Summary
Associations between multilocus heterozygosity and fitness traits, also termed heterozygosity and fitness correlations (HFCs), have been reported in numerous organisms. These studies, in general, indicate a positive relationship between heterozygosity and fitness traits. We studied the association between genome-wide heterozygosity at 706 non-synonymous and synonymous SNPs and 19 quantitative traits, including morphological, biochemical and fitness traits in the Framingham Heart Study. Statistically significant association was found between heterozygosity and systolic and diastolic blood pressures as well as left ventricular diameter and wall thickness. These results suggest that heterozygosity may be associated with traits, such as blood pressure that closely track environmental variations. Balancing selection may be operating in the maintenance of heterozygosity and the major components of blood pressure and hypertension. Genome wide SNP heterozygosity may be used to understand the phenomenon of dominance as well as the evolutionary basis of many quantitative traits in humans.
Keywords: Genome wide heterozygosity, Single Nucleotide Polymorphisms, Balancing selection, inbreeding, association, plastic traits, dominance
1. Introduction
Heterozygous individuals in many organisms are known to exhibit greater levels of vigor, disease resistance, growth rate, longevity and overall Darwinian fitness (Britten, 1996; Lynch & Walsh, 1998), and possess superior buffering capacity to environmental fluctuations-a phenomenon known as genetic homeostasis (Lerner, 1954) - as well as to environmental stress (Parsons, 1971). The concept of homeostasis was first introduced by Cannon (1932), to elucidate mechanisms underlying optimal maintenance of physiological changes in humans, and suggests that biological systems “trigger physiological responses to maintain the constancy of internal environment in face of disturbances of external surroundings” (Cowley 2003). Lerner (op. cit) offered a genetic explanation for Cannon’s hypothesis and suggested that heterozygosity may be essential toward maintaining homeostasis against environmental variations. Following Lerner numerous investigators have studied the relationship between heterozygosity and quantitative traits in a number of organisms and under a wide spectrum of environmental conditions (see Mitton, 1997). Studies investigating the relationship between viability and reproductive fitness and levels of heterozygosity are grouped under the title “heterozygosity and fitness correlations” (HFCs; David, 1998; Bierne, et al. 2000). The HFC concept suggests that heterozygote superiority is maintained in populations due to its inherent selective advantage (also called overdominance; Charlesworth & Charlesworth, 1987; Hansson & Westerberg, 2002) over homozygotes. Selection against homozygosity due to inbreeding (Hansson & Westerberg, 2008) could reduce viability and reproductive fitness, in contrast to heterozygosity; hence the HFCs could also be used as an index of inbreeding depression (Balloux et al. 2004). In a meta-analysis involving 34 data sets, each with a minimum of three populations, 28 out of 34 data sets showed a positive relationship between some measure of fitness and heterozygosity (Reed & Frankham, 2001).
Association studies between levels of heterozygosity, on the one hand, and morphological, physiological, reproductive and longevity traits, developmental stability and disease resistance, on the other, have been attempted in numerous organisms using allozymes (Allendorf et al. 1997; Mitton, 1997; Hansson & Westerberg, 2002), microsatellite markers (Thelen & Allendorf, 2001), and restriction fragment length polymorphisms (RFLPs; Botstein et. al 1980). Among these, allozyme studies have yielded the most comprehensive understanding on the role of heterozygosity in the survival and reproduction of organisms. However, studies involving allozymes and microsatellites also suffer from certain limitations. For example, the number of allozyme markers available for detailed analysis of HFCs is generally limited. At the best, heterozygosity data are available on only a few dozen allozyme markers and on relatively small samples of individuals in each population; this often leads to inaccurate predictions (Eanes, 1999). In contrast, microsatellites are more abundant than allozymes, but they are generally considered neutral; thus it is difficult to determine whether the observed relationship between heterozygosity indicated by microsatellite markers and phenotypic traits is due to direct or indirect effects of the microsatellites studied. Nonetheless, Campbell et al. (2007), measuring associations between microsatellite markers and a panel of cardiovascular traits among Croatian populations, found a positive association between high heterozygosity and low systolic blood pressure. Thus, although microsatellite marker heterozygosity provides some insight on the occurrence and role of HFCs, they fail to provide a comprehensive view of the maintenance of variation in natural populations (see below).
Studies involving either allozymes or microsatellites also suffer from other drawbacks, most notably from the limited availability of phenotypic markers. The latter include metabolic (endo) phenotypes that link the genotype with anatomical, viability and fitness traits. Additionally, three other issues related to HFC, cannot be adequately addressed with either allozyme or microsatellite markers. For instance, dominance, overdominance, and the observed HFC relationships could result from three factors: a) the direct effect of a given locus on the phenotype in question, b) associative overdominance, or the indirect influence of the neighboring locus/allele in linkage disequilibrium with the causal allele, and c) small and cumulative influence of heterozygosity at numerous genome-wide loci (Hansson and Westerberg 2002). Previous studies have not adequately addressed all three hypotheses on the maintenance of HFCs due to the inherent limitations of the markers and the phenotypic traits measured. From this perspective, we suggest that the use of Single Nucleotide Polymorphism (SNP) data in the analysis of HFCs may be more useful for disentangling the direct and indirect effects of polymorphisms than allozyme and microsatellite markers. For example, non-synonymous (NSn) SNPs are known to affect protein products and consequently influence the resulting phenotype. Synonymous (Sn) -SNPs, on the other hand, may have no measurable effect on the phenotype. Hence, if amino acid coding regions (the NSn –SNPs) exert greater influence on the phenotype relative to Sn-SNPs, then the magnitude of HFCs may be used as an index to determine the relative direct or indirect effects of these genomic regions on the phenotype. Furthermore, SNPs are very attractive for HFC studies as they are biallelic, and can be easily assayed at numerous loci (> 6.5 million validated SNPs; http://www.ncbi.nlm.nih.gov/SNP/snp_summary.cgi) across the human genome (DeWoody & DeWoody, 2005). It is important to note that although there is a staggering number of SNPs available in the human genome, the number of Ns-SNPs may be very limited.
