Abstract
Background
Cancer risk prediction tools provide valuable information to clinicians but remain computationally challenging. Many clinics find that Ca Gene or Hughes Risk Apps fit their needs for easy- and ready-to-use software to obtain cancer risks; however, these resources may not fit all clinics’ needs. The Hughes Risk Apps Group and Bayes Mendel Lab therefore developed a web service, called “Risk Service", which may be integrated into any client software to quickly obtain standardized and up-to-date risk predictions for Bayes Mendel tools (BRCAPRO, MMRpro, PancPRO, and MelaPRO), the Tyrer-Cuzick IBIS Breast Cancer Risk Evaluation Tool, and the Colorectal Cancer Risk Assessment Tool.
Findings
Software clients that can convert their local structured data into the HL7 XML-formatted family and clinical patient history (Pedigree model) may integrate with the Risk Service. The Risk Service uses Apache Tomcat and Apache Axis2 technologies to provide an all Java web service. The software client sends HL7 XML information containing anonymized family and clinical history to a Dana-Farber Cancer Institute (DFCI) server where it is parsed, interpreted, and processed by multiple risk tools. The Risk Service then formats the results into an HL7 style message and returns the risk predictions to the originating software client. Upon consent, users may allow DFCI to maintain the data for future research. The Risk Service implementation is exemplified through Hughes Risk Apps.
Conclusions
The Risk Service broadens the availability of valuable, up-to-date cancer risk tools and allows clinics and researchers to integrate risk prediction tools into their own software interface designed for their needs. Each software package can collect risk data using its own interface, and display the results using its own interface, while using a central, up-to-date risk calculator. This allows users to choose from multiple interfaces while always getting the latest risk calculations. Consenting users contribute their data for future research, thus building a rich multi-center resource.
Keywords: Risk Prediction, BayesMendel, BRCAPRO, IBIS Breast Cancer Risk Evaluation Tool, Colorectal Cancer Risk Assessment Tool, Web Service, Risk Service
Introduction
The widely used risk prediction tool BRCAPRO uses family and personal disease history to calculate the risks of carrying mutations in the breast and ovarian cancer-associated genes BRCA1 and BRCA2 and the risk of developing breast and ovarian cancer over time [1]. The authors continually improve the tool as new literature is published regarding the prevalence and penetrance of these genes, and their modifiers [2–7]. BRCAPRO applies the science of Mendelian genetics and Bayesian approaches to calculating risks. The same approach has been also applied to develop tools to evaluate the chance of carrying inheritable mutations associated with pancreatic cancer, melanoma, colorectal cancer, and other cancers associated with mismatch repair genes [8]. The complexity of these tools poses a challenge for practical clinical use, as users are required to enter a potentially large amount of information to evaluate the risk for a given family. This is addressed by implementation tools, which provide user interfaces to simplify data entry, to provide visualizations of results, and to support appropriate interpretation. Many clinics find that the freely available Ca Gene (http://www4.utsouthwestern.edu/breasthealth/cagene/) or Hughes Risk Apps (HRA; http://www.hughesriskapps.com/) applications fit their needs for easy and ready-to-use software to obtain cancer risks; both tools provide a user friendly interface for oncology clinics to collect family and clinical history and both immediately place risk assessments into the clinician’s and/or genetic counselor’s hands [9–11]. Both systems have found a need to continuously upgrade their version of BRCAPRO and other risk assessment tools. HRA has addressed this through a web service known as “Risk Service.”
The Risk Service uses the concept of a web service, which as opposed to a web site, is designed to interact with other software packages, not with humans. The algorithm, guideline or knowledge base of web services (in this case risk prediction algorithms) is installed on a server accessible through the cloud. The Risk Service alleviates from software developers the onus of correctly coding risk prediction tools and allows them instead to develop a convenient software interface to collect, and meaningfully present, information. This report describes the data flow, risk algorithms currently offered, availability, and implementation of the Risk Service (as exemplified through HRA).
