The seasonal cue day length regulates the timing of the floral transition in plants through periodic histone modifications of the FT gene, which encodes a flowering signal in plants. These modifications dampen FT expression at dusk to prevent precocious flowering.
Abstract
The developmental transition from a vegetative to a reproductive phase (i.e., flowering) is timed by the seasonal cue day length or photoperiod in many plant species. Through the photoperiod pathway, inductive day lengths trigger the production of a systemic flowering signal, florigen, to provoke the floral transition. FLOWERING LOCUS T (FT), widely conserved in angiosperms, is a major component of the mobile florigen. In the long-day plant Arabidopsis, FT expression is rhythmically activated by the output of the photoperiod pathway CONSTANS (CO), specifically at the end of long days. How FT expression is modulated at an adequate level in response to the long-day cue to set a proper flowering time remains unknown. Here, we report a periodic histone deacetylation mechanism for the photoperiodic modulation of FT expression. We have identified a plant-unique core structural component of an Arabidopsis histone deacetylase (HDAC) complex. In long days, this component accumulates at dusk, and is recruited by a MADS-domain transcription factor to the FT locus specifically at the end of the day, leading to periodic histone deacetylation of FT chromatin at dusk. Furthermore, we found that at the end of long days CO activity not only activates FT expression but also enables HDAC-activity recruitment to FT chromatin to dampen the level of FT expression, and so prevent precocious flowering in response to the inductive long-day cue. These results collectively reveal a periodic histone deacetylation mechanism for the day-length control of flowering time in higher plants.
Author Summary
The timing of the developmental transition from a vegetative to a reproductive phase is critically important for reproductive success in flowering plants. Plants synchronize the timing of their floral transition with the changing seasons to flower at a suitable time. The change in day length, or photoperiod, is a key seasonal cue, especially at high latitudes. It is through the photoperiod pathway that day lengths trigger the production of a systemic flowering signal, florigen, to induce the transition from vegetative to reproductive growth and flowering. The FT protein is a major component of the mobile florigen signal. In the model flowering plant Arabidopsis, FT mRNA expression is rhythmically activated by the gene CONSTANS, which is the output of the photoperiod pathway, specifically at the end of long days. In this study, we aimed to address how the level of FT expression is modulated in response to the long-day cue to set the appropriate flowering time. We show that on long days a transcription factor recruits histone deacetylase activity to remove acetyl marks from histones at the FT gene specifically at dusk, thereby dampening FT mRNA expression upon its transcriptional activation by CONSTANS. This sets the correct level of FT expression at the end of long days, and thus prevents precocious flowering in response to the long-day cue.
Introduction
The timing of the developmental transition from a vegetative to a reproductive phase is critical for reproductive success in flowering plants. Plants synchronize their timings of floral transition with changing seasons to flower at a suitable time. The change in day length or photoperiod is a key seasonal cue, especially at high latitudes, and is perceived in leaves, leading to the production of florigen in leaf vasculature [1],[2]. Florigen, a systemic signal, is transmitted through phloem from leaf to the shoot apical meristem (SAM), leading to flower formation [1]–[3]. Many plant species respond to day length changes through photoperiod pathways. According to their photoperiodic responses, plants are classified into long-day plants, i.e., flowering occurring only when the day becomes longer than a threshold length or accelerates when the day length increases, short-day plants whose flowering induced upon the day getting shorter, and day-neutral plants [4].
FLOWERING LOCUS T (FT), first identified in Arabidopsis thaliana [5],[6], and FT homologs in other species are the major component of the mobile florigen [7]–[10]. In Arabidopsis, a facultative long-day plant that flowers rapidly under inductive long days, the output of the photoperiod pathway CONSTANS (CO) activates FT expression in leaf veins [2],[11]. The circadian clock sets a high CO mRNA expression in the late afternoon in long days, which coincides with light exposure resulting in CO protein accumulation towards the day's end, because light stabilizes the CO protein [2],[4]. The vasculature-expressed CO protein promotes FT expression activation in the phloem companion cells specifically at the end of long days, resulting in rhythmic FT expression [2],[4]. During night, CO is rapidly degraded by proteasomes and FT expression is repressed. Upon its production in dusk, the FT protein moves from phloem to SAM to induce flowering. FT and its function as florigen are conserved in the flowering plants so far examined; the level of FT expression at the end of long days in Arabidopsis plays a primary determining role for when a plant to flower [1],[2]. To date, beside CO, other factors (if any) that directly regulate FT expression specifically at dusk remain to be identified.
Histone acetylation and de-acetylation can regulate chromatin structure and gene expression. Histone acetyltransferases (HATs) add acetyl groups to core histone tails and typically function to promote transcription of target loci, whereas histone deacetylases (HDACs) remove acetyl groups and are often linked with transcriptional repression [12]. Recent studies of genome-wide binding sites of mammalian HATs and HDACs reveal that both are targeted to actively transcribed genes to control their expression [13]. In addition, rapid and synchronous recruitment of HATs and HDACs to actively transcribed loci have been observed in yeast as well [14],[15]. At these loci HATs act typically to promote transcription, whereas HDACs deacetylate acetylated histones to maintain an adequate level of acetylation and/or reset the acetylation state of a target locus following transcription [13]–[15].
Histone acetylation is involved in plant gene regulation. In Arabidopsis, functional disruption of HATs such as GCN5 and AtHAC1, or HDACs including HDA6 and HDA19, leads to pleiotropic abnormalities in growth and development [16]–[19]. For instance, loss of AtHAC1 or HDA6 function causes upregulation of a potent floral repressor FLOWERING LOCUS C (FLC), resulting in a delay in flowering [17]–[19]. Certain HATs (e.g., GCN5) and HDACs (e.g., HDA6) are involved in genome-wide regulation of gene expression, as revealed by transcript profiling [16],[18], whereas others may control the expression of only a subset of loci. How a HAT or HDAC is recruited to its target loci remains elusive.
HDACs such as the reduced potassium dependency-3 (RPD3) type are often found in multiprotein co-repressor complexes, among which the yeast Sin3-HDAC complex has been well characterized [20],[21]. The core components of this complex typically include the master scaffold protein Sin3, the RPD3 HDAC, and several Sin3-associated structural components including Sin3-Associated Polypeptide 18 (SAP18) and Sin3-Associated Polypeptide 30 (SAP30); homologs of these components are also found in a mammalian Sin3-HDAC like complex [20]. This complex does not bind to DNA directly and is often recruited to specific loci through association with DNA-binding proteins [21]. In Arabidopsis, there are six Sin3 homologs including SNL1–SNL6 (SNL for SIN3-LIKE), and four close RPD3 homologs including HDA19, HDA9, HDA7, and HDA6 [22],[23]. In addition, there is only one SAP18 homolog in Arabidopsis, AtSAP18, that has been shown to play a role in salt stress response and floral organ formation [24],[25]. The Arabidopsis genome does not encode an apparent homolog of the yeast SAP30; hence, whether there are Sin3-RPD3 like co-repressor complexes for histone deacetylation in Arabidopsis remains to be addressed.
Here we report that in Arabidopsis there are two functional relatives of the yeast SAP30, named as SAP30 FUNCTION-RELATED 1 (AFR1) and SAP30 FUNCTION-RELATED 2 (AFR2), acting as part of HDAC complexes (AFR1-HDAC or AFR2-HDAC) to modulate the acetylation level of FT chromatin upon FT activation at the end of long days (LDs) (16-h light/8-h dark). Both AFR1 and AFR2 proteins accumulate at the end of LDs, and bind to FT chromatin at dusk, but not in the middle of the day. Moreover, we found that the MADS-domain transcription factor AGAMOUS LIKE 18 (AGL18) directly interacts with AFR1 and AFR2 and recruits these proteins and presumably AFR1/AFR2-HDAC to FT chromatin specifically at the end of LDs leading to histone deacetylation upon FT activation. The output of the photoperiod pathway CO at the end of LDs not only activates FT expression, but also enables the recruitment of AFRs to the FT locus to dampen FT expression and set it at an adequate level, preventing precocious flowering in response to the day length cue in Arabidopsis.
Results
Two Arabidopsis Functional Relatives of the Yeast SAP30 (ScSAP30) Associate with Histone Deacetylases to Form HDAC Complexes
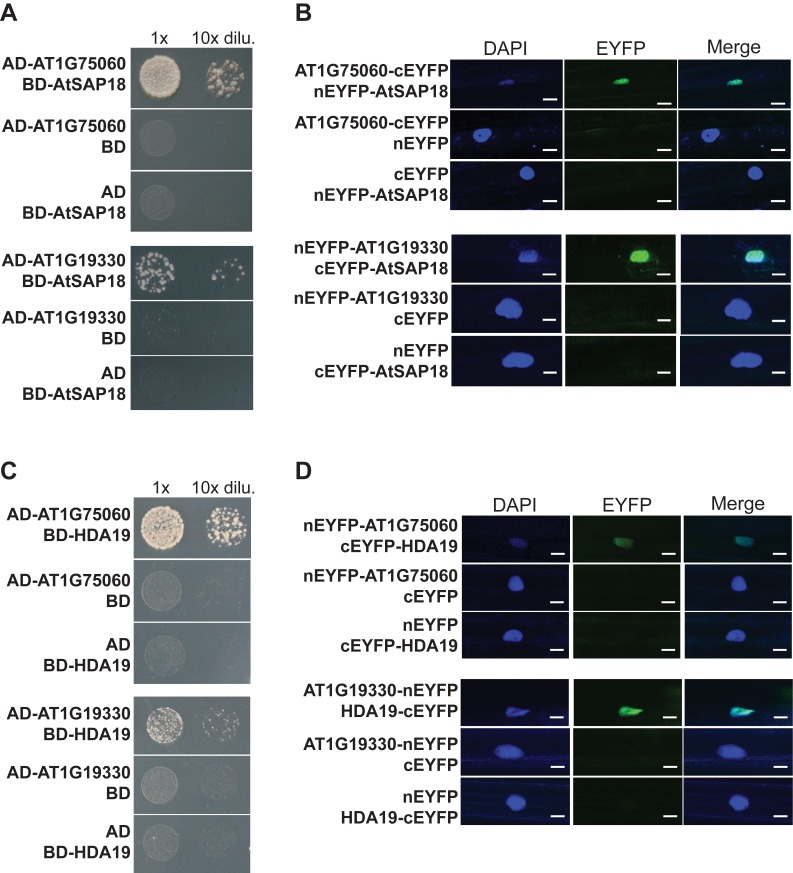
In an effort to explore whether a Sin3-SAP30-RPD3 co-repressor-like complex in Arabidopsis exists, we first examined whether Arabidopsis has any functional relatives of ScSAP30 because it lacks of an apparent homolog of the full-length ScSAP30. In the C-terminal region of ScSAP30 there is a domain important for its function: the 30-amino acid Sin3-binding region (SBR) [26]. We used C-SAP30 to BLAST the Arabidopsis protein database and found that there are nine proteins containing a region with a similarity to SBR, among which AT1G75060 and AT1G19330 are the top two hits (Figure S1A; unpublished data). We reasoned that if these two proteins are functional relatives of SAP30, they would directly associate with SNLs, AtSAP18, and/or RPD3-type HDACs. Yeast two-hybrid assays were performed to check direct interactions of AT1G75060 or AT1G19330 with SNL2 (SIN3-LIKE 2), AtSAP18, HDA9, and/or HDA19. Indeed, these proteins directly interacted with AT1G75060 and AT1G19330 (Figures 1A, 1C, and S2).
