Abstract
Phospholipase D (PLD), a superfamily of signaling enzymes that most commonly generate the lipid second messenger Phosphatidic Acid (PA), is found in diverse organisms from bacteria to man and functions in multiple cellular pathways. A fascinating member of the family, MitoPLD, is anchored to the mitochondrial surface and has two reported roles. In the first role, MitoPLD-generated PA regulates mitochondrial shape through facilitating mitochondrial fusion. In the second role, MitoPLD performs a critical function in a pathway that creates a specialized form of RNAi required by developing spermatocytes to suppress transposon mobilization during meiosis. This spermatocyte-specific RNAi, known as piRNA, is generated in the nuage, an electron-dense accumulation of RNA templates and processing proteins that localize adjacent to mitochondria in a structure also called intermitochondrial cement. In this review, we summarize recent findings on these roles for MitoPLD functions, highlighting directions that need to be pursued to define the underlying mechanisms.
Keywords: Phospholipase D, mitochondria, mouse models, piRNA, spermatogenesis, morphology
1. Introduction
Long known as the powerhouse of the cell, mitochondria are now appreciated to regulate many different cellular functions including apoptosis, intracellular calcium signaling, and lipid synthesis and transport (1, 2). Mitochondrial shape, size and number are regulated by the balance of mitochondrial fusion and fission and affect mitochondrial function, which is important in mammalian health and disease (3, 4). MitoPLD, a member of mammalian phospholipase D (PLD) superfamily, was initially reported to facilitate mitochondrial fusion by hydrolyzing cardiolipin (CL) to generate phosphatidic acid (PA), constituting a novel lipid signaling pathway on the mitochondrial surface (5, 6). More recent work, however, has identified an additional function for MitoPLD in spermatogenesis (7–9).
Zucchini (Zuc), the Drosophila homolog of MitoPLD, was identified through screens for female infertility, which was subsequently determined to ensue from a critical role for Zuc in the PIWI-interacting RNA (piRNA) biogenesis pathway (10). The main function of piRNAs is to defend genomic integrity in germ-line cells by silencing transposable elements (11). Male mice lacking MitoPLD also have decreased piRNA biogenesis and meiotic arrest during spermatogenesis, analogous to the function of Zuc in Drosophila (8, 9). Nuage, an electron dense structure, is the subcellular organelle in which piRNA biogenesis takes place (12). Nuage is normally physically adjacent to mitochondria in spermatocytes; however, this relationship is disrupted in mice lacking MitoPLD (8, 9). Mutations in many of the components found in nuage impair piRNA biogenesis and derepress transposable elements (11, 13–20).
Intriguing questions are raised by the findings reported for MitoPLD and Zuc, centered on the control of mitochondrial shape and trafficking and why lipid-modifying enzymes on the surface of the mitochondria are required for piRNA biogenesis. We review here the current literature and experimental directions immediately apparent.
