Depletion analyses and nanometer-scale mapping of spindle assembly checkpoint proteins reveal how these proteins are integrated within the substructure of the kinetochore.
Abstract
Spindle assembly checkpoint proteins have been thought to reside in the peripheral corona region of the kinetochore, distal to microtubule attachment sites at the outer plate. However, recent biochemical evidence indicates that checkpoint proteins are closely linked to the core kinetochore microtubule attachment site comprised of the Knl1–Mis12–Ndc80 (KMN) complexes/KMN network. In this paper, we show that the Knl1–Zwint1 complex is required to recruit the Rod–Zwilch–Zw10 (RZZ) and Mad1–Mad2 complexes to the outer kinetochore. Consistent with this, nanometer-scale mapping indicates that RZZ, Mad1–Mad2, and the C terminus of the dynein recruitment factor Spindly are closely juxtaposed with the KMN network in metaphase cells when their dissociation is blocked and the checkpoint is active. In contrast, the N terminus of Spindly is ∼75 nm outside the calponin homology domain of the Ndc80 complex. These results reveal how checkpoint proteins are integrated within the substructure of the kinetochore and will aid in understanding the coordination of microtubule attachment and checkpoint signaling during chromosome segregation.
Introduction
The Mad1–Mad2 pathway of the spindle assembly checkpoint in animal cells depends on the protein module of Zwint1, Rod–Zw10–Zwilch (RZZ), Mad1–Mad2, Spindly, and dynein–dynactin (Lara-Gonzalez et al., 2012). In prometaphase, Zwint1 helps recruit RZZ, RZZ helps recruit Mad1–Mad2 and Spindly, and Spindly recruits dynein–dynactin to kinetochores (Starr et al., 2000; Wang et al., 2004; Buffin et al., 2005; Kops et al., 2005; Griffis et al., 2007; Gassmann et al., 2008; Chan et al., 2009; Barisic et al., 2010). Kinetochore-bound Mad1–Mad2 produces a modified Mad2 that binds and inhibits the ability of Cdc20 to activate the APC/C (anaphase-promoting complex/cyclosome; Musacchio 2011). The inhibition of APC/C that prevents anaphase disappears when Mad1–Mad2 becomes depleted from all kinetochores as they acquire a full complement of kinetochore microtubules (kMTs) and come under tension as a result of chromosome biorientation (Musacchio and Salmon, 2007; Maldonado and Kapoor, 2011). Tension is thought to be important for causing loss of kinetochore Mad1–Mad2 by promoting both stabilization of kMT attachment and destabilization of Zw10 through an Aurora B kinase–dependent regulatory system (Famulski and Chan, 2007; Maresca and Salmon, 2010; Kasuboski et al., 2011; Lampson and Cheeseman, 2011). In addition, in animal cells, depletion of Mad1–Mad2 from kinetochores and inactivation of the checkpoint depend critically on microtubule motor activity of the dynein–dynactin complex, which is linked to Spindly (Griffis et al., 2007; Gassmann et al., 2008; Lara-Gonzalez et al., 2012). The formation of kMTs provides MT roadways for dynein motor activity to “strip” Mad1–Mad2, RZZ, and Spindly from kinetochores (Howell et al., 2001; Wojcik et al., 2001; Basto et al., 2004). Zwint1 appears to be stable at kinetochores (Famulski et al., 2008), whereas a full complement of kMTs at metaphase destabilizes RZZ and substantially depletes Mad1–Mad2, Spindly, and kinetochore-associated dynein–dynactin (King et al., 2000; Hoffman et al., 2001; Howell et al., 2004; Karess, 2005; Griffis et al., 2007; Gassmann et al., 2010).
In previous work, we reported nanometer-scale measurements for the positions of 18 kinetochore proteins along the kMT axis in metaphase HeLa cells (Wan et al., 2009). This analysis included the major proteins of the highly conserved Knl1–Mis12–Ndc80 (KMN) network consisting of Knl1 or the Blinkin complex (Knl1 and Zwint1), the Mis12 complex (Mis12, Dsn1, Nsl1, and Nnf1), and the Ndc80 complex (Ndc80 [hsHec1], Nuf2, Spc24, and Spc25). The Ndc80 complex is primarily responsible for robust end-on attachment of kinetochores to the plus ends of kMTs, whereas Knl1 has a major role in regulation of attachment stability and recruiting other outer kinetochore proteins, such as the checkpoint proteins Bub1 and BubR1 and the peripheral coronal protein CENP-F (Varma and Salmon, 2012). From the measurements of Wan et al. (2009), we proposed that the Ndc80 complex and Knl1 form two independent modules of the KMN network that extend along the lattice of kMTs near their plus ends. The kinetochore protein module of RZZ, Mad1–Mad2, and Spindly along with bound dynein–dynactin has traditionally been proposed to reside in the peripheral fibrous corona that is seen by electron microscopy at unattached kinetochores to extend out 100–150 nm from the kinetochore outer plate where the Ndc80 complex is located (Hoffman et al., 2001; DeLuca et al., 2005; Karess, 2005; Griffis et al., 2007; Musacchio and Salmon, 2007; Gassmann et al., 2010; McEwen and Dong, 2010). In this study, we probe the association between the Mad1–Mad2 checkpoint pathway and the KMN network using siRNA-based localization dependency assays and nanometer-scale measurements.
Results and discussion
Knl1 and Zwint1 are partially codependent for their kinetochore localization and recruit RZZ and Mad1 to kinetochores
We first compared the effects of individual depletion of Knl1 and Zwint1 on the localization of the other in nocodazole-treated HeLa cells, where dynein-dependent stripping is prevented. The immunofluorescence procedure we employed involved a brief prefixation followed by detergent extraction and a longer fixation. To ensure that the extraction did not influence the observed localizations, we also fixed cells for a longer period first before extraction. We found that kinetochore labeling of two kinetochore proteins—Zwint1 and Zwilch—was not significantly different between the two procedures (unpublished data). Fixing cells with paraformaldehyde alone without any extraction yielded images of poor quality that were unsuitable for reliable quantification of kinetochore fluorescence (unpublished data). We therefore employed the brief prefixation, extraction, and fixation method, which resulted in the best quality kinetochore labeling, for all subsequent experiments.
