Abstract
Cholesterol is a structural component of the cell, indispensable for normal cellular function, but its excess often leads to abnormal proliferation, migration, inflammatory responses and/or cell death. To prevent cholesterol overload, ATP-binding cassette (ABC) transporters mediate cholesterol efflux from the cells to apolipoprotein A-I (ApoA-I) and to the ApoA-I-containing high-density lipoprotein (HDL)1-3. Maintaining efficient cholesterol efflux is essential for normal cellular function4-6. However, the role of cholesterol efflux in angiogenesis and the identity of its local regulators are poorly understood. Here we show that ApoA-I binding protein (AIBP) accelerates cholesterol efflux from endothelial cells (EC) to HDL and thereby regulates angiogenesis. AIBP/HDL-mediated cholesterol depletion reduces lipid rafts, interferes with VEGFR2 dimerization and signaling, and inhibits VEGF-induced angiogenesis in vitro and mouse aortic neovascularization ex vivo. Remarkably, Aibp regulates the membrane lipid order in embryonic zebrafish vasculature and functions as a non-cell autonomous regulator of zebrafish angiogenesis. Aibp knockdown results in dysregulated sprouting/branching angiogenesis, while forced Aibp expression inhibits angiogenesis. Dysregulated angiogenesis is phenocopied in Abca1/Abcg1-deficient embryos, and cholesterol levels are increased in Aibp-deficient and Abca1/Abcg1-deficient embryos. Our findings demonstrate that secreted AIBP positively regulates cholesterol efflux from EC and that effective cholesterol efflux is critical for proper angiogenesis.
AIBP is a secreted protein discovered in a screen of proteins that physically associate with ApoA-I7. Human APOA1BP mRNA encoding the AIBP protein is ubiquitously expressed7. Although the AIBP binding to ApoA-I implies that AIBP may modulate HDL function7,8, its role in cholesterol efflux has not been experimentally tested.
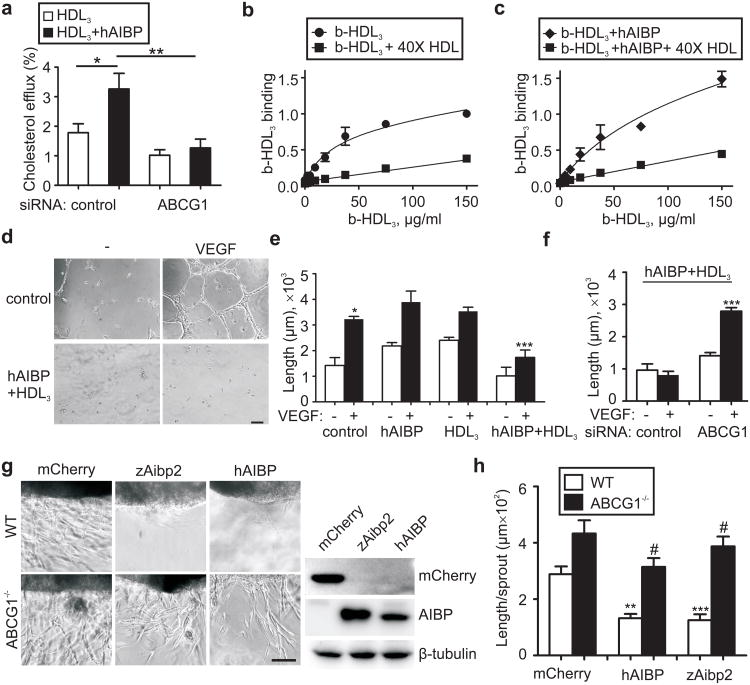
First, we investigated whether human AIBP (hAIBP) had any effect on cholesterol removal from human umbilical vein endothelial cells (HUVEC) in which ABCG1 is a major transporter responsible for cholesterol efflux to HDL9,10. In the presence of hAIBP, cholesterol efflux from HUVEC to HDL3 was increased 2-fold, and the ABCG1 deficiency completely abrogated this effect (Fig. 1a, Supplementary Fig. 2). hAIBP did not promote cholesterol efflux in absence of HDL3 (Supplementary Fig. 2b), but the hAIBP binding to HUVEC (Supplementary Fig. 3) increased the overall HUVEC capacity to bind HDL3 (Bmax=1.5 vs. 0.8) and the constant of HDL3 dissociation from HUVEC (Kd=1.0×10−6M vs. 0.33×10−6M; Figs. 1b, c), thereby creating conditions that would facilitate HDL3–mediated cholesterol efflux.
Figure 1. Role of AIBP in cholesterol efflux from EC and in vitro angiogenesis.
a, hAIBP mediated-cholesterol efflux and effect of ABCG1 knockdown. HUVEC were transfected with control or ABCG1 siRNA, preloaded with 3H-cholesterol and incubated for 1 hour with 50 μg/ml HDL3 in the presence or absence of 0.2 μg/ml hAIBP. Efflux was measured as the 3H counts in the medium divided by the sum of 3H counts in the medium and the cells. Mean±SE; n=6. b and c, Effect of hAIBP on HDL3 binding to HUVEC. HUVEC were incubated on ice with the indicated concentration of biotinyated HDL3 (b-HDL3), in the presence or absence of hAIBP (at a 0.1:50 w/w hAIBP:HDL3 ratio) and 40× excess of unlabeled HDL. Each data point is Mean±SE from 3 to 7 independent experiments. The binding parameters for b-HDL3/HUVEC binding were calculated as Bmax = 0.8±0.1 and Kd = (0.33±0.10)×10−6 M in absence of hAIBP (panel b; R2=0.92, Sy.x=0.1), and Bmax=1.5±0.4 and Kd=(1.03±0.10)×10−6 M in the presence of hAIBP (panel c; R2=0.94, Sy.x=0.1). The differences in Bmax and Kd values were statistically significant (p<0.01 and p<0.05, respectively). d, Effect of hAIBP and HDL3 on EC tube formation. HUVEC were preincubated with or without 50 μg/ml HDL3 + 0.1 μg/ml hAIBP for 4 hours. Cells were then seeded on Matrigel, in the presence or absence of 20 ng/ml VEGF, and imaged following a 12-hour incubation. Scale, 100 μm. e, The length of EC tubes in the experiment illustrated in 1d and Supplementary Fig. 4. Mean±SE; n=5. f, Requirement for ABCG1 in hAIBP inhibition of angiogenesis. HUVEC were transfected with control or ABCG1 siRNA and assayed as in 1d. Mean±SE; n=6. g, Mouse aortic ring angiogenesis assay. Aortic rings from C57BL6 and Abcg1-/- mice were embedded in Matrigel. HEK293 cells transiently expressing mCherry, zAibp2 or hAIBP were inserted approximately 0.5 mm away from the aortic ring, and the plates were incubated with 10 ng/ml VEGF for 7 days. Images show the edge of the aortic rings facing the HEK293 cell clusters. Immunoblots show expression of hAIBP and zAibp2 (both detected with a Flag tag antibody) and mCherry in HEK293 cells. h, The length of aortic ring sprouts. Mean±SE; n=10. In all panels: #, not significant; *, p<0.05; **, p<0.01; ***, p<0.001.
