Abstract
Studies of transcriptome profiles have provided new insights into mechanisms underlying the development of hypertension. Cell type heterogeneity in tissue samples, however, has been a significant hindrance in these studies. We performed a transcriptome analysis in medullary thick ascending limbs of the loop of Henle isolated from Dahl salt-sensitive rats. Genes differentially expressed between Dahl salt-sensitive rats and salt-insensitive consomic SS.13BN rats on either 0.4% or 7 days of 8% NaCl diet (n=4) were highly enriched for genes located on chromosome 13, the chromosome substituted in the SS.13BN rat. A pathway involving cell proliferation and cell cycle regulation was identified as one of the most highly ranked pathways based on differentially expressed genes and by a Bayesian model analysis. Immunofluorescent analysis indicated that just one week of a high salt diet resulted in a several fold increase in proliferative medullary thick ascending limb cells in both rat strains and that Dahl salt-sensitive rats exhibited significantly greater proportion of medullary thick ascending limb cells in a proliferative state than in SS.13BN rats (15.0% ± 1.4% vs. 10.1% ± 0.6%, n=7–9, P<0.05). The total number of cells per medullary thick ascending limb section analyzed was not different between the two strains. The study revealed alterations in regulatory pathways in Dahl salt-sensitive rats in tissues highly enriched for a single cell type, leading to the unexpected finding of a greater increase in the number of proliferative medullary thick ascending limb cells in Dahl salt-sensitive rats on a high-salt diet.
Keywords: Kidney, gene expression, physiological genomics, cell cycle, salt intake
Introduction
Gene or protein expression profiles have been studied in whole organs or partial organs collected from various animal models of hypertension. Examples include analysis of cardiac and renal transcriptomes in the spontaneously hypertensive rat, the Lyon hypertensive rat, and the Dahl salt-sensitive (SS) rat.1,2,3 The SS rat is a well-established model of human salt-sensitive hypertension and renal injury. The consomic SS.13BN rat was derived by substituting Brown Norway chromosome 13 into the SS genome. Salt-induced hypertension and renal injury were substantially attenuated in the SS.13BN rat.4 Our analysis of the transcriptomes and proteomes in the renal medulla and the renal cortex of SS and SS.13BN rats has revealed several novel mechanisms and partial regulatory networks that may contribute to the development of hypertension or renal injury in the SS rat.5,6,7,8,9,10 The novel mechanisms discovered include those involving fumaric acid metabolism and renal 11β-hydroxysteroid dehydrogenase type 1.11,12,13
Cell type heterogeneity, however, has been a significant hindrance in the interpretation of these studies. The renal cortex and the renal medulla contain epithelial, connective, and vascular tissues. Each tissue includes multiple cell types. For example, the nephron epithelia consist of several segments, each with unique physiological and molecular characteristics. Differential expression between animals may in some cases reflect differences in cellular composition of the tissue being analyzed rather than expression changes in a given cell type. Dominant cell types may prevent detection of important changes in gene expression taking place in a minority cell type.
In the present study, we performed a transcriptome analysis in medullary thick ascending limbs (mTAL) of the loop of Henle isolated from SS rats and SS.13BN rats. The mTAL is highly relevant to the pathophysiological mechanisms of hypertension, especially in the SS rat.14,15,16,17,18,19,20,21,22,23,24,25,26 We applied a new method of pathway analysis utilizing a Bayesian modeling approach that incorporated prior knowledge of biological pathways and was independent of any a priori determination of differential expression.27 The study led to an unexpected discovery that mTALs in SS rats on a high-salt diet exhibited increased proliferative activities.
Methods
See the Online Supplement for detailed methods
Animals
Male SS and consomic SS.13BN rats were generated and maintained as described.4,8 Rats were studied at 6 weeks of age while maintained on 0.4% salt diet (AIN-76A, Dyets) or at 7 weeks of age after one week of 8% salt diet. The time points represent an early stage of the development of hypertension.8
Isolation of medullary thick ascending limbs (mTAL)
mTALs were isolated using a bulk dissection method as described previously with modifications.18,28
Tamm-Horshfall protein staining
Samples of isolated mTALs were stained with antibody against Tamm-Horshfall protein.
RNA extraction
RNA was extracted as described8 and assessed by Agilent BioAnalyzer 2100 and spectrophotometry.
Real-time PCR
Real-time PCR analysis of selected mRNAs was performed using the SYBR Green chemistry (Supplemental Table S1).8,29
Affymetrix Expression Array analysis
Affymetrix Rat Expression Array 230 2.0 was hybridized following the manufacturer’s protocols.8 A total of 16 arrays were used to analyze two rat strains and two salt conditions (0.4% and 7 days of 8%) with four individual samples in each experimental group. Signal intensities were normalized by Robust Multi-array Average Expression Measure (RMA). Differentially expressed genes were identified by Rank Product methods. Ingenuity Pathway Analysis was used for pathway analysis.8
Chromosomal representation index
Representation of a chromosome in differentially expressed genes was calculated as we described previously.7
Bayesian model analysis
A pathway-based analysis was performed using a modified version of the Bayesian model described recently.27 The Bayesian method integrates pathway information with the experimental data to identify pathways and genes related to a binary phenotype. Briefly, we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) to retrieve an S matrix indicating memberships of genes to pathways and an R matrix describing relationships between genes within and between pathways. The matrixes were used to define probability distributions of pathways and genes, called priors. A Monte Carlo Markov chain (MCMC) algorithm was applied to explore the posterior space efficiently and find the most probable configurations of genes and pathways. The procedure results in a list of visited models with included pathways and genes and their corresponding relative posterior probabilities. The posterior probability represents the likelihood of a pathway or gene to discriminate two experimental groups.
Defining boundaries of congenic segments using single nucleotide polymorphism (SNP) arrays
We previously identified four non-overlapping congenic regions on rat chromosome 13 that had significant effects on hypertension in the SS rat, designated as lines 1, 5, 9, and 26.30 In the present study, we obtained precise boundaries for lines 5, 9, and 26 using the RATDIVm520813 SNP array (803,484 SNPs genome-wide).
Immunofluorescence analysis of cell proliferation
Proliferative cells were identified by positive staining of Ki-67. Cells in the G2/M phases were identified by positive staining of histone H3 phosphorylated at Ser10 (p-H3 or H3-P). Cell nucleus was visualized by DAPI staining. Ten fields were randomly selected from each kidney. mTALs were identified by morphology. The numbers of all cells (cell nuclei), Ki-67 positive cells, and H3-P positive cells within mTALs were manually counted. The investigator analyzing the images was blinded to the treatment conditions of the rats.
