were searched using Mascot (Matrix Science) against a target-decoy database comprised of forward and reverse sequences from mouse and human proteins (Uniprot v2011_12), as well as common contaminants. Carbamidomethylated cysteine was considered as a fixed modification, oxidized methionine and phosphorylated serine, threonine, and tyrosine, as well as SILAC-labeled lysine (13C615N2; +8.01416 D) and arginine (13C615N4; +10.00822 D) were all considered as variable modifications. Search data were filtered to a 5% peptide false discovery rate using linear discriminant analysis and subsequently identified DLK peptide data were quantified using the VistaQuant algorithm (Bakalarski et al., 2008). To check labeling efficiency of input material, gel regions containing heavy and light SILAC lysate were similarly digested and analyzed. The Ascore algorithm was used to localize and generate confidence scores for each phosphorylation site within the data (Beausoleil et al., 2006). Ascores of 13 and 19 correspond to 95 and 99% confidence in the phosphorylation site localization events. Note on S643 site: because the peptide spectral match reporting S643 has an Ascore of 10.3 (∼90% confidence) and because S643 resides immediately adjacent to proline, we attributed phosphorylation to this site over S642. Note on S295-T306: two phosphorylated residues were detected on this peptide, but because of a lack of site-determining ions a single, doubly modified sequence cannot be unambiguously assigned. Given the number of available permutations, it is possible that several multiply phosphopeptide sequences occur. For this reason, data for this doubly modified sequence are shown in the aggregate, rather than specifically by site (Fig. S4 H).
To accurately determine abundance ratios for individual phosphorylation events between paired SILAC samples, correction factors were calculated to account for differences in total DLK between each pair of conditions. Database search results were used to identify peptides from DLK that were only observed in the unmodified form. Additional searches were performed on the DLK-containing gel region and the regions immediately above the visible DLK band to consider alternate modifications with the potential to confound peptide measurements. After eliminating peptides containing serine or threonine residues as possible sites of phosphorylation, four DLK peptides were selected to use in determining a correction factor; ANNLYM*ELNALM*LQLELK, FHGEEVAVK, GLHHDLLLR, and LEEELVM*R, where M* indicates methionine sulfoxide. The ratio for each of these peptides was determined for each paired SILAC sample and averaged value used to adjust individual phosphopeptide ratios. The log2ratios for the four samples were: WT (light) + okadaic acid (heavy) at +1.29; WT (light) + JNK inhibitor (heavy) at −1.07; WT (light) + DLKS302A (heavy) at −1.99; and DLKS302A (light) + constitutively active JNK (heavy) at 0.77.
Immunoprecipitations
For anti-ubiquitin immunoprecipitations (IPs), DRGs dissected from E12.5 CD-1 mice (Charles River) were lysed as previously stated with the addition of 30 µM MG132 and 5 µM N-ethylmaleimide in the lysis buffer. Lysates were precleared for 30 min with protein G–conjugated Dynabeads (Life Technologies). 6 µg anti-ubiquitin antibody (clone FK2; EMD Millipore) or equivalent mouse IgG1 isotype control were added to the lysates overnight, followed by immunoprecipitation with Protein G–Dynabeads for 1 h. Dynabeads were washed 3× and bound proteins were eluted in sample buffer for Western blotting.
For immunoprecipitations of DLK from retina lysates, the procedure was identical to that given above, except 7.5 µg anti-DLK antibody or equivalent rabbit IgG isotype control were added to 500 µg lysate.
In vitro JNK kinase assay
Flag-tagged DLKS302A was immunoprecipitated from 293T cell lysates using anti–Flag-conjugated magnetic beads (Sigma-Aldrich). After washing, the DLK-bound Flag beads were incubated with 2,000 U lambda protein phosphatase, 1× PMP buffer, and 1× MnCl2 for 30 min at 30°C to remove all phosphate groups from the purified DLK. The beads were then washed with buffer containing phosphatase inhibitors and split into two tubes with kinase reaction buffer (50 mM Hepes, pH 7.2, 10 mM MgCl2, 1 mM EGTA, 0.01% Triton X-100, 2 mM DTT, and 30 µM ATP). 126 ng GST-tagged human recombinant JNK3 (EMD Millipore) was added to one of the two tubes and they were incubated at 30°C for 90 min. DLK was eluted from the Flag beads by heating in sample buffer, and samples were loaded on a gel for blotting.