There are additional concerns, however. While heterozygous superiority (heterosis) is known to affect the entire organism, traits such as blood pressure (plastic traits) in animals, including humans, may be highly modulated by environmental variation relative to other suites of traits, leading us to ask the following questions: Does heterozygosity have differential effects on traits in different pathways? Do the effects of heterozygosity on quantitative traits differ in younger and older individuals? Do these relationships have sex specific effects? Answers to such questions may emerge from a set of association analysis between average heterozygosity and a large number of quantitative traits representing different developmental pathways influencing viability, reproduction and health in human populations.
In the present study, we report the association between average individual heterozygosity of both non-synonymous and synonymous SNPs and 19 quantitative traits, including anthropometric, physiological and anatomical traits in the Framingham Heart Study (FHS). Polymorphisms in the amino acid coding regions (NSn-SNPs) and non-coding regions (Sn-SNPs) of the genome are known to influence traits that show mendelian and quantitative mode of inheritance. Examples include, mutations in the ADAM33 (a disintegrin and metalloprotease domain 33), and NOD2 (nucleotide-binding oligomerization domain containing 2) associated with Asthma (Blakely et. al. 2005) and Crohn’s disease (Ogura et al 2001), respectively. Recent genome wide association studies have focused almost exclusively on establishing associations between SNPs and quantitative traits (Donnelly 2008). Furthermore, a recent report indicates that 44 percent of all the genes in the human genome are heterozygous for one or more loci (i.e. SNPs; Levy et al. 2007); which suggests that the role of SNP heterozygosity and dominance in the human genome cannot be easily discounted. Instead it provides a compelling reason to examine their function in the maintenance of heath and its components, as well as quantitative traits.
Materials and Methods
Cohort, samples and data
The data represented in this study were derived from the first two generations of the Framingham Heart Study (FHS) cohort. Details of the cohort, diversity of phenotypic and biochemical traits measured over the years, as well as genetic analysis conducted using this cohort are presented elsewhere (Cupples et al. 2007; Govindaraju et al. 2008). In brief, the FHS is a longitudinal study, initiated in 1948 with the intent to identify factors that contribute to cardiovascular diseases (CVD). The participants represented in the FHS cohort are of European descent, drawn largely from the Town of Framingham, located 20 miles west of Boston. The Original and Offspring generation cohorts consist of 5209 (2336 men and 2873 women), and 5124 (2483 men and 2641 women), respectively. The participants in the Original and Offspring cohorts have undergone 29 (every two years), and 8 (every four to eight years) cycles of examination, respectively. Clinical data on these participants have been collected under the following six categories in each of the exam cycle: physical exams, life style, medical history, laboratory analysis, non invasive tests and endpoints (dbGaP: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v4.p2).
For the present study, data on 19 traits were assembled from the previously collected panel of data (posted in the dbGaP; see above) on both males and females from examination six of the second generation (offspring) cohort from the FHS. These traits may be grouped roughly into four categories: a) demographic and anthropometric measures (age, sex, age of menopause, height, weight, body mass index (BMI), hip girth, b) physiological (bio) markers [renin, aldosterone to renin ratio (Aldo/Renin), C-reactive protein, fibrinogen, interleukin-6], and c) anatomical [left ventricular diameter-diastolic (LVDD), left atrial diameter – LAD systolic, aortic root diameter (AOR)- diastolic, left ventricular mass (LVM), left ventricular wall thickness (LVWT), d) physical examination variables (systolic and diastolic blood pressure (SBP and DBP), and e) the FHS -risk score for each of the participantin examination cycle. traits were selected from a large number of phenotypic variables available from the FHS data sets (see dbGaP) as many of these traits are known to contribute directly or indirectly toward the development of coronary heart disease (Kannel et al. 1961; Wilson et al. 1998). Traits such as age of menopause and longevity may be considered life history traits, as they contribute to both viability and reproductive fitness (Stearns 1992). Many of the quantitative traits examined in this study show substantial heritability and are also known to be potential risk factors in cardiovascular diseases (reviewed in Govindaraju et al. 2008)., the traits considered for analysis belong to four groups (see above), and traits within each group may show some degree of correlation, indicating their ontogenetic relationships within and among groups. Certain traits, on the other hand, might be independent of each other, showing little or no correlation.
Genotyping and genotype data
The DNA for the study was extracted from whole blood, buffy coat and immortalized cell lines, and was extracted using either phenol-chloroform or salt-precipitation methods. The SNP genotypes reported in the study were derived from the Affymetrix 100K GeneChip (see Cupples et al 2007 for details). A total of 706 SNPs (ALL-SNPs) from the gene-centric regions represented in the 100K chip were employed in the study, of which 313 were non-synonymous (NSn) SNPs and 393 synonymous (Sn) SNPs (from intronic, untranslated and coding regions). Only 313 non-synonymous SNPs were available in the 100K Affymetrix GeneChip panel, and all of them were utilized in the study. The remaining 393 synonymous SNPs were selected randomly from the 110K panel for comparison. There was no statistical difference for the level of heterozygosities between the two panels. It must be noted that a great deal of the SNPs represented in the 100K panel derived from non-amino acid changing exonic regions or non-coding regions of the genome.
Average individual heterozygosity was determined by counting the genotypes as follows: where m is the number of loci examined, and Hij is a binary {0, 1} variable for locus j within ith individual [modified from Weir (1996), Donavan & Welden (2001)] Hij = 1 if the j-th locus is heterozygous, and = 0 otherwise.