To investigate the role of AIBP/HDL-mediated cholesterol efflux in angiogenesis, we incubated HUVEC with hAIBP and/or HDL3 and then stimulated cells with VEGF. hAIBP and HDL3 added separately did not affect EC tube formation, but together they significantly reduced angiogenesis (Fig. 1d, e and Supplementary Fig. 4). Cholesterol depletion by methyl-β-cyclodextrin (MβCD)11 also inhibited angiogenesis, whereas cholesterol-loaded MβCD, which delivers cholesterol to the cell, promoted angiogenesis (Supplementary Fig. 5). If the hAIBP/HDL3 inhibition of angiogenesis is the consequence of accelerated cholesterol efflux, then this effect should depend on the presence of the cholesterol transporter ABCG1. Indeed, knockdown of ABCG1 in HUVEC rescued VEGF-induced angiogenesis from the hAIBP/HDL3 inhibition (Fig. 1f). Further, we tested both hAIBP and zAibp2 (a zebrafish protein, which will be discussed later) in an ex vivo aortic ring angiogenesis assay. A cluster of HEK293 cells producing either hAIBP, zAibp2 or mCherry (negative control) was placed 0.5 mm from the edge of a mouse aortic ring, and VEGF was added to stimulate angiogenesis. Both hAIBP and zAibp2, but not mCherry, significantly reduced neovascularization of aortic rings isolated from a wild type mouse (Figs. 1g, h). Aortic rings from an Abcg1-/- mouse responded to VEGF with a more vigorous angiogenesis, which was not significantly reduced by hAIBP or zAibp2. These results support the hypothesis that cholesterol efflux is necessary for the AIBP-mediated inhibition of angiogenesis.
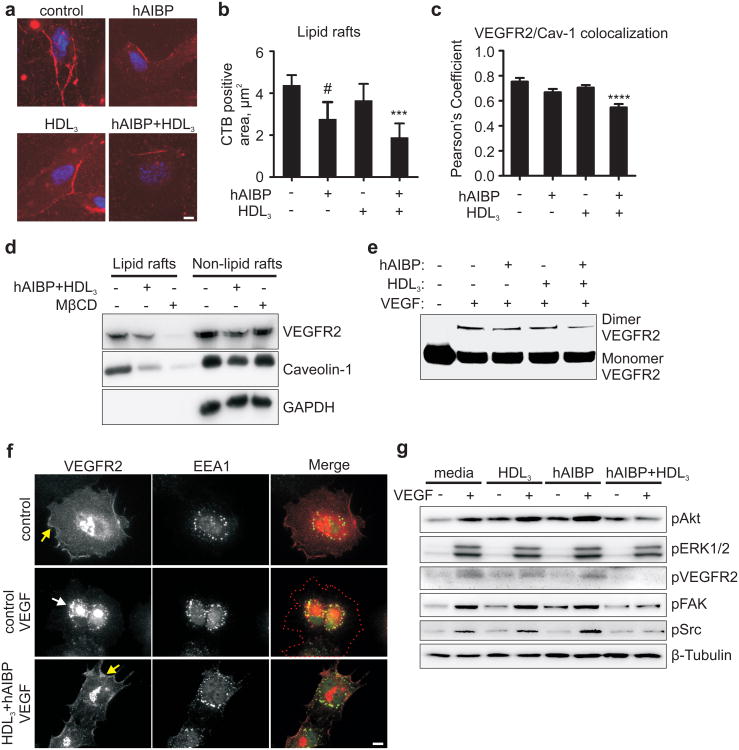
HDL-mediated depletion of cholesterol from plasma membrane disrupts cholesterol- and sphingomyelin-rich membrane microdomains12,13, often designated as lipid rafts, and affects membrane receptor signaling11. We found that hAIBP/HDL3 reduced the lipid raft content in HUVEC and disrupted cell-surface colocalization of caveolin-1 and VEGFR2 (Figs. 2a-c and Supplementary Fig. 6). The hAIBP/HDL3 treatment, similarly to the treatment with MβCD, decreased VEGFR2 and caveolin-1 localization to the lipid raft fraction isolated from cell lysates (Fig. 2d and Supplementary Fig. 7). Many studies suggest that VEGFR2 localization to lipid rafts facilitates VEGFR2 dimerization and endocytosis14-17, the steps required for VEGF-mediated signaling18. In our experiments, the hAIBP/HDL3 treatment reduced VEGF-induced VEGFR2 dimerization and endocytosis as well as phosphorylation of VEGFR2, Akt, FAK, Src and to a lesser degree of ERK1/2 (Figs. 2e-g and Supplementary Figs. 8-10). Importantly, subsequent addition of cholesterol partially reversed inhibition of VEGFR2, FAK and Akt phosphorylation in hAIBP/HDL3-treated cells (Supplementary Fig. 11). Consistent with the effect on VEGF signaling, HUVEC migration toward a VEGF cue was significantly reduced in hAIBP/HDL3-treated cells (Supplementary Fig. 12). These results suggest that hAIBP facilitates cholesterol efflux from HUVEC to HDL and that cholesterol depletion of the plasma membrane disrupts lipid rafts and VEGF signaling and inhibits VEGF-induced angiogenesis.
Figure 2. Effect of AIBP on HUVEC lipid rafts, VEGFR2 localization, dimerization and signaling.
a, Effect of hAIBP and HDL3 on lipid rafts. HUVEC were preincubated with 50 μg/ml HDL3, 0.1 μg/ml hAIBP, or 50 μg/ml HDL3 + 0.1 μg/ml hAIBP for 4 hours. Cells were stained for nuclei (blue, DAPI) and for lipid rafts (red, cholera toxin B (CTB) + anti-CTB antibody). Scale, 10 μm. b, The area of lipid rafts per cell. Mean±SE; n=10; **, p<0.01; #, p=0.08. c, Effect of hAIBP and HDL3 on caveolin-1 and VEGFR2 surface localization. HUVEC were incubated with hAIBP and/or HDL3 as in 2a, fixed and stained with antibodies to caveolin-1 and VEGFR2. Images were captured using TIRF microscopy (Supplementary Fig. 6) and Pearson's coefficient was calculated to assess surface colocalization of VEGFR2 with caveolin-1. Mean±SE; n=38-50; ***, p<0.001. d, VEGFR2 and caveolin-1 localization to lipid rafts. HUVEC were incubated with 20 μg/ml cholesterol-MβCD for six hours, followed by a 1 hour incubation with or without 50 μg/ml HDL3 + 0.1 μg/ml hAIBP, or a 30 min incubation with 10 mM MβCD. HUVEC lysates were separated into lipid rafts and non-lipid rafts fractions by ultracentrifugation, run on SDS-PAGE and blotted with VEGFR2 and caveolin-1 antibodies. e, Effect of hAIBP and HDL3 on VEGFR2 dimerization. HUVEC were preincubated with HDL3 and/or hAIBP as in 1a, followed by a 20 min stimulation with 50 ng/ml VEGF. Cells were treated with a crosslinking reagent, lysed and immunoprecipitated with a VEGFR2 antibody. Monomers and crosslinked dimers of VEGFR2 were visualized on western blot. f, Effect of hAIBP and HDL3 on VEGFR2 endocytosis. HUVEC were preincubated with or without 50 μg/ml HDL3 + 0.1 μg/ml hAIBP for 4 hours, then stimulated with 50 ng/ml VEGF for 20 min, fixed and stained with antibodies to VEGFR2 (red) and the early endosome marker EEA-1 (green). Yellow and white arrows point to the surface and endosomal localization of VEGFR2. Red dotted line traces cell contour. Scale, 10μm. g, Effect of hAIBP and HDL3 on VEGFR2 signaling. HUVEC were preincubated with HDL3 and/or hAIBP as in 2a, followed by a 20 min stimulation with 50 ng/ml VEGF. Total cell lysates were run on SDS-PAGE and probed as indicated.