Methods
Risk Service Architecture and Data Flow
The Risk Service resides on a Dana-Farber Cancer Institute (DFCI) server, receives Health Level 7 (HL7) Pedigree Model [12] XML-formatted family and clinical history inputs, runs all selected risk prediction tools, and returns the risk predictions (Figure 1). There is no human readable interface. For the Risk Service to be practically functional, a software package must create a user interface to collect family and clinical history and to display the results of the calculations; the software interface is not restrictive to operating system, it may be developed for android, iPhone, iPad, and other devices. That developed software communicates, via the HL7 message, with the Risk Service where the calculations are actually performed.
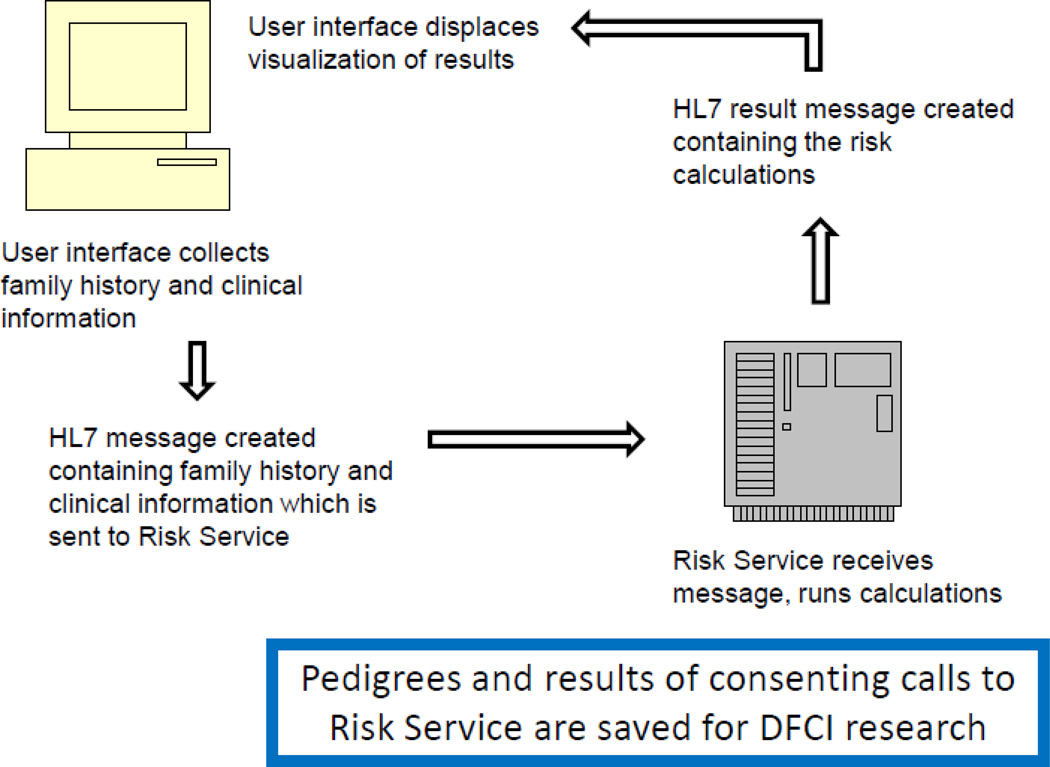
Fig 1.
Risk Service Data Flow. A software client collects data and packages it as an HL7 message which the Risk Service can read and transmit to the Dana-Farber Cancer Institute server. The server runs the prediction tools and returns the results to the software client as an HL7 message. If consented, data sent to the server are kept by DFCI for future research. (Clipart used with permission from Microsoft).
The Risk Service software uses Tomcat, an all Java web server [13]. Within that framework, Apache Axis2 is used as a toolkit for receiving and sending HL7 XML messages to and from the server. HL7 XML input data is interpreted and parsed with XSLT, a specialized XML translation language, and transformed for input into risk tools by an open-source XSLT processor (Saxon, see Figure 2). The risk tools are run allowing for simultaneous calls from different users while preserving each users' data in a safe and separate namespace; they are run in parallel across multiple threads and/or CPUs, even for the same user, to achieve a rapid response time.
