Abstract
Intratumoral transport and binding are important mechanisms that determine the efficacy of cancer drugs. Current drug screening methods rely heavily on monolayers of cancer cells, which overlook the contribution of tissue-level transport and binding. To quantify these factors, we developed a method that couples an in vitro, drug-delivery device containing a three-dimensional cell mass and a mathematical model of drug diffusion, binding to DNA, release from carriers, and clearance. Spheroids derived from LS174T human colon carcinoma cells were inserted into rectangular chambers to form rectangular cell masses (tissue) and subjected to continuous medium perfusion. To simulate drug delivery and clearance, the tissues were treated with doxorubicin followed by drug-free medium. To evaluate the effect of liposome encapsulation, tissues were treated with liposome-encapsulated doxorubicin (Doxil). Spatiotemporal dynamics of drug distribution and apoptosis was measured by fluorescence microscopy. The diffusivity and DNA binding constant of doxorubicin were determined by fitting experimental data to the mathematical model. Results show that an ideal combination of diffusivity, binding constant, clearance rate, and cytotoxicity contribute to the high therapeutic efficacy of doxorubicin. There was no detectable release of doxorubicin from Doxil in the tissues. The rate of doxorubicin release, evaluated by fitting experimental data to the mathematical model, was below therapeutically effective levels. These results show that despite enhanced systemic circulation obtained by liposome encapsulation, the therapeutic effect of Doxil is limited by slow intratumoral drug release. The experimental and computational methods developed here to calculate drug efficacy provide mechanisms to explain poor performance of drug candidates, and enable design of more successful cancer drugs.
Keywords: Mathematical modeling, microfluidics, in vitro models, stealth liposomes, drug binding, drug delivery
Introduction
Many chemotherapeutic drugs suffer from limited penetration and accumulation in solid tumors1-5. Intratumoral transport and binding are important mechanisms that determine a cancer drug's efficacy. Cancer drug development typically starts with screening a large library of compounds for maximum cytotoxicity towards monolayers of cancer cells6, 7. Selected compounds are then tested in mice xenograft models for their ability to shrink tumors8. Only a small percentage of compounds tested in xenograft models successfully regress tumors. There is often no correlation between cytotoxicity to cells in monolayers and ability to shrink solid tumors8. For this reason, cytotoxicity to monolayers alone may not be the best indicator of a cancer drug's therapeutic effect. Lack of experimental systems to effectively determine drug efficacy results in a long and expensive cancer drug development process9. To better predict the therapeutic efficacy of a cancer drug, a method for measuring drug delivery, clearance, intratumoral diffusion and binding is needed.
Several transport processes and reactions affect the efficacy of intravenously administered chemotherapeutic drugs (Fig. 1). Rapid plasma clearance and limited diffusion in tumors, for example, significantly limit the efficacy of cancer drugs10. Immediately after delivery, the plasma concentration of a drug is maximum and decreases over time as a result of absorption and metabolism11. During this period, the drug enters tumors and spreads by diffusion (Fig. 1A). Presence of drug within tissue causes cancer cell death. Some drugs may reversibly or irreversibly bind12, 13 to sites on tumor cells (Fig. 1B). The efficacy of chemotherapeutics like doxorubicin has been attributed to strong ability to intercalate DNA14. After plasma drug levels fall below tumor drug levels, drug is cleared from the tumor (Fig. 1C). The overall therapeutic effect is determined by this interaction between diffusion, binding, cytotoxicity, and systemic clearance rate. Current monolayer-based methods for testing drugs do not measure these interactions. A method to predict in vivo therapeutic efficacy must: a) simulate drug delivery and clearance, and b) mimic tissue-level transport and binding.
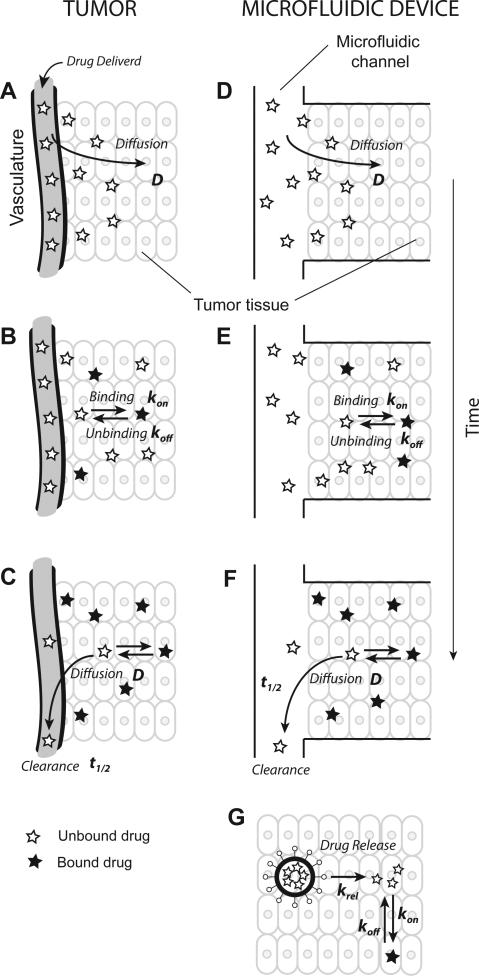
Figure 1. Mechanisms determining therapeutic efficacy.
A. Intravenously injected chemotherapeutic drugs enter tumors by diffusion. B. In tumors, drugs may bind to specific sites. C. Drugs are cleared from plasma causing clearance from tumor tissue. D-F. Tumor tissue positioned alongside a microfluidic channel mimics all of these mechanisms. G. For encapsulated drugs, the drug carrier must first release the drug into tissue before subsequent binding.
Rectangular three-dimensional (3D) cancer cell masses placed next to continuously perfused channels provide a method to capture these interactions (Fig. 1D-F). Several different devices for forming 3D cancer cell masses (hereafter referred to as tissues) in continuously perfused microfluidic devices have been described15-17. In our laboratory, we previously designed a device to grow cancer tissues in a rectangular geometry that forms one-dimensional chemical gradients and can be continuously monitored in time and space under a microscope18-20. Because of its geometry, transport can be described with a simple one-dimensional mathematical model, which enables quantification of the diffusion and binding rates. Coupling to a mathematical model enhances the utility of the device beyond measuring cell death or apoptosis, and enables quantification of the mechanisms of delivery.
An ideal method to test a device to measure transport properties, is comparison of a drug to its liposome-encapsulated form. These two entities would have different tissue transport properties but similar interactions with cells. As a test system, comparison of doxorubicin to encapsulated doxorubicin (Doxil) has the added advantage that doxorubicin is naturally fluorescent. Doxorubicin is a chemotherapeutic drug that is widely used in the treatment of cancer, including cancers of the breast, ovary, and lung. It is an anthracycline compound with wide-spectrum anti-tumor abilities21. In cultured cancer cells, doxorubicin is rapidly taken up and localized in the nucleus22-24. Doxorubicin binds to DNA, disrupts the mitotic process, and causes inhibition of cancer cell growth25, 26. On intravenous administration, the plasma concentration of doxorubicin drops rapidly with a half-life of approximately 5 minutes, as a result of distribution in tissues. After initial rapid clearance of 60 to 80% of the administered drug, the remaining drug undergoes a slow terminal clearance with a half-life of 20-48 hours because of slow elimination from tissues27-30.
Liposome encapsulation of drugs is a method used to overcome sub-therapeutic accumulation levels in tumors and toxicity to other organs. Poly-(ethylene glycol) (PEG) coated liposomes (stealth liposomes) have a reduced uptake by the mononuclear phagocyte system, which results in long plasma circulation times31-33. Doxil (pegylated liposome-encapsulated doxorubicin) is approved for the treatment of AIDS-related Kaposi's sarcoma, ovarian cancer, and multiple myeloma, and has been shown to reduce cardiac toxicity31-33. It has a plasma clearance half-life of 50 hours34. Several other cancer drugs encapsulated in liposomes are currently in advanced stage clinical trials35-37. To be effective, a liposome-encapsulated drug must extravasate into the tumor and release its payload (Fig. 1G). The release of drug from pegylated liposomes is not well-understood31, 38 and there is evidence that the rate of release may be very low39, 40. For example, when a chemotherapeutic agent, cisplatin, and its pegylated liposome-encapsulated variants, SPI-077 and SPI-077 B103, were administered to mice bearing melanoma tumors, the net accumulation of platinum (Pt) in tumors was 2.2 – 5 fold higher for the liposome-encapsulated drugs compared to free cisplatin. However, there was no detectable free active Pt in the tumors injected with the liposome drugs. All tumors injected with cisplatin had detectable free active Pt39. These data suggest that the rate of release of free Pt from pegylated liposomes was low. Further, SPI-077 was found to have no therapeutic efficacy in Phase I/II clinical trials, despite a long circulation time41. Attempts have been made to increase the intratumor rate of release from pegylated liposomes by light activation42, 43.