Figure 1. Direct interactions of AT1G75060 (AFR1) and AT1G19330 (AFR2) with AtSAP18 and HDA19 proteins.
(A) Interactions of AtSAP18 with AT1G75060 and AT1G19330 in yeast. The indicated proteins of full-length were fused with the GAL4-BD or AD domain. Yeast cells harboring the fusion proteins, BD and/or AD (as indicated), were grown on selective synthetic defined media lacking of Trp, Leu, and His. (B) BiFC analysis of the interactions of AtSAP18 with AT1G75060 and AT1G19330 in onion epidermal cells. Onion epidermal cells were co-transformed transiently by a pair of plasmid, as indicated, via biolistic gene bombardment. Yellowish-green signals indicate physical associations of paired proteins in the nuclei. Blue fluorescence from a DAPI (4′,6-diamidino-2-phenylindole) staining indicates a nucleus. Bar = 20 µm. (C) Interactions of HDA19 with AT1G75060 and AT1G19330 in yeast. The indicated full-length proteins were fused with the GAL4-BD or AD domain. Yeast cells were grown on selective synthetic defined media lacking of Trp, Leu, and His. (D) BiFC analysis of the interactions of HDA19 with AT1G75060 and AT1G19330 in onion epidermal cells. Bar = 20 µm.
Next, to confirm these interactions in plant cells, we conducted bimolecular fluorescence complementation (BiFC) assays in which non-fluorescent N-terminal and C-terminal fragments of enhanced yellow fluorescent protein (EYFP) were fused to the full-length AT1G75060, AT1G19330, AtSAP18, or HDA19 proteins. When AT1G75060-cEYFP and nEYFP-AtSAP18 were simultaneously expressed in onion epidermal cells, in the nuclei fluorescence was observed (Figure 1B), demonstrating a direct interaction of AT1G75060 with AtSAP18. Similarly, we also examined and observed direct interactions of AT1G19330 with AtSAP18, AT1G19330 with HDA19, and AT1G75060 with HDA19 in the nuclei of onion cells (Figure 1B and 1D). These results suggest that both AT1G75060 and AT1G19330 function like ScSAP30, namely, as an integral component of Sin3-RPD3 co-repressor like complexes, although there is little similarity of amino acid sequence between these proteins and ScSAP30 outside of the C-terminal SBR (Figure S1A). Hence, the homologous AT1G75060 and AT1G19330 proteins were named as AFR1 and AFR2, respectively. In short, our findings collectively suggest that RPD3-type HDACs and AFRs together with other structural components such as SNLs and AtSAP18 form Sin3-SAP30-SAP18-RPD3 like complexes in Arabidopsis.
AFR1 Acts Additively with AFR2 to Repress the Floral Transition
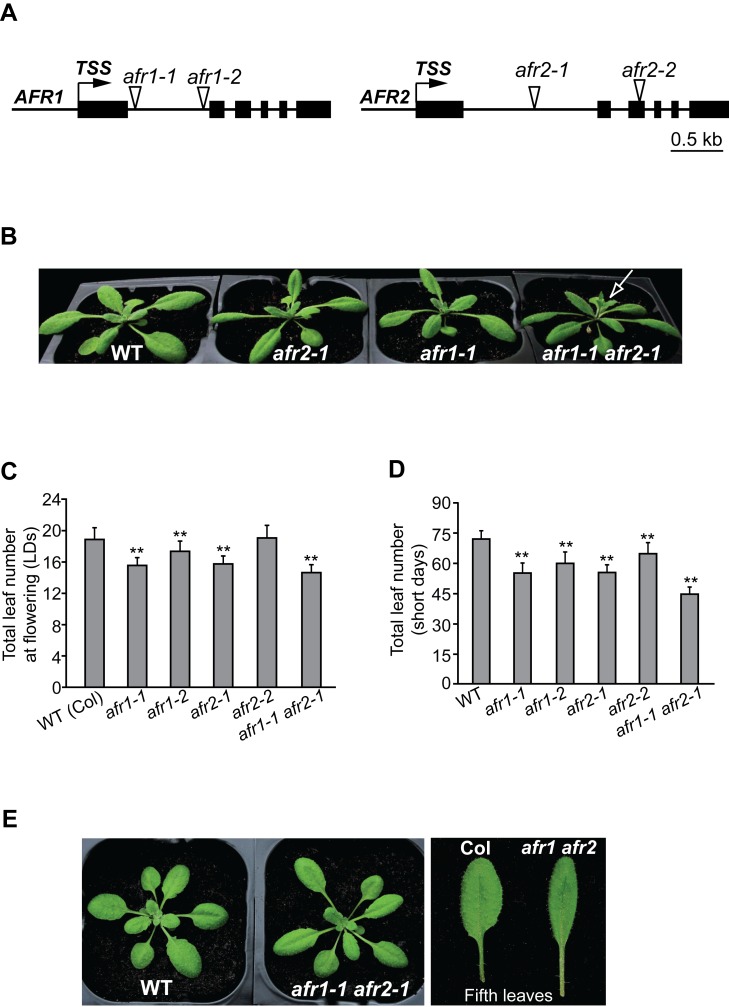
To elucidate the biological functions of AFR1 and AFR2, we identified their loss-of-function mutants, and each carries a transfer DNA (T-DNA) insertion. In afr1-1 and afr1-2, a T-DNA is inserted into the first intron of AFR1, resulting in a great reduction, but not elimination of AFR1 expression (Figures 2A and S3A). In both afr2 alleles, full-length AFR2 transcripts were not detected, but AFR2 transcripts truncated at their 3′ ends were detectable in the afr2-2 mutant, which are predicted to encode a protein containing the first 149 amino acids of AFR2 (Figure S3B). Grown in LDs, afr1-1, afr1-2, and afr2-1 mutants flowered moderately earlier than the wild type (WT) Col, as revealed by the total number of leaves formed prior to flower formation from the primary SAM of an examined line, the developmental standard for Arabidopsis flowering-time measurement (Figure 2B and 2C). In short days (8-h light/16-h dark), all afr1 and afr2 mutants flowered earlier than WT (Figure 2D), and the early-flowering phenotypes of afr1-1 and afr2-1 were fully rescued by complementation with tagged AFR1 and AFR2, respectively (Figure S4). Of note, afr2-2 is a weak allele compared to afr1-1, and it is likely that the truncated AFR2 may retain some function in the afr2-2 mutant. We further constructed the afr1 afr2 double mutant (afr1-1 afr2-1), and found it flowered earlier than either single mutant in both LDs and short days (Figures 2C, 2D, and S5A). Taken together, these results suggest that AFR1 and AFR2 act additively to repress the floral transition in Arabidopsis, indicating that the homologous AFR1 and AFR2 proteins do not function in one complex. In addition to early flowering, the afr1 afr2 mutant exhibited a leaf phenotype: longer petiole, narrower leaf blade, and with a slightly increased leaf initiation rate (Figures 2E and S5C), suggesting that AFR1 and AFR2 may also be required for proper leaf development.
Figure 2. Phenotypes of afr1, afr2, and afr1 afr2 mutants.
(A) AFR1 and AFR2 gene structures. Exons are represented by black boxes, and arrows indicate transcription start sites (TSS); triangles for T-DNA insertion sites. (B) afr1, afr2, and afr1 afr2 mutants grown in LDs. The arrow indicates a main bolt with flowers. (C) Flowering times of the indicated genotypes grown in LDs. 19–23 plants were scored for each line. Double asterisks indicate statistically significant differences in the means between Col (WT) and the indicated mutants, as revealed by two-tailed Student's t test (**, p<0.01). Bars indicate SD (for standard deviation). (D) Flowering times of the indicated genotypes grown in short days. 11–15 plants were scored for each line. Double asterisks indicate statistically significant differences in the means between Col and the indicated mutants. (E) Leaf phenotype of the afr1 afr2 double mutant grown in LDs.
AFR1 and AFR2 Downregulate FT Expression Specifically at the Day's End to Delay Flowering in LDs
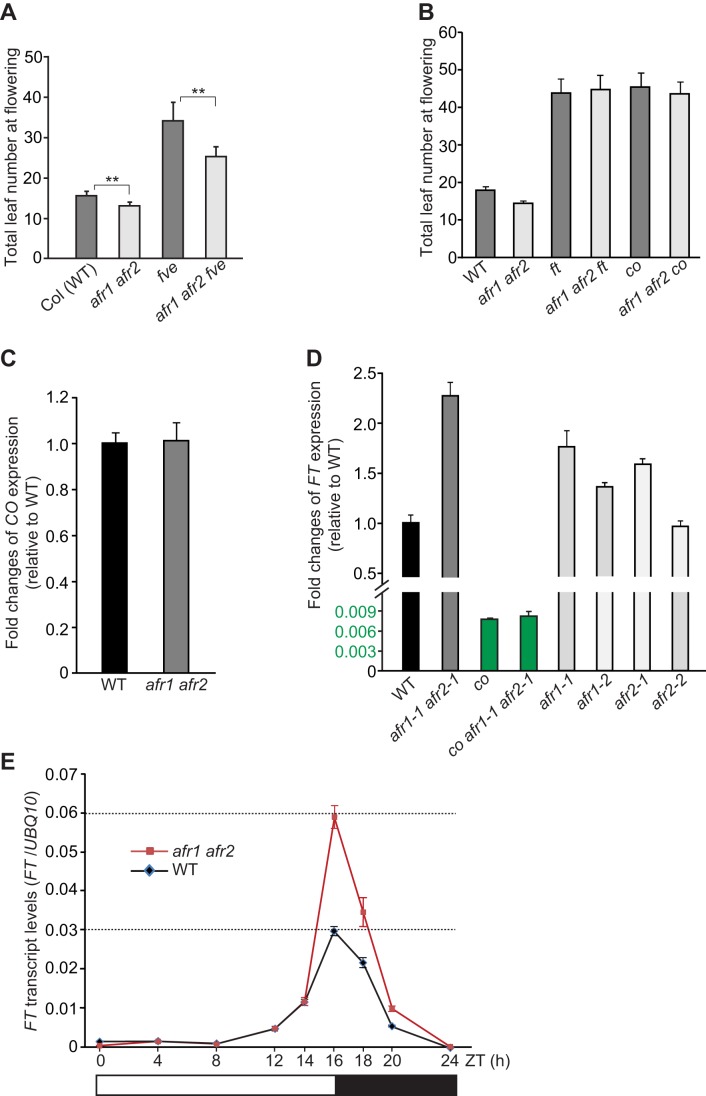
There are several genetic pathways promoting flowering in Arabidopsis, including the autonomous, photoperiod, vernalization, and thermosensory pathways [2],[27],[28]. To explore in which genetic pathway AFR1 and AFR2 act to regulate flowering, we introduced the photoperiod-pathway mutation co or a null mutant of FVE, a component of the autonomous and thermosensory pathways, into the afr1 afr2 double mutant [27],[28]. The late-flowering phenotype of fve was partly suppressed by afr1 afr2, whereas the afr1 afr2 co triple mutant flowered at about the same time as co (Figure 3A and 3B); thus, the co mutation is epistatic to afr1 and afr2. Furthermore, we found that ft, like co, is epistatic to afr1 and afr2 (Figure 3B). These results suggest that AFR1 and AFR2 act through the CO-FT regulatory module of the photoperiod pathway to control flowering.