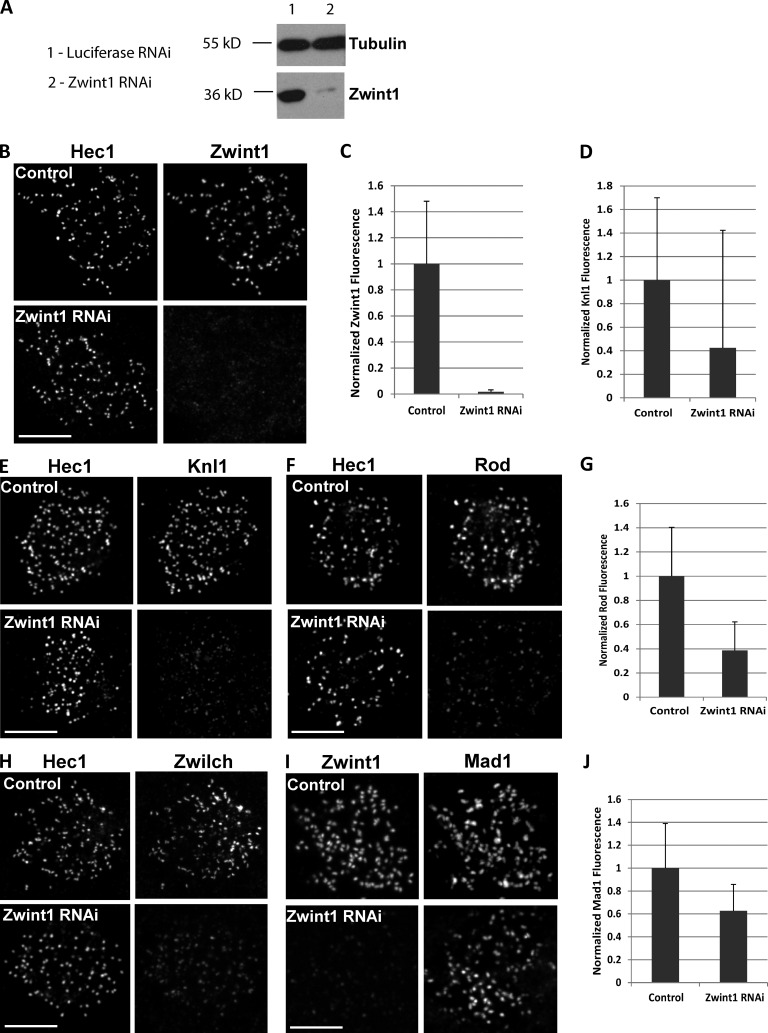
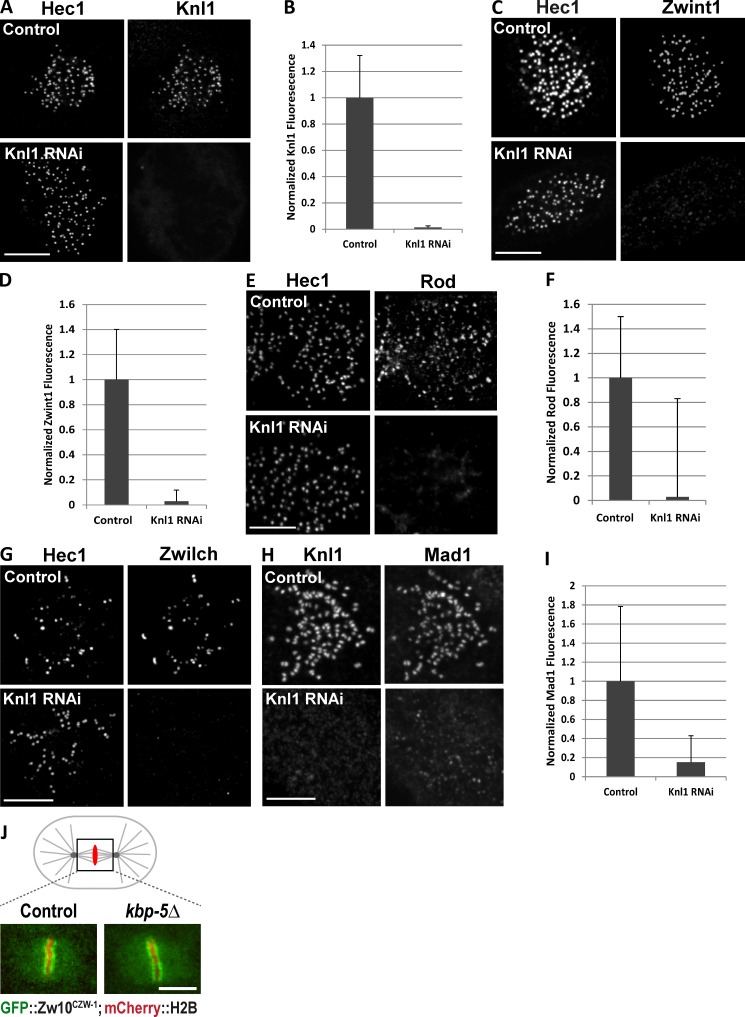
Zwint1 was reduced after siRNA transfection by >98% based on kinetochore immunostaining and immunoblotting (Fig. 1, A–C). Zwint1 depletion also significantly reduced Knl1 levels at kinetochores—Knl1 was present only at ∼40% of the level observed in control cells (Fig. 1, D and E). In the converse scenario, depletion of Knl1 eliminated both Knl1 and Zwint1 kinetochore immunostaining by >97% (Fig. 2, A–D). These results suggest interdependency of Knl1 and Zwint1 kinetochore localization, with Knl1 being required for Zwint1 localization and Zwint1 exerting a partial but significant effect on Knl1 localization.
Figure 1.
Partial loss of Knl1, RZZ, and Mad1 at Zwint1-depleted kinetochores. (A) Immunoblot of endogenous Zwint1 and α-tubulin (loading control) from HeLa cells treated with control or Zwint1 siRNA for 48 h. (B, E, F, H, and I) Nocodazole-treated HeLa cells treated with either control luciferase or Zwint1 siRNA were immunostained using anti-Hec1 9G3 (B, E, F, and H) or anti-Zwint1 (I) and either anti-Zwint1 (B), anti-Knl1 (E), anti-Rod (F), anti-Zwilch (H), or anti-Mad1 (I) antibodies. (C, D, G, and J) Kinetochore fluorescence was normalized relative to Hec1 (C, D, and G) or ACA (J) in control and Zwint1-depleted cells. n = 100 kinetochores; P < 0.001. Error bars are SD from the means. Bars, 5 µm.
Figure 2.
Complete loss of Zwint1, RZZ, and Mad1 at Knl1-depleted kinetochores. (A–I) Nocodazole-treated HeLa cells treated with either control luciferase or Knl1 siRNA were immunostained using anti-Hec1 9G3 (A, C, E, and G) or anti-Knl1 (H) and either anti-Knl1 (A), anti-Zwint1 (B), anti-Rod (E), anti-Zwilch (G), or anti-Mad1 (H) antibodies. Kinetochore fluorescence was normalized relative to Hec1 (B, D, and F) or ACA (I) in control and Knl1-depleted cells. n = 100 kinetochores; P < 0.001. Error bars are SD from the means. (J) Images showing kinetochore localization of GFP-Zw10 in metaphase one-cell-stage embryos of control and kbp-5Δ (ok1358) alleles expressing mCherry-H2B. Bars, 5 µm.
