Abstract
Although it is widely accepted that the regulation of the chromatin landscape is pivotal to conveying epigenetic phenomena, it is still unclear how a defined chromatin domain is reproduced following replication and transmitted from one generation to another. Here we review multiple mechanisms that contribute to the inheritance of epigenetic information with emphasis on the recycling of old histones following replication, the requirement for a positive feedback loop, long-range gene interactions, and the complex network of trans-acting factors.
Proper development of multicellular organisms entails the distinct specifications of disparate cell types. Each of these cellular identities will be conserved during later cell divisions. Despite an identical genomic sequence, these cells exhibit a substantially different profile of gene expression. This highlights the role of “epigenetics” and its stable and inheritable information that is distinct from DNA sequence and fostered by specialized mechanisms. These mechanisms include DNA methylation, small interfering RNA, histone variants, and histone posttranslational modifications (PTMs). To date, however, only DNA methylation has been shown to be stably inherited1. While some histone PTMs are expected to contribute to the transmission of epigenetic information, others actively participate within the process of transcription, the so called “active marks” and others are likely to be restricted to “structural functions”2,3.
In eukaryotes, 1.65 turns of DNA are wrapped around an octamer of histones that include two copies of H2A, H2B, H3, and H4 thereby forming the nucleosome, the basic unit of chromatin. Each core histone contains a well structured globular domain and a more flexible N-terminal tail protruding out from the nucleosome4. These tails are characterized by a high lysine content that gives rise to an overall positive charge and contrasts the surrounding negative DNA charge. DNA folding around octamers is the first step of compaction, forming the 11-nm “beads on a string” fiber. However, additional levels of compaction are required to pack the roughly 2m long DNA filament into the nucleus. It is these higher order chromatin structures that are still poorly understood.
The folding of DNA into chromatin presents a barrier to the machinery that either transcribes, replicates, or repairs DNA. However, it has become evident that the structure of chromatin is not only a protective sheath to guard against these processes occurring in an untimely manner, nor simply a packaging tool, but is also a dynamically adjusted entity that reflects the regulatory cues necessary to program appropriate cellular pathways. Many of these regulatory features are inherited.
Since the first discovery of enzymes that modify histones, numerous laboratories have focused on the identification, the characterization and the role(s) of various histone posttranslational modifications (PTM) such as acetylation, methylation, phosphorylation, and ubiquitinylation5. Genome-wide approaches (ChIP-chip, ChIP-seq) have provided a partial picture of the chromatin landscape including the localization of histone PTMs and histone variants, DNA methylation patterns, and nucleosome occupancy. Moreover, the discovery of protein domains that specifically recognize a defined histone modification, including the chromodomain, bromodomain, PHD, and MBT domains have advanced our understanding of the role of histone PTMs6,7. From these data, histone PTMs were correlated with more or less defined functions such as gene regulation7. However, it also became clear that a single type of histone PTM does not dictate a defined function, as for example in the case of H3K9me3 that is found in silent heterochromatin, but also at some active genes7. In light of this, it seems more prudent to consider chromatin as composite domains (Figure 1). These domains would be characterized by the local enrichment of a histone PTM in combination with other specific PTMs, the presence of histone variant(s), nucleosome occupancy, DNA methylation patterns, and possibly nuclear localization. It is this totality of features that we refer to as chromatin structure or landscape.
Figure 1.
Characteristics of a Chromatin Domain.
Schematic depicting modifications within chromatin that define different chromatin domains. The green dashed lines represent the separation between two adjacent domains. For simplicity, H2A-H2B have been omitted.
Although some key players that regulate chromatin structure are well defined and the functions of certain histone PTMs are clear, exactly how this entire machinery that is composed of the enzymes that modify histones, the histone modifications themselves, and the proteins that recognize these modifications, is localized and restricted to specific loci is currently unclear. Consequently, little is known about how the regulatory information embedded in the chromatin structure is carried over during cell divisions.
We first describe two examples of heritable transcription regulation extracted from two model systems, yeast and Drosophila, in which genetic analyses have contributed importantly to our understanding of chromatin regulation. We will then analyze how histones are segregated during DNA replication and highlight some of the mechanisms required to maintain defined chromatin domains. Finally, we will describe findings that show how chromatin structure acts in concert with trans-acting factors to carry the information understood as being epigenetic.
I) Examples of heritable transcription regulation
Ia) Transcriptional memory in yeast
The inhibition of histone deacetylase activity in cells results in increased histone acetylation and consequently changes in gene expression. Hence, a reporter gene integrated into the centromeric heterochromatin of fission yeast cells switch from a repressed to an active state after exposure to Trichostatin A (TSA, inhibitor of Class I and II histone deacetylases) for 5 generations8. Following this observation, the authors shifted the yeast to media without TSA and monitored the expression of the reporter gene for 200 generations. They observed that the reporter stayed active and hyperacetylated, although at each cell division an average of 2% of the cells reverted to the repressed state8. Interestingly, this retention of an active state required the integration of the reporter within a heterochromatic environment and was maintained through crosses with yeast that had not been exposed to TSA suggesting that this activation was not solely a consequence of transcription factor regulation.
A different example of transcriptional memory in yeast has been described for the GAL regulon. Genes of the yeast GAL regulon are repressed in glucose medium, but strongly induced in the presence of galactose as the sole carbon source. The kinetics of GAL gene activation are dramatically different depending upon prior cell exposure to galactose: whereas galactose induction is slow, requiring up to 2 hrs for full activation, reinduction following a cycle of activation and repression occurs in minutes 9-11. This effect has been referred to as “transcriptional memory.”