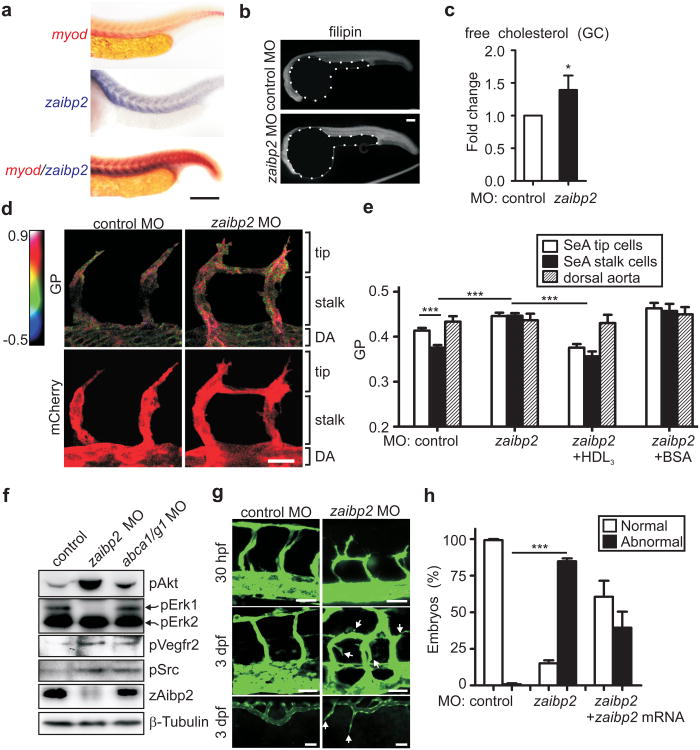
AIBP is evolutionary conserved from Drosophila to zebrafish to mouse and human (Supplementary Fig. 13a). Zebrafish have two genes, apoa1bp1 and apoa1bp2, encoding zAibp1 and zAibp2 proteins, respectively (Supplementary Fig. 13b). The zaibp2 expression in 24-36 hours postfertilization (hpf) zebrafish embryos shows a clear segmental pattern, colocalizing with the somite marker myod (Fig. 3a and Supplementary Fig. 14). By 48 hpf, when segmental angiogenesis is completed, zaibp2 is no longer expressed in somites.
Figure 3. Effect of Aibp deficiency on zebrafish cholesterol, membrane lipid order, Vegfr2 signaling and angiogenesis.
a, Tissue distribution of zaibp2 mRNA in zebrafish embryos. Embryos at 24 hpf were fixed and WISH was performed with antisense myod and zaibp2 probes. Scale, 100 μm. b-c, Free cholesterol levels in zaibp2 morphants. b, Zebrafish embryos were injected with 8 ng of either control MO or zaibp2 MO. Twenty four hpf control and zaibp2 morphants were stained with filipin to detect free cholesterol in embryos. Note the yolks are artificially masked on the images. c, At 24 hpf, the trunk area (without yolk) was dissected, total lipids extracted, and free cholesterol levels determined by gas chromatography (GC). The cholesterol levels were normalized to the protein content and then to the values in control MO embryos. 50-70 embryos were pooled for each sample. Mean±SE; n=4; *, p<0.05. d, Effect of zaibp2 MO on SeA membrane lipid order. Tg(flk1:ras-cherry)s896 embryos were injected with control or zaibp2 MO as in 3b and at 24 hpf were stained with 5 μM Laurdan. In the same embryos, confocal images of mCherry fluorescence (bottom images) and the multiphoton images of Laurdan fluorescence (ex 800 nm, em 400-460 nm and 470-530 nm) were captured. The multiphoton results (top row images) are displayed as pseudocolored GP (generalized polarization, a measure of the membrane lipid order) images, cropped to show only the vasculature, i.e. mCherry-positive areas. Scale, 20 μm. e, The graph shows GP values in the areas corresponding to tip and stalk cells of growing SeA and the dorsal aorta (DA) as indicated in 3d. Some one-cell stage embryos were coinjected with 1 nl of 10 mg/ml human HDL3 or BSA. Note the Y-scale is from 0.2 to 0.5. Mean±SE; n=44-119 SeA in 25-49 embryos; ***, p<0.001. f, Phosphorylation of signaling proteins. Lysates of 24 hpf control (8 ng control MO), zaibp2 (8 ng zaibp2 MO) and abca1/g1 (4 ng abca1 MO + 4ng abcg1 MO) morphants were separated on SDS-PAGE and immunoblotted as indicated. g, Angiogenic defects in zaibp2 morphants. One-cell stage Tg(fli1:egfp)y1 zebrafish embryos were injected with 8 ng of either control or zaibp2 MO. The images are of SeA in 30 hpf embryos (top row), and SeA (middle row) and of SIV (bottom row) in 3 dpf embryos. Arrows point to dysregulated sprouts. Scale, 25 μm. h, Quantification of the number of embryos with normal and abnormal angiogenesis (SeA with ectopic branching). The abnormal angiogenesis was partially rescued by coinjection of 40 pg of zaibp2 mRNA lacking the MO targeting site. Mean±SE; n=100-149. ***, p<0.001.
Both zAibp2 and zAibp1 bound to human ApoA-I and to the HDL in human plasma, but only zAibp2 was effective in promoting cholesterol efflux from HUVEC to HDL3 (Supplementary Figs. 15, 16). Zebrafish embryos injected with antisense morpholino oligonucleotides (MO) targeting zaibp2 translation sites had increased levels of free (unesterified) cholesterol, whereas injections of zaibp1 or scrambled control MO did not result in any changes (Figs. 3b, c and Supplementary Fig. 17). Thus, we focused on zaibp2. Using the polarity-sensitive fluorescent probe Laurdan, we observed a higher membrane lipid order in the areas of growing segmental arteries (SeA) corresponding to tip cells compared to stalk cells (Fig. 3d, e), suggesting a higher content of lipid rafts in tip cells, which may positively regulate Vegfr2 signaling. The membrane lipid order was increased in the SeA of zaibp2 morphants compared to controls, and the difference between tip and stalk cells was lost. To test the hypothesis that zAibp2-mediated cholesterol efflux regulates membrane order in growing SeA, we injected zaibp2 morphants with human HDL3 or with BSA. Adding an excess of HDL3 – to promote cholesterol efflux and to override the zAibp2 deficiency – annulled the increase in membrane order in SeA of zaibp2 morphants, and a spatially indiscriminate HDL3 excess equalized the membrane order in tip and stalk cells. Adding an excess of BSA had no effect on the membrane order in zaibp2 morphants. Lysates of zaibp2 knockdown embryos displayed increased phosphorylation of Vegfr2, Akt and Src, and decreased phosphorylation of Erk1 (Fig. 3f, Supplementary Figs. 18, 19). These results suggest that zAibp2 regulates cholesterol levels, the membrane lipid order, and Vegfr2 signaling and, thus, may control angiogenesis.
Indeed, injection of MOs targeting zaibp2 translation or splicing sites into one-cell stage embryos of Tg(fli1:egfp)y1 zebrafish, which express EGFP in EC19, resulted in remarkable dysregulation of angiogenesis, with profound ectopic branching of both SeA and subintestinal veins (SIV) (Fig. 3g and Supplementary Figs. 20, 21). The zaibp2 knockdown was validated in western blot (Fig. 3f). The ectopic branching of SeAs in zaibp2 morphants was partially rescued by forced expression of zaibp2 mRNA lacking the MO target site (Fig. 3h).
