Abstract
Protein function often depends on the exchange between conformational substates. Allosteric ligand binding or distal mutations can stabilize specific active site conformations and consequently alter protein function. In addition to comparing independently determined X-ray crystal structures, alternative conformations observed at low levels of electron density have the potential to provide mechanistic insights into conformational dynamics. Here, we report a new multi-conformer contact network algorithm (CONTACT) that identifies networks of conformationally heterogeneous residues directly from high-resolution X-ray crystallography data. Contact networks in Escherichia coli dihydrofolate reductase (ecDHFR) predict the long-range pattern of NMR chemical shift perturbations of an allosteric mutation. A comparison of contact networks in wild type and mutant ecDHFR suggests how mutations that alter optimized networks of coordinated motions can impair catalytic function. Thus, CONTACT-guided mutagenesis will allow the structure-dynamics-function relationship to be exploited in protein engineering and design.
INTRODUCTION
Proteins fluctuate between alternative conformations to mediate their biological functions1,2. Perturbations that affect the relative populations of conformations caused by ligand binding3, mutation4, post-translational modification5, and temperature6 can affect biological mechanism. In addition to causing global or local unfolding7, perturbations can affect the structure and dynamics of groups of residues within the folded ensemble8. Proteins can evolve to exploit these perturbations for regulation: binding of ligands or the introduction of mutations at allosteric sites can stabilize transiently populated, but functional, alternative conformations at distant active sites9. However, these low population states are difficult to identify with most biophysical techniques10, making it challenging to characterize how structural fluctuations regulate protein activity by changing the populations of different conformers.
Traditionally, X-ray crystallography has provided a single static model of a “ground state”, which is assumed to represent the lowest energy conformation in the crystal lattice. More “dynamic” interpretations of crystallographic data have historically involved modeling an ensemble of independent conformations11–17. Recently developments in time-averaged refinement select a representative ensemble of 10s–100s of structures18. However, the relationship between individual ensemble members and the representation of conformational heterogeneity in the crystal remains the subject of debate15. In contrast, our recent work identifies conformational heterogeneity from high resolution X-ray diffraction data at levels of electron density (below 1σ) that are commonly ignored by manual model building efforts and are inadequately represented by harmonic atomic displacement parameters19,20 (Supplementary Fig. 1). Throughout this work, our analyses are based on such a single, sparse multi-conformer model built by qFit19.
Multi-conformer and ensemble analyses of X-ray data can be complementary to NMR8,21,22, simulations23, and co-evolutionary analyses24 to reveal how interactions between distant sites enable proteins to respond to perturbations. However, tools to uncover and interpret conformational diversity from X-ray crystallography data and to connect structural mechanisms for biomolecular dynamics remain underdeveloped. We developed an algorithm, COntact Networks Through Alternate Conformation Transitions (CONTACT), which automatically identifies conformationally heterogeneous residues that can connect functional sites, propagate chemical shift differences, and reveal the structural mechanisms of mutations that affect redistributions of the conformational ensemble.
RESULTS
Identifying conformational coupling between residues
To identify interacting residues that can respond collectively to perturbations, we used our robotics-inspired algorithm qFit19 to optimally fit up to four alternative conformations per residue into electron density features derived from experimental X-ray data that are consistent with anharmonic disorder (Fig. 1a). CONTACT analyzes the repulsive van der Waals interactions across all alternative conformations in the qFit multi-conformer model (Supplementary Fig 2.). The goal of this analysis is to define networks of conformationally coupled residues, in which movement between alternative conformations of one residue likely influences the conformations of all other residues in a network.
Figure 1.