Data analysis
The traits used in the study were tested for normality, and the deviations from normality were corrected using log transformations on variables such as renin, CRP, fibrinogen and interleukin-6 to approach a normal distribution. Furthermore, some individuals were treated with blood pressure reducing drugs, hence these traits were adjusted for medications, and only the residuals were used for association studies. Because heterozygosity is known to respond to environmental stresses (Parsons 2007), we used a composite trait, cardiovascular disease risk (the FHS-CVD risk score), to explore the relationship between environmental stress and the average heterozygosity. The FHS-CVD risk score has been constructed using biological (cholesterol, blood pressure etc.,) and environmental indices (smoking) that are known to contribute to coronary heart disease (Wilson et al. 1998); hence the risk score could be viewed as an overall indicator of stress influencing cardiac health.
Descriptive statistics were obtained on all variables. Association studies were performed using regressions, accounting for age and sex. All statistical analyses were conducted in the SAS package, version 9.1 (SAS Institute Inc., Cary, NC). The age specific relationship between heterozygosity and SBP and DBP was carried out by dividing the cohort into two age groups: ≤50 and >50 years. This cut-off was used as most white women in North America reach the age of menopause around 50 years of age (McKinlay 1996). Similarly, gender differences in relation to heterozygosity for all the traits were also tested. Statistical significance of the association was assessed individually for each trait, and using Bonferroni correction, which controls the family wise error rate (FWER), as well as applying the Guo and Rao (2008) procedure, which controls the false discovery rate (FDR). The latter approach is a modified “step-down” procedure, in which the variables are arranged in descending order of importance, and can be applied irrespective of the strength of correlation (degree of dependency) among traits.
Results
Sample sizes, mean, standard deviations for biomarkers, and quantitative traits are provided in Table 1. The average age of individuals in the second generation cohort was 56 years at exam 6, with limits of 29– 85 years of age. Mean heterozygosity for ALL-SNPs was 0.247 (SD 0.189; ranging from0.170 to 0.308); corresponding values for NSn-SNPs and S-SNPs were 0.239 (SD 0.250, ranging from 0.134 to 0.320) and 0.254 (SD 0.240, ranging from 0.170 to 0.328), respectively. The sample size for various traits used in the study varied from 253 to 1017. Except for the age of menopause, sample size for all the other traits ranged from 782 to 1017 (Table 2).
Table 1.
Characteristics of the quantitative variables used in the analysis.
| Trait | Sample size | Mean ± SD | Limits |
|---|---|---|---|
| Age (years) | 1017 | 56 ± 10 | 29 – 85 |
| Height (Cms) | 1007 | 168 ± 10 | 142 – 197 |
| Weight (Kg) | 1014 | 80 ± 18 | 18 – 41 |
| Body mass index (kg/m2) | 1007 | 28.2 ± 5.5 | 16.6 – 53.3 |
| Hip girth (cms) | 993 | 104 ± 10 | 58.4 – 152 |
| Age of menopause (years) | 253 | 48.6 ± 5.3 | 28.0 – 62.0 |
| Systolic blood pressure (mm. Hg) | 1017 | 126 ± 18 | 87 – 203 |
| Diastolic Blood Pressure (mm. Hg) | 1017 | 76 ± 9 | 50 – 118 |
| Renin (log-transformed) | 1000 | 2.57 ± 0.98 | 0.69 – 8.27 |
| Aldosterol/Renin ratio (log-transformed) | 974 | − 0.27 ± 1.02 | − 5.97 – 2.25 |
| C-reactive protein (log-transformed) | 997 | 0.67± 1.15 | −1.83 – 4.67 |
| Fibrinogen (log-tranformed) | 1000 | 5.79 ± 0.19 | 5.23 – 6.56 |
| Interleukin – 6 (log – transformed) | 965 | 1.04 ± 0.72 | −0.56 – 4.65 |
| Left ventricular diameter-end diastole (cm) | 782 | 4.81 ± 0.50 | 3.39 – 7.46 |
| Left atrial diameter – end systole (cm) | 974 | 3.99 ± 0.56 | 2.71 – 5.99 |
| Aortic root diameter – end diastole (cm) | 977 | 3.22 ± 0.40 | 2.17 – 4.82 |
| Left ventricular mass (grams) | 779 | 167 ± 47 | 78 – 414 |
| Left ventricular wall thickness (cm) | 801 | 1.91 ± 0.27 | 1.31 – 3.80 |
| The FHS - Coronary Heart Disease risk score | 996 | 6.73 ± 4.92 | −15 – 22 |
| Mean heterozygosity for all the 706 SNPs | 1017 | 0.247 ± 0.019 | 0.17 – 0.31 |
| Mean heterozygosity for the 313 NSn-SNPs | 1017 | 0.239 ± 0.025 | 0.13 – 0.32 |
| Mean heterozygosity for the 393Sn-SNPs | 1017 | 0.254 ± 0.024 | 0.17 – 0.33 |
Table 2.
Association between heterozygosity and quantitative traits adjusted for age and sex.