The Set1 histone H3 lysine-4 (H3 K4) methylase is targeted to transcriptionally active genes. H3 K4 methylation persists at the GAL10 gene through a cycle of activation and repression, leading to the suggestion that H3 K4 methylation provides a “memory” of recent transcriptional activity 12. However, the enzymes responsible for H3K4me, H3K79me, H2B123Ub and H3Ac were shown to be dispensable for the rapid re-induction 10. GAL gene memory requires the histone variant H2A.Z and the SWI/SNF chromatin remodeling complex 9, 10. A possible explanation for SWI/SNF requirement came from the observation that the GAL1 gene is relocated to the nuclear periphery after the first induction, in a manner dependent upon the histone variant H2A.Z10. Moreover, SWI/SNF deletion prevents H2A.Z deposition onto DNA13. Interestingly, a transcription dependent physical interaction between promoter and terminator (sequences regulating poly A addition) was reported and this “gene looping” is suggested to facilitate transcription re-initiation 14. Independently Hampsey, Proudfoot and their colleagues reported that gene looping is critical for the re-induction of the Gal10 and HXK1 genes respectively (Laine and Hampsey, 09; Wong and Proudfoot, 09). These observations suggest that retention of components of the transcription machinery following active transcription might play an important role in the rapid re-induction of transcription in yeast. In contrast, Tzamarias and colleagues argue that the cytoplasmic level of the Gal1 galactokinase is critical to transcription memory in yeast. This interpretation is supported by the finding that formation of heterokaryons (cells with two nuclei that share the same cytoplasm) in which naïve cells respond quickly to galactose when fused with the cytoplasm of cells induced with galactose prior to fusion11.
Whichever mechanism(s) for “transcription memory” is operational, the above experiments demonstrate the existence of this phenomenon and that the histone variants (H2AZ), the chromatin remodeling complex Swi/Snf, and nuclear localization are important players. Moreover, these studies suggest that the actual structure of the transcription machinery and/or a cytoplasmic kinase can affect the process of “transcription memory”. Most importantly to the studies described in this review, the effects observed were found to be independent of histone modifications, and since the process involved the maintenance of a transcription permissive environment, we conclude that histone modifications that function in transcription (“active marks” H3(K4/K36/K79)me3 are irrelevant to the process of “transcription memory”. The important question then is whether other histone modifications such as those involved in maintaining a repressed state (H3K9/27me and/or H4K20me depending on the model) are important players in the establishment of a chromatin domain that is inherited.
Ib) Position effect variegation
Position Effect Variegation (PEV), a phenomenon initially described in Drosophila but also reported in yeast15, reflects either the silencing of a euchromatic gene when artificially moved in proximity to a heterochromatic region or gene activation in the reverse case16, 17. Using genes whose transcriptional status can be easily monitored (such as eye color), this phenomenon was explored by screening for mutations that enhanced or suppressed variegation; up to 150 loci encoding modifiers of PEV were identified. Importantly PEV is established early in development (by the end of the first larval instar) although eye cells undergo further divisions indicating this regulation is inherited17. Some of the modifiers of variegation display a dosage-dependent effect, meaning that they have opposite effects on variegation whether they are up- or downregulated through genetic manipulation, as reported for Su(var)2-5, Su(var)3-7 or Su(var)3-918. Also, the extent of the heterochromatin involved is critical to PEV regulation as illustrated by the enhanced variegation observed in XO males as compared to XY, considering that the Y chromosome in somatic cells is heterochromatic16. Of note, distinct regions of chromatin appear to be regulated differently. Some of the modifiers of variegation that were identified upon rearrangement of a euchromatic gene at pericentric heterochromatin were ineffectual when this same gene was positioned at subtelomeric heterochromatin17.
Characterization of the modifiers of variegation revealed that many are involved directly or indirectly in the regulation of chromatin structure19. Hence, reduced levels of several histone methyltransferases such as Su(var)3-9, E(z), dSETDB1, Pr-SET7, Suv4-20, or histone demethylases such as Su(var)3-3 were shown to suppress variegation17, 18, 20. The modifications placed by these enzymes are all associated with a repressed state of chromatin and can work in the same pathway or target distinct regions of heterochromatin as shown by the selective loss of heterochromatin on the fourth chromosome upon deletion of dSETDB117, 21 or pericentric heterochromatin upon deletion of Su(var)3-918. Conversely, loss of function of the kinase JIL-1 that phosphorylates H3S10, and thus prevents the methylation of H3K9, acts as an enhancer of variegation at pericentric heterochromatin22. An interesting case of PEV has been reported for the Brown gene. Although PEV is mostly recessive, insertion of a block of centromeric heterochromatin at the Brown locus was reported to have a dominant effect. This trans-inactivation of the second Brown allele is a consequence of chromosome pairing that brings the wild type allele to a heterochromatic region of the nucleus during the interphase period of the cell cycle23. Although chromatin regulation plays a critical role in PEV, we will see throughout this review that the regulation of heterochromatin establishment and maintenance is also regulated by the RNAi machinery and transcription factors.
From these two examples, it is evident that chromatin plays a critical role in the inheritance of transcriptional regulation. However for successful cell division, DNA must be replicated and implicit in this is that the inherent disruption of chromatin is followed by the incorporation of newly synthesized histone. To understand how the modifications of chromatin control its structure and can be propagated, we will briefly summarize how replication occurs and then consider a few models of histone segregation.
II) Histone deposition following replication
The regulation of DNA replication starts in the late M phase of the cell cycle when replication initiation sites are targeted by the ORC (Origin Recognition Complex)24, 25. This is followed by the loading of the MCM complex as well as other proteins to form the preRC (preReplication Complex). This complex is then activated at the G1/S boundary under conditions that prevents the production of new preRCs, ensuring that replication occurs only once per cell cycle. The activation step leads to the production of large complex containing multiple proteins (Replisome Protein Complex) required for the formation of the MCM helicase activity that catalyze DNA unwinding. 26. The last step is the formation of the replisome that entails loading of the replicative DNA polymerases and auxiliary proteins including the clamp loader (RFC), the clamp and the single-strand DNA binding protein RPA. Importantly, the initiation of DNA synthesis requires the synthesis of RNA primers by DNA primase which are extended by DNA polymerase α. Following the loading of PCNA (Proliferating Cell Nuclear Antigen) by Replication Factor C, DNA polymerase δ and ε take over27. PCNA interacts with the DNA polymerases (Figure 2A) and markedly increases their processivity. Of note, the leading and the lagging strands are replicated in a different manner due to the 5′→3′ directionality of the polymerase activities such that the leading and lagging strands are synthesized continuously and discontinuously respectively. Consequently, multiple priming and PCNA loading events are required on the lagging strand.