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL)
TUNEL staining of kidney sections was performed using DeadEnd™ Fluorometric TUNEL System from Promega following the manufacturer’s instructions.
Statistical analysis
Affymetrix array data were analyzed as described above. Data from other experiments were analyzed using student’s t-test or analysis of variance, and reported as mean ± SEM. P<0.05 was considered significant.
Results
Quality control of mTAL isolation and RNA extraction
More than 100 SS or SS.13BN rats were used for mTAL isolation and RNA extraction. Samples from 16 rats were selected for Affymetrix expression array analysis based on substantial enrichment of mTALs, high RNA quality, and sufficient RNA amounts.
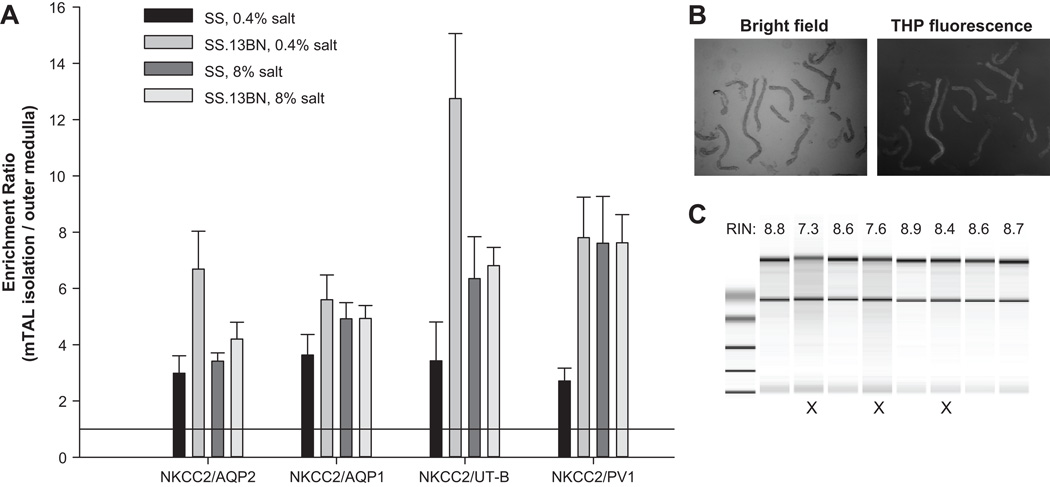
Enrichment of mTALs was assessed by real-time PCR quantification of Na-K-2Cl cotransporter (NKCC2), a marker of thick ascending limbs, aquaporin 1, a marker of proximal tubules, aquaporin 2, a marker of collecting ducts, urea transporter B, a marker of descending Vasa recta, and plasmalemma vesicle protein-1, a marker of endothelial cells. Compared to renal outer medulla tissues, the ratio of NKCC2 abundance to each of the other marker genes was substantially increased in isolated mTALs, indicating a 4 to 7 fold enrichment of mTALs relative to other nephron and vascular segments (Figure 1A). The purity of mTAL isolations was further assessed by staining of Tamm-Horshfall protein, a protein specifically expressed in mTALs (Figure 1B). From 600 tubular segments in 10 images from three isolations, we determined that 93.2% of the segments were mTALs.
Figure 1. Quality control of mTAL isolation and RNA extraction.
A. Real-time PCR analysis of marker genes. Enrichment of mTALs in the mTAL isolations compared to the outer medulla was assessed by examining abundance ratios of an mTAL marker (NKCC2) to markers of other cells (see Results). The ratios were artificially set as 1 for the outer medulla. B. The vast majority of the collected nephron segments (bright field) were stained positive for Tamm-Horshfall protein (THP), a mTAL-specific protein. C. Bioanalyzer analysis of RNA samples. Samples marked with X were discarded.
All 16 samples had RNA Integrity Numbers (RIN) greater than 8.5 (Figure 1C), 260/280 nm ratios of 1.9–2.0, and a total amount of RNA greater than 4µg.
Differentially expressed genes
Of 31,099 EST probe sets on the Affymetrix Expression Array, 14,677 were considered detectable in all mTAL samples. Rank Product analysis identified 217 and 291 ESTs as differentially expressed between SS and SS.13BN rats on 0.4% salt and 7 days of 8% salt, respectively, based on the criteria of FDR < 0.05 and log2 ratio greater than 0.5 or less than −0.5. The differentially expressed ESTs and associated gene annotations are shown in Supplemental Table S2.
Real-time PCR was performed to analyze 3 and 16 of the genes differentially expressed on 0.4% and 8% salt diets, respectively, as well as two genes not considered differentially expressed on the 0.4% salt diet. Several of the genes analyzed by qPCR, including Cdc73, Cdc2, Cenpf, E2f8, and Elk4, are involved in cell cycle regulation, a pathway we chose to focus on in the later part of the current study. Real-time PCR was done in the samples used in the Affymetrix Expression Array study as well as several additional mTAL samples. Nine of the 19 cases of differential expression were confirmed statistically (Supplemental Figure S1). Four cases [Il1b, Elk4, Dars2, Col1a1(LS)] approached statistical significance (P values between 0.05 and 0.1). The remaining 6 cases had P values greater than 0.1, but were still directionally consistent with the array data, although some of them showed smaller fold changes than the array data suggested.