2. Mammalian MitoPLD
2.1 Molecular and Biochemical characterization
Phospholipase D superfamily members are found in prokaryotes, eukaryotes, and some viruses (reviewed in 21). The canonical PLD enzymatic activity is to hydrolyze the lipid phosphatidylcholine to yield choline and the signaling lipid PA, but some family members can hydrolyze other phospholipids or perform lipid synthetic or other actions involving hydrolysis or transfer at the phosphodiester bond linking the phosphate group to the headgroup. PA, a lipid second messenger, plays multiple roles in cellular functions including promoting cytoplasmic membrane fusion, and acting as a lipid anchor to recruit proteins to membrane surfaces and in some cases alter their functional activity (22). PA can also be converted through dephosphorylation by PA phosphohydrolases to diacylglycerol (DAG), another important signaling lipid in many cellular processes (23, 24). The classical isoforms of mammalian PLD, PLD1 and PLD2, were reported in the 1990’s (25, 26). Blast searches of the completed human genome, however, later revealed additional genes encoding the PLD catalytic domain, H(X)K(X4)D (HKD). One of these genes, MitoPLD (later also denoted pld6), was found to be located on the mitochondrial surface (5). However, in contrast to classical mammalian PLD family members which encode two half-catalytic HKD domains (26) and fold together to form the functional enzymatic unit (27), MitoPLD has only a single HKD half-catalytic site, requiring it to dimerize to create an active enzymatic complex (5). Interestingly, having only one copy of the HKD domain makes MitoPLD quite distinct from the classical PLD genes, and in fact closer in similarity to a prokaryotic branch of the superfamily known as “Nuc” (28). The bacterial Nuc genes function as endonucleases, hydrolyzing the phosphodiester bond in the backbone of DNA that is analogous to the phosphodiester bond in phospholipids. Nonetheless, no group to date has been able to demonstrate any type of nuclease activity for MitoPLD (5, 9). The next most similar branch of the PLD superfamily to MitoPLD is a prokaryotic group of enzymes that synthesize cardiolipin (CLS); however, CLS activity also could not be demonstrated for MitoPLD (5, 29). What became apparent, though, was that alterations in lipid content in mitochondria were observed when MitoPLD was overexpressed, leading to the identification of a capability for MitoPLD to hydrolyze CL to generate PA (5).
In contrast to classical PLDs that have lipid-binding domains such as PX, PH, or PIP2-binding motifs, MitoPLD is noteworthy for an N-terminal sequence that both localizes the enzyme to mitochondria and functions as a transmembrane domain to anchor MitoPLD into the mitochondrial surface. MitoPLD overexpression triggers mitochondrial aggregation (discussed below); replacing the key catalytic Histidine residue, H156N, with Asparagine, prevents this mitochondrial aggregation, indicating that it is MitoPLD’s enzymatic activity that mediates the change in morphology observed with overexpression.
2.2 MitoPLD and mitochondrial dynamics
Mitochondrial dynamics, including both fusion and fission, are very important in maintaining mitochondrial and cellular function, which are crucial for mammalian development and health (1, 30). Mitochondrial shape, size and number are determined by the rate and balance of mitochondrial fusion and fission. Increased fusion or decreased fission lead to elongated mitochondria, while decreased fusion or increased fission result in mitochondrial fragmentation. These morphological changes are linked to pathological states including cell apoptosis and neurodegenerative disease.
Mitofusin / fuzzy onion (MFN, fzo) was the first gene discovered to mediate mitochondrial outer membrane fusion (31). The mammalian MFN homologs, MFN1 and MFN2, are transmembrane GTPases located in the mitochondrial outer membrane. Mutations in MFN2 have been linked to the inherited peripheral neuropathy Charcot Marie Tooth disease (30). The C-terminal tail of MFN functions to tether opposing mitochondria as they approach each other during a fusion event, forming a dimeric, antiparallel coiled-coil structure (32). Electron microscopy revealed that the mitochondrial tethering effect creates an interface in which the adjacent mitochondria are separated by 16 nm (32). The GTPase is thought to mediate the fusion event subsequent to tethering.
Overexpression of MitoPLD also results in the juxtaposition of mitochondria. However, the mitochondria are separated by only 6 nm (5), and the ability of MitoPLD to create this mitochondria interface requires Mfn proteins to be present. Thus, it was proposed that the MFN-driven tethering event leads to MitoPLD production of PA on the closely apposed mitochondrial outer membranes, triggering even closer apposition of the membranes to facilitate the MFN-GTPase fusion event. Supporting this hypothesis, overexpression of a catalytically-inactive (dominant-negative) MitoPLD mutant allele or use of MitoPLD RNAi led to mitochondrial fragmentation (5), and mouse embryo fibroblasts (MEFs) isolated from mice lacking MitoPLD exhibit shortened mitochondria (8). These findings indicate that MitoPLD facilitates mitochondrial fusion through a mechanism dependent on its enzymatic activity, although it is not absolutely required for fusion events to occur.