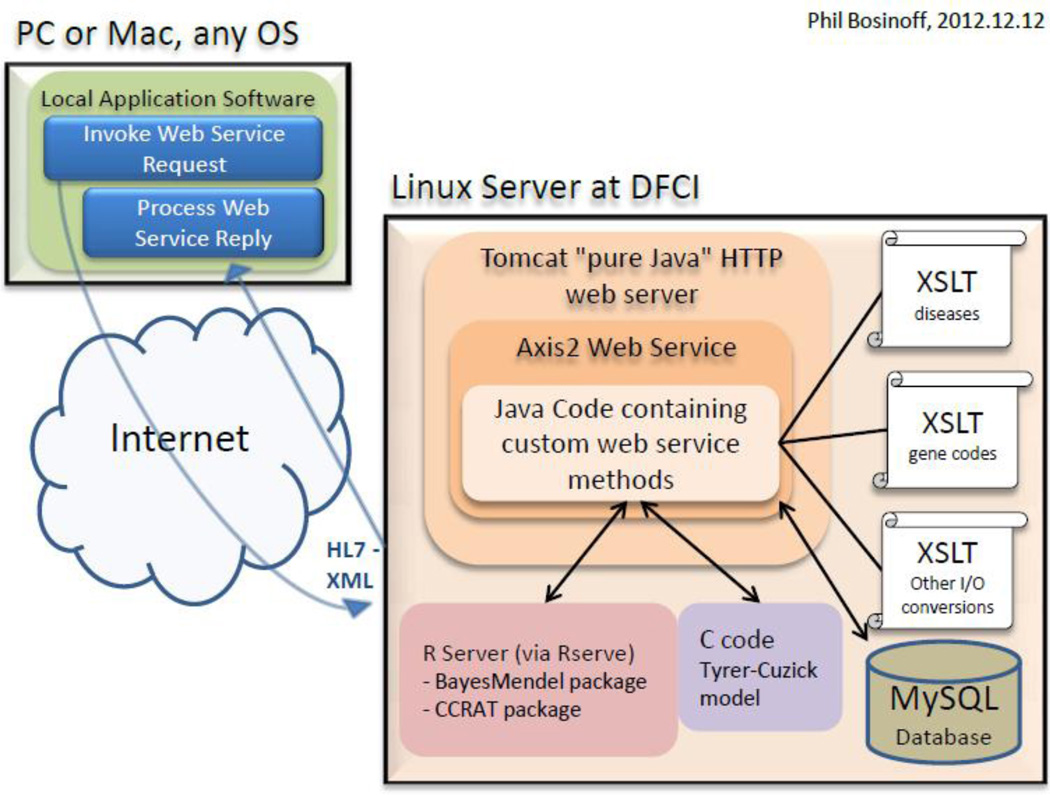
Fig 2.
Risk Service Data Architecture. The Risk Service uses Tomcat and Apache Axis2 to send and receive HL7 XML messages to and from the Dana-Farber Cancer Institute Server. HL7 XML input is interpreted and parsed by XSLT and then transformed to be used by the risk prediction tools.
The Risk Service requires users to obtain a license and valid user agreement (http://bcb.dfci.harvard.edu/bayesmendel/riskservice.php) [14]. All academic institutions, research institutions, and individual health care providers may use the Risk Service free of charge; however, inclusion of the Risk Service into commercial products requires a commercial license. In calling the Risk Service, the user either consents or declines consent for the family history, clinical history, and results to be retained in a local MySQL database, in de-identified form, and used for future research. At minimal risk, the de-identified family history data, collected from multiple institutions, may provide an enormous benefit to the scientific community by helping algorithm developers improve current risk prediction tools. Each user is identified by a user license for the dual purpose of accounting for effects in research and for disseminating the Risk Service.
Risk Prediction Tools
The Risk Service is tailored to clinicians and genetic counselors concerned with hereditary cancer risk. Bayes Mendel tools – including BRCAPRO, MMRpro, PancPRO, and MelaPRO – predict the risk of carrying highly penetrant genetic mutations associated with disease. They use Mendelian laws explicitly and incorporate detailed family history information (race, age, vital status, disease diagnoses, and age of diagnoses, and genetic testing results for each relative), cancer specific data (ER, PR, Her2, CK14, CK5/6) and some behavioral risk factors. BRCAPRO, for example, evaluates the risk of carrying a deleterious BRCA1 and/or BRCA2 mutation and the risk of developing breast or ovarian cancer over time [15–17]. Validations of these tools have been described [18–21].