Surgical procedures, tissue processing, immunocytochemistry, and immunohistochemistry
All experiments with mice were performed under animal protocols approved by the Animal Care and Use Committee at Genentech Inc. Animals (∼12 wk old) were deeply anesthetized using i.p. injection of 0.5 mg/g Avertin. The left eye of each animal was subjected to optic nerve crush injury via an intra-orbital approach. An incision was made on the superior conjunctiva of the surgery eye and the optic nerve was exposed. The crush injury was inflicted for 1 s using a 45° angled fine-tip forceps 1–2 mm from the eyeball and un-operated right eyes were used as controls.
For AAV-Cre studies, 8-wk-old Dlklox/lox;Cre− or DLK+/+;Cre− littermate control mice were injected intravitreally with 2 µl of AAV2-GFP-T2A-iCre (custom vector driving the expression of both EGFP and codon-optimized Cre recombinase under the control of the neuron-specific enhanced human Synapsin I promoter, produced by Vector Laboratories as described previously; Watkins et al., 2013) at a titer of 1013/ml under isoflurane anesthesia using a 2.5-µl Hamilton syringe fitted with a custom 33-gauge, 3/8-inch needle. Needle was inserted just behind the ora serrata at an angle to avoid lens injury, partially removed briefly to allow for drainage of vitreous fluid, reinserted for vector delivery, and held for 10 s after injection before removal. 2 wk after vector injection, optic nerve injury was induced by 5-s retro-orbital nerve crush under isoflurane anesthesia.
For stainings, mouse eyes or optic nerves were collected and fixed in 4% PFA at room temperature for 1 h. After two washes in PBS, tissues were processed for cryosectioning or the retinas were dissected out for whole-mount staining. For immunostaining on retinal sections and optic nerves, 20-µm-thick cryosections were incubated in blocking buffer (2% [vol/vol] donkey serum and 0.2% [vol/vol] Triton X-100 from Sigma-Aldrich in PBS) at room temperature for 1 h, followed by overnight incubation at 4°C in the same blocking buffer containing primary antibodies. The next day, the sections were washed and incubated with DyLight-488–, -549–, and -649–conjugated secondary antibodies (1:400, Jackson ImmunoResearch Laboratories, Inc.) for 1 h and then coverslipped with DAPI–Fluoromount G (SouthernBiotech).
For whole-mount stainings, retinas were blocked in blocking buffer (5% donkey serum and 0.5% Triton X-100 in PBS) at 4°C for 2 h, followed by overnight incubation at 4°C in primary antibodies diluted in blocking buffer. After washes in 0.5% PBST at 4°C for 6 h (six washes), the retinas were incubated overnight at 4°C with DyLight-488–, -549–, and -649–conjugated secondary antibodies (1:400, Jackson ImmunoResearch Laboratories, Inc.). Each retina was cut with four slits along the radial axes from the edge to ∼2/3 of the radius to flatten the retina, and then mounted on slides.
DRG explants to be imaged were fixed in 4% paraformaldehyde and 3% sucrose in PBS for 30 min, followed by permeabilization with 0.2% Triton X-100. They were blocked in 2% BSA in PBS for ≥1 h and incubated with anti–β-tubulin or anti–p-JNK in block overnight at 4°C. After washing 3× in PBS, Alexa 568–conjugated donkey anti–mouse and/or Alexa 488–conjugated donkey anti–rabbit secondary antibodies (1:500; Molecular Probes) were added. The explants were then washed 3× in PBS and mounted in Fluoromount-G containing DAPI stain (SouthernBiotech).