Support for a role for MitoPLD in fusion has recently been described in Drosophila (33). Mitochondrial fission plays a critical role in cellular apoptosis upstream of cytochrome c release. Dynamin-related protein 1 (Drp 1) is the major mitochondrial fission mediator and is required for apoptosis (reviewed in 34). This pathway is relevant during dorsal closure, a complex morphogenetic movement and essential stage in Drosophila embryogenesis: apoptosis in cells in the amnioserosa triggers delamination of the cells which is the driving force for the dorsal closure (35). As reported by Muliyil and colleagues, increasing Drp1 expression or decreasing MitoPLD expression which promotes mitochondrial fragmentation, led to increased delamination, whereas decreasing Drp1 decreased delamination (33). This study showed that MitoPLD, by opposing mitochondrial fragmentation, has a functional role in the regulation of apoptosis and morphogenetic movements during embryogenesis in Drosophila.
Finally, PA is a dynamic lipid capable of being converted into other bioactive signaling lipids, one of which is diacylglycerol (DAG) (23). The family of cytoplasmic enzymes that mediate this activity is known as Lipin (23). Mutations in Lipin 1 cause fatty liver dystrophy and peripheral neuropathy in mice (36, 37). MitoPLD-generated PA recruits Lipin 1b to the mitochondria; Lipin 1b then converts the PA to DAG on the mitochondrial surface, terminating the fusion-promoting function of PA and shifting mitochondrial morphology towards increased fragmentation (8). Many mitochondrial fusion events are followed by a fission event (38); the MitoPLD–PA-lipin1-DAG signaling cascade may play a role in the temporal-spatial changes of mitochondrial dynamics, in particular in mitochondrial dynamic-sensitive tissues such as fat, muscle and neurons. Further studies are needed to show how PA production regulate mitochondrial fusion, such as through recruiting proteins that have PA binding domains, or via the negative charge and membrane curvature-altering properties of PA.
3. Zucchini, the Drosophila homolog of MitoPLD
3.1 Molecular characterization
The Drosophila homolog of MitoPLD, Zuc, was genetically uncovered in a mutagenesis screen for female sterile mutations (39). The molecular characterization of Zuc was reported in 2007 (10), subsequent to the identification of mammalian MitoPLD (5), in a study in which Zuc was shown to be required for transposon suppression during meiosis, a process that involves production of a specialized form of small RNAs known as piRNA (11). Based on the similarity of the MitoPLD / Zuc genes to the prokaryotic branch of the PLD superfamily that exhibits endonuclease activity, Zuc was proposed to be the nuclease that generates piRNA, but no biochemical evidence has yet been generated to support this function. Instead, as discussed above, MitoPLD / Zuc has been shown to hydrolyze CL to generate PA; hence, either that MitoPLD/Zuc posses both (CL) hydrolase and endonuclease activities, or the role for Zuc in piRNA biogenesis is an indirect one. Elegantly confirming the requirement for catalytic activity for Zuc in transposon suppression, the Histidine residue of the HKD domain is critical for the function of PLD superfamily enzymes (26, 28, 40) as discussed earlier, and one of the mutant alleles of Zuc that results in loss of transposon suppression consists of a point mutation in which the HKD His residue was converted to Tyrosine, ZucSG63 (10). Zuc protein is expressed in Drosophila ovary nurse cell and somatic supportive cells (10, 41, 42). Zuc was initially reported to be a cytoplasmic protein and / or in the nuage, a germ cell-specialized electron-dense perinuclear structure that functions as the site for piRNA biogenesis (10, 43). However, these studies were conducted using an allele fused at the N-terminus of the protein to a myc-tag or EGFP, which masked the mitochondrial localization sequence and transmembrane domain. More recent studies using alleles tagged at the C-terminus reported that it localizes to mitochondria (8, 44), similar to that reported for MitoPLD (5, 8, 9).