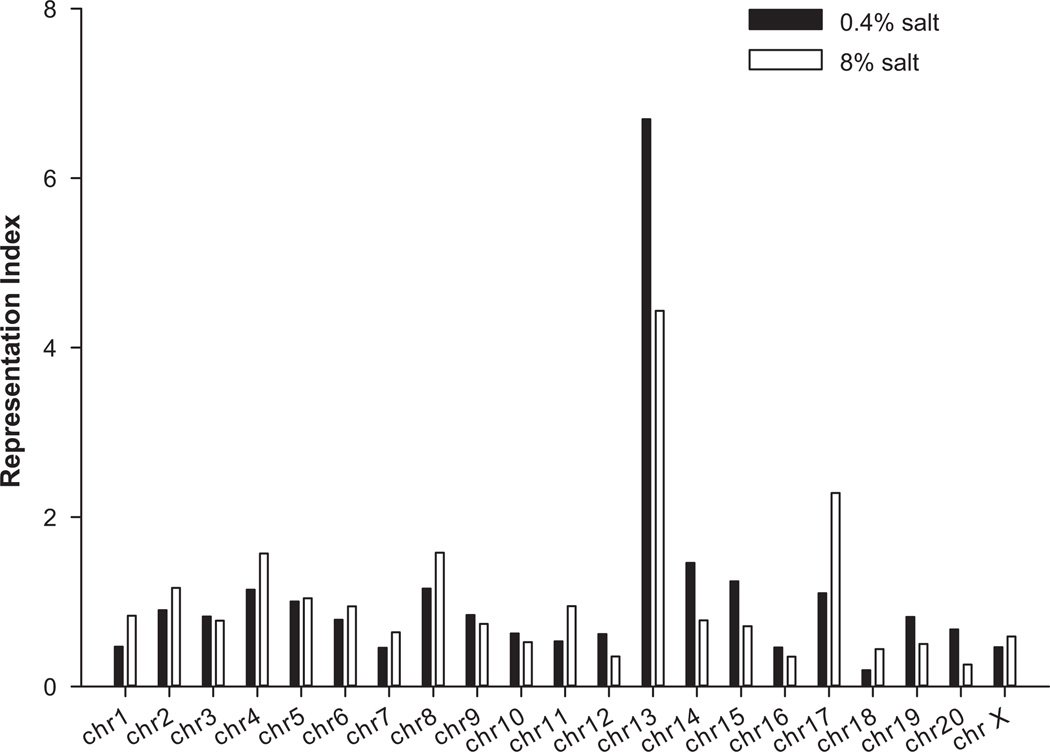
Genes differentially expressed in mTALs between SS and SS.13BN rats were highly enriched for genes located on chromosome 13, the chromosome substituted in SS.13BN (Figure 2). The representation index for genes on chromosome 13 was 6.7 and 4.4 on 0.4% and 8% salt diets, respectively, compared to an average of 1.1 for all chromosomes. Several of the differentially expressed genes located on chromosome 13 fall in the four non-overlapping congenic regions that we previously identified as having significant effects on hypertension in the SS rat (Supplemental Table S2) (Moreno 2007). The genomic boundaries of three of the four congenic regions were newly defined using a rat SNP array analysis (Supplemental Table S3). Several of the genes chosen for real-time PCR verification are located in these congenic regions. These included Elk4 and Slc45a3 in the line 5 region, Plekha6 and Golt1a in line 9, and Fam129a and Dars2 in line 26.
Figure 2. Genes differentially expressed in mTALs between SS and SS.13BN rats were enriched for genes located on chromosome 13.
See Supplemental Methods for an explanation of how “representation index” was calculated.
Ingenuity Pathway Analysis
Ingenuity Pathway Analysis of the genes differentially expressed between SS and SS.13BN rats identified several canonical pathways as highly represented. The top ten pathways identified at each salt diet condition were listed in Supplemental Table S4. The top five pathways identified on the 8% salt diet were hepatic fibrosis / hepatic stellate cell activation, mitotic roles of polo-like kinase, atherosclerosis signaling, cell cycle: G2/M DNA damage checkpoint regulation, and ataxia telangiectasia-mutated gene (ATM) signaling.
Note that the name of a canonical pathway may be uninformative or even misleading in some cases. For example, the “hepatic fibrosis” pathway includes elements of fibrosis in general and should not be considered irrelevant to the kidney tissue. Similarly, the “atherosclerosis signaling” pathway involves several elements of inflammation and fibrosis that are applicable to non-vascular tissues. The “ATM signaling” pathway is involved in the regulation of DNA repair, the cell cycle, and apoptosis.
Pathways and genes identified by the Bayesian model analysis
Ingenuity Pathway Analysis nominates pathways based on enrichment of differentially expressed genes. It does not take into account detailed patterns of changes in mRNA abundance or biological relationships known to exist among genes. Differential expression is typically determined by arbitrary criteria, the limitations of which are exacerbated by small number of replicates in a typical gene expression profiling study.
We therefore performed a Bayesian model analysis to identify biological pathways and genes that were best at discriminating the two rat strains. The Bayesian analysis was performed without a priori determination of differential expression. The intensity of gene expression was directly used in the model to nominate pathways and genes. Moreover, the Bayesian analysis considered known relationships among genes.
2,781 genes were included in the Bayesian model analysis. These genes were selected because they exhibited variations among samples and were members of at least one of the 244 pathways in the KEGG pathway database. Variations among samples were assessed by a K-means clustering algorithm31 (K=6) applied to the standard deviation of each gene among the samples. Table 1 shows the pathways with posterior probability greater than 0.5 in the comparison of SS and SS.13BN rats on the 8% NaCl diet. Also shown in Table 1 are member genes of a pathway that had posterior probability greater than 0.5 given the set of selected pathways and were ranked in the top 10 genes within the pathway. Pathways and genes identified on the 0.4% NaCl diet are shown in Supplemental Table S5. Note that the Bayesian analysis does not unequivocally exclude a pathway or a gene. The analysis ranks pathways or genes by their posterior probabilities, and the ranking is more informative than absolute values of posterior probability.
Table 1. Top pathways (posterior probability > 0.5) and top genes given pathways identified by Bayesian model analysis of data from rats fed the 8% NaCl diet.
The pathways and genes were identified as being best able to discriminate SS and SS.13BN rats fed the high-salt diet. See Methods for detail.