Figure 2.
Models of histone deposition during replication.
Here we illustrate the means by which old and new histones could be deposited following replication. (A) General simplified view of the replication fork. The MCM helicase complex is shown at the fork. The possible interaction of MCM with the histone chaperone ASF1 is shown. The leading strand shows the replication fork, simplified by depicting DNA polymerases ε (Pol), its interaction with PCNA and the interaction of PCNA with ASF1. On the lagging strand, ASF1 and PCNA could interact at three different steps, first during Okazaki fragments replication by the DNA polymerase δ, second during DNA ligation and third with PCNA at the chromatin following replication. Underneath, we present what we believe are important questions that need to be addressed. (B) Random model of histone segregation. In this model, we do not discriminate between dimer or tetramer deposition. The histones segregate randomly between the leading and lagging replicated DNA strands. (C) Semi-conservative model of histone segregation. Left, the scheme is based on the assumption that the H3-H4 tetramer is divided during DNA replication and the parental H3-H4 segregate as dimers onto the newly replicated DNA strands. The parental histones associate with naïve dimers to reconstitute the tetramer. Right, this scheme posits that parental H3-H4 histones segregate as tetramers resulting in the joint deposition of recycled histones and newly deposited naïve histones.
In mammals, different variants of histone H3 exist: the highly related H3.1, H3.2, and H3.3, and the less conserved centromeric Cenp-A histone. Although H3.1 and H3.3 differ by only 5 amino acids, their incorporation into chromatin is regulated distinctly. H3.1 and H3.2 are deposited during DNA replication (S-phase) while H3.3 is deposited during transcription7. Whether in the cytoplasm or during their subsequent import into the nucleus, histones are not free but bound by histone chaperones and histones H3 and H4 are always co-associated. Interestingly, histones are modified posttranslationaly prior to their deposition, hence their K5 and K12 residues are acetylated on most of the cytoplasmic H4 from metazoans to humans28. These marks are required for histone deposition and are removed during chromatin maturation. A sub-fraction of human cytoplasmic histone H3 displays monomethylation of H3K9 and also some acetylation, but the role of these modifications are unclear29. In yeast, H3K56 acetylation plays an important role in nucleosome assembly as a recent study reported that this mark increases the affinity of the CAF-1 chaperone for histone H330. Although this mark was recently detected in mammals31-33, its function is still not clear.
Histones are removed during the passage of the replication fork and in vitro replication experiments have shown that the DNA is refolded into chromatin close behind the replication fork with a slightly different timing for the leading and lagging strands7. It is now accepted that old histones are deposited on both strands of the newly replicated DNA, first H3-H4 and later H2A-H2B7. For a long time it was thought that H3-H4 would be deposited as a tetramer, but recent reports showed that free H3-H4 exists in cells as a dimer7, 34. This observation was supported by the structure of the histone chaperone ASF1 which revealed that it interacts with a dimer of H3-H435, 36.
Histone segregation could follow two different scenarios: First, chromatin can be formed from a pool of new and old histones that are randomly deposited on the newly replicated DNA; this is the “random model” (Figure 2B). This model infringes on the importance of chromatin domain inheritance. Indeed, not only would histone PTMs be diluted by the incorporation of new histones, but also their distribution relative to the DNA sequence is likely to be modified. It is therefore expected that only a defined or a combination of histone PTMs enriched on several adjacent nucleosomes and possibly on each copy of the histones would be transmitted effectively (Figure 2B, red dot). In contrast, histone PTMs present on only a few copies would get diluted and might lose their significance (Figure 2B, yellow dot). In view of this model, whether segregation of H3-H4 occurs as dimers or tetramers is a relatively moot point as the overall enrichment of the domain for a histone PTM takes precedence.
Second is the semi-conservative model suggesting that H3-H4 dimers or tetramers are distributed equally between both strands following DNA replication (Figure 2C). Propagation of epigenetic information would then require machinery that duplicates the histone marks between the corresponding tails of a nucleosome if the segregation occurs as dimers or between adjacent nucleosomes if the segregation occurs as tetramers. Although this hypothesis is attractive in its simplicity with respect to propagation of histone PTMs, it is difficult to conceive how each H3-H4 dimer/tetramer would be deposited, after replication, in an ordered manner on the leading and lagging strands since both strands are not simultaneously synthesized. It was shown that H3-H4 exists as dimers in vivo, but this does not require that they are deposited as dimers considering that H3-H4 are not free outside of the nucleosome, but are instead carried by histone chaperones. ASF1 binds histone H3-H4 as dimers, the trimeric CAF137 complex and it has been suggested that ASF1 also interacts with the MCM complex 37. How the ASF1-H3-H4 complex is transferred across the replication fork and whether ASF1 is the only chaperone that supports this function is unclear. Moreover, it is not known whether ASF1 itself is monomeric or dimeric, and the stoichiometry of the complexes that contain ASF1 (i.e. CAF1-ASF1-H3-H4) and whether PCNA, which is a homotrimer interacts with one or three CAF1 molecules is also unknown (Figure 2A).