Mechanisms for conformational exchange in Cyclophilin A (a) X-ray electron density map contoured at 1 σ (blue mesh) and 0.3 σ (cyan mesh) of CYPA is fit with discrete alternative conformations using qFit. Alternative conformations are colored red, orange, or yellow, with hydrogen atoms added in green. (b) Visualizing a pathway in CYPA: atoms involved in clashes are shown in spheres scaled to van der Waals radii and clashes between atoms highlighted by cyan dashes. This pathway originates with the OG atom of Ser99 conformation A and the CE1 atom of Phe113 conformation B, which clash to 0.8 of their summed van der Waals radii. The pathway progresses from Phe113 to Gln63 and after moving Met61 to conformation B introduces no new clashes the pathway is terminated. A 90 degree rotation of the final panel is shown to highlight how the final move of Met61 relieves the clash with Gln63. (c) Networks identified by CONTACT are displayed as nodes connected by edges representing contacts that clash and are relieved by alternative conformations. The pathway in b forms part of the red contact network in CYPA and is highlighted by the dark purple edges. (d) The six contact networks comprising 29% of residues are mapped on the three dimensional structure of CYPA. The contact network shown in red overlaps with the dynamic network identified by NMR chemical shift perturbation and relaxation dispersion experiments.
CONTACT first identifies ‘pathways’ of van der Waals overlaps. Each residue (e.g. residue i) in turn is moved to an alternative conformation, and overlaps of van der Waals radii with any neighboring residues (e.g. residue j) are identified (Fig. 1b). If neighboring residue j can be moved to an alternative conformation to reduce the steric overlap with residue i, the pathway is continued to neighboring residues of residue j (e.g. residue k, l, etc) until no new clashes are introduced. The relative frequency with which residues can and cannot reduce steric overlap is also recorded (Supplementary Fig. 3).
Pathways can include overlapping or nearly overlapping sets of residues using different combinations of alternative conformations. Thus, pathways that share common members indicate conformational coupling even if the residues are not directly linked in a single pathway. Any pathways that share residues are grouped into a single contact network (Fig. 1c). Thus, a pathway is a single sequence of residues that can be moved between alternative conformations such that van der Waals overlaps are reduced after each move. A contact network is a set of residues that are linked by common pathways.
CONTACT identifies multiple pathways in Cyclophilin A
To test whether CONTACT can automatically identify networks of residues from experimental X-ray data, we first examined the human proline isomerase Cyclophilin A (CYPA). Previous NMR experiments have demonstrated that CYPA undergoes conformational exchange both during its catalytic cycle and in the absence of substrate25. Room temperature X-ray electron density maps revealed extensive alternative side chain conformations for residues extending from the active site into the core, providing a structural rationale for the collective motion inferred by NMR4.
A large contact network of nine residues connects the hydrophobic core of the protein to the active site (Fig. 1d). This “red” contact network generally agrees with findings obtained from a visual analysis of room temperature X-ray data in providing a structural mechanism for collective conformational exchange detectable by NMR relaxation dispersion experiments25. Consistent with the idea that perturbations within the contact network will stabilize alternative conformations of other residues in the contact network, a mutation (S99T) of a core residue leads to chemical shift changes that spread across the red contact network and results in a kcat/KM that is ~0.3% of wild type (WT)4.
Notably, the red contact network consists of many independent pathways that connect contact network residues as reflected in the weights of the edges of the network graph (Fig. 1c). The large number of pathways suggests that the transition between the major and minor forms of the enzyme can occur by multiple structurally distinct mechanisms. The idea of multiple transition paths in CYPA is supported by recent NMR studies that incorporate evidence from the conformational dynamics of several mutants26. The contributions of these transitions are difficult to separate in the collective exchange fitting procedures of the NMR experiment26. Major and minor form end-states distinguished by alternative side chain conformations may dominate the conformational dynamics in the crystal4 and solution25; however, CONTACT identifies multiple plausible transition paths.