| Trait | N | ALL-SNPs (N=706) | NSn-SNPs (N=313) | SN-SNPs (N=393) | |||
|---|---|---|---|---|---|---|---|
| Regression coefficient |
p-value | Regression coefficient |
p-value | Regress ion coeffici ent |
p-value | ||
| LVDD | 782 | 0.019 | 0.021 | 0.012 | 0.06 | 0.011 | 0.09 |
| LAD | 974 | −0.012 | 0.15 | −0.007 | 0.30 | −0.008 | 0.23 |
| AOR | 977 | −0.004 | 0.46 | −0.001 | 0.89 | −0.004 | 0.35 |
| LVM | 779 | 0.099 | 0.89 | −0.32 | 0.55 | 0.384 | 0.49 |
| LVWT | 801 | −0.009 | 0.04 | −0.008 | 0.013 | −0.003 | 0.41 |
| SBP | 1017 | 0.702 | 0.011 | 0.416 | 0.046 | 0.418 | 0.054 |
| DBP | 1017 | 0.428 | 0.0052 | 0.24 | 0.038 | 0.267 | 0.027 |
| HGT | 1007 | −0.001 | 0.26 | −0.001 | 0.09 | 0 | 0.87 |
| WGT | 1014 | −0.326 | 0.22 | −0.275 | 0.17 | −0.123 | 0.56 |
| BMI | 1007 | −0.075 | 0.42 | −0.054 | 0.43 | −0.036 | 0.62 |
| LogRenin | 1000 | −0.005 | 0.77 | −0.007 | 0.58 | 0.001 | 0.96 |
| LogARR | 974 | 0.014 | 0.41 | 0.009 | 0.49 | 0.008 | 0.56 |
| LogCRP | 997 | −0.009 | 0.62 | −0.011 | 0.43 | −0.001 | 0.95 |
| logfib | 1000 | −0.005 | 0.09 | −0.004 | 0.07 | −0.002 | 0.38 |
| Logil6 | 965 | 0.000 | 0.99 | −0.004 | 0.64 | 0.003 | 0.72 |
| Age menopause | 253 | 0.140 | 0.45 | −0.105 | 0.46 | 0.206 | 0.13 |
| hipgirth | 993 | −0.050 | 0.46 | −0.044 | 0.30 | −0.017 | 0.75 |
| CHD risk score | 996 | 0.076 | 0.21 | 0.12 | 0.80 | 0.074 | 0.12 |
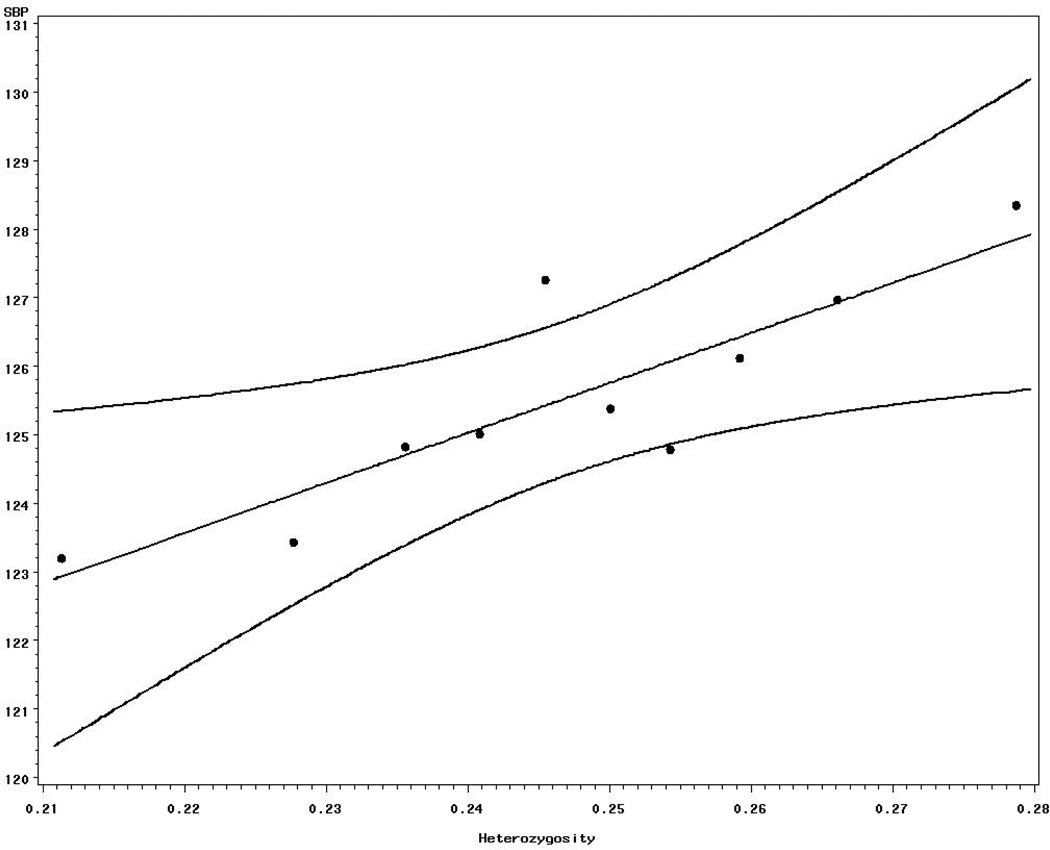
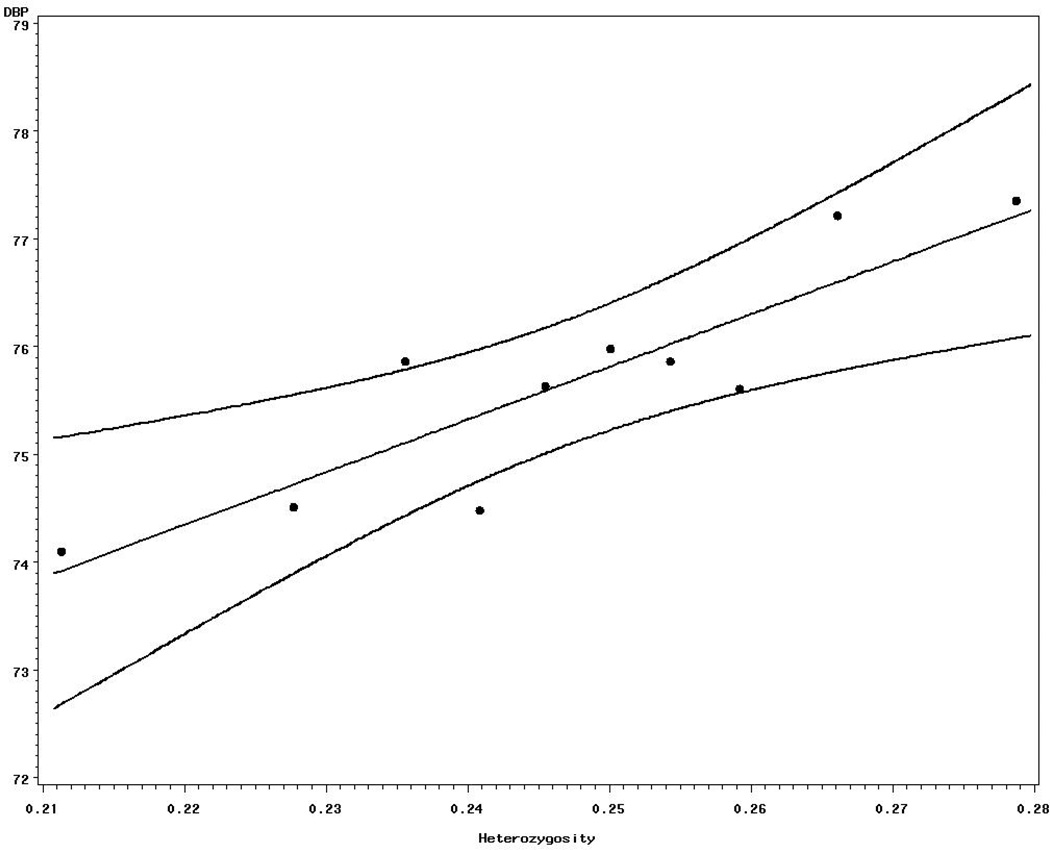
Heterozygosity at ALL-SNPs showed a positive association with left ventricular diameter and wall thickness, systolic and diastolic blood pressure. Associations between heterozygosity at ALL-SNPs on the one hand, and SBP and DBP on the other are presented in Fig 1 and 2, respectively. However, only three (LVWT, SBP, and DBP) traits showed significance with heterozygosity at NSn-SNPs; although LVDD approached significance (P= 0.06). Only diastolic pressure showed statistically significant association with heterozygosity of Sn-SNPs. Weak associations were found between the levels of heterozygosity at NSn-SNPs and height and fibrinogen. Interaction between age and heterozygosity as well as between sex and heterozygosity were not detected (data not shown). In general, heterozygosity at NSn-SNPs tended to be associated with more quantitative traits than either ALL or Sn-SNPs . Influence of sex and age on SBP and DBP with heterozygosity was not significant (data nor shown).
Figure 1.
Relationship between heterozygosity and mean SBP. (For display purposes, decile groups of heterozygosity classes, with equal number within each class, were created and used in the graph)
Figure 2.
Relationship between heterozygosity and mean DBP. (For display purposes, decile groups of heterozygosity classes, with equal number within each class, were created and used in the graph)
Discussion
We extended the well-known heterozygosity and fitness concept (HFCs; Thelen and Allendorf, 2001) to study the influence of SNP heterozygosity on a battery of quantitative traits in an outbred human population. Three hypotheses have been advanced to explain the correlation between multilocus heterozygosity and fitness traits: direct, local and general effects (Hansson and Westerberg, 2002). While direct effects result from functional overdominance of the scored loci (Houle 1989), local effects arise from an association between a neutral marker and the loci under selection (Lynch and Walsh 1998). General effects, on the other hand, arise due to identity disequilibria (correlation of homozygosity or heterozygosity between loci across the entire genome; Tsitrone et al 2001) generated by partial inbreeding