Zwint1 depletion resulted in ∼60% reduction at kinetochores of the RZZ complex subunits Rod and Zwilch in both prometaphase and nocodazole-treated cells compared with controls (Fig. 1, F–H; and Fig. S2 A). By comparison, there was an essentially complete loss of Rod and Zwilch in Knl1-depleted cells in both conditions (Fig. 2, E–G; and Fig. S2 A). The partial versus near-complete effect on RZZ localization of Zwint1 versus Knl1 depletion was mirrored in analysis of Mad1 kinetochore targeting, which requires RZZ (Buffin et al., 2005; Kops et al., 2005). RNAi of Zwint1 caused only ∼35% loss of Mad1 from kinetochores of both prometaphase and nocodazole-treated cells (Fig. 1, I and J; and Fig. S2 A), whereas RNAi of Knl1 caused ∼99% loss of Mad1 from prometaphase kinetochores and ∼85% after nocodazole treatment (Fig. 2, H and I; and Fig. S2 A).
These results show that Knl1 and Zwint1 both contribute to the proper kinetochore recruitment of RZZ and Mad1–Mad2 to unattached kinetochores. Zwint1, initially identified as a yeast two-hybrid interaction partner for Zw10, has been suggested to be the major recruitment factor for the RZZ complex (Starr et al., 2000; Wang et al., 2004). Work in multiple species, including yeasts that completely lack RZZ, has shown that Zwint1 family proteins are conserved interaction partners of the Knl1 protein family (Pagliuca et al., 2009; Petrovic et al., 2010). To explore the relationship between Zwint1 and Knl1 in targeting of RZZ in a system other than HeLa cells, we analyzed the Zwint1 orthologue KBP-5 in Caenorhabditis elegans embryos. C. elegans RZZ, like human RZZ, requires KNL-1 for its kinetochore localization (Gassmann et al., 2008). A mutant allele of kbp-5 in C. elegans (which deletes 75% of the KBP-5 coding sequence; Fig. 2 J and Fig. S1 A) is viable, enabling analysis of RZZ localization in a genetic loss of function of a Zwint1 orthologue. We crossed an integrated transgene expressing GFP-Zw10CZW-1 into the kbp-5 mutant strain and analyzed kinetochore localization of GFP-Zw10CZW-1 in early embryos, which showed no significant effect of the kbp-5 deletion (Fig. 2 J and Fig. S1 B). Thus, in C. elegans, RZZ localization is not dependent on the Zwint1 orthologue KBP-5. Whether the partial loss of RZZ and Mad1 in Zwint1-depleted HeLa cells is caused by a direct contribution from Zwint1 to RZZ recruitment that has been lost in C. elegans KBP-5 or is a result of an indirect effect of the reduction in kinetochore-localized Knl1 in Zwint1-depleted cells remains to be resolved.
Hec1/Ndc80 is not required to recruit Zwint1 or RZZ to kinetochores in nocodazole
Previous work has proposed that Hec1 recruits Zwint1, which in turn recruits RZZ to kinetochores (Lin et al., 2006). This claim appears difficult to reconcile with the tight interaction between Knl1 and Zwint1 observed in biochemical analysis (Petrovic et al., 2010) and the close relationship between Knl1 and Zwint1 in their kinetochore localization dependency (Fig. 1, D and E; and Fig. 2, B and C). We therefore reexamined the relationship between Hec1/Ndc80 and Knl1/Zwint1 as well as the effect of Hec1 depletion on RZZ localization. For HeLa cells, we found that Zwint1 was recruited to kinetochores before Hec1 (Fig. S2 B). Depletion of Hec1 in nocodazole-treated and prometaphase HeLa cells (Guimaraes et al., 2008) did not produce any appreciable decrease in Zwint1 staining at kinetochores (Fig. S2 C and not depicted); the reason for the discrepancy with the previous work (Lin et al., 2006) is unclear. Moreover, depletion of Hec1 from nocodazole-treated HeLa cells (Fig. S2, A, D, and E) or PtK1 cells (Fig. S2 F) resulted in only a minor reduction of RZZ and Mad1 at kinetochores. Thus, Hec1 is not required to recruit Zwint1 or RZZ to kinetochores although it is required to retain these proteins at unattached prometaphase kinetochores in the presence of dynein stripping (Fig. S2 A; DeLuca et al., 2003; Guimaraes et al., 2008; Sundin et al., 2011).
A dynein binding–deficient Spindly motif mutant enables mapping Zwint1, RZZ, and Mad1 location within the substructure of the metaphase kinetochore