The Risk Service also provides estimates for the IBIS Breast Cancer Risk Evaluation Tool [22, 23] and the Colorectal Cancer Risk Assessment Tool (CCRAT) [24]. The IBIS Breast Cancer Risk Evaluation Tool was developed to predict breast cancer risk and risk of carrying a BRCA1 or BRCA2 mutation using family history information, in the presence of high risk lesions in the proband (Atypia, LCIS) and additional behavioral and environmental factors. It accounts for well-established risks of breast cancer including age at menarche, parity, age at first childbirth, age at menopause, atypical hyperplasia, lobular carcinoma in situ, height, and BMI. CCRAT focuses on predicting the risk of colorectal cancer; risks are modified by family history and epi-environmental factors including health and wellness factors, hormone therapy in women, screening for colorectal cancer, and past history of polyps.
Implementation
As an example, we will discuss the implementation of the Risk Service through HRA. HRA provides a user interface for the input of family history, genetic and clinical data (Figure 3), stores the patient’s data into an SQL database, and then packages the data into a message using the HL7-XML coding standard. That message is transmitted to the Risk Service.
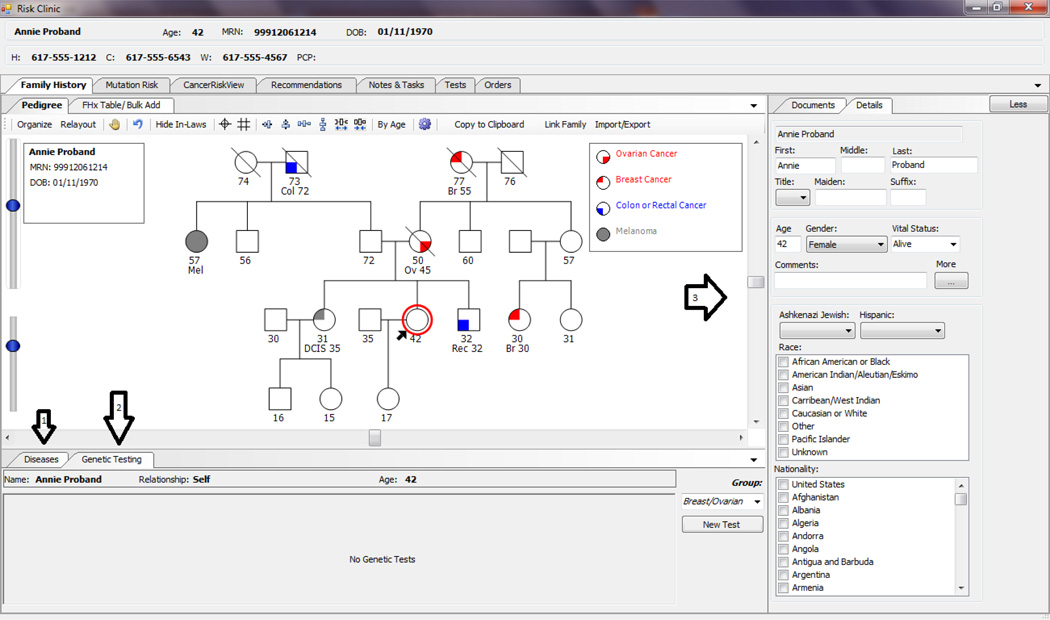
Fig 3.
Example User Interface (HRA) to Enter Family History Data. Arrow 1 shows tab to enter disease data for the selected relative, arrow 2 shows tab to enter genetic testing data for the selected relative, and arrow 3 shows area to enter vital status, age and other data for the selected relative
Receipt of the message triggers a basic call to the Risk Service – get Risk HL7() – which calls each risk tool for each family member having sufficient data for the given risk tool. For example, the counselee may be the only family member with sufficient information to call CCRAT, whereas each family member in the pedigree may have sufficient data to run BRCAPRO. A more succinct set of risk predictions may be requested by calling get Risk HL7 Selected() (Figure 4). The Risk Service User Guide (Online Resource) provides the codes to be used with the HL7 Family History input data. The Risk Service recognizes multiple coding schemas, allowing software packages to choose which best fit their needs, though the most commonly used is the Systematized Nomenclature of Medicine (SNOMED). For a rudimentary example outside of HRA, a minimalistic client calling the Risk Service is online and can be used to test HL7 input family history (http://bayesmendel.dfci.harvard.edu/risk) [25].