and the cumulative effects of multilocus heterozygosity (David 1998). Our results provide an opportunity to make some generalizations on the above three hypotheses. While few traits, particularly the ones related to blood pressure, showed a positive association with NSn-SNPs and ALL-SNPs, suggesting that amino acid coding regions may be influencing the studied traits directly or in concert with the loci in cis or trans positions, only one trait (DBP) showed significant association with the heterozygosity of Sn-SNPs. Synonymous SNPs are in the non-coding regions of the genome, and hence may be indirectly influencing many of the traits studied through NSn-SNPs. Alternatively, correlations arising due to partial inbreeding as a possible explanation for the observed HFC as suggested by Hansson and Westerberg (2002) cannot be ruled out. Possibilities of cryptic homozygosity in the contemporary Massachusetts populations due to consanguinity in the recent past exist. For instance, Danubio & Pettener (1997) reported that between 1880–1920, endogamous marriages among three Italian communities from three parishes in Boston varied from 93.9 to 97.3 percent, of which 2.33 and 6.38 percent were consanguineous and isonymous marriages, respectively. Nearly 19 percent of the individuals in the FHS cohort claim Italian ancestry (Govindaraju et al 2008). Hence, it may not be surprising to find evidence for cryptic homozygosity in the FHS cohort. Cryptic homozygosity in the genome may be important even in modern populations. For instance, Keightley et al (2005) convincingly argued that very old inbreeding traditions might be still influencing the genetic expression in the human genome. As a corollary, long homozygosity stretches, that mimic inbreeding, have been reported even in outbred human populations (Gibson et al. 2006). Hence, the associations between various classes of SNPs and quantitative traits found in this study are consistent with all the three hypotheses (i.e., direct, local and general effects; Hansson and Westerberg, 2002).
Systolic and diastolic pressures as well as left ventricular mass are complex traits and are the major components of blood pressure (Hill and Olsen 2008). From a physiological perspective, Cowley (1992) indicated that optimal “moment-to-moment and long-term stability of arterial pressure are necessary for survival” of all mammals, and “complex mechanisms have evolved to provide a constant level of this pressure throughout a life time of daily stresses and activities.” Either normal or chronically elevated blood pressure (hypertension) is a highly variable trait, and responds to slightest variations in environmentally and physiologically induced changes. Moreover, blood pressure and vascular walls are essential components of hypertension, and show plastic responses to salt intake and other environmental factors (Timarco et al. 1991; Gonzalez et al.). Left ventricular mass and blood pressure are correlated among individuals with hypertension (Mule et al 2003), and left ventricular mass is highly plastic and contributes to “cardiac plasticity” as well as left ventricular hypertrophy (Hill and Olsen op. cit.). From an evolutionary perspective, sustained changes in blood pressure outside the normal bounds in response to environment could lead to hypertension, which in turn could also affect both health and fitness of individuals (Danziger 2001). Thus arterial pressure may be considered as an important fitness trait. Like all fitness traits, systolic and diastolic blood pressure as well as left ventricular mass show moderate heritability (Rijn et al. 2007; Post et al. 1997).