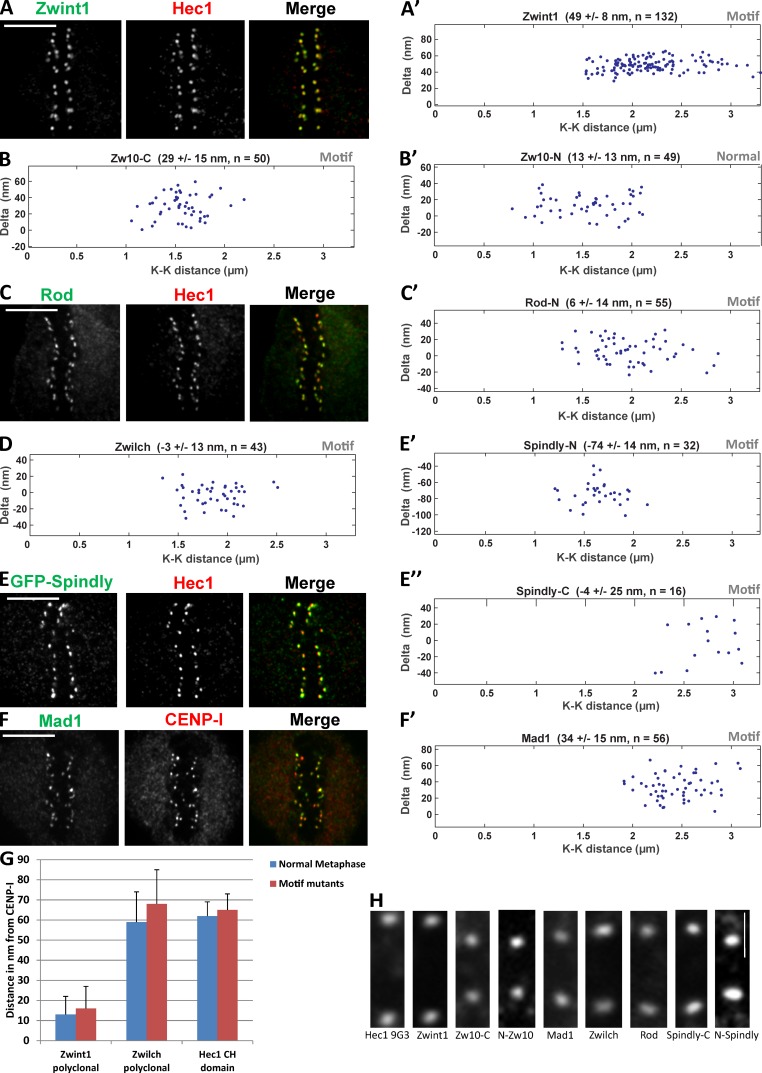
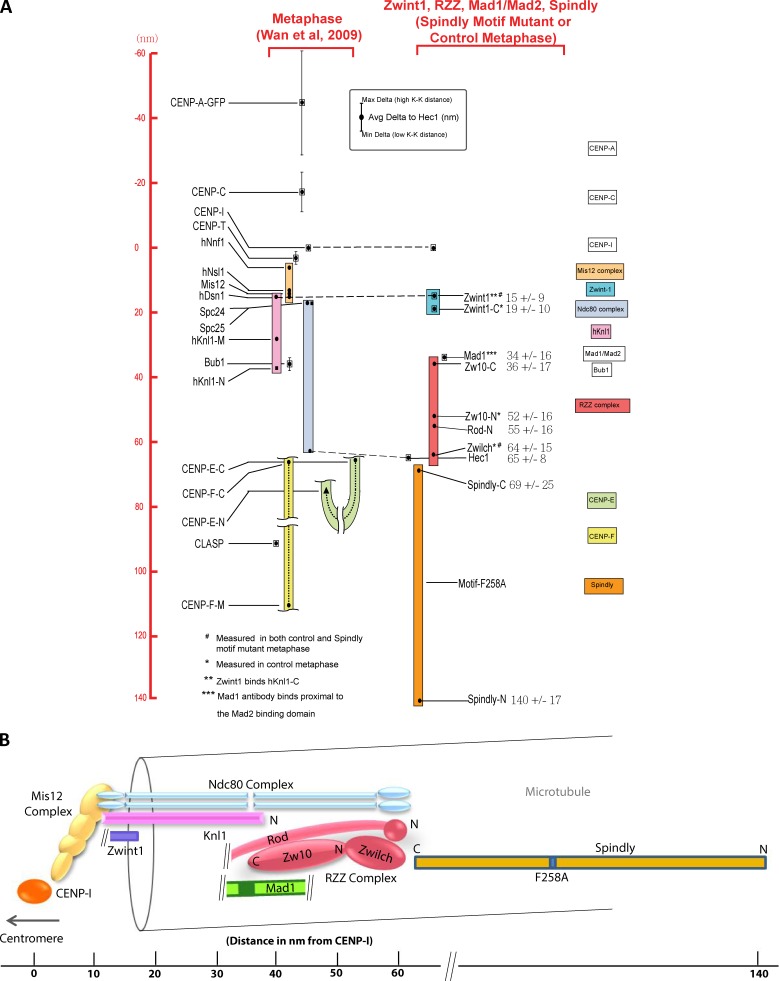
The aforementioned data predict a close relationship between Knl1/Zwint1, the RZZ complex, Mad1–Mad2, Spindly, and dynein–dynactin. To test this prediction, we used our previously described Delta analysis method (Wan et al., 2009) to map the location of Zwint1 and Zwilch, the N terminus of Rod, and the N and C termini of Zw10. The Delta method currently requires paired metaphase kinetochores, which poses a challenge for analyzing proteins such as Mad1–Mad2 and RZZ that are completely or partially removed from kinetochores after microtubule attachment (Hoffman et al., 2001). To circumvent this challenge, we used cells expressing a point mutant (F258A) in a conserved motif of Spindly, which prevents dynein recruitment to kinetochores and causes checkpoint proteins to persist at metaphase kinetochores (Gassmann et al., 2010). Notably, the metaphase persistence of checkpoint proteins is associated with active checkpoint signaling, indicating that the location analysis performed in the Spindly motif mutant reflects a functional state. Previous Delta analysis showed that the distance between the outer, MT-binding head domain of Hec1 and the inner kinetochore protein CENP-I is unchanged in the Spindly mutant, suggesting that its presence does not alter kinetochore architecture (Gassmann et al., 2010). To confirm this, we mapped the location of Zwint1 and the RZZ component, Zwilch, relative to the Hec1 head domain (9G3 antibody epitope) in both the control metaphase and Spindly motif mutant–expressing HeLa cells. Zwint1 is not appreciably depleted between prometaphase and metaphase cells (Famulski et al., 2008), whereas Zwilch is partially depleted after attachment (Gassmann et al., 2010). There were no significant differences in Delta between 9G3 and Zwint1 or 9G3 and Zwilch for the two states (see Fig. 4 G and Table 1; Gassmann et al., 2010), indicating that use of the Spindly mutant state accurately reports physiological kinetochore architecture.
Figure 4.
Kinetochore location of Zwint1, RZZ complex, Mad1, and Spindly in metaphase HeLa cells expressing Spindly containing the motif mutation (GFP-SpindlyF258A). (A, C, E, and F) Example red and green immunofluorescence image pairs used for Delta analysis. Bars, 5 µm. (A′–B′, C′, D, E′, E″, and F′) Example plots of measured Delta values as a function of interkinetochore (K-K) stretch. (G) Comparison of distances measured between CENP-I and three different epitopes in control and Spindly motif mutant metaphase cells. The Hec1–CENP-I separation measurements are replotted from Gassmann et al. (2010). Error bars are SD from the means. (H) Images of a typical sister kinetochore pair for each of the epitopes whose centroids have been measured (indicated by the labels at the bottom) using Delta analysis. Bar, 1 µm.
Table 1.
Summary of Delta values
| Epitope | To 9G3 | To CENP-I | Average to CENP-I |
| nm | nm | nm | |
| Zwint1 (normal) | 51 ± 4 | 13 ± 9 | 15 ± 9 |
| Zwint1 (motif mutant) | 49 ± 8 | 16 ± 11 | |
| Zwint1-C (normal) | 48 ± 4 | 17 ± 9 | 19 ± 10 |
| Zwint1-C–GFP (normal) | 44 ± 6 | 21 ± 10 | |
| Zwilch (normal) | 6 ± 11 | 59 ± 14 | 64 ± 15 |
| Zwilch (motif mutant) | −3 ± 13 | 68 ± 15 | |
| Zw10-C (motif mutant) | 29 ± 15 | 36 ± 17 | 36 ± 17 |
| GFP–Zw10-N (normal) | 13 ± 14 | 52 ± 16 | 52 ± 16 |
| Rod-N (motif mutant) | 10 ± 14 | 55 ± 16 | 55 ± 16 |
| GFP–Spindly-N (Motif mutant) | −74 ± 14 | 140 ± 17 | 140 ± 17 |
| 9G3 (motif mutant) | NA | 65 ± 8 | 65 ± 8 |
| Mad1 (motif mutant) | NA | 34 ± 15 | 34 ± 15 |
| Spindly-C (motif mutant) | −4 ± 25 | 69 ± 25 | 69 ± 25 |
| Hec1-C–tdTomato (normal) | 40 ± 5 | 25 ± 9 | 25 ± 9 |
Summary of Delta values obtained from measurements of 13 different epitopes spanning the Zwint1–RZZ complex, Mad1, and Spindly made relative to either the Hec1 CH (head) domain or the constitutive centromere-associated network protein CENP-I. The average distance of an epitope to CENP-I in the rightmost column refers to the average of the measurements between normal and Spindly motif metaphase kinetochores. NA, not applicable.