Fig 4.
Inputs For Requesting Predictions From Risk Service. get Risk HL7() is the most comprehensive call to the Risk Service. It obtains risk predictions for all family members with sufficient predictors for each given model. get Risk HL7 Selected() allows for obtaining selected risk predictions and output.
Most model inputs into the risk tools are required, though some tool’s model inputs allow for a value of “Unknown”, “missing”, or “NA”. That is, a value of NA is an allowed input value, built into the design of the model. However, if a model input is required, and it is not available, for certain reasonable situations the Risk Service may attempt to use (impute) an appropriate value. For example, if a relative’s age of onset of breast cancer is missing, the current age, if alive, or age at death is used. If the relative’s current age or age at death is unknown, the age of onset of breast cancer is assumed to be age 50. All assumptions made by the risk service about imputation of information appear in the output message section.
The output of the risk models are packaged into an HL7 XML results message, which is returned to the originator of the message, in this case, the HRA instance that originated the message. HRA will store these results in its SQL database and display the results to the user in an intuitive visualization (Figure 5). User developed software will need to develop its own display of the results.
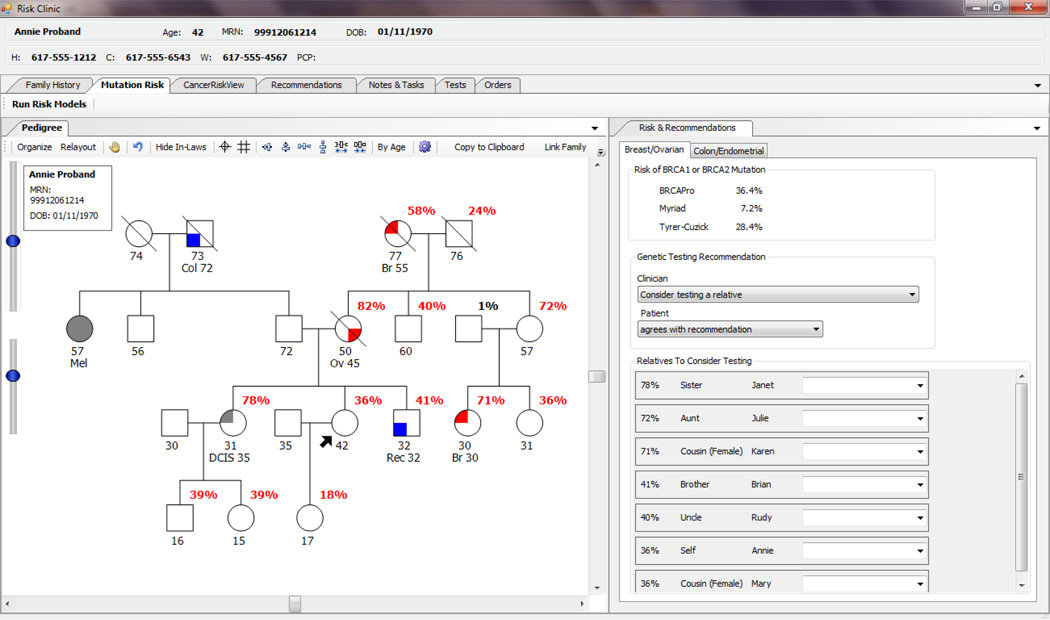
Fig 5.
Example User-Interface Displaying Risk Service Results. Using the data from BRCAPRO via the Risk Service, HRA displays the risk of mutation for each family member.
To illustrate the power of this approach in March 2013, a new version of BRCAPRO that now allows mastectomy as data inputs and families to have mixed ethnicities was installed on the Risk Service. Immediately the 38 installations of HRA began using the latest model. This occurred without any need for local upgrades of the software and was transparent to the user.
Discussion
The Hughes Risk Apps Group and Bayes Mendel Lab jointly developed the Risk Service with the intent to make freely available risk prediction tools for research and clinical use [10, 26]. Running these tools via the Risk Service has the advantage of always using the most up-to-date models without the need for local upgrades and re-installations. HRA exemplifies a client that integrates the Risk Service to provide immediate risk predictions to clinicians. Its user interface allows the collection of data by dynamically asking appropriate sequential questions to collect relevant clinical, genetic and family history information and produces intuitive visualizations that make it easy for the user to understand the information. Ca Gene is another option to obtain risk predictions, but it does not currently integrate with the Risk Service.