| pathway_id | pathway_names | Posterior Probability |
Top 10 genes with posterior probability > 0.5 and in order of ranking |
|---|---|---|---|
| hmr04144 | Endocytosis | 1.0000 | Igf1r, Pdgfra, Hras, Egfr, Met, Src, Cblb, Kdr, Kit, Flt1 |
| hmr04010 | MAPK signaling pathway | 1.0000 | Mapk1, Mapk3, Kras, Pdgfra, Nras, Hras, Egfr, Prkca, Prkcb, Mapk12 |
| hmr04080 | Neuroactive ligand-receptor interaction | 0.8848 | Grm2, Gabbr1 |
| hmr05200 | Pathways in cancer | 0.7966 | Pik3cd, Pik3r1, Pik3r2, Pdgfra, Mapk1, Kras, Pik3r3, Pik3ca, Igf1r, Mapk3 |
| hmr04062 | Chemokine signaling pathway | 0.6943 | Gnai2, Pik3cd, Pik3r2, Pik3ca, Pik3r1, Pik3r3, Mapk1, Mapk3, Gnai3, Gnai1 |
| hmr04060 | Cytokine-cytokine receptor interaction | 0.6521 | Pdgfra, Egfr, , Met, Kdr, Flt1, Kit, Csf1r, Il2rg, Vegfc, Flt4 |
| hmr04910 | Insulin signaling pathway | 0.6111 | Pik3cd, Pik3r2, Mapk1, Pik3r1, Pik3r3, Nras, Kras, Pik3ca, Pik3cb, Mapk3 |
| hmr04141 | Protein processing in endoplasmic reticulum | 0.5987 | Mapk10, Mapk9 |
| hmr03013 | RNA transport | 0.5522 | (none) |
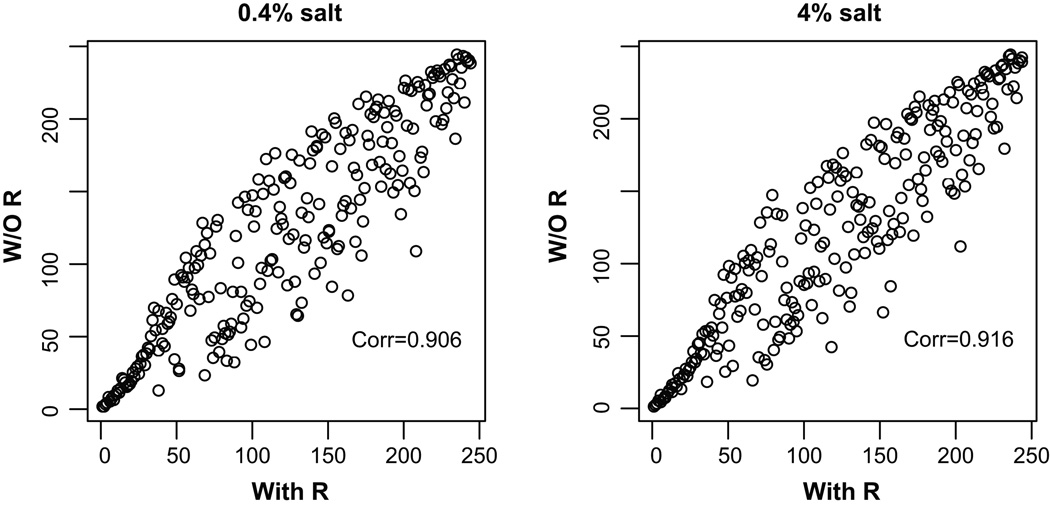
We assessed whether expression data and known relationships among genes (the R matrix) both contributed to the ranking of pathways. Pathways were ranked based on posterior probabilities obtained from analyses considering expression data only or both expression data and the R matrix. Analysis incorporating the R matrix generally yielded higher posterior probabilities. The assessment, however, focused on the ranking of pathways. The correlation of pathway rankings under the two analytical conditions was plotted in Figure 3. The spread of the plot (deviations from the line of identity) suggests that known relationships among genes contributed to the ranking. The high degree of correlation, on the other hand, suggests that expression data or other properties of the pathways, such as pathway size, contributed importantly to the ranking of pathways. As expected, there was a correlation between pathway size and posterior probability. However, there were also pathways of the same size that had very different posterior probabilities. Therefore, it appears that, although larger pathways had a higher probability to be selected, it was necessary that the data supported the selection.
Figure 3. Both existing knowledge of gene-gene relationships and the expression profile data contributed to the ranking of pathways in the Bayesian analysis.
Rankings of 244 pathways were plotted. A higher ranking indicates the pathway is more likely to discriminate the two rat strains. R, R matrix describing known relationships between genes.
Increased proliferative cells in mTALs of SS rats on the 8% salt diet
A “cell cycle” pathway was one of the most highly ranked pathways according to the Ingenuity Pathway Analysis of genes differentially expressed between SS and SS.13BN rats on the 8% salt diet (Supplemental Table S4). A pathway entitled “Pathways in cancer”, which is largely related to cell cycle and proliferation regulation, was one of the most highly ranked pathways according to the Bayesian model analysis (Table 1). Based upon this prediction, we performed studies to specifically examine the status of cell proliferation in mTALs in SS and SS.13BN rats.
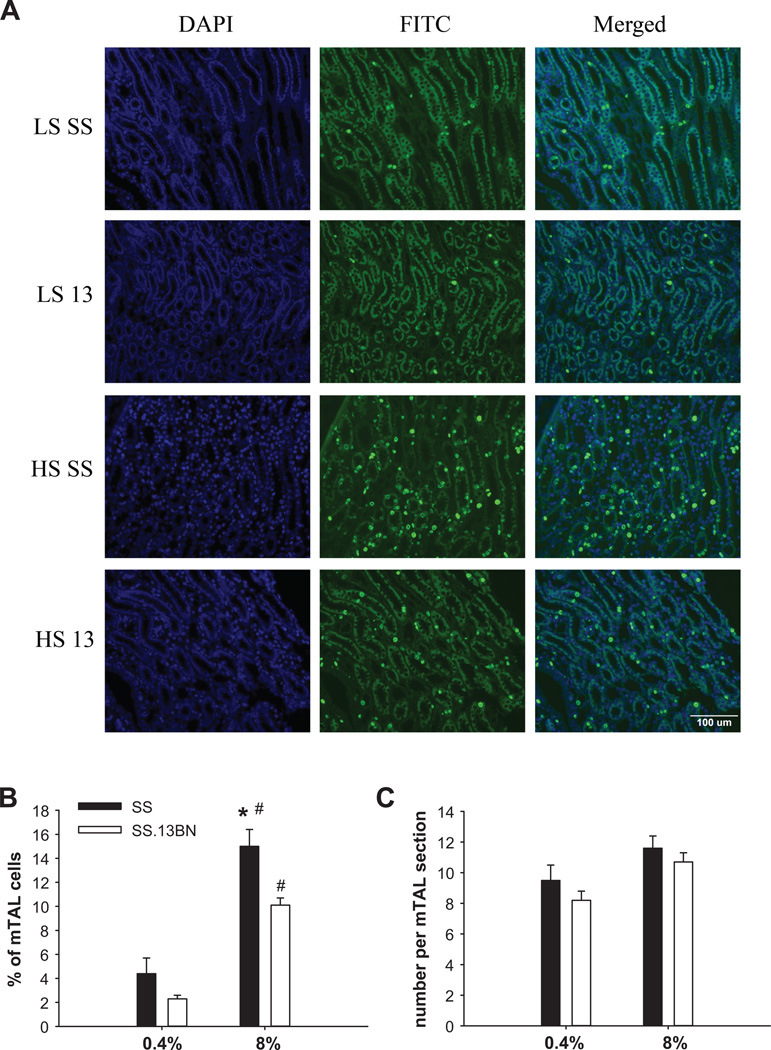
As shown in Figure 4, 2.3% to 4.4% of all mTAL cells were in a proliferative state (positive for Ki-67 staining) when the rats were on the 0.4% salt diet (not significantly different between SS and SS.13BN rats). Exposure to the 8% salt diet for 7 days substantially increased the number of proliferative mTAL cells in both strains. In SS rats on the 8% salt diet, 15.0% of mTAL cells analyzed were in a proliferative state, which was significantly higher than the 10.1% in SS.13BN rats (n=7–9, P<0.05) (Figure 4).