III) Requirements for epigenetic inheritance
Based on of the nucleosome modifications that occur at the silent mating-type region in yeast, Thon and colleagues developed a mathematical model of histone PTM regulation in the context of a two state process38. The authors assumed that the model is applicable for bistable conditions meaning that transition from one state to another occurs but at a relatively low frequency. The bistability is efficiently maintained through replication assuming that histone PTMs are randomly segregated during replication and that newly deposited nucleosomes are naïve (i.e. devoid of the histone modifications regulating the transition from one state to another and are therefore potential targets for these modifications). We mentioned in the previous section that newly incorporated histones do not carry the PTMs potentially involved in transmitting epigenetic information. The bistability requires cooperativity between the histone modifications and positive feedback for the spreading of a defined mark. Furthermore this positive feedback should propagate the mark not only linearly, i.e. to adjacent nucleosomes, but also reach targets several nucleosomes away, thus highlighting the importance of higher order chromatin structure. In the next sections, we will consider known mechanisms that could be critical for the propagation of epigenetic information in light of these theoretical predictions, which we believe to be of great importance in understanding transmission of histone marks and the establishment of chromatin domains that are inherited.
Importantly, while we have some idea about the propagation of DNA methylation and models are emerging for the duplication of histone PTMs, very little is known about inheritance of nucleosome occupancy or histone variants. It was reported that the silencing of the MLH1 promoter in cancer is associated with specific changes in nucleosome occupancy39. Considering that this promoter is also specifically DNA methylated in cancer cells and that changes in nucleosome occupancy are reversed by inhibition of DNA methylation, nucleosome occupancy in this case might be regulated by DNA methylation39. Regarding histone variants, a better characterization of the histone deposition machinery will be required to understand how they might be stably maintained at a defined locus.
IIIa) Duplication of chromatin structure coupled to replication
Following DNA replication the newly formed chromatin must carry the epigenetic information. This step is critical as underrepresentation of the epigenetic information might render that chromatin more likely to switch from one state to another. To insure the stability of the chromatin structure, cells should rapidly duplicate this epigenetic information. In this section, we will discuss the current models for the chromatin-mediated propagation of epigenetic information.
In the case of histone PTMs and considering that old histones are recycled, each chromatin domain could carry a blueprint of the information to be duplicated. It has been suggested that PCNA plays a crucial role in orchestrating the various enzymes required to modify the newly replicated chromatin40, 41. In eukaryotes, the PCNA homotrimer forms a ring shaped structure that slides along the DNA following the DNA polymerase. PCNA not only follows the polymerase, but has been shown to remind bound to the lagging strand chromatin after replication has terminated41. Accordingly, the ratio of trimeric PCNA loaded on the DNA to the replication fork increases markedly during S-phase42. PCNA interacts with many proteins though a hydrophobic pocket situated between its two globular domains27. These proteins usually have a conserved motif called the PIP box and in theory each PCNA ring could contact three different interactors simultaneously.
Cytosine methylation is required for development in large-genome eukaryotes43, knockout of DNA Methyl Transferase 1 (DNMT1) in mice results in early embryonic lethality44. At the cellular level, condition that interfere with the expression of DNMT1 result in cell cycle arrest and cell death45-47. Three DNA methyltransferases have been characterized in mammals, Dnmt1, Dnmt3A and -3B. A simple model is that Dnmt3A and -3B are involved in de novo methylation while Dnmt1 acts on the maintenance of DNA methylation and recognizes specifically hemimethylated DNA. Dnmt1 localizes to replication foci in mid/late S phase48 and was reported to interact with PCNA through its BMAP domain49. However deletion of this domain does not prevent DNMT1 association with the replication foci and it is still able to propagate DNA methylation46, 50. One explanation for this observation came from recent reports of another protein, Np95, that interacts with Dnmt1 and PCNA51, 52. Np95-/- ES cells exhibit a global loss of DNA methylation and a diffuse staining for Dnmt1, instead of its being localized to the replicative heterochromatin region. Moreover Np95 binds methylated CpG through its SET and RING finger-associated domain (SRA)40.
Importantly the distribution of DNA methylation might be a major determinant of the chromatin landscape by controlling histone variant deposition and histone PTMs. In accord with this notion it was shown that H3K4me3 and DNA methylation are inversely correlated53 and, at least in plants, DNA methylation and H2A.Z are also mutually exclusive54 while H3K9me2/3 and DNA methylation are highly coincident55. The link between DNA methylation and H3K9me2/3 is further illustrated by the decreased levels of H3K9me2/3 upon knockdown of DNMT156. Conversely, in N. Crassa, inactivation of a gene related to Su(var)3-9 or mutation of lysine 9 of histone H3 leads to decreased DNA methylation57, 58. In mammals, there are at least five enzymes that catalyze H3K9 methylation including Suv39h1/2, G9a and ESET. Knockdown of Suv39h1/h2 or of G9a impacted DNA methylation at defined loci59, 60. Interestingly, it has been reported that G9a and ESET/SETDB1 interact with PCNA40. G9a could form a complex with DNMT1 and PCNA, while SETDB1 may be associated with the methyl CpG binding protein MBD1 and interact with PCNA through CAF1. The exact mechanisms connecting replication to DNA methylation and H3K9 methylation are still not completely elucidated and might depend on the genomic locus and time of replication. However they appear to be tightly associated to replication through PCNA.
PR-Set7, the only histone methyltransferase that catalyzes H4K20me1 has been reported to interact with PCNA61-63. PR-Set7 and H4K20me1 are cell cycle regulated and accumulate during mitosis64. H4K20me1 levels then decrease possibly due to its conversion to H4K20me2/3 and dilution upon incorporation of naïve histones during DNA replication. PR-Set7 is degraded subsequent to its ubiquitination by SCF/Skp2 in the early G1 phase of the cell cycle65. Genome-wide approaches revealed that H4K20me1 tends to be enriched at active genes66, 67, however H4K20me1 is also found in compact chromatin structures such as the inactive X chromososme68, 69. Finally, this mark has been associated with chromatin maturation70. Considering that the Drosophila homolog of PR-Set7 was reported to be a suppressor of variegation20, it is expected that H4K20me1 should convey some epigenetic information3. Consequently, it was proposed that PR-Set7 propagates H4K20me1 during S-phase through its interaction with PCNA. However, this hypothesis is difficult to reconcile with the low levels of H4K20me1 in S-phase and the finding that PR-Set7 is present only in the G2-M phase of the cell cycle69. Instead, we envision that PR-SET7 would function mostly during mitosis and could be tethered to specific loci through proteins that specifically interact with H4K20me1, such as L3MBTL1 (71 and H. Oda, P. Trojer and D.R., unpublished data). The interaction between PCNA and Pr-Set7 might be important to bring Pr-Set7 to DNA damage site where it can cooperate with the Suv4-20h1/2 enzymes. A role for Pr-Set7 in DNA damage is consistent with increased γ-H2A.X observed upon deletion of Pr-Set7 61, 62, 69,72.