Figure 3. AFR1 acts additively with AFR2 to downregulate FT expression specifically at the day's end in LDs.
(A,B) Flowering times of the indicated genotypes grown in LDs. 11–17 plants were scored for each line. The afr1 afr2 double mutant is afr1-1 afr2-1. Double asterisks indicate statistically significant differences in the means between the indicated genotypes, as revealed by two-tailed Student's t test (**, p<0.01). Bars indicate SD. (C,D) Relative CO (C) and FT transcript levels (D) in the seedlings of indicated genotypes at the end of LDs (ZT16), quantified by qRT-PCR. The transcript levels were normalized first to the endogenous control UBQ10, and relative fold changes to WT are presented (note that the FT transcript level in co was less than 1% of that in WT). Bars indicate SD of triplicate measurements. One of two biological repeats with similar results is shown. (E) FT mRNA levels in Col and afr1 afr2 seedlings over a 24-h LD cycle, as quantified by qRT-PCR. FT transcript levels were normalized to UBQ10; bars indicate SD of triplicate measurements. A biological repeat of this analysis is included as Figure S8A. White and dark bars below the x-axis indicate light and dark periods, respectively.
Next, we examined CO expression upon loss of AFR1 and AFR2 function at the end of LDs (ZT16, 16 h after light on or dawn; Zeitgeber time [ZT]), because CO is highly expressed to activate FT expression at this time point. CO expression remained unchanged in the afr1 afr2 double mutant compared to WT (Figure 3C); additionally, loss of CO function didn't affect AFR1 or AFR2 expression (Figure S6). We further examined FT expression in afr1, afr2, and afr1 afr2 seedlings at the end of LDs. FT transcript levels were increased moderately in afr1 and afr2 single mutants and in the double mutant (Figure 3D), consistent with that AFR1 and AFR2 act additively to repress flowering. In addition, FT expression was slightly upregulated in afr1 afr2 in short days, though its transcript levels were very low in both WT and afr1 afr2 plants (Figure S7). Taken together, these results show that AFR1 and AFR2 function additively to downregulate FT expression and so delay flowering. We further measured FT transcript levels in co and afr1 afr2 co seedlings at ZT16, and found that co completely suppressed FT up-regulation in the afr1 afr2 background (Figure 3D). Hence, CO is required for FT downregulation at ZT16 by AFR1 and AFR2.
FT expression is activated by CO specifically at the end of LDs, and repressed at other times of the day, resulting in rhythmic FT mRNA expression at dusk in LDs. Several histone modifiers including the histone H3 lysine-27 (H3K27) methytransferase CLF and the PKDM7B H3 lysine-4 demethylase (also known as JMJ14 or AtJMJ4), are required for continuous FT repression along day/night cycles [29]–[32]. We explored whether AFR1 and AFR2 could repress FT expression in other times of LDs beside ZT16. Upon loss of AFR1 and AFR2 function, FT expression in LDs was not affected from ZT0 through ZT14, but upregulated only from ZT14 to ZT20 (Figures 3E and S8A). Thus, in LDs AFR1 and AFR2 downregulate FT expression specifically in the presence of CO activity at the day's end, to delay flowering.
AFR1 and AFR2 Downregulate FT Expression in Leaf Vasculature
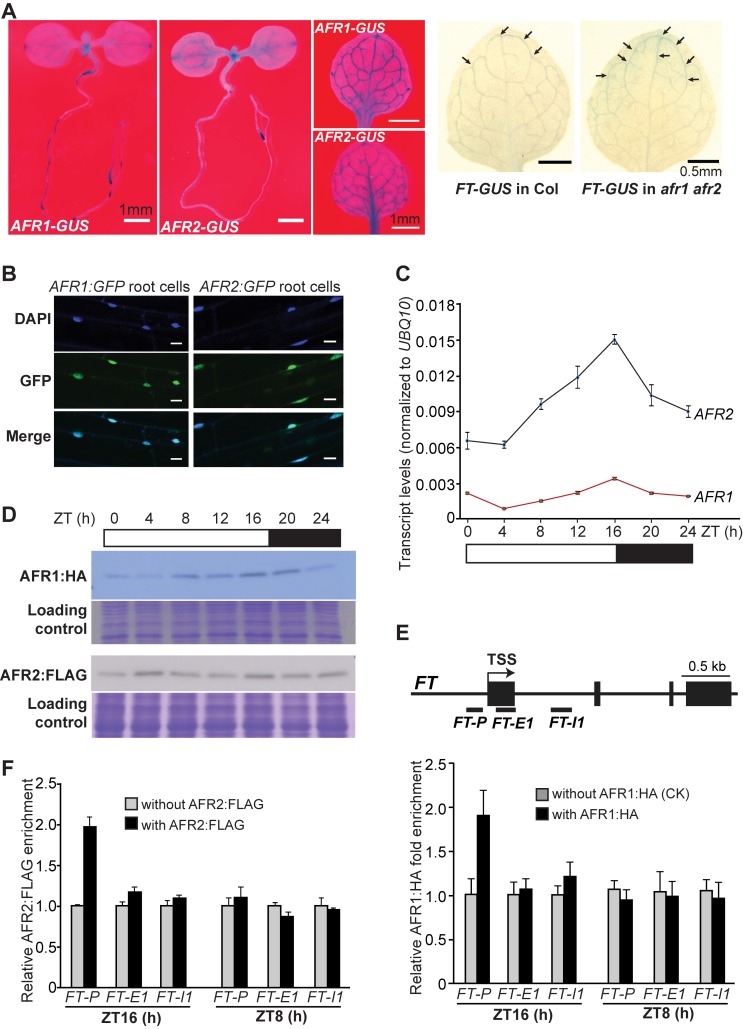
Day length is perceived in leaves, leading to FT activation specifically in the phloem companion cells of leaf veins [1],[2]. We asked whether AFR1 and AFR2 are expressed in leaf veins to downregulate FT expression. To this end, genomic AFR1 and AFR2 fragments (including 5′ promoter plus part of the coding region of each gene), were fused to the β-GLUCURONIDASE (GUS) gene in frame and the fusions were introduced into Arabidopsis by transformation. Histochemical staining revealed that both AFR1 and AFR2 were strongly expressed in leaf vasculature in addition to root tips and shoot apices (Figure 4A). To examine in which tissues AFR1 and AFR2 downregulate FT expression, we introduced afr1-1 and afr2-1 into an FT-GUS reporter line [33], and found that the GUS expression directed by a FT promoter was upregulated only in leaf veins (Figure 4A). Thus, AFR1 and AFR2 downregulate FT expression in the leaf vasculature. Next, we examined the subcellular localization of both AFR1 and AFR2 proteins by fusing them with green fluorescence protein (GFP). Upon their expression in Arabidopsis roots, these fusion proteins were specifically localized in the nuclei (Figure 4B), consistent with that AFR1 and AFR2 act as part of nuclear AFR1/AFR2-HDAC complexes in leaf veins to downregulate FT expression.
Figure 4. Analyses of AFR1 and AFR2 expression patterns and their bindings to FT chromatin.
(A) Spatial expression patterns of AFR1-GUS, AFR2-GUS, and FT-GUS. LD-grown Col seedlings or rosette leaves were stained for 6 h except for AFR1-GUS staining with 8.5 h. Arrows indicate stained veins. (B) Nuclear localization of the AFR1:GFP and AFR2:GFP fusion proteins in Arabidopsis root cells. Scale bars are 50 µm. The blue DAPI staining indicates nuclei. (C) AFR1 and AFR2 mRNA levels in Col (WT) seedlings over a 24-h LD cycle. The mRNA levels were normalized to UBQ10; bars indicate SD of triplicate measurements. A biological repeat of this analysis is included as Figure S8B. White and dark bars below the x-axis indicate light and dark periods, respectively. (D) AFR1:HA and AFR2:FLAG protein levels in Col seedlings over a 24-h LD cycle. Total proteins loaded in a duplicated SDS-PAGE gel were stained with Coomassie Blue, serving as loading controls. (E) ChIP analysis of AFR1:HA enrichment at the FT locus. Amounts of the immunoprecipitated genomic fragments were measured by qPCR, and normalized first to the endogenous control TUBULIN8 (TUB8). The fold enrichment of AFR1:HA in each examined region (at each time point) was calculated by dividing the TUB8-normalized amount of examined region from the AFR1:HA-expressing line, by that of WT (without AFR1:HA) at each time point. Error bars indicate SD of triplicate quantifications (technical replicates). A biological repeat of this analysis is presented as Figure S10A. (F) ChIP analysis of AFR2:FLAG enrichment at the FT locus. The fold enrichments of AFR2:FLAG were calculated in a way similar to those of AFR1:HA. Error bars indicate SD of triplicate quantifications. A biological repeat of this analysis is presented as Figure S10B.