Figure 4. More mTAL cells were in a proliferative state in SS rats on the 8% salt diet than in SS.13BN rats.
LS, 0.4% NaCl diet; HS, 8% NaCl diet for 7 days; 13, SS.13BN rats. N=4 for LS groups, n=7–9 for HS groups, *, P<0.05 vs. SS.13BN on HS, #, P<0.05 vs. LS of the same strain.
Despite a higher proportion of cells that were in a proliferative state, SS rats did not have more total cells in each mTAL section analyzed compared to SS.13BN rats. The total number of cells per mTAL section analyzed was 11.6 in SS rats on the 8% salt diet, which was not significantly different from 10.7 cells per mTAL section in SS.13BN rats (n=7–9, Figure 4). The number of mTAL sections analyzed was lower in SS rats on the 8% NaCl diet than in SS.13BN rats (24.4 ± 1.3 vs. 33.0 ± 1.6 mTAL sections per field, n=7–9, P<0.05), likely because mTALs with overt damage, which were more prevalent in SS rats, were not included in the counting because it was difficult to obtain accurate cell counts from damaged segments.
The number of cells that were in the G2/M phases (positive for p-H3 staining) was very small in all treatment groups. The number tended to be higher in SS rats on the 8% salt diet (0.60 ± 0.20 cells/field) than in SS.13BN rats (0.28 ± 0.13 cells/field), but the difference did not reach statistical significance. The p-H3 staining procedure appeared to have worked properly as a significant number of cultured Hela cells stained positive (Supplemental Figure S2), although we could not directly compare staining in cultured cells and kidney sections.
In SS rats on the high-salt diet, 0.68% ± 0.36% of cells in morphologically normal mTAL segments stained positive in the TUNEL analysis of apoptosis, which tended to be higher than in SS.13BN rats (0.11% ± 0.05%), but the difference did not reach statistical significance. The proportions of positive cells were 0.23% ± 0.19% in SS rats and 0.15% ± 0.06% in SS.13BN rats on the 0.4% salt diet (not significantly different).
Discussion
The present study was the first to characterize the transcriptome in a specific nephron segment in a model of hypertension and one of the first to study the transcriptome in tissues highly enriched for a single cell type in hypertension. Transcriptome analyses have been reported in nephron segments isolated from humans or animal models under physiological conditions.32,33,34,35 The mTAL is particularly relevant to the SS rat model of hypertension. Impairment of pressure natriuresis in SS rats has been attributed to elevated NaCl reabsorption in the loop of Henle.19,25 Dopamine and nitric oxide inhibits Na+/K+-ATPase activity or chloride reabsorption in the thick ascending limb less effectively in SS rats than in Dahl salt-resistant rats.21,22 Production of 20-hydroxyeicosatetraenoic acid, which inhibits chloride transport in mTALs, is diminished in mTALs in SS rats.18,26 NKCC2, which mediates the bulk of NaCl reabsorption in the thick ascending limb, is up-regulated in SS rats and possibly in humans with elevated blood pressure salt-sensitivity.14,15,16,17 mTALs contribute to increased production of reactive oxygen species in SS rats, which may contribute to the impairment of medullary blood flow regulation that is important for pressure natriuresis.20,23,24
In the differentially expressed genes involved in the identified Ingenuity Canonical Pathways, Cxcr4, F5, Fcgr3a, Pla2g4a, and Ptgs2 are located on rat chr. 13. These genes are involved in inflammation, fibrosis, and arachidonic acid metabolism, which are known to contribute to or exacerbate the development of hypertension. Much more work, however, would need to be done before any causal contribution of these genes to hypertension could be established. Importantly, differentially expressed genes not mapped to the consomic chromosome could still be highly relevant to the hypertension phenotype. It is unlikely that causal genes located on the consomic chromosome are the only genes involved in the development and progression of the hypertension phenotype. Instead, it is likely that the causal genes contribute to the hypertension phenotype by influencing, directly or indirectly, biological pathways involving genes located on other chromosomes, forming a tree-like regulatory network that we proposed previously.11
We previously performed a transcriptome analysis of the renal outer medulla, using the same rat strains, dietary conditions, and array platform as the current study of mTALs.8 Several inflammation-related pathways were identified as highly represented in the differentially expressed genes by Ingenuity Pathway Analysis in both studies. Some pathways, however, were identified in only one of the two studies. For example, β-adrenergic signaling and nitric oxide signaling pathways were identified only in the outer medulla study, while pathways related to fibrosis and cell cycle regulation were identified only in the mTAL study.
A novel and unexpected finding of the current study was that SS rats fed the high-salt diet had more mTAL cells that were in a proliferative state, compared to SS.13BN rats. Moreover, the high-salt diet increased proliferative mTAL cells by several fold in both strains of rats. The vast majority of terminally differentiated cells in the kidney, including mTAL cells, are in the quiescent state of G0. Proximal tubular cells can re-enter the cell cycle following massive cell death that occurs in conditions such as severe ischemic-reperfusion injury.36 But even in ischemic-reperfusion injury, mTALs are largely spared. It is, therefore, rather unexpected that we found increased proliferative mTAL cells in rats fed the high-salt diet, especially SS rats. Interestingly, alterations of cell cycle regulation in the kidneys of SS rats were suggested not only by the current mTAL transcriptome analysis but also by our previous transcriptome analysis of the renal cortex.8 The cell cycle regulation pathway, however, was not identified in the analysis of homogenized renal outer medulla tissue.8 In the current mTAL study, the cell cycle regulation pathway was prominent in strain comparisons under both 0.4% and 8% salt conditions according to the Bayesian analysis and the top 10 ranking genes largely overlapped. Yet SS rats on the 0.4% salt diet did not show a significant difference in mTAL proliferative states compared to SS.13BN rats. The ranking of the top genes differs between the two dietary salt conditions. Expression patterns for some of the specific genes also differ between the two conditions. In addition, Ingenuity Pathway Analysis identified cell cycle related pathways from the high-salt dataset only.