Imaging and quantification
Images of retinas and optic nerves were acquired under a confocal microscope (LSM710 with an LSM-TPMT camera, Zen 2010 software; Carl Zeiss) using a 20×/0.4 NA objective (Carl Zeiss) for whole-mount and optic nerve images and a 40×/1.4 NA oil objective (Carl Zeiss) for images of retina sections. To quantify the number of BRN3 and caspase-3–labeled cells from whole-mount retinas, one area covering a 10× field of view was imaged at the center of each quadrant/lobe of the retina on an AxioImager M2 (Carl Zeiss) using Slidebook acquisition software (Intelligent Imaging Innovations). Images were analyzed using Matlab R2011b (MathWorks), except p-c-Jun, in which confocal images were quantified using an IN Cell analyzer (GE Healthcare) due to high density of labeled cells. Images of DRG explants were acquired using a fluorescent microscope (DM5500, Advanced Fluorescence Application Suite software; Leica) with a DFC360 camera using a 20×/0.70 NA objective (Leica) for images in Fig. S3 C and a 63×/1.40 NA oil objective (Leica) for images in Fig. 5 H. All images were acquired at room temperature. Images shown were processed in Adobe Photoshop to convert to grayscale and to enhance visibility by adjusting brightness, contrast, and levels, and pseudocoloring where appropriate. Within an individual figure, all images were subjected to the same post-acquisition processing.
Online supplemental material
Fig. S1 shows blots and notations to clarify background bands in the blots shown in Fig. 3 of the main text. Fig. S2 shows a characterization of DLK levels, cell viability, and axonal structure in retinas after optic nerve crush in DLK+/+ and DLK+/− mice. It also shows a time course of DLK, JNK, and c-Jun activation in DLK+/+ and DLK+/− DRG cultures. Fig. S3 shows that the DLK doublet observed with ubiquitin IP is not due to phosphorylated DLK, that USP9X activity does not change with trophic factor withdrawal and that Phr1mag DRG explants do not have altered neurodegeneration after NGF withdrawal. Fig. S4 shows peptide spectral matches demonstrating phosphorylation on DLK. Fig. S5 shows the strategies used to generate Jnk3 knockout mice and Usp9x conditional knockout mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201303066/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201303066.dv.
Supplementary Material
Acknowledgments
We thank E. Naik for reagents and assistance with the Usp9x conditional knockout mice. Phrmag mutant mice were a gift from the Salk Institute for Biological Studies. All authors are employees of Genentech, Inc.
Footnotes
Abbreviations used in this paper:
- 293T
- human embryonic kidney 293T cells
- ALS
- amyotrophic lateral sclerosis
- DLK
- dual leucine zipper-bearing kinase
- DLK-LZ
- a protein containing only the leucine zipper domains of DLK
- DRG
- dorsal root ganglion
- DUB
- de-ubiquitinating enzyme
- GCL
- ganglion cell layer
- INL
- inner nuclear layer
- JNK
- Jun N-terminal kinase
- MAP3K
- mitogen-activated protein kinase kinase kinase
- MKK4/7
- mitogen-activated protein kinase kinase 4 or 7
- MLK
- mixed lineage kinase
- ONL
- outer nuclear layer
- PHR
- PAM/Highwire/RPM-1 family of E3 ubiquitin ligases
- qRT-PCR
- quantitative reverse-transcription PCR
- RGC
- retinal ganglion cell
- SILAC
- stable isotope labeling by amino acids in cell culture
- USP9X
- ubiquitin-specific peptidase 9, X chromosome
- UbVS
- ubiquitin vinyl sulfone
- WT
- wild type
References
- Babetto E., Beirowski B., Russler E.V., Milbrandt J., DiAntonio A. 2013. The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 3:1422–1429 10.1016/j.celrep.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagowski C.P., Ferrell J.E., Jr 2001. Bistability in the JNK cascade. Curr. Biol. 11:1176–1182 10.1016/S0960-9822(01)00330-X [DOI] [PubMed] [Google Scholar]