3.2 Function of Zucchini in the Drosophila Germline Cell
In Drosophila germline cells, Zuc has been well-demonstrated to have a critical role in biogenesis/maturation of piRNAs (Fig. 1). PIWI-interacting RNAs, or piRNAs, initially named as repeat-associated small interfering RNAs (rasiRNAs) (45), together with microRNA (miRNA) and small interfering RNAs (siRNAs) are the three classes of small RNA molecules that mediate RNA interference action in eukaryotic organisms (11, 46). piRNAs are 24–30nt long RNAs produced from piRNA clusters and repeat sequences by a Dicer endonuclease-independent process. The Piwi family proteins Piwi, Aubergine and Argonaute3 associate with piRNAs during piRNA biogenesis and then mediate silencing of transposons. Transposons are genetic elements that move from site to site within the genome causing genetic mutations. piRNA-mediated transposon silencing is thus critical for maintaining genome stability, in particular in germline cells when transposons are mobilized as a consequence of wide-spread genomic demethylation.
Fig. 1. Components of the piRNA biogenesis complex in Drosophila melanogaster.
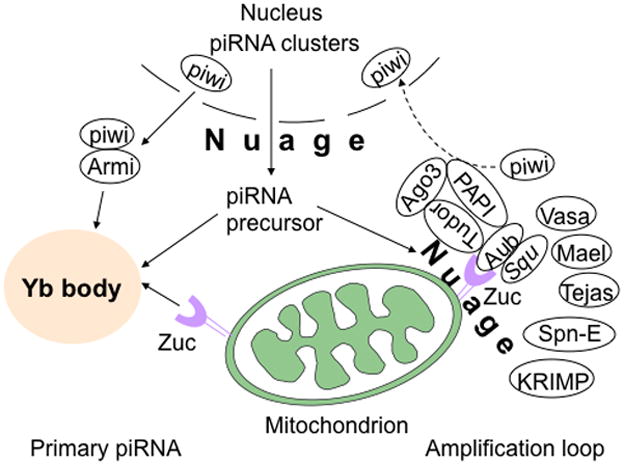
In Drosophila ovarian somatic cells, primary piRNAs are generated at the Yb body. Piwi associates with Armi and translocates to the Yb body, where piRNA precursors are loaded onto piwi-Armi complex and processed to form mature piRNAs in the presence of Zuc, the Drosophila homolog of MitoPLD, which is expressed on the mitochondrial surface. In the absence of Zuc, piwi fails to be released from Yb body and accumulates there. In Drosophila germ cells, piRNA biogenesis occurs in the nuage via an amplification loop, also called the ping-pong cycle. Zuc and Squ have been reported to interact physically with Aub, and Tudor to associate with Aub and Ago3, Vasa, Mael, and Tejas. SpnE and KRIMP are also components of the nuage and may play roles in piRNA biogenesis. See text for additional details.
Genetic mutation of Zuc causes sterility in Drosophila females characterized by dorsal-ventral patterning defects during oogenesis (10). piRNA biogenesis in fly ovaries and testes is impaired in Zuc mutants as indicated by the decreased expression levels of many types of these small RNAs (10, 47), leading to upregulation of retrotransposons. Increased expression of the transposons leads to DNA damage, triggering a meiotic checkpoint that activates the kinase Chk2, that in turn then suppresses translation of Grk, a protein involved in axial patterning of the oocyte. Functional disruption of many of the RNAi pathway proteins similarly leads to increased Chk2, decreased Grk, and dorsal-ventral patterning defects. However, whereas the dorsal-ventral patterning defects can be rescued in most of these cases by eliminating Chk2, the defects persist in embryos lacking Zuc, suggesting that Zuc undertakes morphogenetic functions independent of its role in piRNA biogenesis (10). The recent report that Zuc / MitoPLD plays a role in dorsal closure through regulating mitochondrial morphology (33) is intriguing in connection to the dorsal-ventral patterning defects reported by Pane and colleagues (10).