AFR1 and AFR2 Proteins Accumulate and Directly Interact with the FT locus at the End of LDs
We hypothesized that the downregulation of FT expression by AFR1 and AFR2 in the presence of CO activity at the end of LDs may be attributed to AFR-HDAC binding to FT chromatin only at this time in LDs. To test this hypothesis, we first examined the diurnal expression patterns of AFR1 and AFR2 in LDs, and found that the expression of both genes peaked at the day's end (Figure 4C). Next, using the functional AFR1:HA-expressing line and the functional AFR2:FLAG-expressing line (driven by their native promoters; see Figure S4), we measured the abundance of both proteins every 4 h over a 24-h LD cycle. The levels of AFR1 and AFR2 proteins changed diurnally in LDs, and both proteins accumulated at the end of the day (Figures 4D and S9). This indicates that AFR1/AFR2-HDAC may accumulate at the end of LDs.
We further explored whether AFR1 could bind to FT chromatin, and if so, at what time of the day. Chromatin immunoprecipitation (ChIP) assays were performed using the AFR1:HA-expressing seedlings. We found that at the end of LDs (ZT16), AFR1 bound to the proximal FT promoter region, but not in the first exon or the first intron (Figures 4E and S10A). Unlike at ZT16, AFR1 was not enriched at all in the FT locus in the middle of the day (ZT8) (Figure 4E). Moreover, using ChIP with the AFR2:FLAG-expressing seedlings, we uncovered that AFR2, like AFR1, bound to the proximal FT promoter region at ZT16, but not at ZT8 (Figures 4F and S10B). The bindings of AFR1 and AFR2 to FT chromatin at the end of LDs coincide well with that both proteins accumulate at this time. Taken together, these results suggest that in LDs AFR1/AFR2-HDAC binds to FT chromatin specifically at the day's end to modulate FT expression in the presence of CO activity.
AFR1 and AFR2 Mediate Periodic Histone Deacetylation at FT at the End of LDs
The recruitment of AFR1 and AFR2 to FT chromatin at the day's end, may cause periodic histone deacetylation in LDs. To test this, we examined steady-state levels of histone H3 acetylation of FT chromatin in WT and afr1 afr2 seedlings (and rosette leaves) at ZT8 and ZT16. Loss of AFR1 and AFR2 function led to an increase of steady-state level of acetylated H3 in the proximal FT promoter region, but not in the first exon or the first intron at ZT16, whereas at ZT8 there was no noticeable change in all three regions examined (Figures 5 and S11). This is consistent with the binding of AFR1 and AFR2 to FT chromatin in the proximal promoter at the end, but not the middle of LDs (Figure 4E). Together, these results show that AFR1 and AFR2 (presumably as part of AFR1/AFR2-HDAC) mediate histone deacetylation at the FT locus to downregulate its expression at the end of LDs. Interestingly, there is no apparent increase of the steady-state acetylation levels of FT chromatin in WT at ZT16 relative to ZT8, although FT expression is activated at ZT16 (Figure 5; note that the relative acetylation level of each sample was normalized to the WT at ZT8). Therefore, there must be active histone acetylation by HAT(s) on FT chromatin to “counteract” the AFR-mediated histone deacetylation at the end of LDs.
Figure 5. ChIP analysis of levels of acetylated histone H3 in Col and afr1 afr2 rosette leaves.
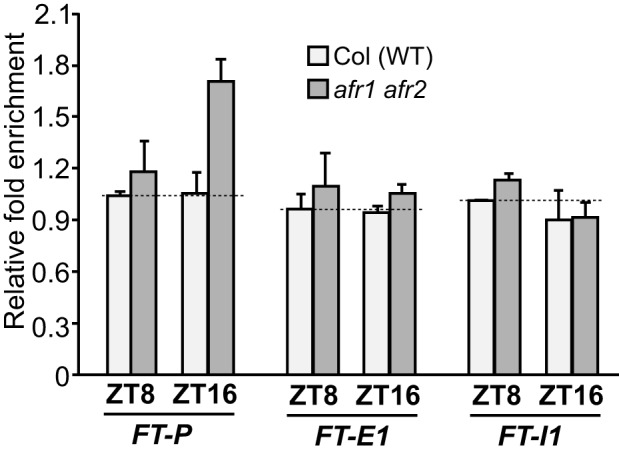
Amounts of the immunoprecipitated genomic fragments were quantified by qPCR. The fold enrichments were calculated as follows: for each examined FT region, the amount of DNA fragments from WT or afr1 afr2 at each time point (ZT8 or ZT16) was first normalized to the constitutively expressed TUBULIN2 (TUB2) in each sample, and subsequently, the TUB2-normalized values for the afr1 afr2 at ZT8, the afr1 afr2 at ZT16, or the WT at ZT16 were divided by the value for the WT at ZT8 to obtain fold enrichments. Shown are the means and SD of two ChIP experiments. An analysis of H3 acetylation on FT chromatin in Col and afr1 afr2 seedlings is presented in Figure S11.
The Transcription Factor AGL18 recruits AFRs to the FT Locus Specifically at the End of LDs
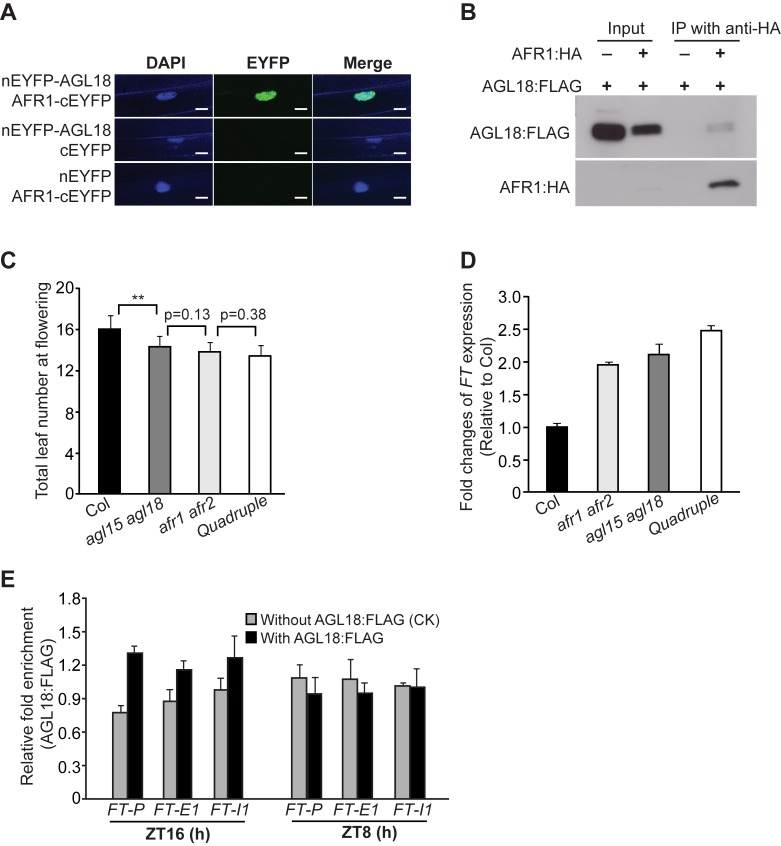
In yeast SAP30 is involved in the recruitment of SAP30-Sin3-RPD3 co-repressor complex to a target locus [21]. We hypothesized that transcription factors may directly interact with AFR1 and AFR2 to recruit AFR1/AFR2-HDAC to the FT locus at the end of LDs. Previous studies have revealed that several MADS-domain transcription factors including FLC, SHORT VEGETATIVE PHASE (SVP), AGL18, and AGAMOUS LIKE 15 (AGL15), are involved in FT repression in the vasculature to inhibit flowering [28],[34],[35]. Using a candidate gene approach, we found that the AGL18 protein directly interacted with AFR1 and AFR2 in the nuclei of onion epidermal cells, as revealed by the BiFC experiments (Figures 6A and S12A). AGL18, like AFR1 and AFR2, are also expressed in leaf veins [35]. To further confirm the in vivo association of AGL18 with AFR1, we conducted co-immunoprecipitation experiments using the seedlings expressing AFR1:HA and a functional AGL18:FLAG (Figure S13A), and found that anti-HA (recognizing AFR1:HA) immunoprecipitated AGL18:FLAG from the seedlings (Figure 6B). Thus, indeed, AGL18 directly interacts with AFR1 and presumably AFR1-HDAC in Arabidopsis.
Figure 6. AGL18 directly interacts with AFR1 and binds to FT chromatin at the day's end in LDs.
(A) BiFC analysis of the interaction of AGL18 with AFR1 in onion epidermal cells. Yellowish-green signals indicate the physical association of AGL18 with AFR1 in the nuclei (indicated by the blue fluorescence from DAPI). Bar = 20 µm. (B) Co-immunoprecipitation of AFR1 with AGL18 in Arabidopsis seedlings. Total protein extracts from F1 seedlings of the doubly hemizygous AGL18:FLAG and AFR1:HA, were immunoprecipitated with anti-HA agarose; subsequently, the precipitates were analyzed by western blotting with anti-FLAG (recognizing AGL18:FLAG) and anti-HA (recognizing AFR1:HA). (C) Flowering times of the indicated genotypes grown in LDs. 12–16 plants were scored for each line. Double asterisks indicate a statistically significant difference in the means between Col and agl15 agl18, as revealed by two-tailed Student's t test (**, p<0.01). Bars indicate SD. (D) Relative FT transcript levels in the seedlings of indicated genotypes at ZT16, quantified by qRT-PCR. The transcript levels were first normalized to UBQ10, and relative fold changes to Col are presented. Bars indicate SD of triplicate measurements. One of two biological repeats with similar results is shown. (E) ChIP analysis of AGL18:FLAG enrichment at the FT locus. Amounts of immunoprecipitated genomic fragments were measured by qPCR, and normalized first to the endogenous control TUB8. The fold enrichment of AGL18:FLAG in each examined region (at each time point) was calculated by dividing the TUB8-normalized amount of examined region from the AGL18:FLAG-expressing line, by that of WT (without AGL18:FLAG) at each time point. Error bars indicate SD of triplicate measurements. A biological repeat of this analysis is presented as Figure S13B.
AGL18 acts redundantly with its homolog AGL15 to repress flowering [35]. We further found that AFR1 directly interacted with AGL15 in yeast and plant cells (Figure S12B and S12C). Of note, it has been previously shown that AGL15 could interact moderately with the conserved HDAC domain of HDA19 in yeast cells [36], which can directly associate with AFR1 and AFR2 (Figure 1C and 1D). Next, we examined the genetic interaction of afr1 afr2 with agl15 agl18 in flowering-time regulation. Grown in LDs, the afr1 afr2 agl15 agl18 quadruple mutant flowered at a similar time to either double mutant (Figure 6C), suggesting that these four genes act in the same flowering-regulatory pathway. Further FT expression analysis revealed that FT transcript levels (at ZT16) in both afr1 afr2 and agl15 agl18 seedlings increased to about 2-fold of the WT level, and that in the quadruple, FT levels were slightly higher than either double mutant (Figure 6D). This slight increase is not surprising given that neither double mutant may not be null as there are low levels of full-length AFR1, and AGL15 and AGL18 transcripts in afr1 and agl15 agl18, respectively (Figures S3 and S14). Taken together, these data are in line with that the DNA-binding AGL15 and AGL18 may act to recruit AFR1 and AFR2 to downregulate FT expression.