Chronic changes in dietary salt intake as well as other environmental variables impose physiological stress (Meng et al 2003) on the vascular system, which ultimately results in sustained high blood pressure or hypertension (Handel & Collister 2005). In our study, systolic and diastolic blood pressure as well as left ventricular diameter showed a positive relationship with heterozygosity. It is argued that heterozygosity may be advantageous under stressful conditions as heterozygotes may have lower energy requirements than homozygotes (Parsons 2007). Note that arterial pressure is a consequence of many factors that “shift continuously depending on the circumstances” (Cowley 1992). Hence, the relationship between heterozygosity and blood pressure related traits may not be surprising, as arterial pressure shifts in relation to environmental variation. Using microsatellite markers, Campbell et al. (2007), reported an inverse relationship between heterozygosity and SBP and DBP in Croatian populations, and interpreted that greater heterozygosity may be advantageous. Furthermore, in a recent study, Ji et al. (2008) screened the FHS cohort for three genes involved in salt handling, and thus contributing to variation in blood pressure and hypertension. They reported that the “causal” genotypes in all the three genes associated with blood pressure were heterozygous, suggesting that heterozygosity may be an important component in the blood pressure pathway.
Scheiner (2006) proposed an interesting hypothesis on the relationship between heterozygosity and quantitative traits, which states that heterozygosity or overdominance may be associated with increased plasticity to traits, irrespective of the direction of association. Hawkins and Day (1999) suggested that the genetic components of fitness respond to environmental variation, in relation to stages of life history and physiological conditions, and multilocus heterozygosity may confer physiological and evolutionary benefits of genome diversity. The present study along with the other two studies (Campbell et al. 2007; Jin et al 2008), points toward a consistent role of heterozygosity in modulating blood pressure in humans. Following Scheiner (op.cit), it may be suggested that plasticity associated with cardiovascular traits may be governing the responses of heterozygosity relation to components of blood pressure. Blood pressure trait and its correlates (left ventricular wall) provide an excellent example of temporally and spatially varying environments, which are in turn conducive for the maintenance of balancing selection (Gillespie 1994; Charlesworth, 2006). Indeed, Young et al (2005) postulated that differential susceptibility to hypertension in modern human populations be the result of their “differential exposure to selection pressure.” In the light of previously reported trait specific relationships between heterozygosity and environments (Lerner 1954, Hoffman & Parsons 1993; Yampolsky and Scheiner 1994), it may be suggested that balancing selection may be operating in the maintenance of heterozygosity in relation to variation among the components of blood pressure. Indeed, balancing selection could maintain genetic variation of complex traits under varying environmental conditions (Turelli and Barton 2004).
This study provided interesting insights on the influence of SNP-heterozygosity and quantitative traits in an outbred White North American population, but there are drawbacks. First, using the “algebraic approach” of Chakraborty (1982), DeWoody & DeWoody (2005) indicated that about 3000 loci may be necessary to achieve a modest correlation of about 0.4 in the HFC studies in humans. Aparicio et al (2007), however, questioned Chakraborty’s (op. cit) assumptions, as well as results obtained by Dewoody & DeWoody (op.cit) employing the algebraic approach. They showed that with more realistic assumptions and in “real populations,” only 200 – 1000 loci representing 100 individuals would provide the same magnitude of correlation as predicted by the algebraic approach. Our study consists of 313 and 393, Ns-SNPs and Sn-SNPs, respectively, and the sample size varied from 253–1017. Hence, the number of SNPs used in the study approach the lower limits of the sample size used by Aparicio et al (op. cit) in their simulation study. Hence, it may be still inadequate to provide a global view of the association between heterozygosity and fitness traits in humans. Nonetheless, the sample size used in this study far exceeds the number employed in most HFC studies reported in the literature. Second, in principle, all the traits examined in the study are correlated as they show genetic, developmental and environmental relationships. Hence, it is necessary to correct for these dependencies among traits. Although a few traits showed statistical significance with heterozygosity, application of Bonferroni correction , and Guo & Rao’s (2008), procedure for multiple testing failed to reveal significance between them. Note that both Bonferroni correction and other multiple testing procedures are known to be conservative, and hence may underestimate the significance. Therefore, we propose that these results found in this study must be treated as provisional. Third, SNPs employed in the study were enriched for greater heterozygosity and hence suffer from ascertainment biases. We suggest that a majority of the genome-wide association studies have shown an overwhelming evidence for additive genetic variance. However, dominance variance may be important toward maintaining traits that are modulated by environmental variation. Our results, although preliminary, point toward studying the importance of dominance variance in the maintenance and evolution of human traits. Detailed phenotypic, biochemical and dense SNP data sets are becoming available from many genome-wide association studies. These rich and diverse data sets would enable to study questions surrounding HFCs in humans in greater detail than has been previously attempted on other organisms, and could potentially illuminate the role of heterozygosity and dominance in human health.
Acknowledgements
DRG is grateful to Dr. Stephen Stearns for comments, and the NESCent working group on “Measuring evolutionary change in modern human populations, using cohort data” for discussions. The working group is supported by the National Evolutionary Synthesis Center (NESCent), NSF #EF-0423641.