Most clinics find Ca Gene and HRA fit their needs for a ready and easy-to-use interface to obtain immediate risk prediction estimates; however, these resources will not fit all clinics’ needs. The Risk Service is available, with a signed user agreement and license, to be integrated with any client software developed to send and receive HL7 XML-formatted family and clinical history. The HL7 format required by the Risk Service is consistent with the health care industry standard and provides the broadest availability for the tools. While clinicians and patients benefit greatly from readily available risk predictions, a large pool of multi-institutional family history data will be continually gathered. This again benefits clinicians and patients as risk prediction tools are further developed and improved.
It is vital that we move toward the web service approach in clinical medicine. Medicine is becoming more and more dependent on knowledge bases and Clinical Decision Support (CDS). The current approach of having 600 Electronic Health Record vendors each develop their own rendition of the knowledge bases and a CDS systems for each aspect of medical care is an approach doomed to failure. We need Specialty Societies and Government to start developing their guidelines, algorithms, and knowledge bases as machine readable web services accessible to any CDS system. This approach is vital if we are to constrain the costs of medical care while simultaneously improving quality care for all. The Risk Service was developed as a prototype to show the value and utility of the web service approach.
Supplementary Material
Acknowledgements
Work partly supported by the Susan G. Komen Foundation and by NCI award 2 P30 CA006516-47 (DF/HCC comprehensive cancer center core grant).
Abbreviations
- HRA
Hughes Risk Apps
- HL7
Health Level 7
- DFCI
Dana-Farber Cancer Institute
- CCRAT
Colorectal Cancer Risk Assessment Tool
- SNOMED
Systematized Nomenclature Of Medicine
- CDS
Clinical Decision Support
Footnotes
Competing Interests
Jonathan Chipman, Brian Drohan, and Amanda Blackford declare they have no conflict of interest. Giovanni Parmigiani, Kevin Hughes, and Phil Bosinoff may receive royalties from the commercial licensing of the Risk Service.
Contributor Information
Jonathan Chipman, Dana-Farber Cancer Institute: 450 Brookline Avenue, Boston MA, 02115, chipmanj@jimmy.harvard.edu, Phone: (617) 582-9619, Fax: (617) 632-2444.
Brian Drohan, Massachusetts General Hospital: 55 Fruit Street, Yawkey 7, Boston, Massachusetts 02114.
Amanda Blackford, Sidney Kimmel Comprehensive Cancer Center: 550 N Broadway, Suite 1111, Baltimore, Maryland 21205.
Giovanni Parmigiani, Dana-Farber Cancer Institute: 450 Brookline Avenue, Boston MA, 02115.
Kevin Hughes, Massachusetts General Hospital: 55 Fruit Street, Yawkey 7, Boston, Massachusetts 02114.
Phil Bosinoff, Massachusetts General Hospital: 55 Fruit Street, Yawkey 7, Boston, Massachusetts 02114.