To explore the role of AGL18 for AFR1 recruitment, first we examined whether and when AGL18 could directly interact with the FT locus. Using ChIP with the AGL18:FLAG-expressing seedlings, we found that at the end of LDs AGL18:FLAG, like AFR1, was apparently enriched in the proximal FT promoter region, in addition to a slight enrichment in the first exon (Figure 6E). In the middle of LDs, AGL18 was not associated with FT chromatin (Figure 6E). Thus, AGL18 directly interacts with the FT locus specifically at the end of LDs. Next, we investigated whether AGL18 is required for AFR1 recruitment to FT chromatin at the end of LDs. The line expressing the functional AFR1:HA (in afr1-1 background) was crossed to agl15 agl18, and the seedlings of afr1 AFR1:HA and agl15 agl18 afr1 AFR1:HA harvested at ZT16 were subjected to ChIP assays with anti-HA. We found that loss of AGL15 and AGL18 function nearly eliminated AFR1 binding to FT chromatin at the end of LDs (Figure 7). These results, together with the direct interactions of AGL18 and AGL15 with AFR1 and AFR2, led us to infer that these two MADS-domain transcription factors recruit AFRs (presumably AFR-HDAC) to the FT locus specifically at the end of LDs to dampen FT expression in the presence of CO activity.
Figure 7. AFR1 binding to FT chromatin at the end of LDs requires CO, AGL15, and AGL18.
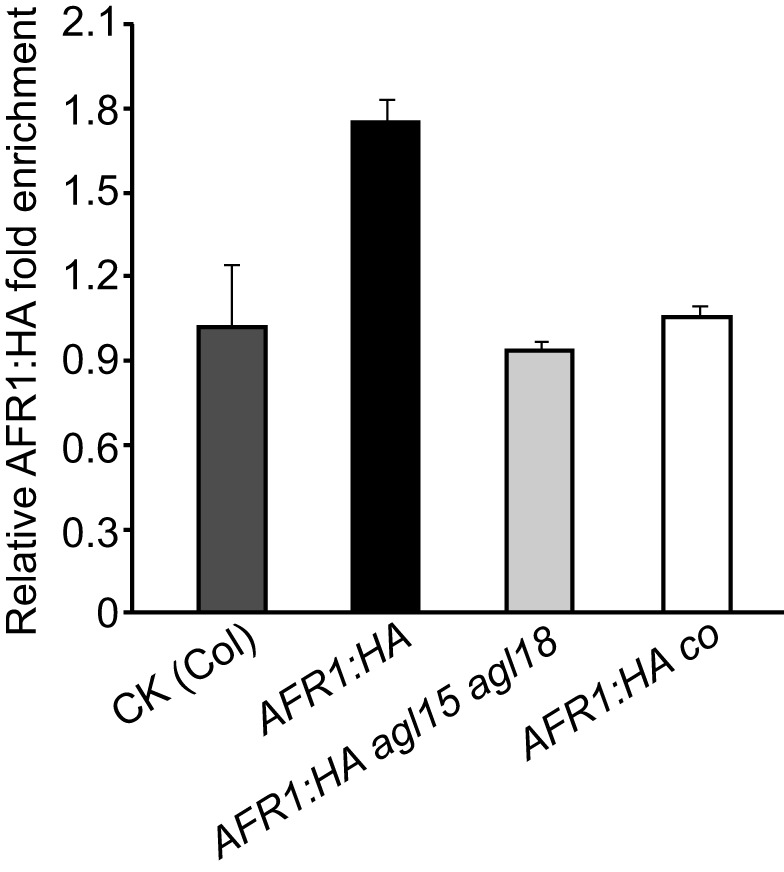
Seedlings of afr1 AFR1:HA, agl15 agl18 afr1 AFR1:HA, co afr1 AFR1:HA, and WT (negative CK) were harvested at ZT16 and subjected to ChIP assays with anti-HA. The fold enrichments of AFR1:HA in FT-P (a proximal promoter region) in the AFR1:HA-expressing lines over CK, are presented. Error bars indicate SD of triplicate measurements. A biological repeat of this analysis is presented as Figure S10C.
CO Activity Is Required for the AFR1 Recruitment to FT Chromatin at the End of LDs
As we showed earlier, CO is required for AFR1- and AFR2-mediated FT downregulation at the end of LDs. We reasoned that the CO activity at the FT locus at the end of LDs may be required for AGL18/AGL15 recruiting AFRs to FT. To test this, first we crossed the AFR1:HA-expressing line (in afr1-1) to co and created co afr1 AFR1:HA. Next, we carried out ChIP assays with the AFR1:HA-expressing seedlings harvested at ZT16, and found that CO, indeed, is required for AFR1 binding to FT chromatin at ZT16 (Figure 7). Upon its accumulation towards dusk in LDs, CO directly binds to the FT proximal promoter to promote its expression [37]. This raises a possibility that CO might directly interact with AGL18 and/or AFRs to recruit these proteins to the FT proximal promoter. Using the yeast-two-hybrid approach we found that CO didn't directly interact with AGL18, AFR1, or AFR2 (Figure S15); hence, we conclude that the CO protein is not directly involved in the recruitment of AFRs to FT chromatin. Taken together, these results suggest that the CO-mediated transcriptional activation of FT at the end of LDs enables or gates the recruitment of AFR-HDAC by AGL18/AGL15 to the FT locus, leading to the dampening of FT expression and so preventing precocious flowering in response to the day-length cue.
Discussion
In this study, we have revealed that the plant-unique AFR1 and AFR2 mediate periodic histone deacetylation on the rhythmically expressed florigen gene FT to dampen its expression specifically at dusk in LDs in Arabidopsis. The output of the photoperiod pathway CO at the end of LDs not only activates FT expression, but also enables the recruitment of AFR-HDAC to the FT locus to dampen FT expression and set it at an adequate level, conferring a proper flowering time in response to the day length cue. The MADS-domain transcription factor AGL18 directly interacts with the AFR1 and AFR2 proteins that accumulate at dusk, and recruits AFR1 (presumably AFR1/AFR2-HDAC) to the FT locus at the end of LDs to modulate FT expression. Our findings collectively uncover a temporal chromatin mechanism, namely periodic histone deacetylation, for the day-length regulation of flowering time in higher plants.
Dynamic Cycles of Histone Acetylation and Deacetylation at the FT Locus
The histone acetylation mark added by HATs can be rapidly removed by HDACs. Both HATs and HDACs have been found to associate with actively transcribed genes in yeast and human, and are often synchronously recruited to certain target loci, leading to dynamic cycles of acetylation and deacetylation [13]–[15]. HATs can bind transiently to and add acetyl groups to certain loci where HDACs transiently bind to erase the acetylation mark, resulting in constant acetylation and de-acetylation cycles at these loci [13]–[15]. Our study reveals that at the FT locus, accompanying rhythmic CO-mediated FT activation at the end of LDs, AFR1 and AFR2 (presumably AFR-HDAC) are recruited to the actively transcribed FT chromatin for histone deacetylation. Given that disruption of AFR1 and AFR2 function leads to an increase of steady-state level of acetylation on FT chromatin at the end of LDs, there must be a HAT (or HATs) to add acetyl groups, suggesting that constant cycles of acetylation and de-acetylation occur at the FT locus upon its activation by CO at dusk. AFR1/AFR2-HDAC is expected to act antagonistically to a HAT to regulate FT expression. Of note, the HAT(s) has not been identified yet.
The dynamic histone acetylation and deacetylation cycles in the proximal FT promoter may result from transient binding of HATs and AFR1/AFR2-HDAC to FT chromatin. Consistent with the transient or unstable AFR1-HDAC binding, our ChIP analysis shows that AFR1 is moderately enriched at the FT promoter (Figures 4E and 7). The increase of steady-state acetylation level at FT and FT upregulation upon disruption of AFR1/AFR2 function, indicate that at the end of LDs histone acetylation promotes FT transcription, whereas histone deacetylation executed by AFR-HDAC acts to downregulate FT expression. It is very likely that when FT chromatin is actively transcribed upon transcriptional activation by CO at the end of LDs, AFR-HDAC may remove acetyl groups following each cycle of HAT activities associated with FT transcription to reset the acetylation state of FT chromatin, and functions to dampen FT expression at dusk. The AFR-HDAC activities at dusk may also serve to reset FT chromatin rapidly to a silent state with a low level of acetylation at night. Consistent with this notion, we have observed that upon loss of AFR1 and AFR2 function, it takes several hours longer to reset FT expression to a silent state at night (Figures 3E and S8A).
Interestingly, the steady-state acetylation levels of FT chromatin, as measured by ChIP, remain largely unchanged from the middle to the end of LDs (Figure 5), although FT expression is switched on at dusk. Not surprisingly, this may reflect the dynamic cycles of histone acetylation and deacetylation upon FT expression induction by CO. In the budding yeast, during transcriptional induction of ARG1 (encoding an argininosuccinate synthase), the Gcn5 HAT and several HDACs are synchronously recruited to ARG1 chromatin, and these opposing activities result in no noticeable changes of steady-state acetylation levels at the ARG1 locus, as measured by ChIP [14]. A similar situation has been observed during the transcriptional induction of several heat-responsive genes by heat shock in yeast [15].
Role of CO in Gating the Recruitment of AFR1/AFR2-HDAC to FT Chromatin at the End of LDs
In this study we have revealed that the MADS-domain transcription factor AGL18 and presumably AGL15 act to recruit AFR1/AFR2-HDAC to the FT locus at the end of LDs. AGL18 is well expressed from ZT8 to ZT16 (unpublished data), but the AGL18 protein binds to FT chromatin (in the proximal promoter) only at ZT16. This indicates that the chromatin state of the FT locus may play a role for the AGL18 binding to it; for instance, the state may determine whether a cis-element such as a CArG motif, recognized typically by a MADS domain, in FT promoter is accessible to AGL18.