- Bakalarski C.E., Elias J.E., Villén J., Haas W., Gerber S.A., Everley P.A., Gygi S.P. 2008. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J. Proteome Res. 7:4756–4765 10.1021/pr800333e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil S.A., Villén J., Gerber S.A., Rush J., Gygi S.P. 2006. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24:1285–1292 10.1038/nbt1240 [DOI] [PubMed] [Google Scholar]
- Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., Kessler B.M. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9:1149–1159 10.1016/S1074-5521(02)00248-X [DOI] [PubMed] [Google Scholar]
- Cavalli V., Kujala P., Klumperman J., Goldstein L.S. 2005. Sunday Driver links axonal transport to damage signaling. J. Cell Biol. 168:775–787 10.1083/jcb.200410136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Jones Y., Ellisman M.H., Goldstein L.S., Karin M. 2003. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell. 4:521–533 10.1016/S1534-5807(03)00094-7 [DOI] [PubMed] [Google Scholar]
- Chen X., Rzhetskaya M., Kareva T., Bland R., During M.J., Tank A.W., Kholodilov N., Burke R.E. 2008. Antiapoptotic and trophic effects of dominant-negative forms of dual leucine zipper kinase in dopamine neurons of the substantia nigra in vivo. J. Neurosci. 28:672–680 10.1523/JNEUROSCI.2132-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdary P.D., Che D.L., Cui B. 2012. Neurotrophin signaling via long-distance axonal transport. Annu. Rev. Phys. Chem. 63:571–594 10.1146/annurev-physchem-032511-143704 [DOI] [PubMed] [Google Scholar]
- Coffey E.T., Smiciene G., Hongisto V., Cao J., Brecht S., Herdegen T., Courtney M.J. 2002. c-Jun N-terminal protein kinase (JNK) 2/3 is specifically activated by stress, mediating c-Jun activation, in the presence of constitutive JNK1 activity in cerebellar neurons. J. Neurosci. 22:4335–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.A., Wairkar Y.P., Johnson S.L., DiAntonio A. 2006. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 51:57–69 10.1016/j.neuron.2006.05.026 [DOI] [PubMed] [Google Scholar]
- Eilers A., Whitfield J., Shah B., Spadoni C., Desmond H., Ham J. 2001. Direct inhibition of c-Jun N-terminal kinase in sympathetic neurones prevents c-jun promoter activation and NGF withdrawal-induced death. J. Neurochem. 76:1439–1454 10.1046/j.1471-4159.2001.00150.x [DOI] [PubMed] [Google Scholar]
- Erkman L., McEvilly R.J., Luo L., Ryan A.K., Hooshmand F., O’Connell S.M., Keithley E.M., Rapaport D.H., Ryan A.F., Rosenfeld M.G. 1996. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 381:603–606 10.1038/381603a0 [DOI] [PubMed] [Google Scholar]
- Fan G., Merritt S.E., Kortenjann M., Shaw P.E., Holzman L.B. 1996. Dual leucine zipper-bearing kinase (DLK) activates p46SAPK and p38mapk but not ERK2. J. Biol. Chem. 271:24788–24793 10.1074/jbc.271.44.27237 [DOI] [PubMed] [Google Scholar]
- Fernandes K.A., Harder J.M., Fornarola L.B., Freeman R.S., Clark A.F., Pang I.H., John S.W., Libby R.T. 2012. JNK2 and JNK3 are major regulators of axonal injury-induced retinal ganglion cell death. Neurobiol. Dis. 46:393–401 10.1016/j.nbd.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J.E., Jr 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14:140–148 10.1016/S0955-0674(02)00314-9 [DOI] [PubMed] [Google Scholar]
- Gagliardini V., Fernandez P.A., Lee R.K., Drexler H.C., Rotello R.J., Fishman M.C., Yuan J. 1994. Prevention of vertebrate neuronal death by the crmA gene. Science. 263:826–828 10.1126/science.8303301 [DOI] [PubMed] [Google Scholar]