Pharmacokinetic/pharmacodynamic (PK/PD) models are used in drug development to measure and predict the time course of a drug's therapeutic effect44, 45. Pharmacokinetics (PK) measures the concentration of drugs in the body as a function of time. Pharmacodynamics (PD) measures the rate and extent of the antitumor effect induced by the drug as a function of drug concentration. A PK/PD model typically consists of two well-mixed compartments – a plasma compartment describing PK, and a tissue compartment describing PD. Because tissues are not well mixed compartments and the mechanism of therapeutic effect in tissues may be complex, there is a time lag between changes in plasma concentration and its effects on the tissue. To account for this, additional hypothetical compartments are introduced between the plasma and tissue compartments46. Different strategies are utilized for linking the effects of the compartments46, 47. The tissue compartments in all these models, however, are assumed to be homogeneous, disregarding the effects of drug concentration gradients, which are known to contribute to limited therapeutic effect in tumors48, 49. In order to more accurately predict tumor tissue drug concentrations and therapeutic effects, PK/PD models must integrate transport processes that govern drug distribution in tumors.
To quantify drug delivery to tumor tissue, we developed an in vitro device that simultaneously measures intratumoral drug diffusion, drug binding, release from carriers, and tissue retention. Tumor tissues in devices were treated with doxorubicin and Doxil and the dynamics of intratumoral accumulation and induced apoptosis was measured using time-lapse fluorescence microscopy. To determine the contribution of individual mechanisms towards the drug distribution, we developed a mathematical model incorporating five mechanisms: i) plasma clearance of drugs (PK), ii) drug diffusion iii) release from drug carriers, iv) drug binding to DNA, and v) cancer cell death (PD). Results from the model were combined with experimental data to determine the contribution of each mechanism towards the efficacy of doxorubicin and Doxil. Perturbing parameters in the mathematical model showed that interactions between the mechanisms are complex and their effects on drug efficacy are non-obvious. Results from these experiments provide reasons for why many drugs perform poorly in mice experiments and why it is difficult to predict drug efficacy. The methods developed in this paper will enable measurement of the mechanisms that contribute to drug efficacy, and more accurate prediction of in vivo drug response.
Results
Device for mimicking drug delivery and clearance
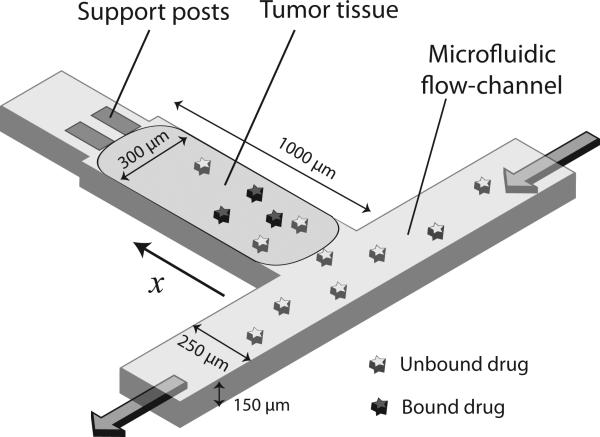
A microfluidic device was designed to mimic the delivery and clearance of drugs through blood vessels of tumor tissues18-20. The device consists of a rectangular tissue culture chamber, 1000 μm × 300 μm × 150 μm, and a microfluidic channel, 250 μm wide and 150 μm tall (Fig. 2). Spheroids prepared from human colon carcinoma cells were introduced into the culture chambers, forming rectangular tissues, which were subjected to continuous medium perfusion through the channels (Fig. 2). The device mimicked regions of tumors adjacent to blood vessels. Drug delivery and clearance was mimicked by adjusting the concentration of drugs in the channels. Construction of devices with a transparent material enabled visualization of drug diffusion and apoptosis induction in tissues.
Figure 2. The drug delivery device.
The drug delivery device is constructed out of PDMS and consists of rectangular tumor tissue next to continuous perfusion through a microfluidic channel along one face. The tumor tissue is held in place by posts at the rear end. Drugs are delivered through the microfluidic flow channel and their concentration is adjusted to mimic clearance. Drugs diffuse into tissue and may undergo binding.
Doxorubicin tissue uptake, clearance, and apoptosis induction
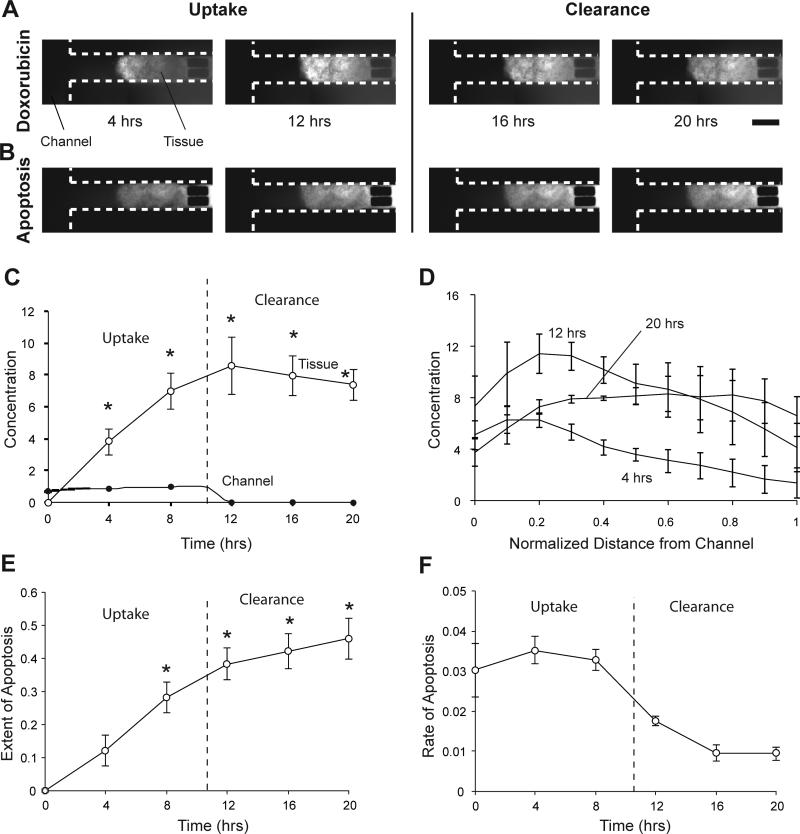
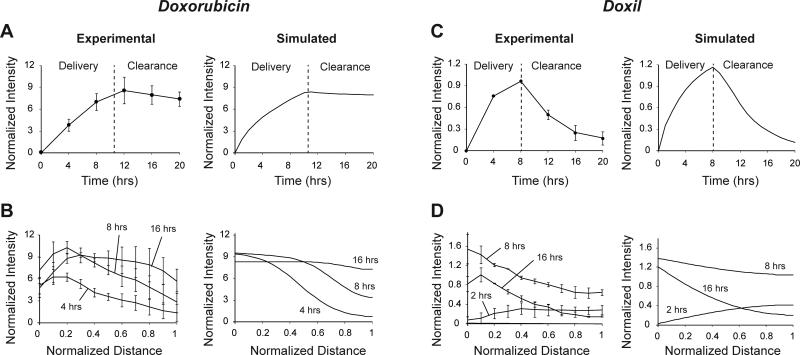
To measure the uptake and clearance of doxorubicin and to quantify apoptosis induced, tumor tissues in devices were treated with cell culture medium containing 5 μM doxorubicin and 0.25μl/ml green fluorescent active caspase-3 stain for 10.5 hours followed by drug-free medium. Doxorubicin delivered through the channels was taken up rapidly by tissue (Fig. 3A). Accumulation of drug in tissue increased apoptosis (Fig. 3B). The amount of drug in tissue increased continuously till 12 hours (Fig. 3C). The drug concentration in tissue was significantly higher (P<0.05, n=3) than in the channels (Fig. 3C). After drug was cleared from channels, the amount of drug in tissue did not decrease rapidly. At 20 hours, the average amount of drug in tissues was 86% of the maximum amount (at 12 hours); only 14% of the drug cleared from tissue in 8 hours (Fig. 3C). Apoptosis increased continuously after administration of drug and continued to increase after drug clearance (Fig. 3E). The rate of apoptosis induction decreased as the concentration of doxorubicin plateaued (Fig. 3F). The extent of apoptosis at 8, 12, 16, and 20 hours was significantly higher (P<0.05; n=3) compared to 0 hrs (Fig. 3E). For tissues not treated with drugs, we have previously shown that the mean extent of apoptosis decreases over time, which is a result of increase in the number of live and proliferating cells19. From 12 to 20 hours, the concentration of doxorubicin in the front region of tissue decreased (Fig. 3D). At the rear end, concentration profiles flattened out and the concentration at 20 hours was higher than at 12 hours (Fig. 3D).