We found that the output of the photoperiod pathway CO is required for, but not directly involved in the recruitment of AFR1 to FT chromatin at dusk. The CO protein directly binds to FT proximal promoter and also associates with NY-F transcriptional factors that bind to FT distal promoter, to promote FT expression specifically at the end of LDs [37]–[40]. Given the indirect role of CO for AFR1/AFR2 recruitment, we infer that at the end of LDs the binding of CO to the FT proximal promoter and/or consequent transcriptional activation of FT expression enable AGL18 recruiting AFRs to the FT locus. It is likely that CO activity may cause a change in FT chromatin state so that the cis-element(s) such as a CArG motif or its variant in the FT promoter become accessible by AGL18. In short, in the photoperiodic flowering regulation CO plays a dual role: activating FT expression and enabling AFR-HDAC-mediated FT downregulation to set a proper level of FT expression at the end of LDs.
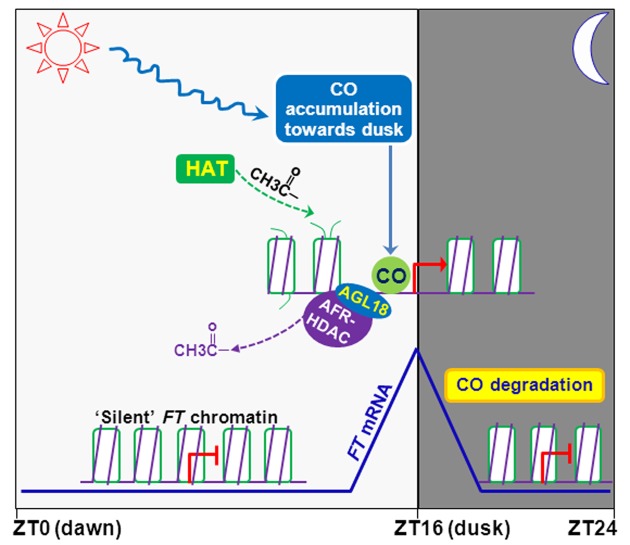
The accumulation of both AFR1 and AFR2 at the end of LDs indicates that AFR1/AFR2-HDAC may accumulate at dusk, which may enable or enhance their recruitment by AGL18 to the FT proximal promoter. CO activity not only enables AGL18-AFR-HDAC binding to FT chromatin to dampen FT expression, but may also facilitate dynamic recruitment of a HAT to the FT locus to promote its expression given the dynamic nature of histone acetylation and deacetylation on FT chromatin at dusk (Figure 8). The opposing activities of HAT and AFR-HDAC at the end of LDs conceivably modulate the acetylation dynamics of FT chromatin and set FT expression at an adequate level at the right time.
Figure 8. A working model for control of FT expression by the dynamic cycles of histone acetylation and deacetylation at the end of LDs.
The coincidence of high CO mRNA expression with light exposure at the day's end leads to the CO protein accumulation towards dusk. CO directly binds to the FT proximal promoter, and CO activity at the FT locus may change the chromatin state and enables/gates AGL18 (and presumably AGL15) binding to the FT proximal promoter. AGL18 recruits AFR1/AFR2-HDAC to FT chromatin at dusk. In addition, the CO activity may also enable the recruitment of a HAT to FT chromatin. The opposing activities of HAT and AFR-HDAC on FT chromatin at the end of LDs conceivably modulate the acetylation dynamics of FT chromatin and set FT expression at an adequate level at dusk. At night, CO is rapidly degraded by proteasomes, which prevents the actions of HAT and AFR-HDAC on FT chromatin, resulting in a “silent” chromatin state. In early day, FT chromatin remains ‘silent’ due to lack of the CO protein. Day and night are indicated with white and gray shadings, respectively.
Role of AFR-HDAC for Photoperiodic Flowering-Time Regulation
We show here that AFRs act in the photoperiod pathway to dampen FT expression in response to inductive long days and so prevent precocious flowering in Arabidopsis. Interestingly, only a moderate early-flowering phenotype has been observed in the afr1 afr2 mutant in which AFR1 is still expressed at a low level (Figure S3A). It is likely that a complete loss of function of AFR1 and AFR2 may cause a strong early-flowering phenotype. In short days, the CO protein does not accumulate and thus FT is expressed at a very low level [2]. We have observed that in short days FT expression is relatively high at ZT4 and ZT20-ZT24 in WT, and that loss of AFR1 and AFR2 function leads to a moderate FT upregulation specifically at these time points (Figure S7B). In addition, it has been shown that AGL18 also moderately represses FT expression in short days [35]. Taken together, these findings suggest that AGL18-AFR-HDAC acts to downregulate FT expression to prevent early flowering in non-inductive short days as well, conferring an accurate photoperiodic response.
CO and FT are widely conserved among angiosperms from the monocotyledonous rice grass to the dicotyledonous poplar tree [1],[2]. In the plants so far examined, the function of FT or FT homologs as a major inducer of flowering (florigen) is largely conserved, and the CO-FT regulatory module underlying the photoperiodic flowering-time control in the dicotyledonous LD plant Arabidopsis, also plays a central role for flowering-time control in the monocotyledonous short-day plant rice [2]. Homologs of the core components of AFR-HDAC including AFR1/AFR2, AtSAP18, RPD3-type HDACs, and SNLs are highly conserved in angiosperms (Figure S1B) [22],[23],[25]. These findings collectively indicate that the temporal chromatin mechanism of periodic histone deacetylation underlying FT expression control in Arabidopsis may modulate the expression of FT homologs in other plant species and so confer an optimal flowering time in response to photoperiodic changes.
Materials and Methods
Plant Materials and Growth Conditions
The co, ft, fve, agl15-3, and agl18-1 were described previously [6],[27],[35],[41]. afr1-1 (Salk_110828), afr1-2 (Salk_059944), afr2-1 (Salk_032653), and afr2-2 (CS842844) were obtained from the Arabidopsis Biological Resource Center and directly used in this study. Plants were grown in long days (16-h light/8-h dark) or short days (8-h light/16-h dark) under cool white fluorescent light at 22°C [41].
Yeast Two-Hybrid Assay
The Matchmaker GAL4 Two-Hybrid System 3 (Clontech) was adapted for this assay. Full-length coding sequences for AFR1, AFR2, AtSAP18, HDA9, HDA19, and SNL2 were cloned into the pGADT7 and/or pGBKT7 vectors, which were subsequently introduced into the yeast strain AH109 according to the manufacturer's instructions (Clontech). Yeast cells were spotted on selective media lacking of leucine, tryptophan, and histidine.
BiFC Assay
The full-length coding sequences for AFR1, AFR2, AtSAP18, HDA19, AGL15, and AGL18 were fused in frame with either the coding sequence for an N-terminal EYFP fragment in the pSAT1A-nEYFP-N1/pSAT1-nEYFPC1 vectors and/or for a C-terminal EYFP fragment in the pSAT1AcEYFP-N1/pSAT1-cEYFP-C1-B vectors [42]. Plasmid pairs were used to transiently transform onion epidermal cells via bombardment by the Helium Biolistic Gene Transformation System (Bio-Rad). Within 24 h following the bombardment, the EYFP fluorescence emitted from the onion cells was imaged with a Zeiss LSM 5 EXCITER upright laser scanning confocal microscopy (Zeiss) [43].
RNA Analysis by Real-Time Quantitative PCR
Total RNAs were extracted from aerial parts of 10- to 11-d-old seedlings grown in long days (at various time points) or 21-d-old seedling grown in short day using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's instructions. cDNAs were reverse transcribed from the total RNAs. Real-time quantitative PCR (qRT-PCR) was performed on an ABI Prism 7900HT sequence detection system using a SYBR green PCR master mix; PCR was conducted as follows: 50°C (2 min), 95°C (10 min), and 40 cycles of 95°C (15 s) and 60°C (60 s). Each sample was quantified at least in triplicate, and the constitutively expressed UBQ10 (its expression is not influenced by day-night cycles) [44], was used for normalization. Ratio of the transcript level of a gene of interest to that of UBQ10 is calculated as 2−ΔCt [ΔCT = CT (gene of interest) − CT (UBQ10)]. The primer pair for CO amplification has been described previously [44], and primer pairs for AFR1, AFR2, FT, and UBQ10 amplifications are specified in Table S1.
Plasmid Construction
To construct AFR1-GUS and AFR2-GUS, genomic fragments of 3.0-kb AFR1 (1.0-kb 5′ promoter plus 2.0-kb genomic coding sequence) and 3.9-kb AFR2 (1.2-kb 5′ promoter plus 2.7-kb genomic coding sequence) were inserted upstream of GUS in the pMDC162 vector [45] via gateway technology (Invitrogen). For AFR1 and AFR2 subcellular localization, the full-length AFR1 and AFR2 coding sequences (except the stop codons) were inserted between the 35S promoter and GFP in the pMDC85-GFP vector [45] via gateway technology (AFR1 and AFR2 are in frame with GFP).
To generate the pAFR1-AFR1:HA plasmid, a 3.0-kb AFR1 genomic fragment (1.0-kb 5′ promoter plus the 2.0-kb full-length genomic coding sequence except the stop codon) was first fused with a 3xHA tag, and cloned into the pHGW vector [46] via gateway technology. For pAFR2-AFR2:FLAG construction, a 3.9-kb AFR2 genomic fragment (1.2-kb 5′ promoter plus the 2.7-kb full-length genomic coding sequence except the stop codon) was first fused with a 3xFLAG tag, and cloned into pHGW. To construct p35S-AGL18:FLAG, the full-length AGL18 coding sequence without the stop codon (772 bp) was first fused with a 3xFLAG tag, and the AGL18:FLAG fusion was subsequently placed downstream of the 35S promoter in the pB2GW7 vector [46] via that gateway technology. The sequences of primers used for plasmid construction are specified in Table S1.
Co-immunoprecipitation
Co-immunoprecipitation experiments were carried out as previously described [43]. Briefly, total proteins were extracted from 10-d-old seedlings and immunoprecipitated with anti-HA M2 affinity gel (Sigma, catalog number A2220). AGL18:FLAG in the immunoprecipitates was detected by western blotting with anti-FLAG (Sigma, catalog number A8592).
Chromatin Immunoprecipitation
ChIP experiments were carried out as described previously with minor modifications [47]. Briefly, total chromatin was extracted from 10-d-old seedlings or the first pair of rosette leaves from 13-d-old seedlings grown in LDs, and immunoprecipitated with Rabbit polyclonal anti- acetylated histone H3 (with acetyl K9+K14+K18+K23+K27; abcam, catalog number ab47915), anti-HA (Sigma, catalog number H6908), or anti-FLAG (Sigma, catalog number F7425). Quantitative PCR was conducted to measure the amounts of FT and the constitutively expressed TUB2 and TUB8 fragments on an ABI Prism 7900HT sequence detection system using a SYBR Green PCR master mix. The primers used are specified in Table S1.