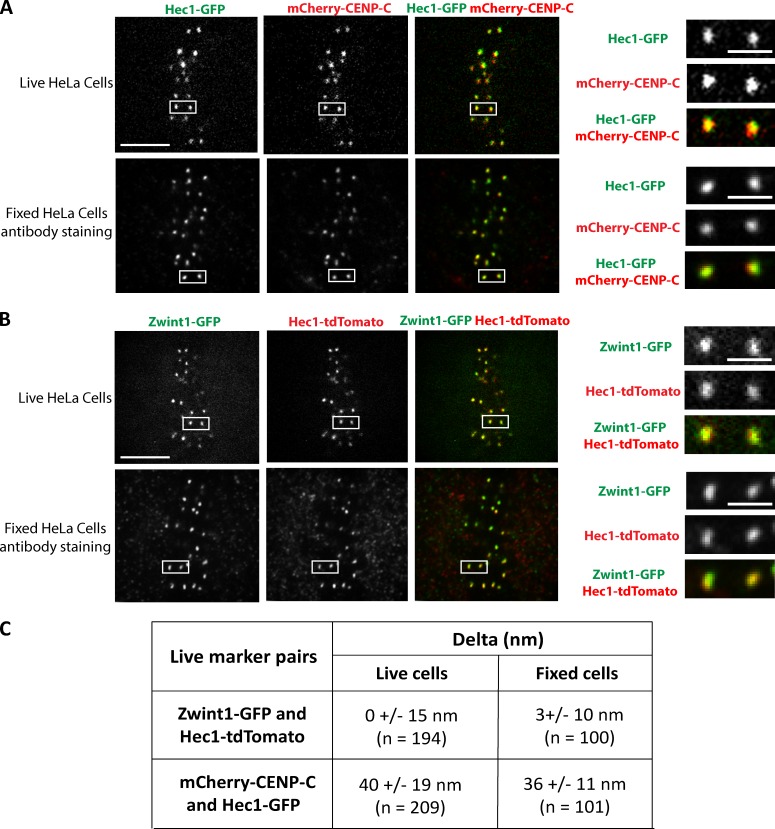
To demonstrate that the methods used to extract and fix the cells before Delta analysis did not interfere with the measurements, we performed live-imaging experiments in HeLa cells transfected with two different pairs of fluorescent-labeled kinetochore protein markers and measured Delta for both pairs. The mean Delta measured for the mCherry–CENP-C/Hec1-GFP pair was 40 ± 19 nm (n = 209) for live cells and 36 ± 11 nm (n = 100) for fixed cells (Fig. 3, A and C; and Video 1). The mean Delta measured for the Zwint1-GFP/Hec1-tdTomato pair was 0 ± 15 nm (n = 194) for live cells and 3 ± 10 nm (n = 100) for fixed cells (Fig. 3, B and C; and Video 2). Neither of these measurements exhibited a statistically significant difference between live and fixed cells (paired Student’s t test).
Figure 3.
Comparison of mean separation distances measured using Delta analysis in live and fixed metaphase HeLa cells. (A and B) HeLa cells transfected with cDNA plasmid pairs encoding Hec1-GFP and mCherry–CENP-C (A) or Zwint1-GFP and Hec1-tdTomato (B) were subjected to two-color live imaging (A and B, top) or indirect immunofluorescence using the anti-GFP antibody (A and B, bottom) and either the anti-mCherry antibody (A, bottom) or the anti-RFP antibody (B, bottom). Bars, 5 µm. (right) Images are boxed regions within the cells in the top and bottom of A and B magnified to show representative sister kinetochore pairs used for Delta analysis. Bars, 1 µm. (C) Mean Delta values measured from live and fixed cells in A and B.
In Fig. 4 (A–F′), we show example Delta measurements of RZZ, Mad1, and Spindly relative to the Hec1 head domain. We note that the diffraction-limited images of kinetochores analyzed for each protein label are typically wider than their thickness and are occasionally tilted relative to the sister–sister kinetochore axis (Fig. 4 H). Table 1 lists the final average Delta values for each fluorescent label pair after minor correction based on kinetochore tilt (Wan et al., 2009). In the summary map in Fig. 5 A, the new measurements are integrated with previous measurements of inner and outer kinetochore proteins (Wan et al., 2009). The origin along the inner–outer kinetochore axis is set to zero at the average CENP-I label position of metaphase control or Spindly motif mutant cells (Fig. 5 A and Table 1).
Figure 5.
Summary of protein label position along the kMT axis for 26 epitopes in 23 kinetochore proteins in control and Spindly motif mutant (SpindlyF258A)–expressing cells. (A) The positions of the newly measured proteins/epitopes obtained from mean values of Delta measurements in Table 1, whereas the previously published measurements are from Wan et al. (2009). Scale on the left side is set to 0 at the position of CENP-I; positive values are outward (toward the spindle microtubules), whereas negative values are inward (toward the centromeric chromatin). Color-coded boxes indicate complexes and large proteins. Black dots indicate the average Delta values that have been corrected for tilt. The vertical lines indicate the minimum and maximum Delta values measured across the variation in centromere stretch. (B) A schematic view of kinetochore protein architecture for the RZZ and Mad1–Mad2 checkpoint protein module proteins relative to the KMN network for metaphase spindly motif mutant cells based on measurements in A. K-K, interkinetochore; N, N terminus; C, C terminus.
None of the proteins tested (Zwint1, Rod, Zwilch, Zw10, Mad1, and Spindly) showed any dependence of Delta on centromere tension, based on the near zero slopes of plots of Delta as a function of interkinetochore separation (Fig. 4); for comparison, see previous data on GFP-CENP-A and CENP-C located in the chromatin (Fig. 5 A; Wan et al., 2009). Next, we describe in detail the Spindly motif mutant enabled mapping of RZZ, the Mad1–Mad2 complex, and Spindly positions within the substructure of kinetochores, and the resulting measurements are summarized in Table 1 and Fig. 5 A.
Zwint1 is located close to the junction between the Mis12 complex and the Spc24/Spc25 end of the Ndc80 complex
Zwint1 is a small (∼30 kD and 277 aa) protein. For both control metaphase and Spindly motif mutant cells, a polyclonal antibody to Zwint1 was located on average 15 ± 9 nm outside of CENP-I label and near the positions of the Spc24/Spc25 end of the Ndc80 complex (Fig. 5 A). The mean positions of an antibody specific for the Zwint1 C terminus or GFP fused to the C terminus of Zwint1 were ∼19 ± 10 nm outside of CENP-I (Fig. 5 A). The polyclonal antibody for Zwint1 likely provides a good measure of the mean position of the C terminus of Knl1 based on in vitro binding experiments (Petrovic et al., 2010). Thus, the position of the C-terminus of Knl1 and its bound Zwint1 is ∼30 nm on average inside the N terminus of Knl1 along the kMT axis (Fig. 5 A). This also indicates that Knl1 extends along the axis of kMTs in metaphase human kinetochores as does the Ndc80 complex (Wan et al., 2009). Knl1 is a large protein of ∼300 kD, and its 3D structure is yet to be elucidated.