IIIb) Positive feedback loop and cooperativity
Following replication, newly formed chromatin is likely subjected to the action of various chromatin modifiers. To insure the faithful propagation of information by the end of mitosis, a defined modification should not only serve as a template but also promote its further deposition on newly incorporated nucleosomes. This requires cooperativity between the mark and the enzyme that catalyzes it, constituting a positive feedback loop. We will discuss a few examples of such a regulatory scheme that have been described thus far.
The hallmark of heterochromatin from fission yeast to mammals is the methylation of H3K9. Although several histone methyltransferases target this residue in mammals, it was shown that Suv39h1/2 are responsible for H3K9me3 at constitutive heterochromatin73. Heterochromatin Protein 1 (HP1) was first identified because of its association with heterochromatin74 and shown to modulate Position Effect Variegation in Drosophila75. This protein is highly conserved with homologues present from fission yeast to humans76. A link to histone methylation was first shown in yeast where it was reported that H3K9 methylation mediated by Clr4, was crucial for the recruitment of Swi6/HP177. Indeed, the chromodomain of Swi6/HP1 mediated the recognition of H3K9me2/378, 79. Furthermore, HP1 also directly interacts with Suv39h1/218, 80. H3K9me3 and heterochromatin could spread through the positive loop constituted by HP1 recognition of H3K9me3 and its subsequent recruitment of Suv39h1/2. The role of non-coding RNA in this positive loop will be discussed below.
Another example of such positive feedback involves the Polycomb Repressive Complex, PRC2, whose enzymatic component Ezh2 catalyzes H3K27 methylation. The literature describing how Ezh2 maintains transcriptional repression is somewhat controversial. Studies using Gal4 DNA-binding domain chimeras to artificially recruit Ezh2, or a component of the PRC2 complex, or an associated polypeptide (PHF1) to a reporter transgene (Gal4-TK-Luciferase) showed that it leads to transcriptional repression81,82. One study concluded that this repression was stable through cell divisions82. In this case, the ratio of Gal4-Ezh2 (or Eed) to H3K27me3 levels within the reporter was monitored as a function of different cell generations after the withdrawal of tetracycline that controls the expression of the chimeras. Yet, a previously published study81 and unpublished data (R.M. and D.R.) contrast this observation. Such differences might reflect the difficulty in ascertaining that 100% of Gal4-Ezh2 is removed after washing away the tetracycline and/or they might be a consequence of the multiple reporter integration sites. Of note, similar discrepancies were reported with the artificial recruitment of Gal4-HP183, 84.
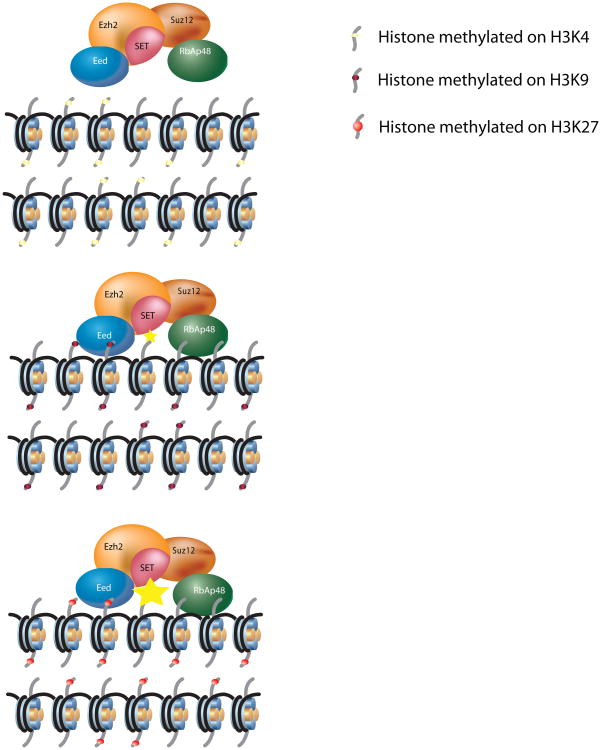
A recent study using a multidisciplinary approach including biochemistry, structural biology, and genetics revealed an important mechanism for the propagation of H3K27me3. This study demonstrated that the PRC2 component EED, a protein with seven β-propeller folds, forms an aromatic cage that allows Eed or the PRC2 complex to bind to repressive histone marks (H1K26me3, H3K9/27me3 and H4K20me3) within nucleosomes. The specificity toward repressive histone marks resides in the hydrophobic interaction between the alanine two residues N-terminal of the methylated lysine (H1K26, H3K9/27) or the leucine two residues C-terminal of the methylated lysine (H4K20me3) and a small pocket on the surface of Eed. Eed was found to be solely responsible for the specific recognition of trimethylated repressive lysines, although other PRC2 components contribute to the overall affinity for nucleosomes85. More importantly, this specific Eed binding leads to an allosteric stimulation of PRC2 activity, and this stimulation was optimal when Eed made specific side-chain interaction with amino acids preceding lysine-27 of the H3 tail. Hence, while Eed displays a slightly higher binding affinity for H3K9me3 relative to H3K27me3, H3K9me3 stimulates PRC2 activity poorly (Figure 3). Most importantly, using the structure of Eed protein as a guide, mutant strains of Drosophila were generated in which the aromatic residues important for cage formation and binding to repressive marks were altered. These mutant strains gave rise to Polycomb phenotypes and reduced levels of H3K27me, thereby underscoring the biological relevance of the Eed cage residues. Together, these results demonstrate that the ability of a histone PTM to promote its own duplication is required for its maintenance85. Furthermore, the observations that several histone repressive marks can support binding of PRC2 and that the co-presence of H3K27me3 results in the allosteric stimulation of PRC2-mediated formation of further H3K27me3, highlight the importance of chromatin domains and further stress that histone marks do not function in isolation.