CONTACT analysis of long-range perturbations in G121V ecDHFR
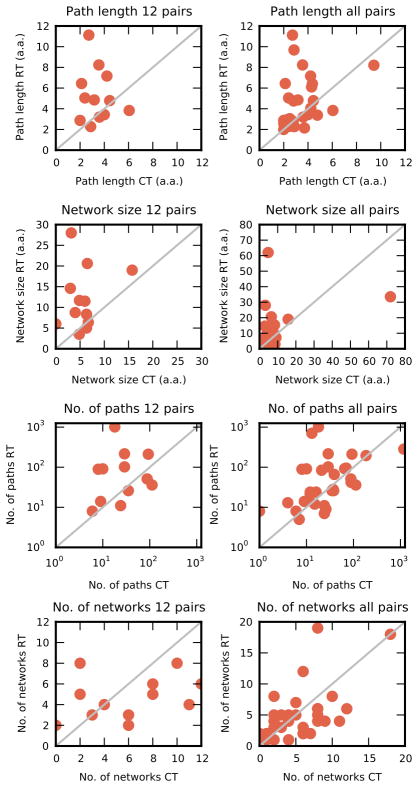
Dihydrofolate reductase (DHFR) is a model system for studying the relationship between conformational dynamics and catalytic activity27. The solution-state dynamics of the E.coli DHFR are dominated by the interconversion of the Met20 loop between closed and occluded conformations, which allow optimal substrate flux through the catalytic cycle28. We obtained X-ray diffraction data sets at cryogenic (100 K - 1.15 Å) and room temperature (~273 K - 1.35 Å) for the model Michaelis complex (E:NADP+:FOL) (Supplementary Table 1). Consistent with the trends observed in a larger dataset of 35 proteins (Figure 2, Supplementary Table 2, Supplementary Figs. 4–6, and Supplementary Note), the room temperature dataset exhibits both more (157 vs. 70) and longer (5.5 vs. 4.3) pathways than the cryogenic dataset.
Figure 2.
Characteristics of pathways and contact networks are sensitive to temperature. Changes to average pathway lengths, contact network sizes, number of pathways, and number of contact networks across 12 closely matched high resolution (left column) or all matched (right column) room temperature (RT) and cryogenic datasets (CT). Each data point represents paired data sets, with values corresponding to room temperature along the vertical axis, and cryogenic temperature along the horizontal axis. Data points are expected to lie along a 45° degree line if there were no differences between the room and cryogenic temperature pairs.
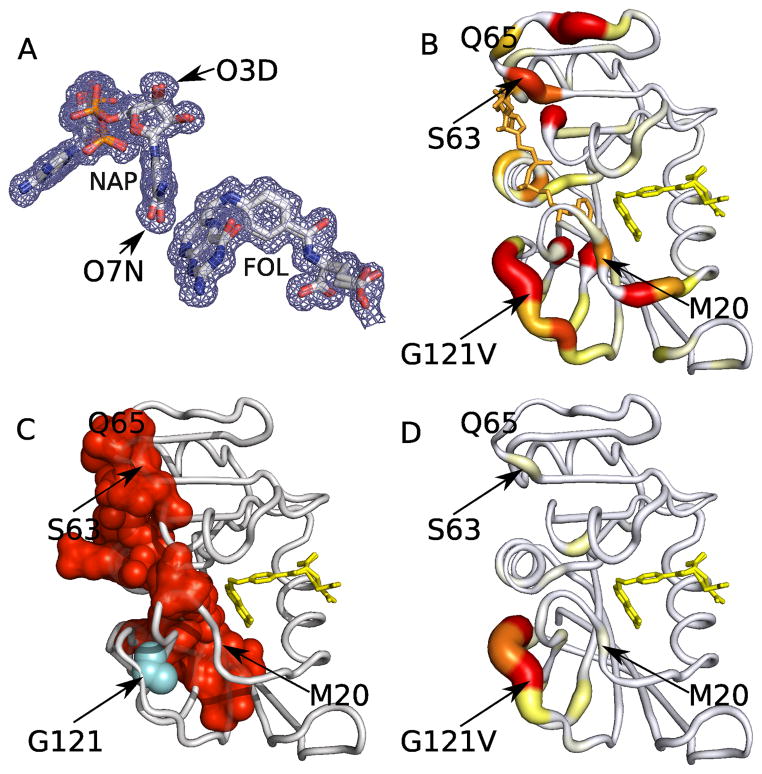
The largest contact network from the room temperature dataset connects the functionally important FG loop to the NADP binding pocket and the adenosine binding domain (Fig. 3) This long-range connection is mediated by the cofactor NADP molecule, which uniquely connects the two subdomains (Fig. 3a and Supplementary Table 3). The electron density map is consistent with discrete disorder around the cofactor (Fig. 4a), providing further evidence that the bound NADP molecule is a dynamic hub. To test this model of conformational coupling of the FG loop to the adenosine-binding domain, we examined the chemical shift perturbations of a mutation in the FG loop (G121V). The NMR data for G121V is consistent with this hypothesis29. Changes in 15N and/or 1H chemical shifts between WT and the G121V mutant (E:NADP+:FOL) complexes propagate from the FG loop to the adenosine binding domain (Fig. 4b). However, no G121V X-ray structure has been published, leaving the structural origin of these long-range effects unclear.