RZZ subunits localize in proximity to the N termini of Knl1 and Hec1
In biochemical reconstitutions, RZZ forms a tight complex (Civril et al., 2010). Rod is a large protein (242 kD and 2,200 aa) with a β-propeller structure at its N terminus, a Sec39p domain in its middle, and repeated α-helical domains from the middle to the C terminus, making it an elongated structure (Civril et al., 2010). An antibody directed against the N-terminal region of Rod was on average located 55 ± 16 nm outside of CENP-I, which is ∼10 nm inside of the 9G3 label on Hec1 calponin homology (CH) domain (Fig. 5 A).
Zwilch is a slightly elongated globular protein (∼65 kD) that binds to the N-terminal region of Rod (Civril et al., 2010). A polyclonal antibody to Zwilch was located at ∼64 ± 15 nm from CENP-I, ∼9 nm outside of the N-terminal of Rod, and coincident with the 9G3 on Hec1 (Fig. 5 A).
Zw10 is a slightly larger globular protein (∼86 kD) than Zwilch. The C terminus of Zw10 binds the Sec39p domain in the middle of Rod (Civril et al., 2010). We measured in Spindly motif mutant cells the mean position of a peptide antibody directed against the C terminus of Zw10 at 36 ± 17 nm outside of CENP-I and ∼29 nm inside of 9G3 on Hec1 (Fig. 5 A). Thus, the C terminus of Zw10 (and possibly the middle of Rod) is ∼20 nm on average inside of the mean position of the N terminus of Rod.
We used a HeLa cell line stably expressing GFP-Zw10 (Kops et al., 2005) to measure the location of N terminus of Zw10. Although partially depleted at metaphase kinetochores, sufficient levels of Zw10 persist in normal cells for accurate measurements (Fig. S3 C; Famulski and Chan, 2007). The mean position of antibodies to GFP was 13 ± 14 nm inside of Hec1 9G3 and ∼33 nm distal to the Zwint1 C terminus. This significant separation between Zw10 and Zwint1 measured at metaphase may be produced by the release of the N terminus of Zw10 from Zwint1 between prometaphase and metaphase as proposed previously (Kasuboski et al., 2011). If so, this must occur independently of force generated by dynein anchored to the RZZ complex via Spindly, as the same results are observed for both control and Spindly motif mutant–expressing cells (Table 1).
The Mad2-binding domain of Mad1 is in close proximity to the C terminus of Zw10, the N terminus of Knl1, and Knl1-associated Bub1
Mad1 (718 aa) forms a homodimer and has a globular C-terminal domain and an α-helical coiled-coil domain that extends from about 600 aa to the N terminus (Kim et al., 2012). The Mad2-binding domain is located before the C-terminal domain at aa 530–560. To label the Mad2 binding site of Mad1, we used a monoclonal antibody whose epitope is located in the region encompassing aa 578–589 (Musacchio, A., personal communication), which is close to the Mad2-binding domain. In Spindly motif mutant–arrested metaphase cells, the average position of the centroid of this antibody was 34 ± 15 nm outside of CENP-I and ∼31 nm inside Hec1 9G3 (Fig. 5 A). This places the Mad2-binding domain on Mad1 near the position measured previously for Bub1 bound to the N terminus of Knl1 along the kMT axis (Wan et al., 2009) and in close proximity to the C terminus of Zw10.
Spindly extends from the RZZ complex into kinetochore domains peripheral to the outer plate
Spindly is a largely coiled-coil and predicted elongated protein. In Spindly motif mutant cells that fail to recruit dynein–dynactin, the average position of antibodies generated against the C terminus of Spindly was 4 ± 25 nm outside the position of 9G3, which is close to the measured position of Zwilch (Fig. 5 A). In contrast, GFP fused to the N terminus of Spindly was 74 ± 14 nm outside of 9G3, extending well into the kinetochore domain peripheral to the outer plate (Fig. 5 A). This places the attachment site on Spindly for dynein–dynactin well outside of the KMN network, potentially to help accommodate the large size of dynein–dynactin. These measurements also suggest Zwilch and its bound N terminus of Rod as the potential recruitment site on RZZ for Spindly.
Summary and conclusions
The depletion and mapping experiments for human kinetochores implicate Knl1 as the key scaffold for checkpoint protein recruitment, a finding supported by results for C. elegans embryos. We also find that Zwint1, RZZ, Mad1–Mad2, and the C terminus of Spindly are in close proximity to and extend along the core kMT attachment site at metaphase when dynein stripping is blocked and the spindle assembly checkpoint is not silenced (Fig. 5, A and B). If the checkpoint proteins are associated with the components of the fibrous corona that remain in metaphase, such as CENP-E and CENP-F (Wan et al., 2009), these coronal fibers overlap with the KMN network.
Our data also suggest that control of Mad1 levels at kinetochores is likely more complicated than release of binding of Zw10 to Zwint1 as proposed recently (Kasuboski et al., 2011). How Mad1–Mad2, Spindly, and dynein–dynactin are substantially depleted from normal metaphase kinetochores but leave moderate levels of RZZ with Zw10 located ∼33 nm or more outside of Zwint1 is an important unresolved issue (Fig. S3, A–F; Chan et al., 2000; Famulski and Chan, 2007; Musacchio and Salmon, 2007; Gassmann et al., 2010). The answer to this puzzle likely depends on contributions from multiple low affinity binding sites for Mad1 provided by proteins associated with the KMN network, including RZZ, Bub1 bound to Knl1, the Hec1 CH domain (Fig. S2 A; Lara-Gonzalez et al., 2012), and in C. elegans, Spindly (Yamamoto et al., 2008). In addition, the activities of kinases, such as Mps1 and Aurora B, are involved (Lara-Gonzalez et al., 2012). The results described here should aid development of mechanistic insights into this critical aspect of checkpoint signaling and silencing.
Materials and methods
Cell culture and transfections
HeLa cells (normal and stably expressing a GFP-tagged protein of interest) were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Sigma-Aldrich), 100 U/ml penicillin, and 100 mg/ml streptomycin. Spindly motif mutant HeLa cells were analyzed after endogenous Spindly depletion and tetracycline induction of the motif mutant as in Gassmann et al. (2010). cDNA transfections were performed using Effectene (QIAGEN) as per the manufacturer’s instructions. siRNA transfections were performed with a total of 100 nM of each siRNA duplex or SMARTpools of oligonucleotides using either DharmaFECT 1 reagent (Thermo Fisher Scientific) or Oligofectamine (Invitrogen) according to the manufacturer’s guidelines. Synthetic duplexed RNA oligonucleotides for Zwint1 (Lin et al., 2006) and Hec1 depletion (DeLuca et al., 2005) were synthesized by Thermo Fisher Scientific. A separate SMARTpool of siRNA oligonucleotides was also used for Zwint1 depletion (catalog no. L-006830-00-0005; Thermo Fisher Scientific). Stealth siRNA oligonucleotides for Knl1 depletion (Kiyomitsu et al., 2007) were synthesized by Invitrogen, and an siRNA for Spindly (5′-GAAAGGGUCUCAAACUGAA-3′) was synthesized by Thermo Fisher Scientific.