Figure 3.
Propagation of H3K27me3 by PRC2.
This scheme shows how pre-existing histone methylation marks regulate PRC2 mediated spreading of H3K27 methylation. For simplicity, only one type of histone methylation is presented for each domain although in vivo a combination of them should be taken into consideration. Importantly this scheme does not consider the recruitment of PRC2. Three examples are envisioned: Top, a chromatin domain enriched for an “active mark” such as H3K4me3 that is not recognize by PRC2 and for this reason does not methylate H3K27. Middle, a chromatin domain is enriched for repressive marks such as H3K9me3, H1K26me3 or H4K20me3 that are recognized by PRC2 recognizes but its enzymatic activity is modestly increased (small yellow star). Bottom, a chromatin domain enriched for H3K27me3 that is recognized by PRC2 recognizes this mark and its enzymatic activity is robustly stimulated (large yellow star).
Cooperativity between chromatin modifiers is a likely consequence of the mutual exclusivity of some PTMs at a defined residue. For example, acetylation and methylation cannot occur on the same residue. Thus histone deacetylases have been found associated with histone methyltransferases as observed with Su(var)39 and either HDAC186 or SirT187. However, cooperativity might also occur for the same type of modification. Hence, the histone methyltransferases Suv39h1/2 and Suv4-20h1/2 di- and tri-methylate H3K9 and H4K20, respectively, but each requires a monomethylated substrate73, 88. Consistent with this, deletion of PR-Set7 that targets H4K20me1 is associated with a loss of H4K20me2/369. More work is required to understand how monomethylation is targeted and whether this substrate contributes to the regulation of the propagation of the di- and tri- methylated versions. Histone PTMs present on distinct residues were also reported to be exclusive as in the case of H3S10-phosphorylation and H3K9me389 indicating that one modification must be erased in order to establish the other. It is likely that cooperativity between chromatin modifiers is a general mechanism.
IIIc) Nuclear architecture and long-range interactions
Transcriptional regulation is usually considered to result from a competition between active and repressive activities that control the level of compaction of a defined gene locus. However it has become evident that a larger picture should be considered, one that takes into account transcription factors, transcription machinery, and chromatin modifying enzymes that are not randomly distributed in the nucleus, but organized in subnuclear domains and bodies90. For instance, heterochromatin in mammalian cells is associated with the nucleolus and nuclear periphery, and defined chromosomes are organized into territories91, 92. With the exception of early G1 phase, chromatin mobility is limited although targeted movements are observed upon gene activation, for instance91. For example, the artificial recruitment of the transcriptional activator VP16 to a transgene is associated with increased mobility of the transgene toward the nucleoplasm93; such movements were also reported for endogenous genes (for review see91). However, it has also been shown that active transcription occurs at the nuclear periphery when genes are localized in the vicinity of nuclear pores94. The link between nuclear organization and chromatin regulation is further supported by the observation that mutation in the lamin A, a component of the nuclear lamina, is associated with reduced levels of H3K9me3 and H3K27me395, 96. Other studies have described relationships between the nuclear architecture and chromatin regulation, but our current knowledge is mostly correlative and little is known about the underlying mechanism.
In contrast, the development of technologies such as chromosome conformational capture has led to a better characterization of long-range chromatin interactions. These interactions can occur in trans as described for the case of the Fab-7 transgene, the insertion of which leads to a variegated repression of neighboring genes97. Fab-7 contains a Polycomb Response Element that is recognized by Polycomb Group proteins in Drosophila. Fab-7 transgene silencing is enhanced in homozygous female flies and requires the endogenous Fab-7 locus. It was shown that the Fab-7 transgene and endogenous loci were physically associated in a manner dependent on the Polycomb Group proteins. Of note, although no clear evidence support the involvement of small non-coding RNA in Polycomb Group protein regulation98, the RNAi machinery was required for the long-range interaction between transgenic Fab-7 copies99. Physical interactions between chromatin regions whose silencing is maintained by Polycomb proteins were reported to be a general mechanism100, 101. Given that Polycomb proteins form subnuclear structures called polycomb bodies102, it is tempting to speculate that higher order chromatin structure could promote the spreading of chromatin-mediated gene silencing. The imprinted Igf2 and H19 mouse genes is a well-documented example of long-range chromatin interaction occurring in cis. The CCCTC-binding factor, CTCF, forms a complex with the Imprinting Control Region (ICR) upstream of H19 and regions proximal to the Igf2 gene103-106. This interaction leads to the formation of a loop which insulates Igf2 from its enhancer and results in its silencing. Importantly, at this loci CTCF also recruits PRC2 that contributes to Igf2 silencing through H3K27 methylation106.
IV) The role of trans-acting factors
Sequencing of the human genome revealed that the number of protein-coding genes does not dramatically increase from C.Elegans to humans, although the size of their respective genomes differs significantly. Consequently, the proportion of the human genome that encodes proteins is limited (1.2%)107. Yet a large part of the genome is being transcribed108 and the functional analysis of 1% of the human genome (ENCODE project) revealed that 70-90% of genome sequences are found in primary transcripts in various cells or tissues and about 15% in any given cell type109. Advances in genome-wide analysis of RNA uncovered a growing family of non-coding RNA (ncRNA); even in the cytoplasm a majority of the polyadenylated RNAs do not correspond to annotated exons from protein coding genes110. In addition to the well-recognized ncRNAs (tRNA, rRNA and snRNA), several classes of small and long ncRNAs have been characterized. Among the small versions, the best characterized are the siRNAs, miRNAs, and piRNAs. SiRNAs and miRNAs are produced from dsRNAs or precursor hairpin RNAs by the ribonuclease Dicer111 while piRNAs are derived from single strand precursors by a mechanism not yet clearly established107. Typically, these small ncRNAs regulate gene expression through targeting RNAs for degradation or by translation inhibition (miRNA and siRNA)111. Small ncRNAs were also reported to regulate DNA methylation in plants112 or DNA elimination in tetrahymena113.