Figure 3. Doxorubicin treatment and apoptosis induction A-B.
Fluorescent images of colon carcinoma tissue in microfluidic devices treated with 5 μM doxorubicin for 10.5 hours, showing the uptake and clearance of the drug (A) and the extent of induced apoptosis (B) measured using an active caspase-3 stain. Scale bar is 300 μm. C. Average normalized fluorescence intensities of doxorubicin from channels and tissues. The fluorescence intensities in tissue were significantly higher (*, P<0.05, n=3) than in channels. The clearance of doxorubicin from tissue was gradual. D. Spatial profiles of doxorubicin concentration. The concentration in the front of the tissue was higher initially. During clearance, the concentration profiles flattened. E. The average normalized fluorescence intensity of the apoptosis-marking stain increased continuously after drug delivery and was significantly greater at 8, 12, 16, and 20 hours (*, P<0.05, n=3) than at 0 hours. F. The rate of increase in apoptosis was positive for 20 hours. This rate of increase was less after doxorubicin was cleared from the system.
Doxil tissue uptake and clearance
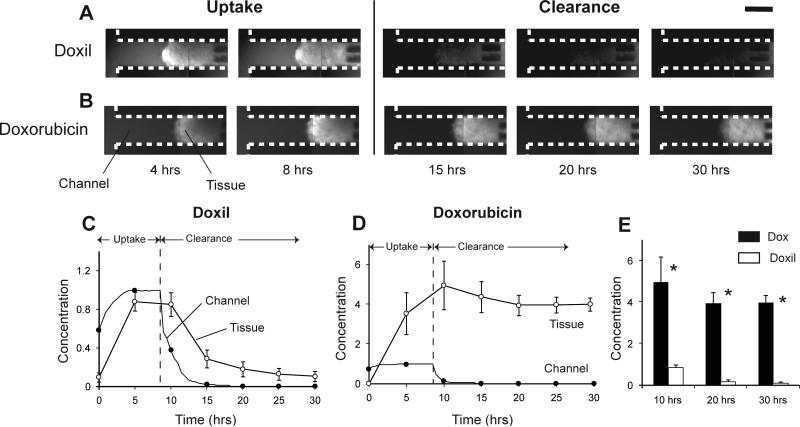
Tumor tissues on devices were treated with 50 μM Doxil for 8 hours followed by drug-free medium to measure uptake and clearance. An 8-hour treatment time was used because it provided sufficient time to measure concentration profiles and diffusion, and it enabled direct comparison with 8-hour doxorubicin treatment. The amount of Doxil taken up in tissue increased when it was delivered through the channels (Fig. 4A). Unlike doxorubicin, the average concentration in tissue was not significantly higher than in the channels (Fig. 4A,C). Only regions close to the channels had higher concentrations than channels (Fig. 4A). Clearance of Doxil from channels after 8 hours resulted in rapid clearance of drug from tissue (Fig. 4A,C). At 30 hours, the average amount of drug in tissue was 12% of the maximum; 88% of the drug was cleared from the tissue (Fig. 4A,C). For direct comparison of the performance of Doxil with doxorubicin, the latter was delivered at 50 μM for 8 hours to tissues. Similar to delivery of doxorubicin above (Fig. 3), the concentration of doxorubicin in tissue was significantly higher than in the channels, and the clearance of doxorubicin from tissues was gradual (Fig. 4B,D). Doxorubicin concentration in tissue was significantly higher (P<0.05, n=3) than Doxil throughout the experiment (Fig. 4E).
Figure 4. Doxil and doxorubicin treatment A-B.
Fluorescent images of colon carcinoma tissue in microfluidic devices treated with 50 μM Doxil (A) and 50 μM doxorubicin (B) for 8 hours, showing the uptake and clearance of the two drugs. Scale bar is 300 μm. C-D. Average normalized fluorescence intensities from channels and tissue for Doxil (C) and doxorubicin (D) over 20 hours. Doxil cleared from tissue rapidly and doxorubicin cleared gradually. E. Comparison of normalized tissue fluorescence intensities of doxorubicin and Doxil. Doxorubicin treated tissues had significantly greater (*, P<0.05, n=3) fluorescence than Doxil treated tissues.
Mathematical model for drug diffusion, release, binding, and clearance
A mathematical model was developed to model the delivery, diffusion, binding, and clearance of drug from tumor tissue. The model incorporates three important mechanisms: a) diffusion in tissue, b) binding, and c) clearance.
| (1) |
| (2) |
| (3) |
This system of equations balances the concentration of three entities: free drug, A (Eq. 1), bound drug, B (Eq. 2), and density of binding sites, S (Eq. 3). The model contains four parameters: i) diffusivity of drug, D; ii-iii) forward and reverse rates of drug binding, kon and koff, respectively; and iv) maximum concentration of drug binding sites in tissue, Smax. The diffusivity, D, describes transport through cellular material, which has been shown to be orders of magnitude slower than in aqueous solution50. Parameter perturbation analysis showed that the solution of the model is an exclusive function of the binding constant, R = kon/koff, reducing the model to three parameters: D, R, and Smax. Drug binding was assumed to be a reversible reaction with 1:1 drug:site stoichiometry, A + S ↔ B, where S is the concentration of available drug binding sites. To fit device results, the drug concentration in the channel was modeled as a step function. Gradients in drug concentration from the flow channels to the boundary of the tissue and along the width of chambers were experimentally observed to be less than 4% in all cases and assumed to be negligible in the model.
For a liposome-encapsulated drug, three additional parameters were introduced to the model: i) rate of drug release from liposomes, krel, ii) diffusivity of liposomes, Dlip, and iii) partition coefficient of the liposomes across the medium-tissue interface, P. Release from liposomes was assumed to be irreversible with first order kinetics. Equations 2 and 3 are valid for liposome-encapsulated drugs. The concentration of liposome-encapsulated drug, LA, and released drug, A, was balanced using the following equations:
| (4) |
| (5) |
Experimental data from doxorubicin and Doxil treatments were fit to the mathematical model to determine model parameters. For doxorubicin, experimental data (Fig. 3) was fit to Eqns. 1-4 to determine the diffusivity of doxorubicin, D, binding constant, R, and the maximum concentration of binding sites, Smax (Table 1). For the determined parameters, the experimental data and model predictions showed good agreement (Fig. 5A,B). The model predicted minimal clearance of doxorubicin from tissue after clearance from channels (Fig. 5A). It also predicted the flattening of the spatial concentration profile at 16 hours (Fig. 5B). For Doxil, experimental data (Fig. 4) was fit to solutions of Eqns. 2-6 to determine the rate of doxorubicin release from liposomes, krel, the diffusivity of liposomes, Dlip, and the liposome partition coefficient, P (Table 1). Experimental and simulated profiles for determined parameters showed good agreement (Fig. 5C,D). The model predicted the rapid clearance of drug from tissue (Fig. 5C) as well as shapes of spatial concentration profiles at all times (Fig. 5D).
Table 1.
