Abstract
In addition to canonical cis splicing, which joins exons from a single pre-mRNA, various forms of trans splicing have been described, whereby two separate precursor transcripts are linked with each other. A new study by Hu et al. in Cell Research characterizes a novel and unusual splicing variation, called exo-endo trans splicing.
A recent study from Bo-Liang Li's group published in Cell Research1 describes a novel type of trans splicing: an exogenous plasmid-derived RNA transcript is joined in trans with an endogenous mRNA, generating a chimeric mRNA that encodes a functional protein. Therefore, the authors have termed this unusual mRNA processing event exo-endo trans splicing. They describe and characterize in great detail this first example, in which a 56-kDa functional splice isoform of the acyl-CoA:cholesterol acyltransferase 1 (ACAT1) was produced, an enzyme making fatty-acid esters of cholesterol, which is physiologically important in cholesterol transport, metabolism and homeostasis.
Trans splicing was initially discovered in the protozoan parasite Trypanosoma brucei, in which a 39-nucleotide, non-coding “mini-exon” derived from the so-called Spliced Leader (SL) RNA is joined to the 5′ end of every protein-coding sequence2,3. Thereby, trans splicing constitutes an essential step in gene expression in trypanosomes. This SL-type trans splicing also appears in a multitude of different organisms, including metazoans like Nematoda and Platyhelminthes, and mostly coexists together with the “normal” cis splicing (i.e., the accurate removal of non-coding intron sequences between the protein-coding exons of a pre-mRNA).
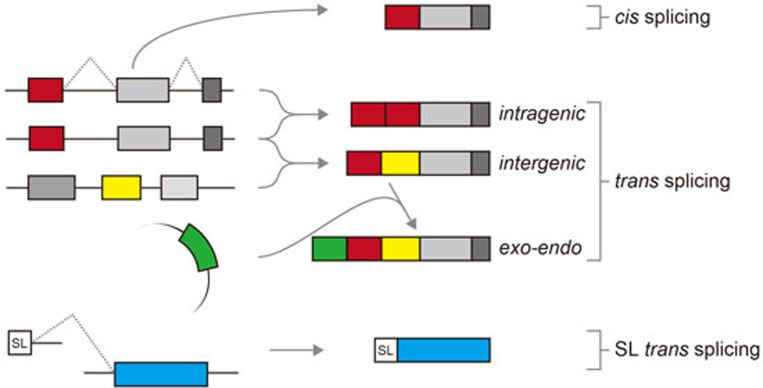
Even in Drosophila and mammals such as rats and humans, trans splicing has been shown to occur between independently transcribed pre-mRNAs4,5,6,7. In contrast to the canonical SL-type trans splicing, which is a prerequisite for generating all mature mRNAs in trypanosomes, different variations of trans splicing patterns have been described in higher eukaryotes (Figure 1): (1) intragenic trans splicing, in which two identical pre-mRNA molecules from the same gene are spliced together to generate an mRNA with duplicated exon sequences; (2) intergenic trans splicing, whereby an mRNA is generated from two pre-mRNAs that are derived from different genes, which can be located even on different chromosomes (interchromosomal trans splicing). A relatively well-characterized example for the latter case is the human ACAT1 gene, as previously described by Bo-Liang Li and colleagues8; importantly, characterization of trans splicing has not only been performed at the mRNA level, as in many other studies, but also been extended to the protein level and the effects on biochemical activities. In this case, exons of the ACAT1 mRNA are transcribed from two different chromosomes, namely 1 and 7, and the resulting 4.3-kb human ACAT1 mRNA encodes two functional protein isoforms (50- and 56-kDa in size) with differential enzymatic activities8.
Figure 1.
Alternative splicing modes in eukaryotes. Different types of pre-mRNA splicing in eukaryotes are schematically represented: canonical cis splicing, three types of trans splicing, and the SL-type trans splicing. Exons including the SL RNA-derived mini-exon (SL) are represented by boxes, and introns as lines.
Now, Hu et al.1 continued their molecular analyses of ACAT1 trans splicing en detail by combining the characterization of both mRNA and protein products of ACAT1. They reveal a new form of trans splicing of the ACAT1 mRNA: an antisense transcript derived from an exogenous ampicillin resistance gene (Ampr) is joined with the endogenous ACAT1 transcript, generating one of the ACAT protein isoforms. This 56-kDa isoform includes additional 46 amino acids at its amino-terminus, which is encoded by the antisense strand of Ampr. By using mutational analysis, Hu et al.1 demonstrate that the antisense transcript of Ampr that is attached to the endogenous ACAT1 mRNA by trans splicing provides the real start codon of the ACAT1 56-kDa isoform, which is different from the start codon previously proposed9.
What is the significance of trans splicing of an antisense transcript in human cells? Surprisingly, Hu et al.1 detected the DNA sequences of Ampr with an upstream cryptic promoter and antisense transcripts of Ampr in human blood cells and a diverse set of human cell lines by PCR, and also in a variety of higher eukaryotes by computational analyses, suggesting that such endo-exo trans splicing events may be more common than expected. Moreover, using both intact cells and an in vitro splicing system, Hu et al.1 demonstrated that the exo-endo trans splicing can occur in different backgrounds of transfected constructs. Finally, they provide evidence based on mutational analysis that the exo-endo trans splicing utilizes the normal spliceosome machinery, as standard splice-site signals are required.
Previously the same group has demonstrated that the 4.3-kb ACAT1 mRNA is produced through interchromosomal trans splicing8. Now they show that the chimeric mRNA derived from chromosomes 1 and 7 is required for the exo-endo trans splicing event (Figure 1). This is the first report in the mammalian system, in which a functional protein is generated through trans splicing of an exogenous transcript and an endogenous cellular RNA. It remains to be investigated how common and widespread this phenomenon is in mammalian gene expression, but it may, in combination with the “normal” alternative splicing patterns, have the potential to further increase the proteome complexity. Moreover, in this special case, this phenomenon might help to explain how plasmid-derived DNA sequences, in particular antibiotic resistance genes, integrate into eukaryotic genomes and utilize the cellular mRNA-processing machinery to modulate protein function.
References
- Hu GJ, Chen J, Zhao XN, et al. Cell Res. 2013. pp. 1007–1024. [DOI] [PMC free article] [PubMed]
- Boothroyd JC, Cross GA. Gene. 1982. pp. 281–289. [DOI] [PubMed]
- van der Ploeg LH, Liu AY, Michels PA, et al. Nucleic Acids Res. 1982. pp. 3591–3604. [DOI] [PMC free article] [PubMed]
- Caudevilla C, Serra D, Miliar A, et al. Proc Natl Acad Sci USA. 1998. pp. 12185–12190. [DOI] [PMC free article] [PubMed]
- Dorn R, Reuter G, Loewendorf A. Proc Natl Acad Sci USA. 2001. pp. 9724–9729. [DOI] [PMC free article] [PubMed]
- Li H, Wang J, Mor G, et al. Science. 2008. pp. 1357–1361. [DOI] [PubMed]
- Takahara T, Tasic B, Maniatis T, et al. Mol Cell. 2005. pp. 245–251. [DOI] [PubMed]
- Li BL, Li XL, Duan ZJ, et al. J Biol Chem. 1999. pp. 11060–11071. [DOI] [PubMed]
- Yang L, Lee O, Chen J, et al. J Biol Chem. 2004. pp. 46253–46262. [DOI] [PubMed]