The involvement of Zuc in the germline piRNA pathway was further supported by studies on retrotransposons located in euchromatin and heterochromatin, I and P elements, respectively. The production of piRNAs involves an amplification loop called the “ping-pong” cycle. The initial antisense piRNAs are produced by heterochromatic loci, after association with Piwi and Aub; these antisense piRNAs target the transcripts of euchromatic elements to generate sense piRNAs, and then the sense piRNAs generate more new antisense piRNAs from heterochromatin. The I element belongs to class I retrotransposons, which lacks the long terminal repeats. Mutation of piRNA pathway genes, including aub, Squ, Zuc, induce derepression of euchromatic I element in a partially post-transcriptional manner (48). Heterochromatic P element is a class II transposon; its movement within the genome depends on a transposase. Mutation in Zuc and Squ showed either complete or partial loss of repression of trans-silencing effect, an effect characterized by repressing a p element-lacz copy in trans due to the presence of another one or two copies of p element-lacz in telomeric-associated sequences in the Drosophila female germline (49, 50). The loss of repression of trans-silencing is likely due to the strong inhibition of accumulation of lacZ small RNAs, which is effected by the piRNA pathway in ovaries.
3.3 Function of Zucchini in Drosophila Somatic Cells
Drosophila ovarian follicle cells are specialized somatic cells. In these cells, piwi is the only PIWI protein present; the other two PIWI proteins, aub and AGO3, are not expressed. Thus, follicle cells possess only a primary piRNA pathway (47), providing a simplified piRNA pathway in comparison to that found in germ cells. The somatic piRNAs derive mainly from piRNA clusters such as flamenco and traffic jam. Mutation in Zuc decreases not only the production of somatic and germline piRNAs from the flamenco locus, but also the somatic piRNAs from the traffic jam locus (41, 43).
Screening of genes involved in the Drosophila piRNA pathway by in vivo RNAi assay identified Zuc, Armitage and Yb as the essential factors for primary piRNA biogenesis in ovarian somatic cells (42, 44). Armitage and Yb are RNA helicases. In contrast to the nuclear localization of piwi, Armitage and Yb co-localize in cytoplasmic foci in the Yb body, an electron-dense cytoplasmic foci involved in the maintenance of germline and somatic stem cells that is associated with the mitochondria, whereas Zuc localizes to the mitochondria (44). Loss of Zuc also affects the localization of piwi. Piwi is no longer seen in the nucleus, and instead exhibits a perinuclear accumulation co-localized with Armitage at the Yb body (42, 51). These findings suggest that a role for Zuc is in the regulation of transit of cytosolic piwi through Armitage and the Yb body, involving biogenesis or loading of piRNAs onto piwi or its release form the Yb body.
Taken together, Zuc plays a critical role in piRNA biogenesis in both ovarian germline cells and somatic cells.
4. MitoPLD and spermatogenesis in mice
Mice lacking MitoPLD have provided insight into the expression, distribution and function of endogenous MitoPLD. As cited above, the Drosophila MitoPLD homologue, Zuc, plays a vital role in piRNA biogenesis; mammalian MitoPLD is also essential for germline nuage formation and spermatogenesis.
MitoPLD−/− mice are grossly normal in appearance and the major phenotype is male mice infertility (8, 9). MitoPLD is strongly expressed in the testis. In the absence of MitoPLD, mitochondrial dynamics in somatic cells are altered with more centralized localization of shorter mitochondria; in fetal germ cells, mitochondria re-localize to the peri-centrosomal region. These findings indicate an altered relationship of mitochondria to the microtubule network.