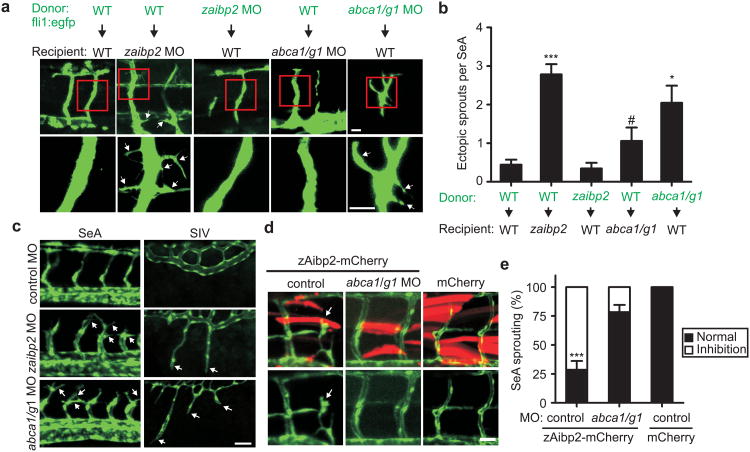
The zaibp2 expression pattern (Fig. 3a) resembles that of type 3 semaphorins, non-cell autonomous repellent cues that guide the patterning of developing SeAs via endothelial-specific PlexinD1 receptors20. To determine the cell autonomy of the zAibp2 effect on angiogenesis, we performed cell transplantation experiments, using Tg(fli1:egfp)y1 donors. Fluorescent EC from wild type donors found in non-fluorescent zaibp2 morphants displayed excessive branching and filopodial projections, while fluorescent EC from zaibp2 morphant donors found in wild type recipients had normal morphology (Figs. 4a, b). In a gain-of-function experiment, overexpression of zAibp2 inhibited SeA sprouting from the dorsal aorta and normal growth of sprouted SeA (Supplementary Fig. 22). These results suggest a role for zAibp2 as a repellent molecule whose function depends on the milieu surrounding EC but not on zAibp2 expression in the EC themselves.
Figure 4. Effect of Aibp and Abca1/Abcg1 deficiency on zebrafish angiogenesis.
a, Mosaic expression analysis of EC branching in control, zaibp2 and abca1/abcg1 knockdown embryos. At 4 hpf, cells were isolated from donor embryos and transplanted into recipient embryos. Recipient embryos were analyzed at 3 dpf. Arrows point to aberrant ectopic branches/sprouts. Scale, 25 μm. b, Numbers of ectopic branches/filopodial projections per SeA. Mean±SE; n=8-16. #, not significant; *, p<0.05; ***, p<0.001. c, Angiogenic defects in abca1/abcg1 morphants. One-cell stage embryos were injected with 8 ng of control MO, 8 ng zaibp2 MO, or 4 ng abca1 MO + 4 ng abcg1 MO. Images of SeA (30 hpf) and SIV (3 dpf) are shown. Scale, 50 μm. d, Knockdown of abca1/g1 cancels the effect of zAibp2 overexpression. One-cell stage embryos were injected with 2 nl of 100 ng/μl myog:zaibp2-mCherry, myog:zaibp2-mCherry + abca1/g1 MO, or myog:mCherry. The arrow points to an aberrant SeA at the site of zAibp2-mCherry expression. Scale, 20μm. e, Abnormal SeA formation was quantified in 8-16 embryos per group. Mean±SE; *** p<0.001.
The loss of zAibp2 resulted in increased expression of genes involved in angiogenesis, such as tie2, vegfr2, vegfr3 and fli1 (Supplementary Figs. 23-24). Thus, in addition to the zAibp2 effect on the membrane lipid order and Vegfr2 signaling (Figs. 3d-f), zAibp2 affects expression of angiogenic genes as well.
To further validate that effective cholesterol efflux is required for normal angiogenesis, we knocked down zebrafish cholesterol transporters abca1 and abcg121,22 and observed higher levels of free cholesterol, increased levels of phosphorylated Akt, Vegfr2 and Src and dysregulated SeA and SIV angiogenesis (Figs. 3f, 4c and Supplementary Figs. 19, 25-27), closely reproducing the angiogenesis defects of zaibp2 morphants. Individual knockdown of each abca1 and abcg1 suggested a dominant role of abca1 in embryonic angiogenesis (Supplementary Fig. 28). In contrast to the zaibp2 non-cell autonomous regulation of angiogenesis, fluorescent EC from abca1/abcg1 morphant donors found in wild type recipients displayed excessive SeA branching (Figs. 4a, b), confirming that cholesterol efflux from EC is required to restrain ectopic angiogenesis. Overexpression of zAibp2-mCherry in somites resulted in inhibition of SeA growth, which was rescued by knocking down abca1/abcg1 (Figs. 4d, e). These results provide additional evidence that expression of zAibp2 limits blood vessel growth and also suggest that zebrafish Abca1- and/or Abcg1-mediated cholesterol efflux is required for the zAibp2 effect on angiogenesis.
Based on our results, we propose that there is an additional level of paracrine regulation of the VEGFR2 pathway in which cholesterol efflux and associated reduction of ordered membrane microdomains/lipid rafts interfere with the VEGFR2 membrane localization, dimerization, endocytosis, and signaling. Because in 24 hpf zebrafish zaibp2 mRNA is highly expressed in somites, but not in the inter-somitic spaces where SeA grow, it is likely that zAibp2-mediated cholesterol efflux inhibits Vegfr2 signaling in a site-specific manner to prevent lateral protrusions from stalk and tip cells and restrains ectopic SeA growth into somites (Supplementary Fig. 1).
The role of cholesterol efflux mechanisms in protecting against endothelial dysfunction, in particular in hypercholesterolemic animals prone to development of atherosclerosis, has been reported10,23. However, our study is the first to demonstrate the role of AIBP in promoting cholesterol efflux from EC to HDL and the importance of this mechanism in regulation of angiogenesis. In contrast to the ApoA-I-containing HDL, ApoB-containing LDL and VLDL deliver cholesterol and other lipids to the cell and, thus, are positioned to promote angiogenesis. Interestingly, a recent paper finds the opposite, that ApoB lipoproteins negatively regulate angiogenesis in zebrafish embryos24. The authors suggest a mechanism in which the ApoB protein, but not the lipid components within ApoB-containing lipoproteins, is responsible for transcriptional regulation of Vegfr1, a soluble decoy receptor for Vegf. Our experiments uncovered a different, lipid-mediated mechanism in which effective cholesterol efflux is a critical process that ensures proper angiogenesis and Aibp secreted by the surrounding tissues serves as an important negative regulator of angiogenesis.