References
- Allendorf FW, Bayles D, Bottom DL, Currens KP, et al. Prioritizing Pacific salmon stocks for conservation. Conserv. Biol. 1997;11:140–152. [Google Scholar]
- Aparicio JM, Ortego J, Cordero PJ. Can a simple algebraic analysis predict markers-genome heterozygosity correlations? J. Hered. 2007;98:93–96. doi: 10.1093/jhered/esl055. [DOI] [PubMed] [Google Scholar]
- Balloux F, Amos W, Coulson T. Does heterozygosity estimate inbreeding in real populations? Molecular Ecology. 2004;13:3021–3031. doi: 10.1111/j.1365-294X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- Bierne N, Tsitrone A, David P. An inbreeding model of associative overdominance during a bottle neck. Genetics. 2000;155:1981–1990. doi: 10.1093/genetics/155.4.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely J, Halapi E, Bjornsdottir US, Wheatley A, et al. Contribution of ADAM33 polymorphisms to the population risk of asthma. Thorax. 2005;60:274–276. doi: 10.1136/thx.2004.027227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnik M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Amer J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Britten HB. Meta-Analyses of the Association Between Multilocus Heterozygosity and Fitness. Evolution. 1996;50:2158–2164. doi: 10.1111/j.1558-5646.1996.tb03606.x. [DOI] [PubMed] [Google Scholar]
- Campbell H, Carothers AD, Rudan I, Hayward C, et al. Effects of genome-wide heterozygosity on a range of biomedically relevant human quantitative traits. Hum Molec Genet. 2007;16:233–241. doi: 10.1093/hmg/ddl473. [DOI] [PubMed] [Google Scholar]
- Cannon WA. Norton, New York, NY: 1932. The Wisdom of the Body. [Google Scholar]
- Charelsworth D. Balancing selection and its effects on sequences in nearby geneome regions. PloS Genetics. 2006;2:379–384. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression. Ann Rev Ecol Syst. 1987;18:237–268. [Google Scholar]
- Cowley AW. Long-term control of arterial blood pressure. Physiol Rev. 1992;72:231–300. doi: 10.1152/physrev.1992.72.1.231. [DOI] [PubMed] [Google Scholar]
- Cowley AW. Genomics and homeostasis. Am J Physiol Regulatory Integrative Comp Physiol. 2003;284:R611–R627. doi: 10.1152/ajpregu.00567.2002. [DOI] [PubMed] [Google Scholar]
- Cupples LA, Arruda H, Benjamin EJ, D’Agostono RB, DeMissie S, et al. The Framingham Heart Study 100K genome-wide association study resource: overview of 17 phenotype working group reports. BMC Medical Genetics. 2007;8(Suppl):1–19. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danubio ME, Pettener D. Marital srtucture of the community of Boston, Massachussets. 1880–1920. J. Biosoc Sci. 1997;29:257–269. doi: 10.1017/s0021932097002575. [DOI] [PubMed] [Google Scholar]
- Danziger RS. Hypertension in an anthropological and evolutionary paradigm. Hypertension. 2001;38:19–22. doi: 10.1161/01.hyp.38.1.19. [DOI] [PubMed] [Google Scholar]
- David P. Heterozygosity-fitness correlations: new perspectives on old problems. Heredity. 1998;80:531–537. doi: 10.1046/j.1365-2540.1998.00393.x. [DOI] [PubMed] [Google Scholar]
- DeWoody YD, DeWoody JE. On the estimation of genome wide heterozygosity using molecular markers. J. Hered. 2005;96:85–88. doi: 10.1093/jhered/esi017. [DOI] [PubMed] [Google Scholar]
- Donavan TM, Weldon CW. Spread Sheet Exercises in Ecology and Evolution. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Shaffer JP, Boldrick JC. Multiple Hypothesis Testing in Microarry Experiments. Statistical Science. 2003;18:71–103. [Google Scholar]
- Eanes WF. Analysis of selection on enzyme polymorphisms. Ann Rev Evol Syst. 1999;30:301–326. [Google Scholar]
- Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. The Causes of Molecular Evolution. Oxford: oxford University Press; 1994. [Google Scholar]
- Gillespie JH, Turelli M. Genotype-environment interaction and the maintenance of polygenic variation. Genetics. 1989;121:129–138. doi: 10.1093/genetics/121.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez W, Fontaine V, Pueyo ME, Laquay N, et al. Molecular Plasticity of Vascular Wall During NG-Nitro-L-Arginine Methyl Ester–Induced Hypertension. Hypertension. 2000;36:103–109. doi: 10.1161/01.hyp.36.1.103. [DOI] [PubMed] [Google Scholar]
- Govindaraju DR, Cupples LA, Kannel WB, O’Donnell, et al. Genetics of the Framingham Heart Study Population. Advances in Genetics. 2008;62:33–65. doi: 10.1016/S0065-2660(08)00602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Rao MB. On control of the false discovery rate under no assumption of dependency. Journal of Statistical Planning and Inference. 2008;138:3176–3188. [Google Scholar]
- Hendel MD, Collister JP. Sodium balance, arterial pressure and the role of the subfornical organ during chronic changes in dietary salt. Am J Physiol Heart Circ Physiol. 2005;289:426–431. doi: 10.1152/ajpheart.01051.2004. [DOI] [PubMed] [Google Scholar]
- Hansson B, Westerberg L. On the correlation between heterozygosity and fitness in natural populations. Molecular Ecology. 2002;11:2467–2474. doi: 10.1046/j.1365-294x.2002.01644.x. [DOI] [PubMed] [Google Scholar]
- Hansson B, Westerberg L. Heterozygosity-fitness within inbreeding classes: local or genome-wide effects? Conserv Genet. 2008;9:73–83. [Google Scholar]
- Hardenberg A, Bassano B, Festa-Bianchet M, Luikart G, et al. Age-dependent genetic effects on a secondary sexual trait in male Alpine ibex, Capra ibex. Mol Ecol. 2007;16:1969–1980. doi: 10.1111/j.1365-294X.2006.03221.x. [DOI] [PubMed] [Google Scholar]
- Hawkins AJS, Day AJ. Metabolic interactions underlying the physiological and evolutionary advantages. Amer Zool. 1999;39:401–411. [Google Scholar]