Another important feature of small ncRNAs is their involvement in heterochromatin assembly. In fission yeast where heterochromatin formation has been extensively studied, it was shown that heterochromatin displays low histone acetylation, high methylation of H3K9 and was disrupted upon interfering with the RNAi machinery114. Similar properties were described for heterochromatin in Drosophila115 and to some extent in mammals, although in the latter system another layer of regulation is provided by DNA methylation116. Importantly, while interference with the RNAi machinery resulted in loss of H3K9 methylation at the centromeres in S. pombe and Drosophila, the knockout of Dicer in mammalian ES cells led to divergent results117, 118. In S. pombe, it was shown that H3K9 methylation is required to stabilize the RNAi machinery at chromatin119.
Considering the interdependence between chromatin modifiers/readers and RNAi machinery, both of which are required for the maintenance of heterochromatin, it is important to understand how these two distinct system are targeted to chromatin in the first place, and how they jointly act. The current model suggests that the RNA-Induced-Transcriptional-Silencing (RITS) complex is recruited to nascent RNA in a RNAi and H3K9 methylation dependent manner119. Artificial recruitment of RISC to nascent transcripts resulted in subsequent methylation of H3K9 and the recruitment of the RNA-Directed-RNA-polymerase Complex (RDRC) and Swi6 which result in heterochromatin silencing120, 121. Increased H3K9 methylation and RNAi production would be expected to generate a positive feedback loop allowing for the spreading of the heterochromatin in cis. Thus, heterochromatin formation could be initiated by an RNA polII-dependent increase in the transcription of the non-coding outer repeats that flank the central centromeric chromatin during late S phase of the cell cycle122, 123. However, how this transcription is regulated remains unclear. Of note, in S. pombe it was shown that sequence specific transcription factors such as Atf1/Pcr1 and Taz1 are required for heterochromatin formation at the mating-type loci and telomeres, respectively119. In Drosophila, the zinc finger protein Suvar(3)7 was also shown to be required for heterochromatin formation124, and binding of transcription factors to DNA could also be crucial for this process in mammals (T. Jenuwein, unpublished data).
Until now, few long ncRNAs have been characterized but recent studies, using specific patterns of histone PTMs as indicator for active promoters, revealed the existence of numerous long intergenic non-coding RNAs125, 126. Long ncRNAs can serve as precursors for small ncRNA while others could have short open reading frames that code for peptides107 yet a number of them directly regulate transcription. Transcription of long ncRNAs represses gene expression by preventing the binding of transcription factors in cis, as reported for the SER3 gene in S. cerivisae127 and the bxd locus in Drosophila128. Some long ncRNAs have been proposed to have structural functions involved in paraspeckle formation, for example, or act as coactivators of nuclear receptor functions as observed with the Steroid Receptor Activator (for review129). More importantly, in the context of epigenetic regulation, long ncRNA expression triggers chromatin mediated silencing of the tumor suppressor p15130 or regulates HOX gene expression by PRC2 recruitment131. Other examples of long ncRNAs that recruit chromatin modifiers have been reported and this may be important in gene imprinting132-134, although the mechanisms by which RNA and chromatin modifier interactions are unclear since their association appears to be cell specific132. In keeping with their role in transcription regulation, ncRNAs are expressed in a developmentally dependent manner107, 135, 136. Specific patterns of histone methylation are associated with their promoters as a function of their expression137 and common transcription factors (such as c-myc and Sp1) are involved in their regulation138.
Clearly, nucleosome occupancy, histone PTMs with their direct and indirect effects on chromatin structure, and DNA methylation are pivotal in determining which response elements which in excess in the genome will be bound successfully by a defined transcription factor. Notwithstanding, we should mention that transcription factors can also form regulatory loops (including positive feedback loops) that can generate epigenetic regulation139. Furthermore, the importance influence that transcription factors have on the chromatin landscape is illustrated by reprogramming experiments. Transforming cells from their fully differentiated state into pluripotent ES cells requires the transient expression of only a few transcription factors which leads to the dramatic restructuring of the chromatin landscape140.
Conclusion
We have tried to emphasize in this review that the transmission of epigenetic information results from the contributions of different mechanisms including recycling of old histones during replication, cooperativity in histone mark duplication, long range interactions, defined chromatin architecture and the complex network of trans-acting factors whether they be proteins or RNAs (Figure 4). We did not include studies concerned with X chromosome inactivation in mammals or dosage compensation in Drosophila and worms since these topics have been reviewed elsewhere 141,142, but they are probably the most striking examples of the complexity inherent to epigenetic regulation. Indeed, in mammals, starting with two chromosomes sharing an identical DNA sequence, only one is silenced and this silencing is inherited through cell divisions. The establishment and maintenance of this silencing program requires the spectrum of agents covered herein: DNA methylation, histone modifications, histone variants, defined nuclear localization, ncRNA, and transcription factors.
Figure 4.
Different phenomena that contribute to propagation of regulatory information.
This figure illustrates the different factors that contribute to the propagation of epigenetic information through the regulation of chromatin structure. Regulatory information is represented as a histone posttranslational modification which is diluted by the incorporation of newly synthesized histones during replication. We distinguish two steps: the first one addresses the targeting of the Chromatin Modifying Complex (CMC) at a defined locus, and the second one the spreading of a putative modification throughout a defined domain. Based on known examples, we have attributed defined mechanisms to one step or the other, however it is clear that this distinction is not a strict one and that each of these mechanisms probably contributes to some extent to both steps.