It remains to be determined what mechanisms cause the increase in proliferative mTAL cells in SS rats on the high-salt diet. Angiotensin II, at doses that do not increase proliferation of proximal tubular cells, stimulates proliferation of a murine mTAL cell line via AT1 receptors.37 The SS rat is a low-renin model of hypertension. Yet intra-renal levels of angiotensin II have been shown to be abnormally high in SS rats fed a high-salt diet.38,39 In addition, reactive oxygen species at moderate concentrations can stimulate cell proliferation.40,41 The renal medulla of SS rats is known to have higher levels of reactive oxygen species including superoxide and H2O2 compared to SS.13BN rats, which contributes to the development of hypertension in SS rats.42,43
The nature of the cell cycle alteration that we observed and their functional consequences remain to be investigated. Nevertheless, the current study has provided several interesting clues. Despite increased proliferative mTAL cells in SS rats, total cells per mTAL section were not significantly different between SS and SS.13BN rats. In fact, total cells per mTAL section were not significantly different between rats on the two salt diets, even though proliferative mTAL cells increased by several fold in rats fed the high-salt diet. There are two possible explanations. One, mTAL cells were proliferating to replace lost cells. Two, the proliferative mTAL cells had exited G0, but were not able to progress through the cell cycle to complete cell division. SS rats fed the high-salt diet had more mTALs that were overtly damaged and filled with casts, as one would expect. However, we only counted cells in mTALs that appeared normal because cells in damaged mTALs were distorted, making it difficult to obtain accurate counts. Therefore, it appears that the more likely scenario is that proliferative mTAL cells were arrested at some point of the cell cycle.
G2/M arrest of proximal tubular cells following injury contributes to the development of tubulointerstitial fibrosis.36 Tubulointerstitial fibrosis in the outer medullary region is a pathological hallmark of SS rats and is significantly attenuated in SS.13BN rats.4,9 It would be consistent with the fibrotic phenotype if proliferative mTAL cells in SS rats were arrested in G2/M. SS rats on the high-salt diet indeed tended to have more mTAL cells that were in G2/M phases. However, very few mTAL cells overall were in G2/M phases in either strain of rats on either salt diet. It suggests that the proliferative mTAL cells that we observed might be in G1 or S phase. G1/S arrest can be a response to mild DNA damage, allowing DNA repair to take place.44,45 Failure of repair could lead to apoptosis. Apoptosis appears to increase in the injured kidneys of SS rats,46 although we were not able to detect a significant difference in apoptotic cells in morphologically normal mTALs between SS and SS.13BN rats. A pro-inflammatory milieu, which exists in the kidneys of SS rats,38,47 could also cause G1/S arrest.48
It remains to be determined if the signal that stimulates mTAL cell proliferation in SS rats is the same signal that prevents mTAL cells from completing the cell cycle. Cellular signals for proliferation and G1/S arrest are often distinct. For example, reactive oxygen species can stimulate proliferation, while anti-oxidant treatments can lead to late-G1 arrest.40,41,49 In some cases, however, a cellular signal can cause proliferation followed by G1/S arrest (Shimi 2011).50
Perspectives
The study provides genome-wide insights into the mechanism of hypertension at the level of a specific nephron segment. The finding of abnormalities in cell proliferation in mTALs may stimulate a new direction of research of hypertension or hypertensive renal injury.
Supplementary Material
NOVELTY AND SIGNIFICANCE (per journal requirement).
What is new?
Novel and unexpected findings on changes in cell proliferation in mTALs.
One of the first to examine the transcriptome in tissues highly enriched for a single cell type in hypertension.
Application of a Bayesian method to hypertension research to identify biological pathways independent of determination of differential expression.
What is relevant?
The study provides genome-wide insights into the mechanism of hypertension at the level of a specific nephron segment.
Summary
We identified several biological pathways in the mTAL that might contribute to hypertension or hypertensive renal injury and provided novel evidence for alterations in cell proliferation in mTALs in SS rats.
Acknowledgments
Sources of Funding
This work was supported by NIH grant HL082798 and Advancing a Healthier Wisconsin Fund FP1701. We thank Glenn Slocum for assistance with microscopy and image analysis, and Dr. Martin Hessner, Mary Kaldunski and Shuang Jia for assistance with microarray equipment and rank-product analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
References
- 1.Joe B, Letwin NE, Garrett MR, Dhindaw S, Frank B, Sultana R, Verratti K, Rapp JP, Lee NH. Transcriptional profiling with a blood pressure QTL interval-specific oligonucleotide array. Physiol Genomics. 2005;23:318–326. doi: 10.1152/physiolgenomics.00164.2004. [DOI] [PubMed] [Google Scholar]
- 2.Malek RL, Wang HY, Kwitek AE, Greene AS, Bhagabati N, Borchardt G, Cahill L, Currier T, Frank B, Fu X, Hasinoff M, Howe E, Letwin N, Luu TV, Saeed A, Sajadi H, Salzberg SL, Sultana R, Thiagarajan M, Tsai J, Veratti K, White J, Quackenbush J, Jacob HJ, Lee NH. Physiogenomic resources for rat models of heart, lung and blood disorders. Nat Genet. 2006;38:234–239. doi: 10.1038/ng1693. [DOI] [PubMed] [Google Scholar]
- 3.Marques FZ, Campain AE, Yang YH, Morris BJ. Meta-analysis of genome-wide gene expression differences in onset and maintenance phases of genetic hypertension. Hypertension. 2010;56:319–324. doi: 10.1161/HYPERTENSIONAHA.110.155366. [DOI] [PubMed] [Google Scholar]