- Gan L., Xiang M., Zhou L., Wagner D.S., Klein W.H., Nathans J. 1996. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc. Natl. Acad. Sci. USA. 93:3920–3925 10.1073/pnas.93.9.3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.S., Wang B., Pozniak C.D., Chen M., Watts R.J., Lewcock J.W. 2011. DLK induces developmental neuronal degeneration via selective regulation of proapoptotic JNK activity. J. Cell Biol. 194:751–764 10.1083/jcb.201103153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V., Levi-Montalcini R. 1949. Proliferation, differentiation and degeneration in the spinal ganglia of the chick embryo under normal and experimental conditions. J. Exp. Zool. 111:457–501 10.1002/jez.1401110308 [DOI] [PubMed] [Google Scholar]
- Hammarlund M., Nix P., Hauth L., Jorgensen E.M., Bastiani M. 2009. Axon regeneration requires a conserved MAP kinase pathway. Science. 323:802–806 10.1126/science.1165527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai S., Kawaguchi A., Hirasawa R., Baba M., Ohnishi T., Ohno S. 2002. MAPK-upstream protein kinase (MUK) regulates the radial migration of immature neurons in telencephalon of mouse embryo. Development. 129:4483–4495 [DOI] [PubMed] [Google Scholar]
- Hirai S., Kawaguchi A., Suenaga J., Ono M., Cui D.F., Ohno S. 2005. Expression of MUK/DLK/ZPK, an activator of the JNK pathway, in the nervous systems of the developing mouse embryo. Gene Expr. Patterns. 5:517–523 10.1016/j.modgep.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Holzman L.B., Merritt S.E., Fan G. 1994. Identification, molecular cloning, and characterization of dual leucine zipper bearing kinase. A novel serine/threonine protein kinase that defines a second subfamily of mixed lineage kinases. J. Biol. Chem. 269:30808–30817 [PubMed] [Google Scholar]
- Hunot S., Vila M., Teismann P., Davis R.J., Hirsch E.C., Przedborski S., Rakic P., Flavell R.A. 2004. JNK-mediated induction of cyclooxygenase 2 is required for neurodegeneration in a mouse model of Parkinson’s disease. Proc. Natl. Acad. Sci. USA. 101:665–670 10.1073/pnas.0307453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Kwon Y.K., Cho K.H. 2007. Coupled positive and negative feedback circuits form an essential building block of cellular signaling pathways. Bioessays. 29:85–90 10.1002/bies.20511 [DOI] [PubMed] [Google Scholar]
- Kim M.J., Futai K., Jo J., Hayashi Y., Cho K., Sheng M. 2007. Synaptic accumulation of PSD-95 and synaptic function regulated by phosphorylation of serine-295 of PSD-95. Neuron. 56:488–502 10.1016/j.neuron.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Kuan C.Y., Yang D.D., Samanta Roy D.R., Davis R.J., Rakic P., Flavell R.A. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 22:667–676 10.1016/S0896-6273(00)80727-8 [DOI] [PubMed] [Google Scholar]
- Lei K., Nimnual A., Zong W.X., Kennedy N.J., Flavell R.A., Thompson C.B., Bar-Sagi D., Davis R.J. 2002. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Mol. Cell. Biol. 22:4929–4942 10.1128/MCB.22.13.4929-4942.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung I.W., Lassam N. 2001. The kinase activation loop is the key to mixed lineage kinase-3 activation via both autophosphorylation and hematopoietic progenitor kinase 1 phosphorylation. J. Biol. Chem. 276:1961–1967 10.1074/jbc.M004092200 [DOI] [PubMed] [Google Scholar]
- Lewcock J.W., Genoud N., Lettieri K., Pfaff S.L. 2007. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 56:604–620 10.1016/j.neuron.2007.09.009 [DOI] [PubMed] [Google Scholar]
- Luo L., O’Leary D.D. 2005. Axon retraction and degeneration in development and disease. Annu. Rev. Neurosci. 28:127–156 10.1146/annurev.neuro.28.061604.135632 [DOI] [PubMed] [Google Scholar]