Calculated parameter values
| Parameter | Description | Value |
|---|---|---|
| Doxorubicin | ||
| D | Diffusivity | (2.86 ± 1.15) × 10−11 m2/s |
| R | Binding constant | 48.6 ± 24.4 μM−1 |
| Smax | Maximum binding site concentration | 42.8 ± 5.6 μM |
| Doxil | ||
| krel | Rate of release from liposome | (6.05 ± 0.18) × 10−11 s−1 |
| Dlip | Diffusivity of liposomes | 3.72 ± 0.57 × 10−12 m2/s |
| P | Partition coefficient | 1.34 ± 0.09 |
Figure 5. Results from experimental data fitting A-B.
Experimental and simulated average tissue intensities (A) and spatial profiles (B) over time for doxorubicin. C-D. Experimental and simulated average tissue intensities (C) and spatial profiles (D) over time for Doxil. Simulated profiles matched experimental profiles well.
Effects of diffusion, binding, and clearance on therapeutic efficacy
The mathematical model was used to determine the contribution of each mechanism – diffusion, binding, and clearance – to the therapeutic effect of doxorubicin. To determine therapeutic effect, the concentration of drug predicted by the mathematical model (Eqns. 1-3) was linked to cell death. Cell death was assumed to be a function of the effective drug concentration, including free (A) and bound (B) drug, E = A + B, and had saturation kinetics with maximum rate of cell death and saturation constant, μd,max and Km, respectively.
| (6) |
where l is the live cell density (0 < l < 1). Parameters μd,max and Km for doxorubicin were calculated from data published by Mantovani51, 52 and were set to 0.14 hr−1 and 1.66 μM, respectively. Clearance of drug or liposomes from the blood was assumed to have a half-life of t1/2. The geometry of this model to calculate therapeutic efficacy matches the shape of the microfluidic device. This geometry has some influence on the predicted efficacy. For example, a very shallow chamber would retain drugs for less time. To produce realistic predictions, the depth of the chamber was chosen to match the diffusion distances that control delivery to tumors13, 53. The linear geometry of the device provides a straightforward means to compare the efficacy of different drugs and compare the effects of model parameters. All experimental parameters measured with the device (e.g. diffusivity, binding, and release) are independent of shape and could be used to calculate efficacy in any geometry.
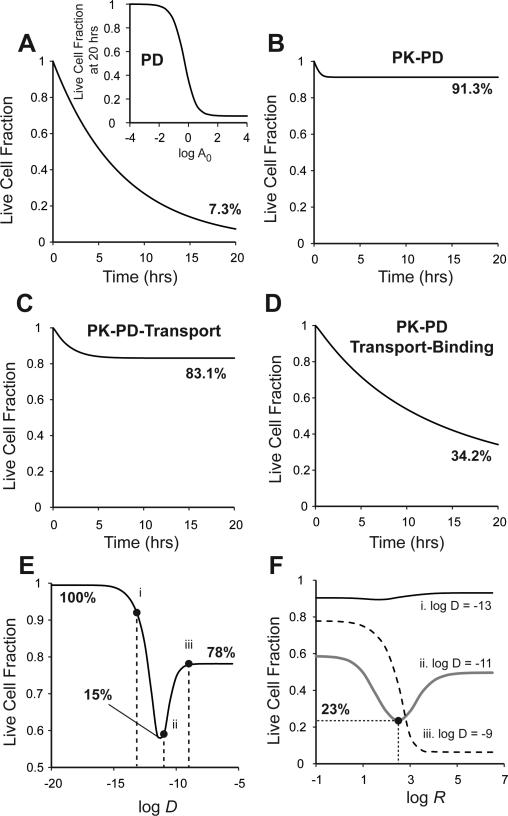
The mathematical model for free drugs (Eqns. 1-3 and Eq. 6) was solved for four limiting cases, introducing the contribution of one mechanism one at a time. Case1: In the absence of transport limitations, clearance, and binding (D→∞, t½→∞, R→0), doxorubicin decreased live cell density at 20 hours to 7.3% (Fig. 6A). A dose response curve for doxorubicin was obtained using these parameters (Fig. 6A, inset) that is identical to standard pharmacodynamic response curves obtained with monolayers54. Case2: To account for the rapid clearance rate of doxorubicin, t1/2 was set to 0.2 hrs27-30. After the initial decrease in live cell density for approximately 1 hour, there was no further decrease (Fig. 6B). At 20 hours, the live cell density was 91.3% (Fig. 6B). Case3: Transport limitations were introduced by setting the diffusivity to the experimentally determined value from above, D = 2.86×10−11 m2/s (Table 1). Contrary to expectation, limited transport enhanced the efficacy of doxorubicin and decreased the surviving cell fraction at 20 hours from 93.1% (Fig. 6B) to 83.1% (Fig. 6C). Case4: Finally, binding was taken into account by setting the binding constant to the measured value, R = 48.6 μM−1 (Table 1). Binding caused a significant increase in efficacy and decreased the surviving cell fraction to 34.2% (Fig. 6D).
Figure 6. Effects of diffusion, binding, and clearance.
The contribution of diffusion, binding, and clearance to doxorubicin efficacy was estimated by solving the mathematical model for limiting cases. A. Pharmacodynamic response was predicted by eliminating transport, binding, and clearance. The drug killed cells at the maximum rate μdmax. Inset: Dose response curve for pharmacokinetic model of doxorubicin without transport or clearance. A0 is normalized doxorubicin dose. B. PK/PD response was predicted by introducing a clearance rate, t1/2 = 0.2 hours. Clearance reduced drug efficacy significantly. C. Effects of transport were predicted by introducing a nominal diffusivity, D = 2.86 × 10−11 m2/s. This improved the therapeutic response marginally by reducing the rate of drug wash out from tissue. D. Effects of binding were predicted by introducing a nominal drug binding constant, R = 48.6 μM−1. Binding improved therapeutic efficacy significantly. E. Limited diffusion can increase as well as decrease therapeutic response. An optimal diffusivity exists that maximizes therapeutic effect. Points i, ii, and iii are at reference diffusivities of 10−13, 10−11 and 10−9 m2/s, respectively. F. The interaction between diffusivity, D, and binding, R, is complex. Binding can enhance as well as reduce drug efficacy, depending on the value of diffusivity. Lines are plotted for diffusivities of (i) 10−13, (ii) 10−11 and (iii) 10−9 m2/s, as in (E).
The interaction between diffusion and binding was complex and their combined effects on drug efficacy were non-obvious. The mathematical model (Eqns. 1-3 and Eq. 6) was used to predict live cell density after treatment for 20 hours for different combinations of diffusivities and binding constants (Fig. 6E,F). In the absence of binding (low binding constant; R = 10−5 μM−1), limited transport both increased and decreased efficacy (Fig. 6E). For very low diffusivities (D < 10−15 m2/s), there was no cell death and the live cell fraction was 100%. For very high diffusivities (D > 10−8 m2/s), the live cell fraction was 78%. There was an optimum diffusivity where the therapeutic effect was at a maximum. At D = 10−11 m2/s, the live cell fraction was 15% (Fig. 6E).
The effect of binding on drug efficacy was dependent on diffusivity. For low diffusivity (D = 10−13 m2/s; point i; Fig. 6E), increasing binding did not significantly affect efficacy (dark line; Fig. 6F). For an intermediate diffusivity (D = 10−11 m2/s; point ii; Fig. 6E), there was a range of binding constants at which the live cell fraction decreased (Fig. 6F). For an optimum binding constant, log R = 2.5, the therapeutic effect was maximum and the live cell fraction decreased to 23% (gray line; Fig. 6F). For high diffusivities (D = 10−9 m2/s; point iii; Fig. 6E), increasing binding considerably decreased the live cell fraction (dotted line; Fig. 6F).