Antibodies
Anti-Zwint1 polyclonal, anti-Zwilch polyclonal, and anti-Mad1 monoclonal rabbit antibodies were provided by A. Musacchio (Max Planck Institute of Molecular Physiology, Dortmund, Germany), anti-Zw10 C-terminal rabbit polyclonal antibody was provided by R. Vallee (Columbia University, New York, NY), anti-Rod N-terminal rabbit polyclonal antibody was provided by G. Chan (University of Alberta, Edmonton, Alberta, Canada), anti–human anti-CREST antiserum (ACA) antibody was provided by B. Brinkley (Baylor College of Medicine, Houston, TX), anti–CENP-I rabbit polyclonal antibody was provided by S.-T. Liu (The University of Toledo, Toledo, OH), anti-Knl1 rabbit polyclonal antibody has been previously described (Cheeseman et al., 2008), and Spindly C-terminal rabbit polyclonal antibody was affinity purified from serum obtained using aa 450–605 of Spindly as the antigen. Other primary antibodies used in this study include anti-Hec1 mouse monoclonal antibody (Abcam), anti-GFP rabbit polyclonal (Invitrogen), and mouse monoclonal (EMD Millipore) antibodies, anti-mCherry (Takara Bio Inc.) and anti-mRFP (MBL) rabbit polyclonal antibodies, and the anti-Zwint1 C-terminal rabbit polyclonal antibody (Bethyl Laboratories, Inc.). Alexa Fluor 488–, Rhodamine Red-X–, or Cy5-labeled donkey secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc.
Immunofluorescence microscopy and Delta analysis
Cells were rinsed briefly in PHEM buffer (120 mM Pipes, 50 mM Hepes, 20 mM EGTA, and 4 mM magnesium acetate, pH 6.9) before fixation. Cells were typically prefixed with 4% paraformaldehyde for 30–60 s, permeabilized with 0.5% Triton X-100 for 5 min, and then fixed for 20 min using 4% paraformaldehyde. We also employed alternate fixation procedures to ensure that the extraction did not influence the observed kinetochore staining pattern, which included fixing cells with 4% paraformaldehyde for a longer period (20 min) first before extraction using 0.5% Triton X-100 and also with 4% paraformaldehyde alone for 20 min without any extraction. For Zw10 antibody staining, the cells were fixed for 6 min at −20°C with ice-cold methanol. All the antibody incubations and washes were also performed in PHEM buffer plus 0.05% BSA. All antibody incubations were at 37°C for 1 h, and washes were performed for 10 min at RT. DAPI staining (0.1 µg/ml) was performed for 10 min, and cells were mounted using either ProLong Antifade (Molecular Probes) or a solution of 0.5% p-phenylenediamine in 90% glycerol and 20 mM Tris, pH 8.8. Nocodazole treatment of cells was for 30 min at 37°C at a final concentration of 10 µM.
For image acquisition, 3D stacks were obtained sequentially at 200-nm steps along the z axis through the cell using MetaMorph software (Molecular Devices) and a high-resolution inverted microscope (TE300; Nikon) equipped with a spinning disk (CSU-10 ; Yokogawa Corporation of America) with image magnification yielding a 65-nm pixel size from the cooled charge-coupled device camera (Orca ER; Hamamatsu Photonics; Maddox et al., 2003) and an 100×/1.4 NA (Plan Apochromat) differential interference contrast oil immersion objective (Nikon). For kinetochore fluorescence quantifications, the average values for integrated fluorescence minus background for control and experimental cell samples were normalized by the average value obtained from the normal cells (Hoffman et al., 2001). Bars are 5 µm in all figures and supplemental figures unless otherwise defined in the figure legends.
For Delta analysis, 3D centroid positions were first measured by 3D Gaussian-fitting function for the fluorescent signals of protein epitopes of different colors (Wan et al., 2009). For each kinetochore pair, the centroids of one color were projected to the axis defined by the centroid of the other color, usually the centroid of either the Hec1 9G3 epitope or CENP-I. The average separation of the projection distance (Delta) between the signals of different colors for that pair was then calculated (Wan et al., 2009). For image acquisition from GFP-Spindly motif mutant–expressing cells, secondary antibodies conjugated with Rhodamine Red-X and Cy5 fluorophores were used to detect the epitopes subjected to Delta analyses, whereas for all other Delta measurements, Alexa Fluor 488– or Rhodamine Red-X–conjugated secondary antibodies were used.
Live-cell imaging
HeLa cells were synchronized using double thymidine treatment and double transfected (using Effectene) with the required live kinetochore marker pairs, Zwint1-GFP/Hec1-tdTomato (Zwint1-GFP was a gift from G. Chan) or mCherry–CENP-C/Hec1-GFP (mCherry–CENP-C was a gift from A. Straight, Stanford University School of Medicine, Stanford, CA), during the first round of thymidine treatment. Live imaging was performed at 9 h after the second thymidine washout, and image stacks of double-transfected metaphase cells with kinetochore labeling of both the markers were obtained using the same imaging set up as for fixed cell image acquisition. A focal plane with maximum number of kinetochore pairs was selected for Delta analysis. Same kinetochore pairs were measured multiple times until they bleached out to average the effect of kinetochore movement and eliminate the difference caused by imaging the two channels at different times. Duplicate dishes treated identically as the live-imaged samples were fixed (using the standard fixation protocol described earlier in this paper) to acquire the stacks for making the same Delta measurements from fixed cells. Both live and fixed sample measurements applied the 2D Gaussian-fitting function to find the centroids of the kinetochore signal (Wan et al., 2012).
C. elegans strains and imaging
The following C. elegans strains were used in this study: OD1254 (unc-119(ed3) III; unc-119(ed3) III; ltIs37 [pAA64; pie-1/mCherry::his-58]; ltIs21 [pIC54; pie-1/GFP-TEV-STag::czw-1; unc-119 (+)]) and OD945 (kbp-5 (ok1358) I; unc-119(ed3) III; unc-119(ed3) III; ltIs37 [pAA64; pie-1/mCherry::his-58]; ltIs21 [pIC54; pie-1/GFP-TEV-STag::czw-1; unc-119 (+)]). For imaging embryos expressing mCherry-H2B and GFP–CZW-10, 5 × 2–µm z stacks were collected with 2 × 2 binning and a 60×/1.4 NA Plan Aprochromat every 20 s at 21°C on a spinning-disk confocal mounted on an inverted microscope (TE2000-E; Nikon) equipped with an interline charge-coupled device (Clara; Andor Technology).
Statistical analyses
All experiments including the RNAi-mediated depletion analysis of protein localization and the nanometer-scale epitope mapping analysis were performed three times to generate the numbers that are reported in the respective figures or figure legends. Test of significance was performed for all the quantifications, and measurements were performed using Student’s t test, the values of which are also reported in the figure legends.
Online supplemental material
Fig. S1 shows additional examples of GFP-Zw10 kinetochore localization in C. elegans one-cell stage metaphase embryos of control and kbp-5Δ alleles. Data in Fig. S2 support the observation that Hec1 is not required for RZZ and Mad1 kinetochore localization in nocodazole-treated cells. Fig. S3 provides data to demonstrate that the components of the RZZ complex are retained at higher levels at metaphase kinetochores as compared with Mad1. Videos 1 and 2 are examples of metaphase HeLa cells expressing Hec1-GFP/mCherry–CENP-C and Zwint1-GFP/Hec1-tdTomato construct pairs, respectively, used to make live Delta measurements. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304197/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201304197.dv.