Supporting Information
Alignments of AFR1 and AFR2 relatives. Numbers refer to amino acid residues. Identical residues among these proteins are shaded black, whereas similar residues are shaded gray. (A) Alignment of the Arabidopsis AFR1 (AtAFR1) and AFR2 (AtAFR2) with the yeast Sap30 (ScSap30). The conserved residues in the putative SBRs are indicated by asterisks (*). (B) Alignment of AtAFR1 with its homologs from other plants including Populus trichocarpa (PtSFR), Picea sitchensis (PsSFR), and Physcomitrella patens (PpSFR). The GenBank accession numbers for PtSFR, PsSFR, and PpSFR are XP_002302473.1, ADE76839.1, and XP_001766227.1, respectively.
(EPS)
Direct interactions of AT1G75060 (AFR1) and AT1G19330 (AFR2) with SNL2 and HDA9 proteins in yeast. The indicated full-length proteins were fused with the GAL4-BD or AD domains. Yeast cells harboring the fusion proteins, BD and/or AD (as indicated), were grown on the selective synthetic defined media lacking of Trp (W), Leu (L), and His (H). (A,B) Interactions of SNL2 with AFR1 and AFR2 in yeast. (C,D) Interactions of HDA9 with AFR1 and AFR2 in yeast.
(TIF)
Analysis of AFR1 and AFR2 expression in afr1 and afr2 seedlings by semiquantitative RT-PCR. The constitutively expressed TUB2 served as a control. (A) Analysis of the full-length AFR1 and AFR2 transcripts. (B) Analysis of 5′- or 3′-truncated AFR2 transcripts in afr2 mutants.
(EPS)
Rescue of afr1 and afr2 mutants by pAFR1-AFR1:HA and pAFR2-AFR2:FLAG , respectively. Plants were grown in short days, and total leaf number for each line (9–11 plants per line) was scored. Bars indicate SD. (A) Flowering times of afr1-1 and a transgenic line of pAFR1-AFR1:HA in afr1-1 (native AFR1 fused with 3x HA; single-locus T3 homozygotes). (B) Flowering times of afr2-1 and a transgenic line of pAFR2-AFR2:FLAG in afr2-1 (native AFR2 fused with 3x FLAG; single-locus T3 homozygotes).
(EPS)
Analysis of flowering time and leaf initiation rate of afr1 afr2 in LDs. 12 plants were scored for each line; bars for SD. (A) Days to flowering of Col and afr1 afr2. (B) Total leaf numbers at flowering of Col and afr1 afr2. (C) Leaf initiation rates of Col and afr1 afr2 plants.
(EPS)
Relative AFR1 and AFR2 transcript levels in Col and co seedlings at the end of LDs, quantified by qRT-PCR. The transcript levels were normalized first to the endogenous control UBQ10, and relative fold changes to Col are presented. Bars indicate SD of triplicate measurements.
(EPS)
AFR1 and AFR2 moderately repress FT expression in short days. (A) Flowering times of the indicated lines grown in short days. Total leaf number for each line (8–14 plants per line) was scored. Error bars indicate SD. (B) FT mRNA levels in Col and afr1 afr2 seedlings over a 24-h short-day cycle. Total RNAs were extracted from 21-d old seedlings every 4 h and quantified by qRT-PCR. Bars indicate SD of three measurements.
(EPS)
Biological repeats of FT , AFR1 , and AFR2 expression analyses. (A) A biological repeat of the FT expression analysis presented in Figure 3E. (B) A biological repeat of the AFR1 and AFR2 expression analysis presented in Figure 4C.
(EPS)
Relative levels of AFR1:HA and AFR2:FLAG proteins over a 24-h LD cycle. The proteins in respective 10- to 11-d-old seedlings were analyzed by western blotting. The intensities of protein bands were quantified by the ImageJ 1.44j program, and relative levels to ZT0 are presented. Note that the time of the day has a significant effect on the levels of both AFR1 and AFR2 proteins in the seedlings, as revealed by single-factor ANOVA tests (for AFR1, p<0.01; for AFR2, p<0.05). (A) Relative AFR1:HA levels over a 24-h LD cycle. Average values of three biological repeats are presented; bars for SD. (B) Relative AFR2:FLAG levels over a 24-h LD cycle. Average values of four biological repeats are presented; bars for SD.
(EPS)
Biological repeats of the ChIP assays on AFR1 and AFR2 binding to FT chromatin. (A) A biological repeat of the ChIP analysis presented in Figure 4E. (B) A biological repeat of the ChIP analysis presented in Figure 4F. (C) A biological repeat of the ChIP analysis presented in Figure 7.
(EPS)
ChIP analysis of the histone H3 acetylation state of FT chromatin in Col and afr1 afr2 seedlings grown in LDs. Amounts of the immunoprecipitated genomic DNA was quantified and normalized first to the constitutively expressed TUB2. Subsequently, the fold enrichments were calculated by dividing the levels of acetylated H3 in afr1 afr2 (at ZT8 or ZT16) or the WT at ZT16, by the level in the WT at ZT8. Shown are the means and SD of two ChIP experiments.
(EPS)
Direct interactions of AGL18 with AFR2 and AGL15 with AFR1. (A) BiFC analysis of the interaction of AGL18 with AFR2 in onion epidermal cells. Yellowish-green signals indicate the binding of AGL18 with AFR2 in the nuclei (indicated by the blue fluorescence from DAPI). Bar = 20 µm. (B) AFR1 interacts with AGL15, but not FLC, in yeast cells. The full-length AFR1, and full-length FLC and the 208-aa AGL15 (without MADS domain) were fused with the GAL4-AD and BD domains, respectively. Yeast cells were grown on the selective synthetic defined media lacking of W, L, and H or lacking of W and L. Of note, FLC directly interacts with SVP and binds to FT chromatin to repress FT expression [48], and serves as a negative control in this experiment. (C) BiFC analysis of the interaction of AGL15 with AFR1 in onion epidermal cells. The full-length AGL15 and AFR1 were fused with nEYFP and cEYFP fragments, respectively. Yellowish-green signals indicate the binding of AGL15 with AFR1 in the nuclei. Bar = 20 µm.
(TIF)
Characterization of the p35S-AGL18:FLAG line and a biological repeat of ChIP analysis of AGL18 binding to FT chromatin. (A) Flowering times of the p35S-AGL18:FLAG line (T3; in the Col background) grown in LDs. Overexpression of AGL18 causes moderate late flowering [35]. Double asterisk indicates a statistically significant difference in the means between Col and the transgenic line, as revealed by two-tailed Student's t test (**, p<0.01). Bars indicate SD. (B) A biological repeat of ChIP analysis of AGL18 binding to FT chromatin presented in Figure 6E.
(EPS)
Analysis of AGL15 and AGL18 expression in Col and agl15 agl18 seedlings by RT-PCR. The constitutively expressed TUB2 served as a control.
(EPS)
Examination of CO interaction with AFR1, AFR2, or AGL18 by the yeast-two-hybrid assay. The full-length CO was fused with GAL4-AD, whereas the full-length AFR1, AFR2, and AGL18 were fused with GAL4-BD. Yeast cells were grown on the selective synthetic defined media lacking of W, L, and H or lacking of W and L.
(TIF)
List of primers used in this study.
(DOCX)
Acknowledgments
We thank Donna E. Fernandez for generously providing agl15 and agl18 seeds, Stanton Gevin for providing the BiFC vectors, and Flanders Interuniversity Institute for Biotechnology (Belgium) for providing the pHGW and pB2GW7 vectors.
Abbreviations
- AFR1
SAP30 FUNCTION-RELATED 1
- AFR2
SAP30 FUNCTION-RELATED 2
- AGL18
AGAMOUS LIKE 18
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
- CO
CONSTANS
- EYFP
enhanced yellow fluorescent protein
- FT
FLOWERING LOCUS T
- GUS
β-GLUCURONIDASE
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- LD
long day
- qRT-PCR
real-time quantitative PCR
- RPD3
Reduced Potassium Dependency-3
- SAM
shoot apical meristem
- SAP30
Sin3-Associated Polypeptide 30
- SBR
Sin3-binding region
- SD
standard deviation
- ,
Sin3-Associated Polypeptide 18 (SAP18)
- ZT
Zeitgeber time
Funding Statement
This work was supported in part by grants from the Singapore Ministry of Education (AcRF Tier 2; MOE2009-T2-1-081), the Singapore National Science Foundation (2010NRF-CRP002-018), and the Temasek Life Sciences Laboratory to YH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wigge PA (2011) FT, a mobile developmental signal in plants. Curr Biol 21: R374–378. [DOI] [PubMed] [Google Scholar]
- 2. Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594. [DOI] [PubMed] [Google Scholar]
- 3. Kobayashi Y, Weigel D (2007) Move on up, it's time for change–mobile signals controlling photoperiod-dependent flowering. Genes Dev 21: 2371–2384. [DOI] [PubMed] [Google Scholar]
- 4. Imaizumi T, Kay S (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558. [DOI] [PubMed] [Google Scholar]
- 5. Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- 6. Kardailsky I, Shukla V, Ahn J, Dagenais N, Christensen S, et al. (1999) Activation tagging of the floral inducer FT . Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- 7. Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis . Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- 8. Tamaki S, Matsuo S, Wong H, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- 9. Mathieu J, Warthmann N, Kuttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis . Curr Biol 17: 1055–1060. [DOI] [PubMed] [Google Scholar]
- 10. Jaeger K, Wigge P (2007) FT protein acts as a long-range signal in Arabidopsis . Curr Biol 17: 1050–1054. [DOI] [PubMed] [Google Scholar]
- 11. Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- 12. Berger S (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412. [DOI] [PubMed] [Google Scholar]
- 13. Wang Z, Zang C, Cui K, Schones D, Barski A, et al. (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Govind C, Zhang F, Qiu H, Hofmeyer K, Hinnebusch A (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25: 31–42. [DOI] [PubMed] [Google Scholar]
- 15. Kremer S, Gross D (2009) SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression. J Biol Chem 284: 32914–32931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Servet C, Conde e Silva N, Zhou D (2010) Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis . Mol Plant 3: 670–677. [DOI] [PubMed] [Google Scholar]
- 17. Deng W, Liu C, Pei Y, Deng X, Niu L, et al. (2007) Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis . Plant Physiol 143: 1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu C, Liu X, Luo M, Chen C, Lin X, et al. (2011) HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis . Plant Physiol 156: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han S, Song J, Noh Y, Noh B (2007) Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J 49: 103–114. [DOI] [PubMed] [Google Scholar]
- 20. Ahringer J (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 16: 351–356. [DOI] [PubMed] [Google Scholar]