Figure 3.
All-atom contact networks in ecDHFR. (a) Contact networks are displayed in surface rendering on the crystal structure of the room temperature E:NADP+:FOL complex (3QL3) (top panels) above the largest contact network graphs (bottom panels). Nine all-atom contact networks comprise 47% of the residues in the room temperature model of ecDHFR. The NADP+ cofactor is part of the red contact network, and shown in red spheres. Folate, part of the yellow contact network, is shown in yellow spheres. The red network connects residue Phe125 in the FG loop to the adenosine binding domain (Ser63–Gln65) exclusively through the NADP cofactor. Residues in the cyan (b) and green (c) contact networks broadly agree with those identified undergoing collective exchanges in CPMG relaxation dispersion experiments in a process that is distinct from the conformational exchange observed near the active site. Consistent with NMR data, we observed that the cyan and green contact networks do not contact active site residues. The salmon and blue contact networks (c) are implicated in hinge motions. The orange contact network is implicated in changing hydrogen bonding patterns during the closed to occluded transition of the M20 loop. The yellow contact network links active site residues I5 and I95 to the folate.
Figure 4.
Contact network analysis and an allosteric mutant (G121V) of ecDHFR. (a) A 2mFo-DFc electron density map around the cofactor and substrate of the room temperature (E:NADP+:FOL) complex contoured at 0.3 σ. Asymmetric density profiles around oxygen atoms (O7N, O3D) of the NADP molecule support multiple conformations. (b) The color (yellow to red) and thickness of the backbone tube represents the magnitude of the weighted chemical shift differences obtained from WT and a G121V mutant (E:NADP+:FOL) complex from 0.1–1ppm. Residues Ser63, Gln65, Gly67, and Thr68 in the adenosine binding domain exhibit large chemical shift changes despite their location over 23 Å away from the mutation site. The G121V mutation stabilizes the occluded conformation of the enzyme. In the WT E:NADP+:FOL complex, the M20 loop is in the “closed” conformation, whereas in the corresponding complex of G121V the M20 loop is in the “occluded” conformation. (c) The red contact network obtained from the room temperature WT (E:NADP+:FOL) complex (also Fig. 3a) in the same orientation. For illustrative purposes the G121V mutation (cyan) is modeled on the WT molecule, abutting the red contact network. Long-range conformational coupling generally corresponds to chemical shift propagation, while local effects owing to increased flexibility of the FG loop in the mutant complex are absent. (d) Chemical shift differences between binary WT and G121V mutant (E:FOL) complexes are localized to the site of mutation, confirming the central role of the NADP cofactor in coupling distant sites in ecDHFR.
Modeling a Val side chain at position 121 causes severe clashes with residues 13–15 and directly impinges upon the red ecDHFR contact network (Fig. 4c). The pattern of contacts in the network includes FG-loop residue 125 and extends to the adenosine-binding domain, resembling the chemical shift perturbations between WT and G121V ecDHFR. These results propose a physical model underlying the long-range chemical shift perturbations: the G121V mutation selectively stabilizes pre-existing conformations that propagate from the FG loop to the adenosine-binding domain. While the distance encompassed by the chemical shift changes is surprising, a single contact network mediates the direction and extent of long-range conformational coupling. In addition to these long-range effects, the “allosteric” mutation G121V destabilizes the catalytically competent closed conformation of the enzyme, stabilizes the occluded conformation, and reduces kcat approximately to ~0.6% of WT.