Effects of encapsulation on therapeutic efficacy
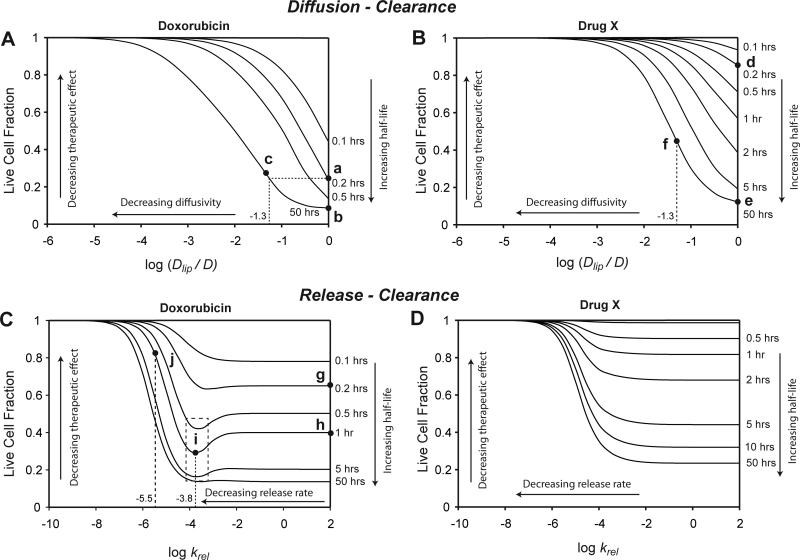
The effects of liposome encapsulation depended on the characteristics of the encapsulated drug, and specifically on the extent of cell binding. To demonstrate these differences, the mathematical model for encapsulated drugs (Eqns. 2-6) was solved for two drugs: a) doxorubicin, and b) a hypothetical drug X that does not bind cells, but is otherwise identical to doxorubicin. Encapsulated drug was assumed to not kill cells. After drug is released from liposomes, it was assumed to bind cells and causes cell death similar to free drug31. For both doxorubicin (Fig. 7A) and drug X (Fig. 7B), efficacy decreased with decreasing diffusivity. The right edge of these maps represents non-encapsulated drugs, where Dlip = D. Encapsulation increases the clearance half-life of doxorubicin from 0.2 hours (Fig. 7A; point a) to 50 hours (point b)34. The therapeutic benefit of this increase would be negated if encapsulation reduced diffusivity to less than 5% of the free drug, i.e. log (Dlip/D) < −1.3 (Fig. 7A; point c). Compared to doxorubicin, the map was shifted upward for non-binding drug X (Fig. 7B). Increasing the half-life of drug X from 0.2 hours (Fig. 7B; point d) to 50 hours (point e) caused a considerably larger improvement (73%) in efficacy compared to doxorubicin (15%). For the same 95% decrease in diffusivity, i.e. log (Dlip/D) = −1.3, the encapsulated drug was still significantly more efficacious than the parent drug (Fig. 7B; point f).
Figure 7. Effects of liposome encapsulation.
The effects of liposomal encapsulation were calculated for doxorubicin and a hypothetical drug X, identical to doxorubicin, except having no ability to bind tissue. Every curve on the maps represents one clearance rate. A-B: Diffusion-Clearance maps show that therapeutic response can be improved by increasing half-life, but decreases as a result of low diffusivity of liposomes, Dlip. The shape of these maps depends on the characteristics of the encapsulated drug. The curves are shifted up (lower efficacy) for the non-binding drug X (B). Points show efficacy at the clearance rate of free drug (a&d, t1/2 = 0.2 hrs), at the clearance rate of encapsulated drug with equal diffusivity (b&e, t1/2 = 50 hrs), and at the clearance rate of encapsulated drug with reduced diffusivity, log (Dlip/D) = −1.3 (c&f, t1/2 = 50 hrs). C-D. Release-clearance maps show a similar trade-off between increasing half-life and decreasing rate of drug release. C. For drugs that bind (doxorubicin), there exists an optimum release rate that can increase therapeutic effect (dotted rectangle). A hypothetical encapsulation system for doxorubicin (g) that increases the half-life to 1 hour (h) would have optimal efficacy at a release rate of krel = 10−3.8 (i). Too fast a release (krel = 10−5.5) would reduce efficacy (j). D. For non-binding drugs, therapeutic effect decreases monotonically with decreasing release rate.
The effects of release and clearance on cell viability was determined for the two drugs (Fig. 7C,D) using the measured value of Doxil diffusivity, Dlip = 3.72 × 10−12 m2/s (Table 1). The right edge of these maps represents non-encapsulated drugs, where release rates are high. Decreasing the release rate of doxorubicin from liposomes, at high and low clearance rates, decreased the therapeutic effect (Fig. 7C). Counterintuitively, at intermediate clearance rates (0.5 to 5 hrs) decreasing the release rate increased the therapeutic effect (Fig. 7C). In this range of clearance rates, the efficacy curves had minima at release rates of approximately 10−4 s−1 (Fig. 7C; dotted box; log krel = −4). To illustrate the benefit of an optimized release rate, consider a hypothetical encapsulation system for doxorubicin (Fig. 7C; point g) that increases the half-life from 0.2 to 1 hour (point h). If the release was optimized (log krel = −3.8; point i; Fig. 7C), the therapeutic effect would increase. However, too slow a release rate (log krel = −5.5; point j; Fig. 7C) would reduce the efficacy beyond that of the parent drug. For the non-binding drug X, decreasing the release rate would decrease the therapeutic effect at all clearance rates (Fig. 7D).
Location of doxorubicin and Doxil on parameter-response maps
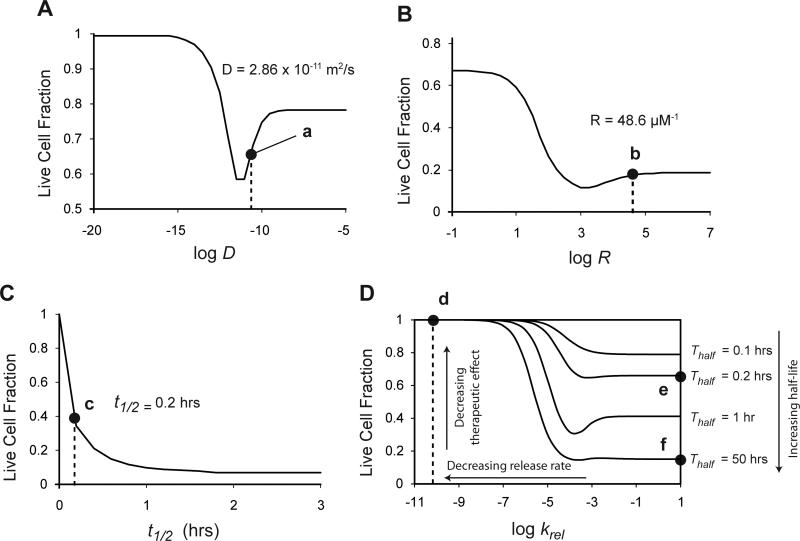
The location of the measured parameter values for doxorubicin and Doxil on the parameter-response maps (Fig. 6,7) may explain their performance (Fig. 8). On the diffusivity-response curve (Fig. 6E), the diffusivity of doxorubicin (D = 2.86×10−11 m2/s; Table 1), was near the bottom of a trough that maximizes therapeutic efficacy (Fig. 8A; point a). On the binding-response curve for this diffusivity, the binding constant of doxorubicin (R = 48.6 μM−1; Table 1) was also near the minimum (Fig. 8B; point b). However, on the clearance-rate-response curve for these values of D and R, the half-life of doxorubicin, t1/2 = 0.2 hours27-30, was not near the maximum response (Fig. 8C; point c). On the Release-Clearance map (Fig. 7C), increasing the half-life of doxorubicin from 0.2 hours (Fig. 8D; point e) to the 50-hour half-life34 of Doxil (Fig. 8D; point f) would increase the therapeutic effect. However, because of its slow release rate (krel = 6.05 × 10−11 s−1; Table 1), Doxil would have a very low therapeutic effect (Fig. 8D; point d) compared to doxorubicin.
Figure 8. Location of doxorubicin and Doxil on performance maps.
A. Diffusivity-response map shows that the diffusivity of doxorubicin, 2.86 × 10−11 m2/s, lies very near to the optimal diffusivity for maximizing therapeutic effect (point a). B. Binding-response map shows that the binding constant of doxorubicin, 48.6 μM−1, lies very near to the optimal binding constant for maximizing therapeutic effect (point b). C. Half-life response map shows that a short half-life, t1/2 = 0.2 hours, limits the therapeutic effect of doxorubicin (point c). D. Release-Clearance response map for Doxil. At half-life of 0.2 hours, doxorubicin lies at point e. Doxil increases the half-life to 50 hours (point f), but has a low tissue drug release rate (point d) that severely limits the therapeutic response.