Supplementary Material
Acknowledgments
We would like to thank Dr. Andrea Musacchio for anti-Zwint1, anti-Zwilch, and anti-Mad1 antibodies, Dr. Gordon Chan for the anti-Rod antibody and Zwint1-GFP construct, Dr. Richard Vallee for the anti-Zw10 antibody, Dr. Bill Brinkley for the ACA, Dr. Don Cleveland for the GFP-Zw10 stable HeLa cell line, Dr. Aaron Straight for the mCherry–CENP-C construct, and Dr. Song-Tau Liu for the anti–CENP-I antibody. We acknowledge the Michael Hooker Microscopy Core Facility at The University of North Carolina-Chapel Hill for their help with imaging. We would also like to thank Drs. Tom Maresca and Ryan O’Quinn for helpful discussions and all members of the Salmon and Desai laboratories for their support of the project. We thank the Caenorhabditis Genetics Center for the kbp-5 deletion strain.
This work was supported by the National Institutes of Health grant R37GM024364 and supplement R37GM024364-31S (to E.D. Salmon) and National Institutes of Health grant GM074215 (to A. Desai).
Footnotes
Abbreviations used in this paper:
- ACA
- anti-CREST antiserum
- CH
- calponin homology
- kMT
- kinetochore microtubule
- KMN
- Knl1–Mis12–Ndc80
- RZZ
- Rod–Zw10–Zwilch
References
- Barisic M., Sohm B., Mikolcevic P., Wandke C., Rauch V., Ringer T., Hess M., Bonn G., Geley S. 2010. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell. 21:1968–1981 10.1091/mbc.E09-04-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T., Karess R. 2004. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 14:56–61 10.1016/j.cub.2003.12.025 [DOI] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Starr D.A., Goldberg M.L., Yen T.J. 2000. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2:944–947 10.1038/35046598 [DOI] [PubMed] [Google Scholar]
- Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H., Gerlich D.W., Nigg E.A., Santamaria A. 2009. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 185:859–874 10.1083/jcb.200812167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F., Wehenkel A., Giorgi F.M., Santaguida S., Di Fonzo A., Grigorean G., Ciccarelli F.D., Musacchio A. 2010. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 18:616–626 10.1016/j.str.2010.02.014 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Howell B.J., Canman J.C., Hickey J.M., Fang G., Salmon E.D. 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13:2103–2109 10.1016/j.cub.2003.10.056 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., McEwen B.F. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531 10.1091/mbc.E04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski J.K., Chan G.K. 2007. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr. Biol. 17:2143–2149 10.1016/j.cub.2007.11.037 [DOI] [PubMed] [Google Scholar]
- Famulski J.K., Vos L., Sun X., Chan G. 2008. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J. Cell Biol. 180:507–520 10.1083/jcb.200708021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R., III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., Desai A. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Stuurman N., Vale R.D. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015 10.1083/jcb.200702062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes G.J., Dong Y., McEwen B.F., Deluca J.G. 2008. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 18:1778–1784 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., Salmon E.D. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., Salmon E.D. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., Salmon E.D. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953–964 10.1016/j.cub.2004.05.053 [DOI] [PubMed] [Google Scholar]
- Karess R. 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15:386–392 10.1016/j.tcb.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Kasuboski J.M., Bader J.R., Vaughan P.S., Tauhata S.B., Winding M., Morrissey M.A., Joyce M.V., Boggess W., Vos L., Chan G.K., et al. 2011. Zwint-1 is a novel Aurora B substrate required for the assembly of a dynein-binding platform on kinetochores. Mol. Biol. Cell. 22:3318–3330 10.1091/mbc.E11-03-0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Sun H., Tomchick D.R., Yu H., Luo X. 2012. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc. Natl. Acad. Sci. USA. 109:6549–6554 10.1073/pnas.1118210109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.M., Hays T.S., Nicklas R.B. 2000. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J. Cell Biol. 151:739–748 10.1083/jcb.151.4.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R., III, Tagaya M., Cleveland D.W. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60 10.1083/jcb.200411118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Cheeseman I.M. 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21:133–140 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Westhorpe F.G., Taylor S.S. 2012. The spindle assembly checkpoint. Curr. Biol. 22:R966–R980 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Lin Y.T., Chen Y., Wu G., Lee W.H. 2006. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene. 25:6901–6914 10.1038/sj.onc.1209687 [DOI] [PubMed] [Google Scholar]
- Maddox P.S., Moree B., Canman J.C., Salmon E.D. 2003. Spinning disk confocal microscope system for rapid high-resolution, multimode, fluorescence speckle microscopy and green fluorescent protein imaging in living cells. Methods Enzymol. 360:597–617 10.1016/S0076-6879(03)60130-8 [DOI] [PubMed] [Google Scholar]
- Maldonado M., Kapoor T.M. 2011. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat. Cell Biol. 13:475–482 10.1038/ncb2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Salmon E.D. 2010. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 123:825–835 10.1242/jcs.064790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.F., Dong Y. 2010. Contrasting models for kinetochore microtubule attachment in mammalian cells. Cell. Mol. Life Sci. 67:2163–2172 10.1007/s00018-010-0322-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. 2011. Spindle assembly checkpoint: the third decade. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3595–3604 10.1098/rstb.2011.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Pagliuca C., Draviam V.M., Marco E., Sorger P.K., De Wulf P. 2009. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS ONE. 4:e7640 10.1371/journal.pone.0007640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A., et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852 10.1083/jcb.201002070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Saffery R., Li Z., Simpson A.E., Choo K.H., Yen T.J., Goldberg M.L. 2000. HZwint-1, a novel human kinetochore component that interacts with HZW10. J. Cell Sci. 113:1939–1950 [DOI] [PubMed] [Google Scholar]
- Sundin L.J., Guimaraes G.J., Deluca J.G. 2011. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol. Biol. Cell. 22:759–768 10.1091/mbc.E10-08-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Salmon E.D. 2012. The KMN protein network—chief conductors of the kinetochore orchestra. J. Cell Sci. 125:5927–5936 10.1242/jcs.093724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., Cimini D., Cameron L.A., Salmon E.D. 2012. The coupling between sister kinetochore directional instability and oscillations in centromere stretch in metaphase PtK1 cells. Mol. Biol. Cell. 23:1035–1046 10.1091/mbc.E11-09-0767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hu X., Ding X., Dou Z., Yang Z., Shaw A.W., Teng M., Cleveland D.W., Goldberg M.L., Niu L., Yao X. 2004. Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279:54590–54598 10.1074/jbc.M407588200 [DOI] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaërou F., Karess R., Hays T. 2001. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007 10.1038/ncb1101-1001 [DOI] [PubMed] [Google Scholar]
- Yamamoto T.G., Watanabe S., Essex A., Kitagawa R. 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 183:187–194 10.1083/jcb.200805185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.