- Hendel MD, Collister JP. Sodium balance, arterial pressure, and the role of the subfornical organ during chronic changes in dietary salt. Am. J. Physiol. Heart Circ Physiol. 2005;289:426–431. doi: 10.1152/ajpheart.01051.2004. [DOI] [PubMed] [Google Scholar]
- Hill JA, Olsen EN. Cardiac plasticity. NEJM. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- Hoffman AA, Parsons PA. Evolutionary Genetics and Environmental Stress. Oxford: Oxford University Press; 1993. [Google Scholar]
- Houle D. Allozyme-associated heterosis in Drosophila melanogaster. Genetics. 1989;123:789–801. doi: 10.1093/genetics/123.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Foo JN, O’Roak BJ, Zhao H, Larson MG, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nature Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, Dawber TR, Kagan A, Revotski N, Stokes J., III Factors of risk in the development of coronary heart disease-six year follow-up experience. Ann Intern Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Lerchner MJ, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. Plos Biol. 2005;3:282–288. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner IM. Genetic Homeostasis. London: Oliver and Boyd; 1954. [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;4:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland: Sinauer Associates; 1998. [Google Scholar]
- McKinlay M. The normal menopause transition: an overview. Maturitas. 1996;23:137–145. doi: 10.1016/0378-5122(95)00985-x. [DOI] [PubMed] [Google Scholar]
- Meng S, Cason GW, Gannon AW, Racusen LC, Davis MR, Jr, et al. Oxidative Stress in Dahl Salt-Sensitive Hypertension. Hypertension. 2003;41:1346–1352. doi: 10.1161/01.HYP.0000070028.99408.E8. [DOI] [PubMed] [Google Scholar]
- Mitton JF. Selection in Natural Populations. Oxford: Oxford University Press; 1997. [Google Scholar]
- Mule G, Nardi E, Andronico G, Cottone S, et al. Pulsatile and steady 24-h blood pressure components as determinants of left ventricular mass in young and middle-aged essential hypertensives. J Hum Hypertens. 2003;17:231–238. doi: 10.1038/sj.jhh.1001542. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Parsons PA. Extreme environmental heterosis and genetic loads. Heredity. 1971;26:579–583. doi: 10.1038/hdy.1971.59. [DOI] [PubMed] [Google Scholar]
- Parsons PA. Energetic efficiency under stress underlies positive genetic correlations between longevity and other fitness traits in natural populations. Biogerentology. 2007;8:55–61. doi: 10.1007/s10522-006-9028-8. [DOI] [PubMed] [Google Scholar]
- Post WS, Larson MG, Myers RH, Galderisi M, Levy D. Heritability of lefet ventricular mass: The Framingham Heart Study. Hepertension. 1997;30:1025–1028. doi: 10.1161/01.hyp.30.5.1025. [DOI] [PubMed] [Google Scholar]
- Reed DH, Frankham R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution. 2001;55:1095–1103. doi: 10.1111/j.0014-3820.2001.tb00629.x. [DOI] [PubMed] [Google Scholar]
- van Rijn MJ, Schut AF, Aulchenko YS, Deinum J, et al. Heritability of blood pressure traits and the genetic contribution to blood pressure variance explained by four blood-pressure-related genes. J Hypertens. 2007;25:565–570. doi: 10.1097/HJH.0b013e32801449fb. [DOI] [PubMed] [Google Scholar]
- Rudan I, Biloglav Z, Vorko-Jovic A, et al. Effects of inbreeding, endogamy, genetic admixture and outbreeding on human health: a ”1000 Dalmatian” study. Croat. Med. J. 2006;47:601–610. [PMC free article] [PubMed] [Google Scholar]
- Sheiner SM. Genotype-environment interactions and evolution. In: Fox CW, Wolf JB, editors. Evolutionary Genetics, Concepts and Case Studies. Oxford: Oxford University Press; 2006. [Google Scholar]
- Stearns S. The Evolution of Life Histories. Oxford: Oxford University Press; 1992. [Google Scholar]
- Thelen CG, Allendorf FW. Heterozygosity-fitness correlations in rainbow trout:Effects of allozyme loci or associative overdominance? Evolution. 2001;55:1180–1187. doi: 10.1111/j.0014-3820.2001.tb00637.x. [DOI] [PubMed] [Google Scholar]
- Timarco B, Lembo G, Ricciardelli B, De Luca N, et al. Salt-induced plasticity in cardiopulmonary baroreceptor reflexes in salt-resistant hypertensive patients. Hypertension. 1991;18:483–493. doi: 10.1161/01.hyp.18.4.483. [DOI] [PubMed] [Google Scholar]
- Tsitrone A, Roussset F, Zull DP. Heterosis, marker mutational process and population inbreeding history. Genetics. 2001;159:1845–1859. doi: 10.1093/genetics/159.4.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Barton NH. Polygenic Variation Maintained by Balancing Selection: Pleiotropy,Sex-Dependent Allelic Effects and G X E Interactions. Genetics. 2004;166:1053–1079. doi: 10.1534/genetics.166.2.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of short tandem repeats. Hum. Mol. Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic Data Analysis II. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- Weiss LA, Pan L, Abney M, Ober C. The sex-specific genetic architecture of quantitative traits in humans. Nature Genet. 2005;38:218–222. doi: 10.1038/ng1726. [DOI] [PubMed] [Google Scholar]
- Wilson PWF, D’Agostino RB, Levy D, Belanger AM, Silberscahtz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- Yampolsky LY, Schiener SM. Developmental noise, phenotypic plasticity and allozyme heterozygosity in Daphnia. Evolution. 1994;48:1715–1722. doi: 10.1111/j.1558-5646.1994.tb02207.x. [DOI] [PubMed] [Google Scholar]
- Young JH, Chang Y-PC, Kim JD-O, Chertien J-P, Klag MJ, et al. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1:730–738. doi: 10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]