Discussion
The drug-delivery devices used for these experiments mimic regions of tumors adjacent to blood vessels. Incorporating three-dimensional tumor tissues mimics intratumoral drug transport. Drug efficacy is a result of a complex interaction between several mechanisms: diffusion, release from carriers, binding, and clearance (Fig. 6, 7). To accurately determine therapeutic efficacy of cancer drugs, all of these mechanisms must be taken into account. Cancer cells attached to flat surfaces cannot quantify them. The drug delivery device developed here provides a platform for measuring the interactions of these critical mechanisms (Fig. 1,2).
Tissues on devices treated with doxorubicin and Doxil displayed important characteristics of the two drugs. Minimal tissue clearance of doxorubicin shows that it binds to tissue strongly (Fig. 3,4). Retention in tissue is a result of DNA intercalation25, 26. Strong tissue binding explains the long terminal half-life of doxorubicin after rapid initial clearance from plasma, as observed in humans27-30. DNA intercalation also explains the higher concentration of doxorubicin in tissue compared to channels (Fig. 3,4). In contrast, the rapid clearance of Doxil from tissue (Fig. 4) shows that there was minimal intra-tissue drug release. There was no drug retention detected in the experiments after clearance indicating that doxorubicin was not released from liposomes (Fig. 4).
The success of doxorubicin as one of the most widely used chemotherapeutic agents may partly be attributed to its optimal diffusivity and binding (Fig. 8A,B). Its diffusivity and binding are both near the maximum theoretical values. The clearance rate of doxorubicin, however, is not near the maximum (Fig. 8C), suggesting that improving this characteristic could substantially improve its efficacy. Doxil was designed to increase the circulation time of doxorubicin. It increased the half-life in humans from 0.2 hours to approximately 50 hours34. Doxil, however, has been successful for only a few cancers, which may be explained by its low release rate (Fig. 8D). Clinically, the benefits of Doxil are due to low toxicity and minimal systemic release31. Limited drug release at the tumor site would be a significant limitation to the therapeutic effect of Doxil. Engineered liposomes that can enhance drug release rates in tissues may radically enhance therapeutic response.
These results with doxorubicin and Doxil show that the effects of diffusion and binding on therapeutic efficacy must be evaluated during the drug development process. The technique demonstrated here, which combines a microfluidic device and a mathematical model, enables quantification of these factors. Monolayer experiments alone cannot measure them. Applying doxorubicin to a monolayer of cells will result in the classic PD model (Fig. 6A and inset). In the absence of transport limitations and clearance, the live cell fraction decreases at the maximum rate (Fig. 6A). Conventional wisdom suggests that accounting for plasma clearance and intratumoral transport would reduce efficacy. As expected, when clearance (i.e. pharmacokinetics) is added to the model, the therapeutic effect is significantly reduced (Fig. 6B). However, when diffusivity is added, there is an increase in the therapeutic response (Fig. 6C). This is caused by increased drug-tissue contact time and reduced clearance from tissue. Binding enhances the drug-tissue contact time and further improves the therapeutic effect (Fig. 6D). Because the effects of diffusion and binding cannot be predicted by monolayer studies, a method that integrates diffusion, binding, and clearance is necessary.
The complex interaction between transport and binding may be the cause for the failure of many cancer drugs in mice experiments and it is important to understand their effects. Limited transport in tumor tissue may enhance as well as reduce therapeutic efficacy (Fig. 6E). There is a limit to the benefit of increasing drug-tissue contact time by decreasing diffusivity. When diffusivity is very low, drugs cannot enter tissues and the therapeutic effect decreases (Fig. 6E). These non-linear effects are compounded by the complex relationship between binding and diffusivity (Fig 6F). For drugs with low diffusivities, stronger binding would have a slightly unfavorable effect, because binding prevents deep penetration (Fig. 6F). For drugs with high diffusivities, strong binding would be favorable because it would increase drug-tissue contact time by preventing rapid clearance (Fig. 6F). For drugs with intermediate diffusivities, there is an optimum binding that would maximize therapeutic effect (Fig. 6F). Both low and high binding rates would limit efficacy.
Encapsulation systems are often created to enhance a drug's delivery properties. However, because these systems are associated with many trade-offs, it is necessary that they be carefully evaluated before beginning development. The enhanced therapeutic effect achieved by increasing plasma circulation can be countered by slow diffusion in tissue (Diffusion-Clearance maps; Fig. 7A,B) and limited release (Release-Clearance maps; Fig. 7C,D). Ideally, increasing the half-life of clearance by liposome encapsulation would increase efficacy (Fig. 7A). However, encapsulation often lowers diffusivity (Table 1), which can offset the benefit of increased clearance (Fig. 7A). The benefit can be much greater for drugs that do not bind cells (Fig. 7B). The improvement in therapeutic effect for encapsulation of a non-binding drug could be large enough to overcome the reduction caused by poor diffusivity (Fig. 7B). This difference suggests that drugs that do not bind to tissue strongly could benefit more from liposome encapsulation.
The rate of drug release from liposomes could also be used strategically to maximize therapeutic efficacy. For example, the efficacy of encapsulated doxorubicin could be improved if the rate of release was optimized (Fig. 7C). An encapsulated drug with an optimum release rate could be more effective than a rapidly releasing system because the liposomes would penetrate deep into tissue before releasing their payload. A fast release rate would limit penetration and reduce efficacy. However, if the rate of release is too low, the benefits of encapsulation are diminished and the resulting drug would be less efficacious than the parent drug (Fig. 7C). In contrast, optima do not exist for non-binding drugs (Fig. 7D). Decreasing the release rate from these liposomes monotonically reduces efficacy and is never beneficial (Fig. 7D).
This drug-delivery device and computational method has many potential applications. It could be used to screen new drugs, revive previously failed drugs, and improve the performance of existing drugs. The method could be used to screen drugs prior to preclinical animal studies and after clearance rates had been measured. Prior to animal studies, exact clearance rates are not known. For these early screens, the method would identify and eliminate drugs that could not be effective at any clearance rate. Eliminating drugs at this point in the drug-development pipeline would considerably reduce the number of animal experiments. Once pharmacokinetic rates had measured, the method could predict efficacy, identify ineffective drugs, and prevent further animal experiments. The ability to identify limiting mechanisms would enable re-investigation of previously failed drugs. In some cases, drugs could be modified to improve performance. For example, liposome encapsulation and attachment to nanoparticles55 could improve circulation, diffusivity or release. The efficacy of approved drugs could be similarly improved by identifying the mechanisms that limit efficacy.
Conclusions
We have developed a method of estimating a cancer drug's efficacy by measuring critical phenomena that determine its efficacy – intratumoral diffusion, release from drug carriers, binding, and clearance – using an in vitro drug delivery device. We developed a mathematical model that incorporates all of these mechanisms and used it to evaluate parameters for doxorubicin and liposome-encapsulated doxorubicin (Doxil). Results from the model show that diffusion, binding, and clearance interact in a complex manner and this complex interaction determines efficacy. Doxorubicin has optimum diffusivity and binding, which maximizes its therapeutic effect and may be the cause of its success as a chemotherapeutic agent. The model shows that the benefits of prolonged circulation achieved by liposome encapsulation may be offset by limited drug release and diffusion in tissue. Doxil did not release doxorubicin in tissues in the experimental system, showing that limited release from liposomes is a major shortcoming of Doxil. Monolayers of cancer cells do not capture these critical mechanisms and fail to determine the efficacy of cancer drugs accurately. The drug-delivery device developed here provides a tool for predicting drug efficacy in vitro before subsequent animal experiments, which will reduce the duration and cost of drug development.
Materials and Methods
Mammalian cell culture
Human colon adenocarcinoma cells LS174T (ATCC, Manassas, VA) were grown in Dulbecco's Modified Eagle's Medium (DMEM, Sigma-Aldrich, St. Louis, MO) containing 1 g/L glucose and 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) at pH 7.4. Cells were incubated at 37°C in ambient air with 5% CO2 and 100% relative humidity. Spheroids were made by plating single cell suspensions of 2.5 × 104 cells/ml on non-adherent poly(2-hydroxyethyl methacrylate)-coated T25 flasks.