Meiotic arrest at the primary spermatocyte stage, more specifically, early in development of pachytene spermatocytes, is observed in MitoPLD−/− mice (8, 9). The germline-specific structure, the nuage, also known as intermitochondrial cement, or chromatoid body, depending on the specific stages, is absent from MitoPLD−/− spermatocytes. Mutation of genes encoding other nuage components, including MILI, MIWI2, TDRD1, TDRD9, MVH, Mael and GASZ (14–20), leads to a similar meiotic arrest phenotype during spermatogenesis (Fig. 2). As nuage is the major site of piRNA biogenesis, derepression of L1 retrotransposons, loss of TDRD1 expression, increased level of genomic damage and reduced methylation of the Rasgrf1 differentially-methylated region in MitoPLD−/− testis all indicate the important role of MitoPLD in the piRNA pathway (8, 9, 52).
Fig. 2. Proposed piRNA biogenesis complex in mice.
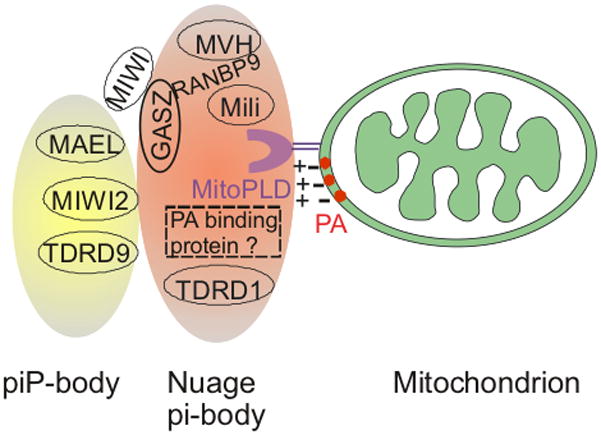
MitoPLD localized on the mitochondrial surface hydrolyzes cardiolipin to generate PA, a negatively-charged lipid, which may promote nuage complex formation either by recruiting the positively-charged nuage via electrostatic attraction, by anchoring a nuage-localized PA-binding protein, or by affecting piRNA precursor or mitochondrial trafficking along microtubules. GASZ co-localizes with mili, MVH, TDRD1, RanBP9, or MIWI depending on the stages of spermatogenesis. The piP-body contains Mael, MIWI2 and TDRD9 and physically interacts with nuage/the pi-body. See text for additional details.
The major biochemical process currently demonstrated for MitoPLD is to hydrolyze cardiolipin to generate PA; the PA recruits Lipin 1 to the mitochondrial surface to convert the PA to DAG. The presumably elevated levels of PA on the mitochondrial surface appear to increase nuage formation in mice lacking Lipin 1 (fld/fld) mice, confirming the presence of the PA and DAG generating pathways on the mitochondrial surface and their influence on nuage formation. However, the fld/fld mice produce motile sperm, indicating that increased nuage formation does not interfere with spermatogenesis. How PA exerts its function in nuage formation and in piRNA complex formation / biogenesis are not currently known. Possibilities include recruitment of PA-binding proteins that promote the complex formation or effects on the microtubule trafficking of mitochondria or piRNA precursor transcripts to the mitochondria. PA is also a highly negatively charged lipid, it could also attract nuage by electrostatic force, since the nuage is enriched in positively-charged histone proteins.
In summary, the discovery of MitoPLD as a protein that regulates mitochondrial dynamics revealed an important lipid signaling pathway on the mitochondrial surface. PA is the only signaling molecule known to be generated by the activity of MitoPLD at present. The establishment of MitoPLD−/− mice uncovered the piRNA biogenesis-regulating role of MitoPLD, which is similar to that of its Drosophila homolog Zuc. Further studies are needed to understand how MitoPLD affects nuage formation as well as piRNA generation.
Finally, since piRNA biogenesis is known to be critical only for spermatogenesis, the requirement for MitoPLD in the process raises the potential of small molecule MitoPLD inhibitors as potential male contraceptive agents. Specific and potent small molecule inhibitors with utility in vivo have been developed for other members of the PLD family (53–58), making it feasible to propose identifying one for MitoPLD (the existing classical PLD1/2 small molecule inhibitor does not block MitoPLD activity (54)). Males account for approximately one third of contraceptive usage in general, making this an attractive therapeutic target that could be highly effective and easily reversible.