Acknowledgments
We thank Drs. D. Beck, R. Bonasio, and E. Campos for their critical reading of the manuscript. We are grateful to Drs. L. Vales and J. Hurwitz for critical reading of this manuscript and active discussions. We apologize to authors whose studies could not be cited due to space limitation. Work in the laboratory of D.R. is funded by NIH (NIH RO1GM064844 and 4R37GM037120) and by HHMI.
References
- 1.Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–3. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–7. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–94. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos EI, Reinberg D. Histones: Annotating Chromatin. Annual Review of Genetics. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 8.Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–32. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 9.Brickner DG, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007;17:2041–6. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 12.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–19. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 13.Lemieux K, Larochelle M, Gaudreau L. Variant histone H2A.Z, but not the HMG proteins Nhp6a/b, is essential for the recruitment of Swi/Snf, Mediator, and SAGA to the yeast GAL1 UAS(G) Biochem Biophys Res Commun. 2008;369:1103–7. doi: 10.1016/j.bbrc.2008.02.144. [DOI] [PubMed] [Google Scholar]
- 14.Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–78. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–62. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 16.Girton JR, Johansen KM. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv Genet. 2008;61:1–43. doi: 10.1016/S0065-2660(07)00001-6. [DOI] [PubMed] [Google Scholar]
- 17.Eissenberg JC, Reuter G. Cellular mechanism for targeting heterochromatin formation in Drosophila. Int Rev Cell Mol Biol. 2009;273:1–47. doi: 10.1016/S1937-6448(08)01801-7. [DOI] [PubMed] [Google Scholar]
- 18.Schotta G, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. Embo J. 2002;21:1121–31. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–92. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 20.Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–5. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brower-Toland B, Riddle NC, Jiang H, Huisinga KL, Elgin SC. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics. 2009;181:1303–19. doi: 10.1534/genetics.108.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao X, Deng H, Johansen J, Girton J, Johansen KM. Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect variegation at pericentric sites in Drosophila heterochromatin. Genetics. 2007;176:1355–8. doi: 10.1534/genetics.107.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 24.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–80. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 26.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–86. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–41. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–16. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–55. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–27. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–7. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. Embo J. 2009;28:1878–89. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 35.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–41. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 37.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–31. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 38.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–22. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 39.Lin JC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–44. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 41.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–85. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 42.Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- 43.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 44.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 45.Egger G, et al. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci U S A. 2006;103:14080–5. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spada F, et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–71. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen T, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–6. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 48.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–73. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 49.Chuang LS, et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 50.Schermelleh L, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–12. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 52.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 53.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–9. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espada J, et al. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J Biol Chem. 2004;279:37175–84. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 57.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–83. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 58.Tamaru H, et al. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–9. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 59.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 60.Dong KB, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. Embo J. 2008;27:2691–701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179:1413–26. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jorgensen S, et al. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–45. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–7. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rice JC, et al. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–30. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin Y, Yu VC, Zhu G, Chang DC. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle. 2008;7:1423–32. doi: 10.4161/cc.7.10.5867. [DOI] [PubMed] [Google Scholar]
- 66.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 67.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kohlmaier A, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oda H, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–95. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scharf AN, et al. Monomethylation of lysine 20 on histone H4 facilitates chromatin maturation. Mol Cell Biol. 2009;29:57–67. doi: 10.1128/MCB.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalakonda N, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293–304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paulsen RD, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–39. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 74.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–72. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eissenberg JC, et al. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990;87:9923–7. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–3. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 78.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 79.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 80.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–38. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–31. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 83.Seum C, Delattre M, Spierer A, Spierer P. Ectopic HP1 promotes chromosome loops and variegated silencing in Drosophila. Embo J. 2001;20:812–8. doi: 10.1093/emboj/20.4.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Y, Danzer JR, Alvarez P, Belmont AS, Wallrath LL. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 2003;130:1817–24. doi: 10.1242/dev.00405. [DOI] [PubMed] [Google Scholar]
- 85.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009 doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 87.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–4. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 88.Schotta G, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–61. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–22. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 90.Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- 91.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–15. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 92.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 93.Tumbar T, Belmont AS. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol. 2001;3:134–9. doi: 10.1038/35055033. [DOI] [PubMed] [Google Scholar]
- 94.Casolari JM, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–39. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 95.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–5. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–20. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 2009;6 doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- 99.Grimaud C, et al. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–71. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 100.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–74. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 101.Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–9. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buchenau P, Hodgson J, Strutt H, Arndt-Jovin DJ. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: impact on models for silencing. J Cell Biol. 1998;141:469–81. doi: 10.1083/jcb.141.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–93. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 104.Kurukuti S, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoon YS, et al. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li T, et al. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol. 2008;28:6473–82. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–92. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 108.Willingham AT, Gingeras TR. TUF love for “junk” DNA. Cell. 2006;125:1215–20. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 109.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 111.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–20. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–76. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 113.Liu Y, et al. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–45. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 115.Pal-Bhadra M, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–72. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 116.Wallace JA, Orr-Weaver TL. Replication of heterochromatin: insights into mechanisms of epigenetic inheritance. Chromosoma. 2005;114:389–402. doi: 10.1007/s00412-005-0024-6. [DOI] [PubMed] [Google Scholar]
- 117.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L. Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol. 2009;53:245–57. doi: 10.1387/ijdb.082691av. [DOI] [PubMed] [Google Scholar]
- 120.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–86. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 121.Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–21. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 122.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–7. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 123.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–5. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Reuter G, et al. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature. 1990;344:219–23. doi: 10.1038/344219a0. [DOI] [PubMed] [Google Scholar]
- 125.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–4. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 128.Petruk S, et al. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–21. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 133.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 134.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sessa L, et al. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. Rna. 2007;13:223–39. doi: 10.1261/rna.266707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ke XS, et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 2009;4:e4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cawley S, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 139.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–6. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 140.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 141.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–66. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 142.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]