Perfused tumor tissue culture
Microfluidic devices were used to continuously perfuse cultures of tumor tissue. Spheroids were introduced into rectangular chambers on devices to form cuboidal tissues (Fig. 2). These tissues were subjected to continuous medium perfusion along one face, and were incubated for 24 hours at 37°C in a customized heated enclosure on a microscope before drug treatment. Humidity and pH was maintained by flowing 25mM HEPES buffered DMEM at 3 μl/min. Devices were constructed using standard soft lithography techniques as described previously18, 20. Molds were made by selectively exposing SU-8 (MicroChem, Newton, MA) coated wafers to UV light through photomasks to produce positive-relief features. Replicates were made using Sylgard® 184 Silicone Elastomer Kit (Dow Corning, Midland, MI). A 1.5 mm biopsy punch was used to make holes for connecting inlet and outlet tubing. We have previously shown that this device can be used to measure the diffusion coefficient of doxorubicin18.
Doxorubicin and Doxil treatment and Quantification of Apoptosis
Doxorubicin was obtained from Sigma-Aldrich (St. Louis, MO) and Doxil was obtained as a kind gift from Dr. T. Emrick (University of Massachusetts Amherst, Amherst, MA). Apoptosis was quantified by using a fluorescent stain from the CaspGLOW Fluorescein Active Caspase-3 Staining Kit (Biovision, Milpitas, CA), which binds irreversibly to active caspase-3, a marker of apoptosis in cells. All experiments were run in multiple replicates (n=3). Two-tailed Student's t-tests were used for statistical comparisons. Errors are reported as standard errors of the mean.
Image acquisition and analysis
All images were acquired using an Olympus (Center Valley, PA) IX71 inverted epifluorescent microscope equipped with a Plan-APO 10x objective. Doxorubicin and Doxil naturally fluoresce red when excited with green light. Transmitted light and red fluorescent images were acquired every 30 minutes immediately after drugs were introduced into the device. To capture an entire culture chamber (1000μm × 300 μm), two adjoining images (867.15μm × 660.68μm each) were obtained and tiled using a macro in IPLab (BD Bioscience, Rockville, MD). Fluorescent images were acquired using a 560/10 nm excitation and a 590 nm long pass emission filter (Chroma, Rockingham, VT). Intensity values obtained from fluorescent images were used to generate space and time profiles. Rectangular regions of interest (ROI) incorporating each tissue were created and background-subtracted linear intensity profiles were generated by averaging intensities along successive widths of the ROIs. Distances were normalized by the length of the ROIs. Doxorubicin concentrations in tissues were determined by normalizing intensity values by the background-subtracted intensities in the flow channels, which had known concentrations. All tissue concentrations are reported relative to these flow channel concentrations. A similar transformation was done for green fluorescent images to calculate normalized intensity profiles corresponding to extent of apoptosis. The rate of apoptosis increase was calculated as the rolling three-point average of the local slope. Because encapsulation in liposomes quenches fluorescence, the fluorescence intensity of equal concentrations of doxorubicin and Doxil was measured. Fluorescence from free doxorubicin was determined to be 3.28 times greater than Doxil. For comparison of overall drug uptake by tissues, average normalized intensity values over entire ROIs were obtained at all time points.
Solution of Mathematical Model and Parameter Estimation
For doxorubicin, the mathematical model was comprised of Eqns. 1-3. The concentration (A) at the front edge (x = 0) was modeled as a time-dependent step function that matched the concentration in the channels of the device. An impermeable boundary was assumed at the rear end (x = 600 μm). Initially, concentrations of free (A) and bound (B) doxorubicin were set to zero. The three equations were discretized using finite differences into a set of non-linear algebraic equations and solved for A and B in space and time using the MATLAB (The Mathworks Inc, Natick, MA) function fsolve. For Doxil, the mathematical model was comprised of Eqns. 2-5. Similar to doxorubicin, the concentration of encapsulated drug, LA, at the front edge was set to match the concentration in the channel. A partition coefficient (P) was included to account for the partitioning across the medium-tissue interface. Because the concentration of free drug, A, at the front edge was unknown, a convective boundary condition was included with a large mass transfer coefficient, Kl = 1.1×10−3 m/s. An impermeable boundary was assumed at the rear end. Similar to the doxorubicin simulations, the Doxil equations (Eqns. 2-5) were discretized using finite differences and solved for LA, A and B.
Normalized fluorescent intensity profiles obtained from experimental data from doxorubicin and Doxil treatment were fit to the mathematical models to determine transport parameters. For doxorubicin, the free drug model (Eqns. 1-3) was solved individually for each experiment (n=3). The least square error between experimental and simulated profiles was minimized using the MATLAB unconstrained nonlinear optimization function, fminsearch, to determine values of D, R and Smax (Table 1). These values were used to solve the mathematical model for Doxil (Eqns. 2-5). To account for increase in fluorescence as a result of doxorubicin release from Doxil, the contribution to fluorescence intensity from released doxorubicin was 3.28 times higher than Doxil. Similar to the doxorubicin experiments, the mathematical model for Doxil was solved individually for each experiment (n=3) and least square minimization was performed to determine values of Dlip, krel, and P (Table 1).
Predictive Modeling
To predict the effect of doxorubicin delivery on therapeutic effectiveness, an equation describing the rate of cell death (Eq. 6) was added to the doxorubicin model. To match the PK, the concentrations of doxorubicin (A) at the front edge (x = 0) was modeled as an exponential decay with half-life t1/2. Initially, concentrations of free (A) and bound (B) doxorubicin were set to zero and the live cell density (l) was set to 1. This system of equations (Eqns. 1-3 and Eq. 6) was solved using fsolve as described above. To simulate the absence of transport limitations, clearance, and binding (Case1), model parameters were set to high diffusivity (D = 102 m2/s), long half-life (t1/2 = 100 hrs), and a low binding constant (R = 10−5 μM−1). To introduce the effects of clearance, diffusion and binding, the model was solved after adjusting three parameters: plasma half-life (t1/2) to 0.2 hrs (Case2); diffusivity (D) to 2.86×10−11 m2/s (Case3); and binding constant (R) to 48.6 μM−1 (Case4). Diffusion and binding profiles were determined by solving the system (Eqns 1-3 and Eq. 6) for ranges of diffusivities and binding constants, with a clearance rate (t1/2) of 0.5 hours. Profiles report live cell fractions (l) at 20 hours.
To predict the effect of Doxil delivery, cell death (Eq. 6) was added to the Doxil model (Eqns. 2-5). The concentrations of Doxil (LA) at the front edge (x = 0) was modeled as an exponential decay with half-life t1/2. Initially, concentrations of Doxil (LA), free (A) and bound (B) doxorubicin were set to zero and the live cell density (l) was set to 1. To model the effects of encapsulating a hypothetical non-binding drug (drug X), the binding constant set to be very low (R = 10−5 μM−1). Diffusion-Clearance maps were generated by solving the system of equations for a range of liposome diffusivities (Dlip) and clearance rates (t1/2), with rapid drug release. The live cell fraction (l) is reported at 20 hours. Release-Clearance maps were generated by solving the equations for a range of liposome release (krel) and clearance rates (t1/2).
Acknowledgements
We gratefully acknowledge financial support from the National Science Foundation (Grant No. 1159689), National Institutes of Health (Grant Nos. R21CA112335 and R01CA120825), and the Eugene M. Isenberg Award for Bhushan J. Toley. We gratefully acknowledge Dr. Todd Emrick at the University of Massachusetts for providing Doxil for experiments.