Acknowledgments
Supported by NIH GM071520 and GM084251 to MAF and a fellowships from NIH (T32-DK07521) to QG.
Footnotes
The authors do not declare any conflicts of interest.
References
- 1.Braschi E, McBride HM. Mitochondria and the culture of the Borg: understanding the integration of mitochondrial function within the reticulum, the cell, and the organism. Bioessays. 2010;32:958–966. doi: 10.1002/bies.201000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frohman MA. Mitochondria as integrators of signal transduction and energy production in cardiac physiology and disease. Journal of molecular medicine (Berlin, Germany) 2010;88:967–970. doi: 10.1007/s00109-010-0662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Liesa M, Palacin M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 5.Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 6.Huang H, Frohman MA. Lipid signaling on the mitochondrial surface. Biochimica et biophysica acta. 2009;1791:839–844. doi: 10.1016/j.bbalip.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravin AA, Chan DC. piRNAs meet mitochondria. Dev Cell. 2011;20:287–288. doi: 10.1016/j.devcel.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, Noce T, Nakano T, Nakatsuji N, Lin H, Sasaki H. MitoPLD Is a Mitochondrial Protein Essential for Nuage Formation and piRNA Biogenesis in the Mouse Germline. Developmental Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature reviews Molecular cell biology. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 12.O’Donnell KA, Burns KH, Boeke JD. A descent into the nuage: the maelstrom of transposon control. Dev Cell. 2008;15:179–181. doi: 10.1016/j.devcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci U S A. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, Hata K, Martin SL, Noce T, Kuramochi-Miyagawa S, Nakano T, Sasaki H, Pillai RS, Nakatsuji N, Chuma S. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, Totoki Y, Shibata T, Kimura T, Nakatsuji N, Noce T, Sasaki H, Nakano T. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, Yan W, Matzuk MM. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang P, Frohman MA. The potential for phospholipase D as a new therapeutic target. Expert opinion on therapeutic targets. 2007;11:707–716. doi: 10.1517/14728222.11.5.707. [DOI] [PubMed] [Google Scholar]
- 23.Brindley DN, Pilquil C, Sariahmetoglu M, Reue K. Phosphatidate degradation: phosphatidate phosphatases (lipins) and lipid phosphate phosphatases. Biochimica et biophysica acta. 2009;1791:956–961. doi: 10.1016/j.bbalip.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazzolli R, Shemon AN, Fang MQ, Hughes WE. Phospholipid signalling through phospholipase D and phosphatidic acid. IUBMB life. 2006;58:457–461. doi: 10.1080/15216540600871142. [DOI] [PubMed] [Google Scholar]
- 25.Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- 26.Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, Morris AJ, Frohman MA. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- 27.Uesugi Y, Hatanaka T. Phospholipase D mechanism using Streptomyces PLD. Biochimica et biophysica acta. 2009;1791:962–969. doi: 10.1016/j.bbalip.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 28.Stuckey JA, Dixon JE. Crystal structure of a phospholipase D family member. Nat Struct Biol. 1999;6:278–284. doi: 10.1038/6716. [DOI] [PubMed] [Google Scholar]
- 29.Choi SY, Gonzalvez F, Jenkins GM, Slomianny C, Chretien D, Arnoult D, Petit PX, Frohman MA. Cardiolipin deficiency releases cytochrome c from the inner mitochondrial membrane and accelerates stimuli-elicited apoptosis. Cell death and differentiation. 2007;14:597–606. doi: 10.1038/sj.cdd.4402020. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Chan DC. Physiological functions of mitochondrial fusion. Annals of the New York Academy of Sciences. 2010;1201:21–25. doi: 10.1111/j.1749-6632.2010.05615.x. [DOI] [PubMed] [Google Scholar]
- 31.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 32.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 33.Muliyil S, Krishnakumar P, Narasimha M. Spatial, temporal and molecular hierarchies in the link between death, delamination and dorsal closure. Development. 2011;138:3043–3054. doi: 10.1242/dev.060731. [DOI] [PubMed] [Google Scholar]