Full Methods
Cloning of human and zebrafish AIBP, recombinant protein expression and purification, and antibody production
Zebrafish aibp2 (Ggene ID:557840) and aibp1 (Gene ID:436891) were cloned from zebrafish brain cDNA using primers: CCGGAATTCCATGTTGGGGGTTCGAGCTCTG (5′) and CGCGGATCCTCAGTTGAGCTGAAACACACACTC (3′) for zaibp1; and CCGGAATTCCGCCACCATGAACCACAGCTCCAACG (5′) and CGCGGATCCCGCAGTTCTATAATACATTCTGTGC (3′) for zaibp2. The fragments were cloned in frame into pFLAG-CMV4 (Sigma). Human APOA1BP (Gene ID: 128240) was cloned from HEK293 cell cDNA using primers: CCGGAATTCCATGTCCAGGCTGCGGGCGCTGCTGGGCCTCG (5′) and CGGGGTACCTCACTGCAGACGATAGACACACTC (3′). For expression of AIBP proteins, the genes were cloned in frame into pHUE vector26 (kindly provided by Tracy Handel), expressed in BL21 DE3 competent cells (Invitrogen) and purified with a Ni-NTA agarose resin column (Qiagen). Deubiquitinase (DUB) expressed in pHUE was used as a negative control in experiments with recombinant AIBP. To produce a zAibp2 antibody, recombinant zAibp2 was mixed with complete Freund's adjuvant (Sigma) and injected subcutaneously into a guinea pig. The guinea pig was boosted 3 more times. Post-immune plasma was compared with pre-immune plasma from the same animal and used in western blot to detect zAibp2 in zebrafish lysates (Supplementary Fig. 18). The specificity of the antibody was confirmed by adding excess of recombinant zAibp2 to the antibody, which prevented its binding to a specific band on the western blot.
Cholesterol efflux
A cholesterol efflux assay was performed as described23,27, with modifications. In brief, HUVEC (ATCC) were loaded with 2 μCi/ml 3H-cholesterol, and cholesterol efflux was initiated by the addition of 0.2% BSA/EBM with 50 μg/ml HDL3 (isolated from normolipidemic human plasma by ultracentrifugation), in the presence or absence of 0.2 μg/ml zAibp2, zAibp1, or hAIBP. DUB, replacing AIBP, was used as a negative control. Background, non-specific release of 3H-cholesterol was measured in absence of HDL or any other protein. After 1 to 6 hours of incubation, the medium was collected and counted in a liquid scintillation counter LS 6500 (Beckman Coulter). The cells were extracted with 2-propanol, and the lipid extract was added to ScintiVerse BD Cocktail (Fisher) and counted. Cholesterol efflux was expressed as a percentage of 3H counts in the medium compared to combined 3H counts in the cells and the medium. Background, non-specific release of 3H from the cells was subtracted.
ABCG1 knockdown
Both negative control and ABCG1 siRNA oligonucleotides were from Ambion. HUVEC were plated in 6-well plates at 5×105 cells/well and transfected with 66.6 nM siRNA using SuperFect Transfection Reagent (Qiagen) as described in the manufacturer's protocol. Two days after transfection, cells were washed and used in an efflux assay. Two additional wells of transfected cells were used to confirm ABCG1 knockdown in western blot using an antibody from Novus Biologicals.
AIBP/HDL3 - HUVEC binding assay
hAIBP and HDL3 were biotinylated with EZ-Link Sulfo-NHS-Biotin (Thermo Scientific) according the manufacture's protocol. Binding of biotinylated hAIBP or biotinylated HDL3 to HUVEC was assessed by a chemiluminescent binding assay as described by Fang et al.28, with modifications. HUVEC (2×104) were seeded into 96-well flat bottom plates in 5% FBS-EBM. After 72 h, plates were blocked with ice-cold 1% BSA-PBS for 30 min on ice, incubated with ice-cold biotinylated proteins for 2 h on ice, washed, and fixed with ice-cold 4% paraformaldehyde (PFA) in PBS for 30 min. HUVEC-bound biotinylated hAIBP or HDL3 were detected with NeutrAvidin-conjugated alkaline phosphatase (Pierce) and LumiPhos 530 (Lumigen, Southfield, MI), using a Dynex luminometer (Dynex Technologies). Data were recorded as relative light units counted per 100 ms. All samples were assayed in triplicates. The parameters of hAIBP and HDL3 binding to HUVEC (Bmax and Kd) were calculated using a total and non-specific binding algorithm within the GraphPad Prism 5.0 software package. The following model was used: H + C ↔ HC, where H is unbound HDL3, C is cells, and HC is HDL3 bound to the cells. The equations used for calculating binding parameters were:
where a is background and b is the slope of the linear fit of nonspecific binding. Goodness of fit of non-linear regression was estimated using R2 and standard deviation of residuals (Sy.x), expressed in the same units as [H] and Bmax. A molecular mass of 80 kDa was used for the HDL protein.
In vitro angiogenesis assay
The angiogenesis assay was carried out as described in reference29. Growth factor reduced Matrigel (BD Biosciences) was thawed at 4°C overnight and diluted with an equal volume of serum-free EBM medium (Lonza). Each well of 96-well plates was coated with 50 μl diluted Matrigel and incubated at 37°C for 1 hour. HUVEC were serum-starved and then pre-incubated with HLD3 and/or hAIBP. Cells were harvested and added to Matrigel-coated plates at 1×104 cells per well in EBM, in the presence or absence of 20 ng/ml VEGF (R&D Systems). Following a 12 hour incubation, tubular structures were imaged with a phase contrast microscope.
Free cholesterol measurements in HUVEC
HUVEC cholesterol levels were measured in cellular lipid extracts using a colorimetric assay (BioVision) as described14.
Aortic ring neovascularization assay
The method was adopted from reference30, with modifications. Thoracic aorta was isolated from a 6-week old male C57BL6 mouse or an age and gender matched Abcg1-/- mouse (kindly provided by Catherine Hedrick, La Jolla Institute for Allergy and Immunology), cleaned from surrounding fat and connective tissue and sliced into 1 mm long rings. The aortic rings were placed in wells of a 48-well plate containing solidified Matrigel and then covered with additional Matrigel. Small wells were made in Matrigel approximately 0.5 mm from aortic rings and 50 μl aliquots of Matrigel containing 1×105 HEK293 cells transfected with mCherry (negative control), zAIBP2 or hAIBP were placed in these wells. After 10 min, each well was filled with EBM medium supplemented with 10 ng/ml VEGF and the plates were incubated at 37°C for 6 days. Media were changed every two days. The rings were photographed in phase contrast using a Nikon Eclipse Ti microscope.
Visualization of lipid rafts with cholera toxin B
HUVEC were plated on glass coverslips and preincubated with 50 μg/ml HDL3, 0.1 μg/ml hAIBP, or 50 μg/ml HDL3 + 0.1 μg/ml hAIBP for 4 hours. Cells were washed once with medium before the addition of 1 μg/ml Alexa Fluor 594-labeled cholera toxin B (CTB, from Invitrogen). Cells were incubated for 15 min at 4°C, washed with PBS, and then incubated for 15 min at 4°C with an anti-CTB antibody (EMD Chemicals) to crosslink CTB and lipid rafts. After washing with PBS, cells were fixed in 4% PFA for 20 min at 4°C, mounted with a Prolong Antifade Kit with DAPI (Invitrogen) and images were captured with a Leica DM IRE2 fluorescent microscope.