References
- 1.Jain RK. Sci Am. 1994;271:58–65. doi: 10.1038/scientificamerican0794-58. [DOI] [PubMed] [Google Scholar]
- 2.Jain RK. Nat Med. 1998;4:655–657. doi: 10.1038/nm0698-655. [DOI] [PubMed] [Google Scholar]
- 3.Tredan O, Galmarini CM, Patel K, Tannock IF. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 4.St Jean AT, Zhang M, Forbes NS. Curr Opin Biotechnol. 2008;19:511–517. doi: 10.1016/j.copbio.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim BJ, Forbes NS. Biotechnol Bioeng. 2007;96:1167–1182. doi: 10.1002/bit.21205. [DOI] [PubMed] [Google Scholar]
- 6.Shoemaker RH. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 7.Grever MR, Schepartz SA, Chabner BA. Semin Oncol. 1992;19:622–638. [PubMed] [Google Scholar]
- 8.Plowman J, Dykes DJ, Hollingshead M, Simpson-Herren L, Alley MC. In: Anticancer Drug Development Guide. 1 edn. Teicher BA, editor. Humana Press; Totowa: 1997. pp. 101–125. ch. 6. [Google Scholar]
- 9.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minchinton AI, Tannock IF. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 11.Lin JH, Lu AY. Pharmacol Rev. 1997;49:403–449. [PubMed] [Google Scholar]
- 12.Bryn SR, Dolch GD. J Pharm Sci. 1978;67:688–693. doi: 10.1002/jps.2600670532. [DOI] [PubMed] [Google Scholar]
- 13.Lankelma J. Curr Pharm Des. 2002;8:1987–1993. doi: 10.2174/1381612023393512. [DOI] [PubMed] [Google Scholar]
- 14.Sartiano GP, Lynch WE, Bullington WD. J Antibiot (Tokyo) 1979;32:1038–1045. doi: 10.7164/antibiotics.32.1038. [DOI] [PubMed] [Google Scholar]
- 15.Hsiao AY, Torisawa YS, Tung YC, Sud S, Taichman RS, Pienta KJ, Takayama S. Biomaterials. 2009;30:3020–3027. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen MC, Gupta M, Cheung KC. Biomed Microdevices. 2010;12:647–654. doi: 10.1007/s10544-010-9417-2. [DOI] [PubMed] [Google Scholar]
- 17.Wu LY, Di Carlo D, Lee LP. Biomed Microdevices. 2008;10:197–202. doi: 10.1007/s10544-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 18.Walsh CL, Babin BM, Kasinskas RW, Foster JA, McGarry MJ, Forbes NS. Lab Chip. 2009;9:545–554. doi: 10.1039/b810571e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toley BJ, Forbes NS. Integr Biol (Camb) 2012;4:165–176. doi: 10.1039/c2ib00091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toley BJ, Ganz DE, Walsh CL, Forbes NS. J. Vis. Exp. 2011:57. doi: 10.3791/2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arcamone F. Doxorubicin: Anticancer Antibiotics. Academic Press Inc.; New York: 1981. [Google Scholar]
- 22.Di Marco A, Massimo L. Minerva Med. 1968;59:3511–3521. [PubMed] [Google Scholar]
- 23.Rusconi A, Dimarco A. Cancer Research. 1969;29:1507–&. [PubMed] [Google Scholar]
- 24.Silvestr R, Gambaruc C, Dasdia T. Tumori. 1970;56:137–&. doi: 10.1177/030089167005600301. [DOI] [PubMed] [Google Scholar]
- 25.Silvestrini R, Di Marco A, Dasdia T. Cancer Res. 1970;30:966–973. [PubMed] [Google Scholar]
- 26.Silvestrini R, Gambarucci C, Dasdia T. Tumori. 1970;56:137–148. doi: 10.1177/030089167005600301. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson PM, Mawer GE. Br J Clin Pharmacol. 1974;1:241–247. doi: 10.1111/j.1365-2125.1974.tb00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamin RS, Riggs CE, Jr., Bachur NR. Clin Pharmacol Ther. 1973;14:592–600. doi: 10.1002/cpt1973144part1592. [DOI] [PubMed] [Google Scholar]
- 29.Arena E, D'Alessandro N, Dusonchet L, Gebbia N, Gerbasi F, Palazzoadriano M, Raineri A, Rausa L, Tubaro E. Arzneimittelforschung. 1971;21:1258–1263. [PubMed] [Google Scholar]
- 30.Mhatre RM, Herman EH, Waravdekar VS, Lee IP. Biochem Med. 1972;6:445–453. doi: 10.1016/0006-2944(72)90089-0. [DOI] [PubMed] [Google Scholar]
- 31.Immordino ML, Dosio F, Cattel L. Int J Nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 32.Lao J, Madani J, Puértolas T, Alvarez M, Hernández A, Pazo-Cid R, Artal A, Antón Torres A. J Drug Deliv. 2013;2013:456409. doi: 10.1155/2013/456409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duggan ST, Keating GM. Drugs. 2011;71:2531–2558. doi: 10.2165/11207510-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 34.Gabizon A, Catane R, Uziely B, Kaufman B, Safra T, Cohen R, Martin F, Huang A, Barenholz Y. Cancer Research. 1994;54:987–992. [PubMed] [Google Scholar]
- 35.Seetharamu N, Kim E, Hochster H, Martin F, Muggia F. Anticancer Res. 2010;30:541–545. [PubMed] [Google Scholar]
- 36.Farhat FS, Temraz S, Kattan J, Ibrahim K, Bitar N, Haddad N, Jalloul R, Hatoum HA, Nsouli G, Shamseddine AI. Clin Breast Cancer. 2011;11:384–389. doi: 10.1016/j.clbc.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Zamboni WC, Ramalingam S, Friedland DM, Edwards RP, Stoller RG, Strychor S, Maruca L, Zamboni BA, Belani CP, Ramanathan RK. Clin Cancer Res. 2009;15:1466–1472. doi: 10.1158/1078-0432.CCR-08-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabizon AA. Cancer Invest. 2001;19:424–436. doi: 10.1081/cnv-100103136. [DOI] [PubMed] [Google Scholar]
- 39.Zamboni WC, Gervais AC, Egorin MJ, Schellens JHM, Zuhowski EG, Pluim D, Joseph E, Hamburger DR, Working PK, Colbern G, Tonda ME, Potter DM, Eiseman JL. Cancer Chemotherapy and Pharmacology. 2004;53:329–336. doi: 10.1007/s00280-003-0719-4. [DOI] [PubMed] [Google Scholar]
- 40.Panwar P, Pandey B, Lakhera PC, Singh KP. Int J Nanomedicine. 2010;5:101–108. doi: 10.2147/ijn.s8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrington KJ, Lewanski CR, Northcote AD, Whittaker J, Wellbank H, Vile RG, Peters AM, Stewart JS. Ann Oncol. 2001;12:493–496. doi: 10.1023/a:1011199028318. [DOI] [PubMed] [Google Scholar]
- 42.Bondurant B, Mueller A, O'Brien DF. Biochim Biophys Acta. 2001;1511:113–122. doi: 10.1016/s0005-2736(00)00388-6. [DOI] [PubMed] [Google Scholar]
- 43.Leung SJ, Romanowski M. Theranostics. 2012;2:1020–1036. doi: 10.7150/thno.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derendorf H, Lesko LJ, Chaikin P, Colburn WA, Lee P, Miller R, Powell R, Rhodes G, Stanski D, Venitz J. J Clin Pharmacol. 2000;40:1399–1418. [PubMed] [Google Scholar]
- 45.Gabrielsson J, Green AR. J Pharmacol Exp Ther. 2009;331:767–774. doi: 10.1124/jpet.109.157172. [DOI] [PubMed] [Google Scholar]
- 46.Derendorf H, Meibohm B. Pharm Res. 1999;16:176–185. doi: 10.1023/a:1011907920641. [DOI] [PubMed] [Google Scholar]
- 47.Meibohm B, Derendorf H. International Journal of Clinical Pharmacology and Therapeutics. 1997;35:401–413. [PubMed] [Google Scholar]
- 48.Jain RK. Cancer Res. 1987;47:3039–3051. [PubMed] [Google Scholar]
- 49.Jain RK. Adv Drug Deliv Rev. 1997;26:71–90. doi: 10.1016/s0169-409x(97)00027-6. [DOI] [PubMed] [Google Scholar]
- 50.Pluen A, Boucher Y, Ramanujan S, McKee TD, Gohongi T, di Tomaso E, Brown EB, Izumi Y, Campbell RB, Berk DA, Jain RK. Proc Natl Acad Sci U S A. 2001;98:4628–4633. doi: 10.1073/pnas.081626898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mantovani A. Cancer Res. 1977;37:815–820. [PubMed] [Google Scholar]
- 52.Erlichman C, Vidgen D. Cancer Res. 1984;44:5369–5375. [PubMed] [Google Scholar]
- 53.Helmlinger G, Yuan F, Dellian M, Jain RK. Nat Med. 1997;3:177–182. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 54.Chen CT, Au JL, Wientjes MG. Clin Cancer Res. 1998;4:277–282. [PubMed] [Google Scholar]
- 55.Kim B, Han G, Toley BJ, Kim CK, Rotello VM, Forbes NS. Nat Nanotechnol. 2010;5:465–472. doi: 10.1038/nnano.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]