- Martin L.J. 1999. Neuronal death in amyotrophic lateral sclerosis is apoptosis: possible contribution of a programmed cell death mechanism. J. Neuropathol. Exp. Neurol. 58:459–471 10.1097/00005072-199905000-00005 [DOI] [PubMed] [Google Scholar]
- Mata M., Merritt S.E., Fan G., Yu G.G., Holzman L.B. 1996. Characterization of dual leucine zipper-bearing kinase, a mixed lineage kinase present in synaptic terminals whose phosphorylation state is regulated by membrane depolarization via calcineurin. J. Biol. Chem. 271:16888–16896 10.1074/jbc.271.28.16888 [DOI] [PubMed] [Google Scholar]
- Miller B.R., Press C., Daniels R.W., Sasaki Y., Milbrandt J., DiAntonio A. 2009. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat. Neurosci. 12:387–389 10.1038/nn.2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy V., Han S., Beauchamp R.L., Smith N., Haddad L.A., Ito N., Ramesh V. 2004. Pam and its ortholog highwire interact with and may negatively regulate the TSC1.TSC2 complex. J. Biol. Chem. 279:1351–1358 10.1074/jbc.M310208200 [DOI] [PubMed] [Google Scholar]
- Nakata K., Abrams B., Grill B., Goncharov A., Huang X., Chisholm A.D., Jin Y. 2005. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 120:407–420 10.1016/j.cell.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Nihalani D., Merritt S., Holzman L.B. 2000. Identification of structural and functional domains in mixed lineage kinase dual leucine zipper-bearing kinase required for complex formation and stress-activated protein kinase activation. J. Biol. Chem. 275:7273–7279 10.1074/jbc.275.10.7273 [DOI] [PubMed] [Google Scholar]
- Pettmann B., Henderson C.E. 1998. Neuronal cell death. Neuron. 20:633–647 10.1016/S0896-6273(00)81004-1 [DOI] [PubMed] [Google Scholar]
- Phu L., Izrael-Tomasevic A., Matsumoto M.L., Bustos D., Dynek J.N., Fedorova A.V., Bakalarski C.E., Arnott D., Deshayes K., Dixit V.M., Kelley R.F., Vucic D., Kirkpatrick D.S. 2011. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell Proteomics. 10:M110.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley H.A., Nickells R.W., Kerrigan L.A., Pease M.E., Thibault D.J., Zack D.J. 1995. Retinal ganglion cell death in experimental glaucoma and after axotomy occurs by apoptosis. Invest. Ophthalmol. Vis. Sci. 36:774–786 [PubMed] [Google Scholar]
- Sabapathy K., Hu Y., Kallunki T., Schreiber M., David J.P., Jochum W., Wagner E.F., Karin M. 1999. JNK2 is required for efficient T-cell activation and apoptosis but not for normal lymphocyte development. Curr. Biol. 9:116–125 10.1016/S0960-9822(99)80065-7 [DOI] [PubMed] [Google Scholar]
- Schwickart M., Huang X., Lill J.R., Liu J., Ferrando R., French D.M., Maecker H., O’Rourke K., Bazan F., Eastham-Anderson J., et al. 2010. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 463:103–107 10.1038/nature08646 [DOI] [PubMed] [Google Scholar]
- Seibler J., Zevnik B., Küter-Luks B., Andreas S., Kern H., Hennek T., Rode A., Heimann C., Faust N., Kauselmann G., et al. 2003. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 31:e12 10.1093/nar/gng012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.E., Cho Y., Beirowski B., Milbrandt J., Cavalli V., DiAntonio A. 2012. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 74:1015–1022 10.1016/j.neuron.2012.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D.J., Weimer R.M., McLaughlin T., Kallop D., Stanger K., Yang J., O’Leary D.D., Hannoush R.N., Tessier-Lavigne M. 2012. A caspase cascade regulating developmental axon degeneration. J. Neurosci. 32:17540–17553 10.1523/JNEUROSCI.3012-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z., Lu K.P., Kwon Y.T., Tsai L.H., Filhol O., Cochet C., Brickey D.A., Soderling T.R., Bartleson C., Graves D.J., et al. 1996. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16:6486–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell D.G., Paredes M.F., Galvao R.P., Jones D.L., Froemke R.C., Sebe J.Y., Alfaro-Cervello C., Tang Y., Garcia-Verdugo J.M., Rubenstein J.L., et al. 2012. Intrinsically determined cell death of developing cortical interneurons. Nature. 491:109–113 10.1038/nature11523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A., Wang X., Siedlak S.L., Perry G., Smith M.A., Zhu X. 2007. c-Jun phosphorylation in Alzheimer disease. J. Neurosci. Res. 85:1668–1673 10.1002/jnr.21298 [DOI] [PubMed] [Google Scholar]