Cell fractionation
Lipid rafts (light membrane fractions) were isolated using a detergent-free, discontinuous gradient ultracentrifugation method14. Briefly, HUVEC were washed twice with ice-cold PBS and cells were scraped from the plate in 0.5 M sodium carbonate buffer (pH 11.0) containing a protease inhibitor cocktail (Sigma), homogenized and sonicated 3 × 10 sec. Samples were adjusted to 45% sucrose by adding a 90% sucrose solution in MBS (25 mM Mes, 0.15 M NaCl, pH 6.5) and placed into ultracentrifugation tubes. A 5-35% sucrose discontinuous gradient was formed above the sample, followed by ultracentrifugation at 35×103 rpm for 18 hours at 4°C in a SW-41 rotor (Beckman). Ten 1 ml fractions were collected from the top to the bottom of each gradient. The lipid rafts fraction (fraction 5) and the non-lipid rafts fraction (fraction 10) were used for further analysis, which included measurements of protein concentration and immunoblotting. Thirty μl of lipid rafts and non-lipid rafts fractions (adjusted to load equal protein concentrations of each sample) were run on SDS-PAGE, transferred to PVDF membranes and blotted with the indicated antibodies.
Caveolin-1 and VEGFR2 colocalization
HUVEC plated on chamber coverglass (Lab-Tek™ II) were incubated for 4 hours with 0.1 μg/ml hAIBP, 50 μg/ml HDL3 or 0.1 μg/ml hAIBP + 50 μg/ml HDL3, and the cells were washed with PBS and fixed with warm 4% PFA for 15min at room temperature. HUVEC were permeabilized, blocked, and incubated with anti-Caveolin-1 (BD Biosciences) and anti-VEGFR2 (Cell Signaling Technology) antibodies, followed by incubation with anti-mouse IgG-Alexa Fluor 488 and anti-rabbit IgG-Cy3 antibodies. Images were captured using a Nikon Eclipse Ti inverted fluorescent microscope operating in TIRF mode. Raw TIFF images of Caveolin-1 and VEGFR2 were analyzed using JACoP algorithm31 in Image J. Colocalization was quantified using Pearson's coefficient.
VEGFR2 endocytosis
HUVEC were incubated for 4 hours with 0.1 μg/ml hAIBP, 50 μg/ml HDL3 or 0.1 μg/ml hAIBP + 50 μg/ml HDL3, followed by a 20 min incubation with 50 ng/ml VEGF. Cells were fixed and stained with antibodies against VEGFR2 and the early endosomal marker EEA-1-FITC (BD Biosciences). Images were captured with a Nikon Eclipse Ti inverted fluorescent microscope. VEGFR2 and EEA-1 colocalization was quantified using Pearson's coefficient with the JACoP plugin loaded to ImageJ31.
VEGFR2 dimerization assay
The assay was carried out as described in reference32. Two days after plating 1 × 106 HUVEC in a 10 cm dish, the cells were starved overnight in 0.5% FBS-EBM. Next day, cells were incubated with hAIBP and/or HDL3, followed by a 20 min incubation with 50 ng/ml VEGF and then crosslinked with 1 mg/ml bis-sulfosuccinimidyl (Thermo Scientific) for 30 min on ice. Cell lysates were immunoprecipitated with an anti-VEGFR2 antibody immobilized on agarose beads. The beads were washed and the eluted samples were run on SDS-PAGE, followed by immunoblotting with the VEGFR2 antibody.
HUVEC migration assay
Serum starved HUVEC were pretreated with 50 μg/ml HDL3, 0.2 μg/ml hAIBP, or 50 μg/ml HDL3 + 0.2 μg/ml hAIBP for 4 hours at 37°C in 5% LPDS/EBM, harvested from the plate, washed, resuspended in 5% LPDS/EBM and added to the transwell (8 μm pore size). VEGF was added to the lower chamber at 20 ng/ml. Following a 4 hour incubation, the transwell membranes were fixed in ice-cold methanol for 10 min and stained with filtered 0.5% Crystal Violet for 10 min, and transmigrated cells were counted.
Zebrafish
Wild type AB and transgenic Tg(fli1:egfp)y1 and Tg(flk1:ras-cherry)s896 zebrafish lines19,33 were kindly provided by Dr. David Traver and Neil Chi (UCSD). Zebrafish were maintained as previously described34, and all experimental procedures were approved by the UCSD IACUC.
Confocal microscopy
Confocal imaging was carried out as previously described35. Briefly, anaesthetized zebrafish embryos (treated at 24 hpf with 0.003% PTU) were housed in a sealed chamber (Invitrogen) in a small drop of 0.02% tricaine (Sigma) containing E3 medium and imaged using a Nikon C1-si confocal microscope. Z-stacks were acquired with a 1-3 μm step, and images were 3D rendered and analyzed using Imaris® software (Bitplane). All 3D reconstructions were performed with the same threshold settings.
Morpholino oligonucleotide injections
To knock down gene expression, 4-8 ng of morpholino antisense oligonucleotides (MO; synthesized by GeneTools) were injected into one-cell stage embryos. A control MO was derived from the zaibp2 sequence, with 5 mismatched oligonucleotides.
Control MO: TGAGCTTCATGTTCATTTATTCCGC; zaibp2 MO (zaibp2 MO1): TGTGGTTCATCTTGATTTATTCGGC; zaibp2 splicing MO (zaibp2 MO2):TGTTGAGTGTCAGACAAACCTTGGT zaibp1 MO: TCTGTATTCAAATCAGACGCTCAGT; abca1 MO: AACCCAACTGAGTGGAGACAGCCAT; abcg1 MO: AAAAGGCTGCCATGAGACATGCCAT.
Quantitative analysis of cell sprouts in segmental arteries (SeA) and subintestinal veins (SIV)
To determine changes in segmental artery cell sprouts in embryos injected with MO targeting zaibp2, zaibp1, abca1 and/or abcg1, we counted abnormal projections in 4 to 6 pairs of segmental arteries in adjacent somite boundaries in each zebrafish. For each set of injections, 15 embryos (i.e. 60-90 sprouts) were examined. Values were expressed as a number of ectopic sprouts per SeA. To examine sprouts in SIV, only the sprouts moving in the ventral direction out of the SIV were counted. Values were expressed as a number of ventral SIV sprouts per zebrafish.
Measuring membrane lipid order with polarity-sensitive probe
The experiments were carried out as described in references36,37. Briefly, live Tg(flk1:ras-cherry)s896 zebrafish embryos were incubated with 5 μM Laurdan (Invitrogen) at 28°C for 30 min. The concentration of a Laurdan stock solution was measured using OD at 365 nm and an extinction coefficient of 19000 cm-1M-1. After incubation with Laurdan, embryos were incubated with E3 medium for additional 30 min, fixed in PFA for 4 hours at room temperature, deyolked, and embedded in 1% low melting temperature agarose for imaging. Images were captured with a Leica SP5 confocal/multiphoton system, using a water immersion 20× objective. The confocal mode was used to capture mCherry fluorescence and the multiphoton mode was used to capture Laurdan images (ex 800 nm, em 400-460 nm and 470-530 nm) in the same embryos. The multiphoton results were displayed as pseudocolored GP (a measure of the membrane lipid order) images, derived from Laurdan ratiometric measurements and using a ImageJ plug-in as described36. The quantitative data were obtained by measuring GP values in the areas corresponding to tip cells (top 1/3 of the SeA length), stalk cells (bottom 2/3 of the SeA length) and the dorsal aorta in several individual, mCherry-masked z-sections. This method ensured that GP values were derived only from EC (e.g. from the areas where mCherry was in focus in each z-section). The GP images in Fig. 3d were composed each from four to five individual z-sections, with a minimal overlap of mCherry-masked GP images.