- 21. Grzenda A, Lomberk G, Zhang JS, Urrutia R (2009) Sin3: master scaffold and transcriptional corepressor. Biochim Biophys Acta 1789: 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pandey R, Muller A, Napoli C, Selinger D, Pikaard C, et al. (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bowen A, Gonzalez D, Mullins J, Bhatt A, Martinez A, et al. (2010) PAH-domain-specific interactions of the Arabidopsis transcription coregulator SIN3-LIKE1 (SNL1) with TELOMERE-BINDING PROTEIN 1 and ALWAYS EARLY2 Myb-DNA binding factors. J Mol Biol 395: 937–949. [DOI] [PubMed] [Google Scholar]
- 24. Liu C, Xi W, Shen L, Tan C, Yu H (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16: 711–722. [DOI] [PubMed] [Google Scholar]
- 25. Song C, Galbraith D (2006) AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis . Plant Mol Biol 60: 241–257. [DOI] [PubMed] [Google Scholar]
- 26. Xie T, He Y, Korkeamaki H, Zhang Y, Imhoff R, et al. (2011) Structure of the 30-kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding. J Biol Chem 286: 27814–27824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ausin I, Alonso-Blanco C, Jarillo J, Ruiz-Garcia L, Martinez-Zapater J (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166. [DOI] [PubMed] [Google Scholar]
- 28. Lee J, Yoo S, Park S, Hwang I, Lee J, et al. (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis . Genes Dev 21: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang D, Wang Y, Wang Y, He Y (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu F, Cui X, Zhang S, Liu C, Cao X (2010) JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis . Cell Res 20: 387–390. [DOI] [PubMed] [Google Scholar]
- 31. Jeong J, Song H, Ko J, Jeong Y, Kwon Y, et al. (2009) Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis . PLoS One 4: e8033 doi:10.1371/journal.pone.0008033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang W, Jiang D, Jiang J, He Y (2010) A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis . Plant J 62: 663–673. [DOI] [PubMed] [Google Scholar]
- 33. Takada S, Goto K (2003) TERMINAL FLOWER 2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Searle I, He Y, Turck F, Vincent C, Fornara F, et al. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis . Genes Dev 20: 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adamczyk B, Lehti-Shiu M, Fernandez D (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis . Plant J 50: 1007–1019. [DOI] [PubMed] [Google Scholar]
- 36. Hill K, Wang H, Perry S (2008) A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J 53: 172–185. [DOI] [PubMed] [Google Scholar]
- 37. Tiwari S, Shen Y, Chang H, Hou Y, Harris A, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66. [DOI] [PubMed] [Google Scholar]
- 38. Kumimoto R, Zhang Y, Siefers N, Holt B (2010) NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana . Plant J 63: 379–391. [DOI] [PubMed] [Google Scholar]
- 39. Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, et al. (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis . Plant Cell 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blackman B, Michaels S (2010) Does CONSTANS act as a transcription factor or as a co-activator? The answer may be–yes. New Phytol 187: 1–3. [DOI] [PubMed] [Google Scholar]
- 41. Michaels S, Amasino R (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee L, Fang M, Kuang L, Gelvin S (2008) Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang D, Kong N, Gu X, Li Z, He Y (2011) Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet 7: e1001330 doi:10.1371/journal.pone.0008033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343. [DOI] [PubMed] [Google Scholar]
- 45. Curtis M, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta . Plant Physiol 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105. [DOI] [PubMed] [Google Scholar]
- 47. Johnson L, Cao X, Jacobsen S (2002) Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367. [DOI] [PubMed] [Google Scholar]
- 48. Li D, Liu C, Shen L, Wu Y, Chen H, et al. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis . Dev Cell 15: 110–120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignments of AFR1 and AFR2 relatives. Numbers refer to amino acid residues. Identical residues among these proteins are shaded black, whereas similar residues are shaded gray. (A) Alignment of the Arabidopsis AFR1 (AtAFR1) and AFR2 (AtAFR2) with the yeast Sap30 (ScSap30). The conserved residues in the putative SBRs are indicated by asterisks (*). (B) Alignment of AtAFR1 with its homologs from other plants including Populus trichocarpa (PtSFR), Picea sitchensis (PsSFR), and Physcomitrella patens (PpSFR). The GenBank accession numbers for PtSFR, PsSFR, and PpSFR are XP_002302473.1, ADE76839.1, and XP_001766227.1, respectively.
(EPS)
Direct interactions of AT1G75060 (AFR1) and AT1G19330 (AFR2) with SNL2 and HDA9 proteins in yeast. The indicated full-length proteins were fused with the GAL4-BD or AD domains. Yeast cells harboring the fusion proteins, BD and/or AD (as indicated), were grown on the selective synthetic defined media lacking of Trp (W), Leu (L), and His (H). (A,B) Interactions of SNL2 with AFR1 and AFR2 in yeast. (C,D) Interactions of HDA9 with AFR1 and AFR2 in yeast.
(TIF)
Analysis of AFR1 and AFR2 expression in afr1 and afr2 seedlings by semiquantitative RT-PCR. The constitutively expressed TUB2 served as a control. (A) Analysis of the full-length AFR1 and AFR2 transcripts. (B) Analysis of 5′- or 3′-truncated AFR2 transcripts in afr2 mutants.
(EPS)
Rescue of afr1 and afr2 mutants by pAFR1-AFR1:HA and pAFR2-AFR2:FLAG , respectively. Plants were grown in short days, and total leaf number for each line (9–11 plants per line) was scored. Bars indicate SD. (A) Flowering times of afr1-1 and a transgenic line of pAFR1-AFR1:HA in afr1-1 (native AFR1 fused with 3x HA; single-locus T3 homozygotes). (B) Flowering times of afr2-1 and a transgenic line of pAFR2-AFR2:FLAG in afr2-1 (native AFR2 fused with 3x FLAG; single-locus T3 homozygotes).
(EPS)
Analysis of flowering time and leaf initiation rate of afr1 afr2 in LDs. 12 plants were scored for each line; bars for SD. (A) Days to flowering of Col and afr1 afr2. (B) Total leaf numbers at flowering of Col and afr1 afr2. (C) Leaf initiation rates of Col and afr1 afr2 plants.
(EPS)
Relative AFR1 and AFR2 transcript levels in Col and co seedlings at the end of LDs, quantified by qRT-PCR. The transcript levels were normalized first to the endogenous control UBQ10, and relative fold changes to Col are presented. Bars indicate SD of triplicate measurements.
(EPS)
AFR1 and AFR2 moderately repress FT expression in short days. (A) Flowering times of the indicated lines grown in short days. Total leaf number for each line (8–14 plants per line) was scored. Error bars indicate SD. (B) FT mRNA levels in Col and afr1 afr2 seedlings over a 24-h short-day cycle. Total RNAs were extracted from 21-d old seedlings every 4 h and quantified by qRT-PCR. Bars indicate SD of three measurements.
(EPS)
Biological repeats of FT , AFR1 , and AFR2 expression analyses. (A) A biological repeat of the FT expression analysis presented in Figure 3E. (B) A biological repeat of the AFR1 and AFR2 expression analysis presented in Figure 4C.
(EPS)
Relative levels of AFR1:HA and AFR2:FLAG proteins over a 24-h LD cycle. The proteins in respective 10- to 11-d-old seedlings were analyzed by western blotting. The intensities of protein bands were quantified by the ImageJ 1.44j program, and relative levels to ZT0 are presented. Note that the time of the day has a significant effect on the levels of both AFR1 and AFR2 proteins in the seedlings, as revealed by single-factor ANOVA tests (for AFR1, p<0.01; for AFR2, p<0.05). (A) Relative AFR1:HA levels over a 24-h LD cycle. Average values of three biological repeats are presented; bars for SD. (B) Relative AFR2:FLAG levels over a 24-h LD cycle. Average values of four biological repeats are presented; bars for SD.
(EPS)
Biological repeats of the ChIP assays on AFR1 and AFR2 binding to FT chromatin. (A) A biological repeat of the ChIP analysis presented in Figure 4E. (B) A biological repeat of the ChIP analysis presented in Figure 4F. (C) A biological repeat of the ChIP analysis presented in Figure 7.
(EPS)
ChIP analysis of the histone H3 acetylation state of FT chromatin in Col and afr1 afr2 seedlings grown in LDs. Amounts of the immunoprecipitated genomic DNA was quantified and normalized first to the constitutively expressed TUB2. Subsequently, the fold enrichments were calculated by dividing the levels of acetylated H3 in afr1 afr2 (at ZT8 or ZT16) or the WT at ZT16, by the level in the WT at ZT8. Shown are the means and SD of two ChIP experiments.
(EPS)
Direct interactions of AGL18 with AFR2 and AGL15 with AFR1. (A) BiFC analysis of the interaction of AGL18 with AFR2 in onion epidermal cells. Yellowish-green signals indicate the binding of AGL18 with AFR2 in the nuclei (indicated by the blue fluorescence from DAPI). Bar = 20 µm. (B) AFR1 interacts with AGL15, but not FLC, in yeast cells. The full-length AFR1, and full-length FLC and the 208-aa AGL15 (without MADS domain) were fused with the GAL4-AD and BD domains, respectively. Yeast cells were grown on the selective synthetic defined media lacking of W, L, and H or lacking of W and L. Of note, FLC directly interacts with SVP and binds to FT chromatin to repress FT expression [48], and serves as a negative control in this experiment. (C) BiFC analysis of the interaction of AGL15 with AFR1 in onion epidermal cells. The full-length AGL15 and AFR1 were fused with nEYFP and cEYFP fragments, respectively. Yellowish-green signals indicate the binding of AGL15 with AFR1 in the nuclei. Bar = 20 µm.
(TIF)
Characterization of the p35S-AGL18:FLAG line and a biological repeat of ChIP analysis of AGL18 binding to FT chromatin. (A) Flowering times of the p35S-AGL18:FLAG line (T3; in the Col background) grown in LDs. Overexpression of AGL18 causes moderate late flowering [35]. Double asterisk indicates a statistically significant difference in the means between Col and the transgenic line, as revealed by two-tailed Student's t test (**, p<0.01). Bars indicate SD. (B) A biological repeat of ChIP analysis of AGL18 binding to FT chromatin presented in Figure 6E.
(EPS)
Analysis of AGL15 and AGL18 expression in Col and agl15 agl18 seedlings by RT-PCR. The constitutively expressed TUB2 served as a control.
(EPS)
Examination of CO interaction with AFR1, AFR2, or AGL18 by the yeast-two-hybrid assay. The full-length CO was fused with GAL4-AD, whereas the full-length AFR1, AFR2, and AGL18 were fused with GAL4-BD. Yeast cells were grown on the selective synthetic defined media lacking of W, L, and H or lacking of W and L.
(TIF)
List of primers used in this study.
(DOCX)