- Vila M., Przedborski S. 2003. Targeting programmed cell death in neurodegenerative diseases. Nat. Rev. Neurosci. 4:365–375 10.1038/nrn1100 [DOI] [PubMed] [Google Scholar]
- Vila M., Jackson-Lewis V., Vukosavic S., Djaldetti R., Liberatore G., Offen D., Korsmeyer S.J., Przedborski S. 2001. Bax ablation prevents dopaminergic neurodegeneration in the 1-methyl- 4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson’s disease. Proc. Natl. Acad. Sci. USA. 98:2837–2842 10.1073/pnas.051633998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlug A.S., Teuling E., Haasdijk E.D., French P., Hoogenraad C.C., Jaarsma D. 2005. ATF3 expression precedes death of spinal motoneurons in amyotrophic lateral sclerosis-SOD1 transgenic mice and correlates with c-Jun phosphorylation, CHOP expression, somato-dendritic ubiquitination and Golgi fragmentation. Eur. J. Neurosci. 22:1881–1894 10.1111/j.1460-9568.2005.04389.x [DOI] [PubMed] [Google Scholar]
- Watkins T.A., Wang B., Huntwork-Rodriguez S., Yang J., Jiang Z., Eastham-Anderson J., Modrusan Z., Kaminker J.S., Tessier-Lavigne M., Lewcock J.W. 2013. DLK initiates a transcriptional program that couples apoptotic and regenerative responses to axonal injury. Proc. Natl. Acad. Sci. USA. 110:4039–4044 10.1073/pnas.1211074110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsbie D.S., Yang Z., Ge Y., Mitchell K.L., Zhou X., Martin S.E., Berlinicke C.A., Hackler L., Jr, Fuller J., Fu J., et al. 2013. Functional genomic screening identifies dual leucine zipper kinase as a key mediator of retinal ganglion cell death. Proc. Natl. Acad. Sci. USA. 110:4045–4050 10.1073/pnas.1211284110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White F.A., Keller-Peck C.R., Knudson C.M., Korsmeyer S.J., Snider W.D. 1998. Widespread elimination of naturally occurring neuronal death in Bax-deficient mice. J. Neurosci. 18:1428–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Wang X., Ewanek R., Bhat P., Diantonio A., Collins C.A. 2010. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J. Cell Biol. 191:211–223 10.1083/jcb.201006039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Hao Y., Sun K., Li J., Li X., Mishra B., Soppina P., Wu C., Hume R.I., Collins C.A. 2012. The Highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol. 10:e1001440 10.1371/journal.pbio.1001440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Maroney A.C., Dobrzanski P., Kukekov N.V., Greene L.A. 2001. The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Mol. Cell. Biol. 21:4713–4724 10.1128/MCB.21.14.4713-4724.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Jin Y. 2012. Regulation of DLK-1 kinase activity by calcium-mediated dissociation from an inhibitory isoform. Neuron. 76:534–548 10.1016/j.neuron.2012.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Nguyen T.V., Pike C.J. 2005. Beta-amyloid-induced neuronal apoptosis involves c-Jun N-terminal kinase-dependent downregulation of Bcl-w. J. Neurosci. 25:1149–1158 10.1523/JNEUROSCI.4736-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Yankner B.A. 2000. Apoptosis in the nervous system. Nature. 407:802–809 10.1038/35037739 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.