References
- 1.Parmigiani G, Berry DA, Aguilar O. Determining Carrier Probabilities for Breast Cancer Susceptibility Genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–158. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tai YC, Domchek S, Parmigiani G, Chen S. Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2007;99(23):1811–1814. doi: 10.1093/jnci/djm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tai YC, Chen S, Parmigiani G, Klein AP. Incorporating tumor immunohistochemical markers in BRCA1 and BRCA2 carrier prediction. Breast Cancer Res. 2008;10(2):401. doi: 10.1186/bcr1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katki HA, Blackford A, Chen S, Parmigiani G. Multiple diseases in carrier probability estimation: accounting for surviving all cancers other than breast and ovary in BRCAPRO. Stat Med. 2008;27(22):4532–4548. doi: 10.1002/sim.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katki HA. Incorporating medical interventions into carrier probability estimation for genetic counseling. BMC Med Genet. 2007;8:13. doi: 10.1186/1471-2350-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Blackford A, Parmigiani G. Tailoring BRCAPRO to Asian-Americans. J Clin Oncol. 2009;27(4):642–643. doi: 10.1200/JCO.2008.20.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas S, Tankhiwale N, Blackford A, Barrera AMG, Ready K, Lu K, Amos CI, Parmigiani G, Arun B. Assessing the added value of breast tumor markers in genetic risk prediction model BRCAPRO. Breast Cancer Res Treat. 2012;133(1):347–355. doi: 10.1007/s10549-012-1958-z. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. Bayes-Mendel: an R Environment for Mendelian Risk Prediction. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1063. Article 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CaGene. [ http://www4.utsouthwestern.edu/breasthealth/cagene/]
- 10.HughesRiskApps. [ http://www.hughesriskapps.com/]
- 11.Ozanne EM, Loberg A, Hughes S, Lawrence C, Drohan B, Semine A, Jellinek M, Cronin C, Milham F, Dowd D, Block C, Lockhart D, Sharko J, Grinstein G, Hughes KS. Identification and management of women at high risk for hereditary breast/ovarian cancer syndrome. Breast J. 2009;15(2):155–162. doi: 10.1111/j.1524-4741.2009.00690.x. [DOI] [PubMed] [Google Scholar]
- 12.Health Level Seven International. [ http://www.hl7.org/]
- 13.Tomcat. [ http://www.apache.org]
- 14.To Obtain Risk Service. [ http://bcb.dfci.harvard.edu/bayesmendel/riskservice.php]
- 15.King MC, Marks JH, Mandell JB New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 16.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 Penetrance. J Clin Oncol. 2007;25(11):1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parmigiani G, Chen S, Iversen ES, Jr, Friebel TM, Finkelstein DM, Anton-Culver H, Ziogas A, Weber BL, Eisen A, Malone KE, Daling JR, Hsu L, Ostrander EA, Peterson LE, Schildkraut JM, Isaacs C, Corio C, Leondaridis L, Tomlinson G, Amos CI, Strong LC, Berry DA, Weitzel JN, Sand S, Dutson D, Kerber R, Peshkin BN, Euhus DM. Validity of models for predicting BRCA1 and BRCA2 mutations. Ann Intern Med. 2007;14(7):441–450. doi: 10.7326/0003-4819-147-7-200710020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, Watson P, Gruber S, Euhus D, Kinzler K, Jass J, Gallinger S, Lindor N, Casey G, Ellis N, Giardiello F, Offit K, Parmigiani G The Colon Cancer Family Registry. Prediction of Germline Mutations and Cancer Risk in the Lynch Syndrome. JAMA. 2007;296(12):1479–1487. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Chen S, Brune K, Hruban R, Parmigiani G, Klein A. PancPRO: Risk Assessment for Individuals With a Family History of Pancreatic Cancer. J Clin Oncol. 2007;25(11):1417–1422. doi: 10.1200/JCO.2006.09.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Niendorf K, Patel D, Blackford A, Marroni F, Sober AJ, Parmigiani G, Tsao H. Estimating CDKN2A carrier probability and personalizing cancer risk assessments in hereditary melanoma using MelaPRO. Cancer Res. 2010;70(2):552–559. doi: 10.1158/0008-5472.CAN-09-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Stat Med. 2004;23(7):1111–1130. doi: 10.1002/sim.1668. [DOI] [PubMed] [Google Scholar]
- 23.Boughey J, Hartmann L, Anderson S, Degnim A, Vierkant R, Reynolds C, Frost M, Pankratz V. Evaluation of the Tyrer-Cuzick (International Breast Cancer Intervention Study) model for breast cancer risk prediction in women with atypical hyperplasia. J Clin Oncol. 2010;28(22):3591–3596. doi: 10.1200/JCO.2010.28.0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freedman AN, Slattery ML, Ballard-Barbash R, Willis G, Cann B, Pee D, Gail MH, Pfeiffer RM. A colorectal cancer risk prediction tool for white men and women without known susceptibility. J Clin Oncol. 2009;27(5):686–93. doi: 10.1200/JCO.2008.17.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risk Service Demo. [ http://bayesmendel.dfci.harvard.edu/risk]
- 26.BayesMendel Lab. [ http://bcb.dfci.harvard.edu/bayesmendel/index.php]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.