Several additional lines of evidence support this model. In molecular dynamics simulations, the dynamics of residue 121 are correlated with the dynamics of residues in the adenosine binding domain30. Sampling of locally unfolded states identified a similar pattern of energetic coupling7. NMR spin relaxation experiments in the fast (ps–ns) timescale found that the majority of residues with fast dynamics affected by G121V mutation are located in the adenosine binding domain29,31.
Based on our contact network analysis, we predicted that removing NADP would disrupt the conformational coupling of the FG loop to the adenosine-binding domain. To assess this idea, we compared 15N and 1H chemical shifts for the binary WT and G121V E:FOL complexes. In the absence of NADP, we observed no major chemical shift perturbations for residues in the adenosine-binding domain in the folate binary complexes (Fig. 4d). Thus, CONTACT provides testable hypotheses about how mutations shift conformational ensembles and induce long-range chemical shift perturbations observed in NMR experiments.
Altered conformational heterogeneity in N23PP/S148A ecDHFR
We used CONTACT to study the N23PP/S148A mutant of ecDHFR, which was designed to destabilize the occluded conformation of the Met20 loop. This mutant populates a nearly wild type structure as defined by a single cryogenic X-ray model32. Interestingly, and somewhat surprisingly, the N23PP/S148A mutant has a reduced rate of hydride transfer (khyd). In contrast to the WT enzyme, the N23PP/S148A mutant displays no evidence of conformational exchange for most active site residues on the millisecond timescale. However, faster (ps-ns) timescale backbone dynamics remained similar to WT32. While previous experiments indicated that the alteration of millisecond conformational dynamics in the mutant influenced the chemical step of catalysis, the underlying mechanism remained elusive. To further investigate the origins of the reduction in hydride transfer rate, we crystallized the N23PP/S148A mutant (E:NADP+:FOL) complex in the same crystal form as WT ecDHFR and collected a new room temperature data set to 1.38 Å resolution (Fig. 5, Supplementary Fig. 7, and Supplementary Tables 1 and 4).
Figure 5.
Increased conformational heterogeneity at the active site of the crystal structure of the E:NADP+:FOL complex of the N23PP/S148A mutant. (a) The red contact network shares similarities with its WT counterpart, but several residues in the Met20 loop now participate in this contact network. The connection between the FG loop and NAP to the adenosine binding domain through residue I14 has been replaced by Met20 loop residues Met20, Pro21, and Trp22. (b) The cyan contact network is largely comprised of three beta strands towards the C terminus of the molecule. (c) An isomorphous difference electron density map of the WT and N23PP/S148A datasets phased with the WT crystal structure. Positive difference density (4.0σ green) in the absence of negative difference density (−4.0σ red) indicates elevated heterogeneity. Mutation sites are shown with spheres, NADP+ in orange and folate in yellow (see also Supplementary Figure 7b).
Given the minimal structural effects of the mutation and the loss of observable millisecond conformational dynamics, we expected that CONTACT would reveal a large reduction in pathways and a decreased number of residues participating in active site contact networks. Surprisingly, we found a ~500% increase in the number of all-atom pathways in N23PP/S148A (806) compared to WT (157). In both WT and N23PP/S148A ecDHFR, a large contact network connects the FG loop to the adenosine-binding domain through the NADP cofactor. However, several residues of the Met20 loop were included in the mutant, but not wild type, contact network (Fig 5a). This result suggests that additional non-productive motions surrounding the active site of N23PP/S148A ecDHFR can influence the relative positions of the NADP and FOL during the reaction cycle. A second large contact network (blue) in the N23PP/S148A mutant reveals an extensive set of connections across the central beta strand of the protein (Fig. 5a,b). While in WT ecDHFR the connections across these beta strands were distributed over several contact networks (Fig. 4b,c), the entire beta sheet forms a single contact network in N23PP/S148A ecDHFR. These results suggest that the mutant enzyme exhibits increased active-site heterogeneity, despite the loss of detectable conformational averaging on the millisecond timescale.