Whole mount in situ hybridization
WISH was carried out as described38-40. Briefly, wild type embryos or morphants at indicated developmental stages were fixed with 4% PFA and the embryos older than 24 hpf were permeabilized with 10 μg/ml proteinase K (Roche). Subsequently, the embryos were pre-hybridized at 70°C for 4-6 hours, and then hybridized with a digoxigenin-labeled zaibp2 antisense probe at 65°C for 2 days. Both the control sense and anti-sense RNA probes were directly synthesized from zaibp2 full length gene using a Roche T7/SP6 RNA or Ambion T3 RNA synthesis kit. After extensive wash, hybridized RNA was detected by immunohistochemistry using an alkaline phosphatase-conjugated antibody against digoxigenin (Roche) and a chromogenic substrate nitro blue tetrazolium (NBT) (Sigma) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP)38 (Sigma). A similar procedure was performed with tie2, vegfr2, vegfr3, fli1 and cdh5 probes. Double WISH was performed as described41. Both digoxigenin-labeled zaibp2 and fluorescein-labeled myod were hybridized with the same embryos. The embryos were then first incubated with alkaline phosphatase-conjugated anti-fluorescein Ab, and fast red (Roche) was used as a substrate. Subsequently, the embryos were washed, fixed with 4% PFA and incubated with anti-digoxigenin Ab conjugated alkaline phosphatase, and then NBT/BCIP was used as a chromogenic substrate.
Transplantation experiments
Cell transplantation was performed as described39, with different combinations of donors and recipients as indicated in Fig. 4a. Donor embryos were of the Tg(fli1:egfp)y1 origin, and recipient embryos were of the wild type origin. At the one-cell stage, donor Tg(fli1:egfp)y1 embryos were injected with rhodamine-labeled dextran (Mini-Ruby, Invitrogen) as a lineage tracer. At the sphere stage (approximately 4 hpf), embryos were dechorionated by 0.4 mg/ml pronase (Sigma) and transferred to agarose wells (Adaptive Science Tools, Worcester, MA). Approximately 20-40 cells from the margin of a donor embryo were transferred to the margin of a recipient embryo. The recipient embryos were subsequently grown at 28°C and imaged at 72 hpf. Endothelial cells in chimeric zebrafish originating from donor embryos were visualized by their green fluorescence using a Leica M165FC fluorescent stereoscope. For detailed analysis, images were captured using a Nikon C1-si confocal microscope. Numbers of ectopic branches in each fluorescent SeA were counted.
Real time PCR
Real time PCR was performed using a Rotor Gene Q qPCR machine (Qiagen). Real time PCR master mix Platinum SYBR Green qPCR SuperMix was from Invitrogen. The primers were synthesized by IDT. The PCR program was: 50°C for 2 minutes (UDG incubation), 95°C for 2 minutes, 40 cycles of: 95°C for 15 seconds, 60°C for 1 minute. Primer sequences: fli1 (F): CTTGGCACGTTGCCTTGATAAG, fli1 (R): CCTTCATATCTGAGAGTGATCCC; tie2 (F): GCGATGGATGGCAATAGAGT, tie2 (R): CGACAGCAGGATCTGAGAGA; vegfr2 (F): TCCACGAGGGTGGGCAGTCA; vegfr2 (R): AGACGGGTGGTGTGGAGTAACGA. kdrb (F): TGCCCACATGGAGCTGCTAGCA; kdrb (R): TGTGGCACATTCAACCACATGAGC. β-actin (F): CTCTTCCAGCCTTCCTTCCT, β-actin (R): GGTTGGTTCGTTCGTTTGAAT.
Immunoblot of zebrafish lysates
Zebrafish were lysed on ice with a lysis buffer (50 mM Tris-HCl, pH 7.5, 4 mM sodium deoxycholate, 1% Triton X 100, 150 mM NaCl, 1 mM EDTA, and a protease inhibitor cocktail from Sigma). Protein content was determined with a DC protein assay kit (BioRad) and equal protein amounts of the cell lysates were run on a 4-12% Bis-Tris SDS-PAGE with MOPS buffer (Invitrogen) and then transferred to a PVDF membrane (Invitrogen). The blots were probed with appropriate antibodies against specific phosphorylated and non-phosphorylated proteins (Cell Signaling Technology), secondary antibodies conjugated with HRP and developed using a Super Signal West Dura substrate (Pierce).
Filipin staining
Filipin staining of embryos was performed as described42. Zebrafish were fixed with 4% PFA overnight at 4°C. The fixed fish were incubated overnight with 0.05% filipin (Sigma) in PBS with 1% sheep serum, and then washed 3 times with PBS. Images were captured immediately with a Leica M165FC fluorescent stereoscope and quantified.
Total lipid extraction and free cholesterol measurements in zebrafish
Total lipid was extracted from zebrafish embryos as we previously described28. In brief, trunk/tail segments were dissected from fifty 24 hpf embryos and pooled together. The tissue was homogenized and supplemented with 50 μg stigmasterol, an internal standard to control for recovery of extracted sterols. Total lipid extraction was performed with 1:2 methanol/dichloromethane. No saponification of cholesterol esters was performed because the goal of this study was to measure free cholesterol, the form of cholesterol transferred from the cells via ABC transporters to ApoA-I/HDL. Cholesterol and stigmasterol were measured with a Shimadzu GC-2014 gas chromatograph using a 30 m × 0.25 mm (i.d.) ZB-5HT inferno capillary column, film thickness 0.2 μm (Phenomenex). Cholesterol levels were normalized to protein and then to the levels in embryos injected with control MO.
Supplementary Material
Acknowledgments
We thank David Traver, Neil Chi, Joseph Witztum, Richard Klemke, Deborah Yelon, Tracy Handel, Konstantin Stoletov, Wilson Clements, Claire Pouget, Zayra Garavito-Aguilar, Ararat Ablooglu, Ruiling Zhang, Xiaohong Yang, Mila Angert, Kersi Pestonjamasp and Jennifer Santini (UC San Diego), Catherine Hedrick, Klaus Ley, Duygu Sag, Prithu Sundd and Amy Wu (La Jolla Institute for Allergy and Immunology), Sean Trzaska (New York University), Shao Jun Du (University of Maryland), Bettina Schmid and Christian Haass (Ludwig-Maximilians-University München, Germany), Dylan Owen and Astrid Magenau (University of New South Wales, Australia), Arndt Siekmann (Max-Planck Institute for Molecular Biomedicine), and Christoph Binder (Medical University of Vienna, Austria) for many helpful discussions, technical assistance and/or for providing reagents and access to equipment for this study. The project was supported by the NIH grants HL093767 (Y.I.M.), HL055798 (Y.I.M.) and HL114734 (L.F.), and the fellowship 18FT-0137 from the UC Tobacco-Related Disease Program (L.F.), as well as the UCSD Neuroscience Microscopy Facility Grant P30 NS047101.
Footnotes
The authors declare no conflicts of interests.
Author Contributions: F.L. and Y.I.M. conceived the project, designed the experiments and wrote the manuscript. J.T.-V. made important intellectual contributions and helped revise the manuscript. F.L. performed the majority of the experiments. S.H.C., J.S.B., C.L., F.A., F.U., P.W., A.T., E.D., J.P., A.C.L. performed experiments and/or provided research assistance.