- 4.Cowley AW, Roman RJ, Kaldunski ML, Dumas P, Dickhout JG, Greene AS, Jacob HJ., Jr Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension. 2001;37:456–461. doi: 10.1161/01.hyp.37.2.456. [DOI] [PubMed] [Google Scholar]
- 5.Liang M, Yuan B, Rute E, Greene AS, Zou AP, Soares P, MCQuestion GD, Slocum GR, Jacob HJ, Cowley AW., Jr Renal medullary genes in salt-sensitive hypertension: a chromosomal substitution and cDNA microarray study. Physiol Genomics. 2002;8:139–149. doi: 10.1152/physiolgenomics.00083.2001. [DOI] [PubMed] [Google Scholar]
- 6.Liang M, Yuan B, Rute E, Greene AS, Olivier M, Cowley AW., Jr Insights into Dahl salt-sensitive hypertension revealed by temporal patterns of renal medullary gene expression. Physiol Genomics. 2003;12:229–237. doi: 10.1152/physiolgenomics.00089.2002. [DOI] [PubMed] [Google Scholar]
- 7.Liang M, Lee NH, Wang H, Greene AS, Kwitek AE, Kaldunski ML, Luu TV, Frank BC, Bugenhagen S, Jacob HJ, Cowley AW., Jr Molecular networks in Dahl salt-sensitive hypertension based on transcriptome analysis of a panel of consomic rats. Physiol Genomics. 2008;34:54–64. doi: 10.1152/physiolgenomics.00031.2008. [DOI] [PubMed] [Google Scholar]
- 8.Lu L, Li P, Yang C, Kurth T, Misale M, Skelton M, Moreno C, Roman RJ, Greene AS, Jacob HJ, Lazar J, Liang M, Cowley AW., Jr. Dynamic convergence and divergence of renal genomic and biological pathways in protection from Dahl salt-sensitive hypertension. Physiol Genomics. 2010;41:63–70. doi: 10.1152/physiolgenomics.00170.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori T, Polichnowski A, Glocka P, Kaldunski M, Ohsaki Y, Liang M, Cowley AW., Jr High perfusion pressure accelerates renal injury in salt-sensitive hypertension. J Am Soc Nephrol. 2008;19:1472–1482. doi: 10.1681/ASN.2007121271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian Z, Greene AS, Usa K, Matus IR, Bauwens J, Pietrusz JL, Cowley AW, Liang M., Jr Renal regional proteomes in young Dahl salt-sensitive rats. Hypertension. 2008;51:899–904. doi: 10.1161/HYPERTENSIONAHA.107.109173. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Singh RJ, Usa K, Netzel BC, Liang M. Renal medullary 11 beta-hydroxysteroid dehydrogenase type 1 in Dahl salt-sensitive hypertension. Physiol Genomics. 2008;36:52–58. doi: 10.1152/physiolgenomics.90283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian Z, Liu Y, Usa K, Mladinov D, Fang Y, Ding X, Greene AS, Cowley AW, Liang M., Jr Novel role of fumarate metabolism in dahl-salt sensitive hypertension. Hypertension. 2009;54:255–260. doi: 10.1161/HYPERTENSIONAHA.109.129528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang M. Hypertension as a mitochondrial and metabolic disease. Kidney Int. 2011;80:15–16. doi: 10.1038/ki.2011.84. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez-Guerra M, Garay RP. Renal Na-K-Cl cotransporter NKCC2 in Dahl salt-sensitive rats. J Hypertens. 2002;20:721–727. doi: 10.1097/00004872-200204000-00031. [DOI] [PubMed] [Google Scholar]
- 15.Aviv A, Hollenberg NK, Weder A. Urinary potassium excretion and sodium sensitivity in blacks. Hypertension. 2004;43:707–713. doi: 10.1161/01.HYP.0000120155.48024.6f. [DOI] [PubMed] [Google Scholar]
- 16.Haque MZ, Ares GR, Caceres PS, Ortiz PA. High salt differentially regulates surface NKCC2 expression in thick ascending limbs of Dahl salt-sensitive and salt-resistant rats. Am J Physiol Renal Physiol. 2011;300:F1096–F1104. doi: 10.1152/ajprenal.00600.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoagland KM, Flasch AK, Dahly-Vernon AJ, dos Santos EA, Knepper MA, Roman RJ. Elevated BSC-1 and ROMK expression in Dahl salt-sensitive rat kidneys. Hypertension. 2004;43:860–865. doi: 10.1161/01.HYP.0000120123.44945.47. [DOI] [PubMed] [Google Scholar]
- 18.Ito O, Roman RJ. Role of 20-HETE in elevating chloride transport in the thick ascending limb of Dahl SS/Jr rats. Hypertension. 1999;33:419–423. doi: 10.1161/01.hyp.33.1.419. [DOI] [PubMed] [Google Scholar]
- 19.Kirchner KA. Greater loop chloride uptake contributes to blunted pressure natriuresis in Dahl salt sensitive rats. J Am Soc Nephrol. 1990;1:180–186. doi: 10.1681/ASN.V12180. [DOI] [PubMed] [Google Scholar]
- 20.Mori T, O’Connor PM, Abe M, Cowley AW., Jr Enhanced superoxide production in renal outer medulla of Dahl salt-sensitive rats reduces nitric oxide tubular-vascular cross-talk. Hypertension. 2007;49:1336–1341. doi: 10.1161/HYPERTENSIONAHA.106.085811. [DOI] [PubMed] [Google Scholar]
- 21.Nishi A, Eklöf AC, Bertorello AM, Aperia A. Dopamine regulation of renal Na+,K(+)-ATPase activity is lacking in Dahl salt-sensitive rats. Hypertension. 1993;21:767–771. doi: 10.1161/01.hyp.21.6.767. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz PA, Garvin JL. Intrarenal transport and vasoactive substances in hypertension. Hypertension. 2001;38:621–624. doi: 10.1161/hy09t1.093361. [DOI] [PubMed] [Google Scholar]
- 23.O’Connor PM, Lu L, Liang M, Cowley AW., Jr A novel amiloride-sensitive h+ transport pathway mediates enhanced superoxide production in thick ascending limb of salt-sensitive rats, not na+/h+ exchange. Hypertension. 2009;54:248–254. doi: 10.1161/HYPERTENSIONAHA.109.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connor PM, Lu L, Schreck C, Cowley AW., Jr Enhanced amiloride-sensitive superoxide production in renal medullary thick ascending limb of Dahl salt-sensitive rats. Am J Physiol Renal Physiol. 2008;295:F726–F733. doi: 10.1152/ajprenal.00137.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roman RJ, Kaldunski ML. Enhanced chloride reabsorption in the loop of Henle in Dahl salt-sensitive rats. Hypertension. 1991;17:1018–1024. doi: 10.1161/01.hyp.17.6.1018. [DOI] [PubMed] [Google Scholar]