To complement the results of CONTACT, we generated an isomorphous difference map between the WT and N23PP/S148A datasets. This map revealed that the most prominent difference features were located immediately adjacent to the site of mutation, corresponding to the change in amino acid identity (N23 to P) or proline insertion (Fig. 5c). However, several other features surrounding the active site suggest that the mutations had shifted the conformational distributions of neighboring residues. The presence of positive difference density without a corresponding negative difference density peak implies increased disorder in the mutant electron density map. A single model would fail to reveal any differences between the WT and mutant enzyme as the mean positions are not altered. Rather, one major effect of the mutation is to broaden the conformational distributions relative to WT, thus increasing frustrated, non-productive motions in the mutant enzyme. Indeed, difference density around the M20 loop residues Met20 and Trp22 revealed elevated conformational heterogeneity, and these residues participate in N23PP/S148A, but not WT, contact networks (Figs. 3 and 5a). Additional difference density is observed across the beta sheet contacting the C terminus. These results provide a further indication that the mutations can alter dynamic properties in distant regions of the molecule and that the N23PP/S148A mutation increases conformational heterogeneity observable by X-ray crystallography.
DISCUSSION
The ability to infer networks of coordinated motion from X-ray data provides a gateway to understanding the structure-dynamics-function relationships in proteins. By analyzing experimental X-ray data, CONTACT complements existing methods for analyzing molecular dynamics trajectories33 Monte Carlo simulations34 or Rosetta-based analyses35. CONTACT automatically identifies coupled conformational heterogeneity, previously called “dynamic close packing”17, present in electron density distributions. These networks can connect allosteric and functional sites, explain the propagation of chemical shift differences, and reveal the structural mechanisms of mutations that have minimal effects on the ground state conformation.
Our analysis reveals that conformations accessible in the crystal are sensitive to perturbations such as temperature and mutations. Recent experiments have postulated that intramolecular pathways of signaling exist within a protein structure24. The physical basis of these pathways and how they are insulated from and supported by the surrounding protein structure remains unclear. While previous studies have largely focused on using the functional response to mutations to delineate the boundaries of intramolecular pathways, our results suggest that changes to contact networks as a function of data collection temperature may reveal new insights. For most proteins, we observed a decrease in the number and lengths of pathways at cryogenic temperature, implying that the broader conformational ensembles of residues at room temperature go beyond ‘filling the voids’ provided by expansion of the lattice6. Rather, networks of residues collectively sample alternative conformational substates. Some cryo-cooling procedures stabilize contact networks into specific conformational substates from those sampled at room temperature. It is important to note that exposure to X-ray radiation can introduce serious radiation damage artifacts that may complicate these analyses. Radiation damage is dramatically reduced at cryogenic temperatures. New advances in serial femto-second crystallography may mitigate some of these concerns, retaining the advantages of non-cryogenic collection with radiation damage protection that exceeds cryogenic techniques36. Thus, new biophysical tools may help circumvent the complex trade-off between collecting data below the glass transition37, which alters conformational heterogeneity, and protecting against radiation damage.
In CYPA, a large contact network consists of residues that collectively sample different conformations during the catalytic cycle. A mutation (S99T) outside of the active site that reduces the extent of the contact network, impairs the kinetics of interconversion, and reduces catalytic efficiency. In contrast, in ecDHFR, a mutant (N23PP/S148A) with a rigidified Met20 loop has the surprising effect of increasing non-productive, frustrated conformational heterogeneity in the active site. Frustration resulting from competing low-free-energy conformations that cannot be mutually satisfied are thought to play a key role in native state dynamics38. The loss of detectable millisecond conformational exchange in the active site is due to a decrease in backbone flexibility of the Met20 loop. However, local anharmonic side chain motions, which do not generate a large enough chemical shift difference or occur on a timescale inaccessible for detection by a relaxation dispersion experiment, have likely increased around the active site and may inhibit progress towards the transition state. Further NMR experiments, particularly for side chain methyl groups, may provide additional insights.