Author Information: Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Bodzioch M, et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 2.Rust S, et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 3.Klucken J, et al. ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc Natl Acad Sci USA. 2000;97:817–822. doi: 10.1073/pnas.97.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yvan-Charvet L, et al. ATP-Binding Cassette Transporters and HDL Suppress Hematopoietic Stem Cell Proliferation. Science. 2010;328:1689–1693. doi: 10.1126/science.1189731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong AJ, Gebre AK, Parks JS, Hedrick CC. ATP-binding cassette transporter G1 negatively regulates thymocyte and peripheral lymphocyte proliferation. J Immunol. 2010;184:173–183. doi: 10.4049/jimmunol.0902372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensinger SJ, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritter M, et al. Cloning and Characterization of a Novel Apolipoprotein A-I Binding Protein, AI-BP, Secreted by Cells of the Kidney Proximal Tubules in Response to HDL or ApoA-I. Genomics. 2002;79:693–702. doi: 10.1006/geno.2002.6761. [DOI] [PubMed] [Google Scholar]
- 8.Jha KN, et al. Biochemical and Structural Characterization of Apolipoprotein A-I Binding Protein, a Novel Phosphoprotein with a Potential Role in Sperm Capacitation. Endocrinology. 2008;149:2108–2120. doi: 10.1210/en.2007-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefulj J, et al. Human Endothelial Cells of the Placental Barrier Efficiently Deliver Cholesterol to the Fetal Circulation via ABCA1 and ABCG1. Circ Res. 2009;104:600–608. doi: 10.1161/CIRCRESAHA.108.185066. [DOI] [PubMed] [Google Scholar]
- 10.Terasaka N, et al. ABCG1 and HDL protect against endothelial dysfunction in mice fed a high-cholesterol diet. J Clin Invest. 2008;118:3701–3713. doi: 10.1172/JCI35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fessler MB, Parks JS. Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. J Immunol. 2011;187:1529–1535. doi: 10.4049/jimmunol.1100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez AJ, et al. Membrane Lipid Domains Distinct from Cholesterol/Sphingomyelin-Rich Rafts Are Involved in the ABCA1-mediated Lipid Secretory Pathway. J Biol Chem. 2001;276:3158–3166. doi: 10.1074/jbc.M007717200. [DOI] [PubMed] [Google Scholar]
- 13.Murphy AJ, et al. High-Density Lipoprotein Reduces the Human Monocyte Inflammatory Response. Arterioscler Thromb Vasc Biol. 2008;28:2071–2077. doi: 10.1161/ATVBAHA.108.168690. [DOI] [PubMed] [Google Scholar]
- 14.Noghero A, et al. Liver X Receptor Activation Reduces Angiogenesis by Impairing Lipid Raft Localization and Signaling of Vascular Endothelial Growth Factor Receptor-2. Arterioscler Thromb Vasc Biol. 2012;32:2280–2288. doi: 10.1161/ATVBAHA.112.250621. [DOI] [PubMed] [Google Scholar]
- 15.Oshikawa J, et al. Novel role of p66Shc in ROS-dependent VEGF signaling and angiogenesis in endothelial cells. Am J Physiol Heart Circ Physiol. 2012;302:H724–H732. doi: 10.1152/ajpheart.00739.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda S, et al. Novel Role of ARF6 in Vascular Endothelial Growth Factor-Induced Signaling and Angiogenesis. Circ Res. 2005;96:467–475. doi: 10.1161/01.RES.0000158286.51045.16. [DOI] [PubMed] [Google Scholar]
- 17.Liao Wx, et al. Compartmentalizing VEGF-Induced ERK2/1 Signaling in Placental Artery Endothelial Cell Caveolae: A Paradoxical Role of Caveolin-1 in Placental Angiogenesis in Vitro. Mol Endocrinol. 2009;23:1428–1444. doi: 10.1210/me.2008-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichmann A, Simons M. VEGF signaling inside vascular endothelial cells and beyond. Curr Opin Cell Biol. 2012;24:188–193. doi: 10.1016/j.ceb.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson ND, Weinstein BM. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 20.Torres-Vazquez J, et al. Semaphorin-plexin signaling guides patterning of the developing vasculature. Dev Cell. 2004;7:117–123. doi: 10.1016/j.devcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Ann Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 22.Archer A, et al. Transcriptional activity and developmental expression of liver X receptor (lxr) in Zebrafish. Dev Dyn. 2008;237:1090–1098. doi: 10.1002/dvdy.21476. [DOI] [PubMed] [Google Scholar]
- 23.Whetzel AM, et al. ABCG1 Deficiency in Mice Promotes Endothelial Activation and Monocyte-Endothelial Interactions. Arterioscler Thromb Vasc Biol. 2010;30:809–817. doi: 10.1161/ATVBAHA.109.199166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avraham-Davidi I, et al. ApoB-containing lipoproteins regulate angiogenesis by modulating expression of VEGF receptor 1. Nat Med. 2012;18:967–973. doi: 10.1038/nm.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carmona G, et al. Role of the small GTPase Rap1 for integrin activity regulation in endothelial cells and angiogenesis. Blood. 2009;113:488–497. doi: 10.1182/blood-2008-02-138438. [DOI] [PubMed] [Google Scholar]
- 26.Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Connell BJ. Cellular Physiology of Cholesterol Efflux in Vascular Endothelial Cells. Circulation. 2004;110:2881–2888. doi: 10.1161/01.CIR.0000146333.20727.2B. [DOI] [PubMed] [Google Scholar]
- 28.Fang L, et al. Oxidized cholesteryl esters and phospholipids in zebrafish larvae fed a high cholesterol diet: macrophage binding and activation. J Biol Chem. 2010;285:32343–32351. doi: 10.1074/jbc.M110.137257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao F, et al. L-5F, an apolipoprotein A-I mimetic, inhibits tumor angiogenesis by suppressing VEGF/basic FGF signaling pathways. Integr Biol (Camb) 2011;3:479–489. doi: 10.1039/c0ib00147c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellacen K, Lewis EC. Aortic ring assay. J Vis Exp. 2009 doi: 10.3791/1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 32.Chung TW, et al. Ganglioside GM3 inhibits VEGF/VEGFR-2-mediated angiogenesis: direct interaction of GM3 with VEGFR-2. Glycobiology. 2009;19:229–239. doi: 10.1093/glycob/cwn114. [DOI] [PubMed] [Google Scholar]
- 33.Chi NC, et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westerfield M. The Zebrafish Book. (University of Oregon Press; Eugene, Oregon: 2007. [Google Scholar]
- 35.Stoletov K, et al. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ Res. 2009;104:952–960. doi: 10.1161/CIRCRESAHA.108.189803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen DM, Rentero C, Magenau A, Abu-Siniyeh A, Gaus K. Quantitative imaging of membrane lipid order in cells and organisms. Nat protocols. 2012;7:24–35. doi: 10.1038/nprot.2011.419. [DOI] [PubMed] [Google Scholar]
- 37.Gaus K, Le Lay S, Balasubramanian N, Schwartz MA. Integrin-mediated adhesion regulates membrane order. J Cell Biol. 2006;174:725–734. doi: 10.1083/jcb.200603034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 39.Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–784. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- 40.Lawson ND, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- 41.Jowett T. Analysis of protein and gene expression. Methods Cell Biol. 1999;59:63–85. doi: 10.1016/s0091-679x(08)61821-x. [DOI] [PubMed] [Google Scholar]
- 42.Schwend T, Loucks EJ, Snyder D, Ahlgren SC. Requirement of Npc1 and availability of cholesterol for early embryonic cell movements in zebrafish. J Lipid Res. 2011;52:1328–1344. doi: 10.1194/jlr.M012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.