- 34.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyama Y, Peralta XG, Wells AR, Kiehart DP, Edwards GS. Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science. 2008;321:1683–1686. doi: 10.1126/science.1157052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langner CA, Birkenmeier EH, Ben-Zeev O, Schotz MC, Sweet HO, Davisson MT, Gordon JI. The fatty liver dystrophy (fld) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J Biol Chem. 1989;264:7994–8003. [PubMed] [Google Scholar]
- 37.Langner CA, Birkenmeier EH, Roth KA, Bronson RT, Gordon JI. Characterization of the peripheral neuropathy in neonatal and adult mice that are homozygous for the fatty liver dystrophy (fld) mutation. J Biol Chem. 1991;266:11955–11964. [PubMed] [Google Scholar]
- 38.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung TC, Roper RL, Zhang Y, Rudge SA, Temel R, Hammond SM, Morris AJ, Moss B, Engebrecht J, Frohman MA. Mutagenesis of phospholipase D defines a superfamily including a trans- Golgi viral protein required for poxvirus pathogenicity. EMBO J. 1997;16:4519–4530. doi: 10.1093/emboj/16.15.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 44.Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 46.Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138:1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 48.Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronsseray S, Josse T, Boivin A, Anxolabehere D. Telomeric transgenes and trans-silencing in Drosophila. Genetica. 2003;117:327–335. doi: 10.1023/a:1022929121828. [DOI] [PubMed] [Google Scholar]
- 50.Josse T, Maurel-Zaffran C, de Vanssay A, Teysset L, Todeschini AL, Delmarre V, Chaminade N, Anxolabehere D, Ronsseray S. Telomeric trans-silencing in Drosophila melanogaster: tissue specificity, development and functional interactions between non-homologous telomeres. PLoS One. 2008;3:e3249. doi: 10.1371/journal.pone.0003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szakmary A, Reedy M, Qi H, Lin H. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol. 2009;185:613–627. doi: 10.1083/jcb.200903034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, Hiura H, Arima T, Fujiyama A, Sado T, Shibata T, Nakano T, Lin H, Ichiyanagi K, Soloway PD, Sasaki H. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su W, Chen Q, Frohman MA. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future oncology (London, England) 2009;5:1477–1486. doi: 10.2217/fon.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su W, Yeku O, Olepu S, Genna A, Park JS, Ren H, Du G, Gelb M, Morris A, Frohman MA. FIPI, a Phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Molecular pharmacology. 2009;75:437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, Armstrong MD, Arteaga CL, Lindsley CW, Brown HA. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nature chemical biology. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishikimi A, Fukuhara H, Su W, Hongu T, Takasuga S, Mihara H, Cao Q, Sanematsu F, Kanai M, Hasegawa H, Tanaka Y, Shibasaki M, Kanaho Y, Sasaki T, Frohman MA, Fukui Y. Sequential Regulation of DOCK2 Dynamics by Two Phospholipids during Neutrophil Chemotaxis. Science. 2009;324:384–387. doi: 10.1126/science.1170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dall’Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB, Yu WH, Robinson KS, Yeku O, Small SA, Duff K, Frohman MA, Wenk MR, Yamamoto A, Di Paolo G. The phospholipase D1 pathway modulates macroautophagy. Nature communications. 2010;1:142. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukahara T, Tsukahara R, Fujiwara Y, Yue J, Cheng Y, Guo H, Bolen A, Zhang C, Balazs L, Re F, Du G, Frohman MA, Baker DL, Parrill AL, Uchiyama A, Kobayashi T, Murakami-Murofushi K, Tigyi G. Phospholipase D2-dependent inhibition of the nuclear hormone receptor PPARgamma by cyclic phosphatidic acid. Mol Cell. 2010;39:421–432. doi: 10.1016/j.molcel.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