- 26.Zou AP, Drummond HA, Roman RJ. Role of 20-HETE in elevating loop chloride reabsorption in Dahl SS/Jr rats. Hypertension. 1996;27:631–635. doi: 10.1161/01.hyp.27.3.631. [DOI] [PubMed] [Google Scholar]
- 27.Stingo FC, Chen YA, Tadesse MG, Vannucci M. Incorporating Biological Information into Linear Models: A Bayesian Approach to the Selection of Pathways and Genes. Annals of Applied Statistics. 2011;5:1978–2002. doi: 10.1214/11-AOAS463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinh-Trang-Tan MM, Bouby N, Coutaud C, Bankir L. Quick isolation of rat medullary thick ascending limbs. Enzymatic and metabolic characterization. Pflugers Arch. 1986;407:228–234. doi: 10.1007/BF00580681. [DOI] [PubMed] [Google Scholar]
- 29.Liang M, Pietrusz JL. Thiol-related genes in diabetic complications: a novel protective role for endogenous thioredoxin 2. Arterioscler Thromb Vasc Biol. 2007;27:77–83. doi: 10.1161/01.ATV.0000251006.54632.bb. [DOI] [PubMed] [Google Scholar]
- 30.Moreno C, Kaldunski ML, Wang T, Roman RJ, Greene AS, Lazar J, Jacob HJ, Cowley AW., Jr Multiple blood pressure loci on rat chromosome 13 attenuate development of hypertension in the Dahl S hypertensive rat. Physiol Genomics. 2007;31:228–235. doi: 10.1152/physiolgenomics.00280.2006. [DOI] [PubMed] [Google Scholar]
- 31.Hartigan JA, Wong MA. A K-means clustering algorithm. Applied Statistics. 1979;28:100–108. [Google Scholar]
- 32.Cheval L, Pierrat F, Dossat C, Genete M, Imbert-Teboul M, Duong Van Huyen JP, Poulain J, Wincker P, Weissenbach J, Piquemal D, Doucet A. Atlas of gene expression in the mouse kidney: new features of glomerular parietal cells. Physiol Genomics. 2011;43:161–173. doi: 10.1152/physiolgenomics.00093.2010. [DOI] [PubMed] [Google Scholar]
- 33.Takenaka M, Imai E, Nagasawa Y, Matsuoka Y, Moriyama T, Kaneko T, Hori M, Kawamoto S, Okubo K. Gene expression profiles of the collecting duct in the mouse renal inner medulla. Kidney Int. 2000;57:19–24. doi: 10.1046/j.1523-1755.2000.00834.x. [DOI] [PubMed] [Google Scholar]
- 34.Virlon B, Cheval L, Buhler JM, Billon E, Doucet A, Elalouf JM. Serial microanalysis of renal transcriptomes. Proc Natl Acad Sci U S A. 1999;96:15286–15291. doi: 10.1073/pnas.96.26.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu MJ, Miller RL, Uawithya P, Rinschen MM, Khositseth S, Braucht DW, Chou CL, Pisitkun T, Nelson RD, Knepper MA. Systems-level analysis of cell-specific AQP2 gene expression in renal collecting duct. Proc Natl Acad Sci U S A. 2009;106:2441–2446. doi: 10.1073/pnas.0813002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Besschetnova TY, Brooks CR, Shah JV, Bonventre JV. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat Med. 2010;16:535–543. doi: 10.1038/nm.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf G, Ziyadeh FN, Helmchen U, Zahner G, Schroeder R, Stahl RA. ANG II is a mitogen for a murine cell line isolated from medullary thick ascending limb of Henle’s loop. Am J Physiol. 1995;268:F940–F947. doi: 10.1152/ajprenal.1995.268.5.F940. [DOI] [PubMed] [Google Scholar]
- 38.De Miguel C, Das S, Lund H, Mattson DL. T lymphocytes mediate hypertension and kidney damage in Dahl salt-sensitive rats. Am J Physiol Regul Integr Comp Physiol. 2010;298:R1136–R1142. doi: 10.1152/ajpregu.00298.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobori H, Nishiyama A, Abe Y, Navar LG. Enhancement of intrarenal angiotensinogen in Dahl salt-sensitive rats on high salt diet. Hypertension. 2003;41:592–597. doi: 10.1161/01.HYP.0000056768.03657.B4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 41.Klein JA, Ackerman SL. Oxidative stress, cell cycle, neurodegeneration. J Clin Invest. 2003;111:785–793. doi: 10.1172/JCI18182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng D, Yang C, Geurts AM, Kurth T, Liang M, Lazar J, Mattson DL, O’Connor PM, Cowley AW., Jr Increased expression of NAD(P)H oxidase subunit p67(phox) in the renal medulla contributes to excess oxidative stress and salt-sensitive hypertension. Cell Metab. 2012;15:201–208. doi: 10.1016/j.cmet.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor NE, Glocka P, Liang M, Cowley AW., Jr NADPH oxidase in the renal medulla causes oxidative stress and contributes to salt-sensitive hypertension in Dahl S rats. Hypertension. 2006;47:692–698. doi: 10.1161/01.HYP.0000203161.02046.8d. [DOI] [PubMed] [Google Scholar]
- 44.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tentner AR, Lee MJ, Ostheimer GJ, Samson LD, Lauffenburger DA, Yaffe MB. Combined experimental and computational analysis of DNA damage signaling reveals context-dependent roles for Erk in apoptosis and G1/S arrest after genotoxic stress. Mol Syst Biol. 2012;8:568. doi: 10.1038/msb.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ying WZ, Wang PX, Sanders PW. Induction of apoptosis during development of hypertensive nephrosclerosis. Kidney Int. 2000;58:2007–2017. doi: 10.1111/j.1523-1755.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- 47.Mattson DL, James L, Berdan EA, Meister CJ. Immune suppression attenuates hypertension and renal disease in the Dahl salt-sensitive rat. Hypertension. 2006;48:149–156. doi: 10.1161/01.HYP.0000228320.23697.29. [DOI] [PubMed] [Google Scholar]
- 48.Penzo M, Massa PE, Olivotto E, Bianchi F, Borzi RM, Hanidu A, Li X, Li J, Marcu KB. Sustained NF-kappaB activation produces a short-term cell proliferation block in conjunction with repressing effectors of cell cycle progression controlled by E2F or FoxM1. J Cell Physiol. 2009;218:215–227. doi: 10.1002/jcp.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Havens CG, Ho A, Yoshioka N, Dowdy SF. Regulation of late G1/S phase transition and APC Cdh1 by reactive oxygen species. Mol Cell Biol. 2006;26:4701–4711. doi: 10.1128/MCB.00303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25:2579–2593. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.