Superficially, the S99T mutation of CYPA and the N23PP/S148A mutation of ecDHFR are similar: both result in losses of catalytic activity and micro- to millisecond timescale conformational dynamics. However, the CONTACT results offer distinct interpretations of these mutations. In CYPA, an overpacked core decreases the kinetics of interconversion between conformational substates and catalytic rate. In contrast, in ecDHFR, the stabilization of Met20 loop dynamics have increased heterogeneity in the active site, which likely decreases the rate of hydride transfer. Enzyme engineering and design efforts could target residues in contact networks for simultaneous sequence optimization based on the principle that network residues exert a substantial influence on the conformations at other contact network sites. As protein design methods are optimized, the conformational dynamics necessary for catalytic cycles (for example, to occlude water or prevent product inhibition) will need to be defined and engineered. However, recently de-novo designed enzymes appear to suffer from packing imperfections that allow too much conformational heterogeneity, suggesting that contact networks can be targeted to improve packing39.
As multi-conformer and ensemble models are more widely adopted, several improvements to this analysis can reveal further links between conformational flexibility and mechanism. Minor improvements in Rfree should be carefully evaluated against model fit, stereo-chemistry and, importantly, a biologist’s ability to develop structure-based hypotheses of bio-molecular function. A particular challenge is to determine the extent of conformational coupling caused by steric mechanisms, as modeling of atomic interactions with a repulsive hard-sphere potential is an egregious simplification. As in ecDHFR, where the NADP ligand connects distant sites, it is likely that non-protein atoms play key roles in coupling distant sites; however, qFit does not currently automatically fit multi-conformer small molecules or waters into the electron density. Despite these simplifications, analysis of a multi-conformer model from a single X-ray experiment complements NMR22, long-timescale molecular dynamics40, Rosetta sampling35 and comparison of multiple independent crystal structures21. Integrated analyses combining these approaches will help to derive structural bases for conformational dynamics of proteins and to develop new hypotheses about how protein motions relate to function.
ONLINE METHODS
qFit
To model heterogeneous features in the electron density qFit computes an optimal fit of one to four conformations per residue together with their occupancies19. No explicit information about non-bonded contacts is included in the initial assignment of conformations or occupancy. These conformations are subsequently connected allowing for backbone heterogeneity into a multi-conformer model. The multi-conformer models can provide evidence of discrete heterogeneity of populations down to ~10%. qFit is available as a web server at: http://smb.slac.stanford.edu/qFitServer/qFit.jsp.
We used Phenix version 1.8–1069 to add hydrogens to qFit models and for subsequent conventional positional and ADP refinement steps 41. The qFit model is prepared for refinement with phenix.ready_set using the default parameters, which add riding hydrogens. The model is refined without manual intervention with phenix.refine using the flags:
“optimize_xyz_weight=true optimize_adp_weight=true main.strategy.individual_sites_real_space=false main.number_of_macro_cycles=5”
All other phenix.refine parameters are set to the default values, which includes occupancy refinement. For models with diffraction to greater than 1.55A resolution, we also include the flag:
“adp.individual.anisotropic=”not water or element H”
Since phenix.refine optimizes the hydrogen placement to the center of X-ray scattering rather than the likely nuclear position, hydrogens are stripped after refinement with phenix.pdbtools using the flags:
modify.remove=”element H”
Hydrogens were then restored to their nuclear positions with phenix.ready_set using the default parameters.
Model building and calculation of pathways
The CONTACT algorithm (available at http://smb.slac.stanford.edu/CONTACT) calculates the most severe van der Waals clash between atoms from different residues that are separated by less than the sum of the van der Waals radii. This calculation takes into account all conformations of all pairs of residues with alternative conformations, excluding main chain hydrogen bonded atoms (identified by MMDB) and pairs of cysteine residues. In evaluating a multi-conformer model, we use the most severe 30% of these van der Waals overlaps to define the threshold value for clashes (Tstress). The 30th percentile of van der Waal’s overlap is an adjustable parameter in CONTACT. Functional biological insights may be obtained at other values of this parameter subject to considering resolution, data quality, and crystal environment.
Starting at each residue in succession, if we have obtained a pathway up to residue i, a residue j is added to the pathway if the following two conditions are satisfied:
Changing conformation ui at residue position i to conformation ri creates at least one substantial (≥ Tstress) overlap on conformation uj at residue position j: E(ui,uj) ≤ E(ri,uj)
Changing conformation rj to conformation uj at residue position j reduces the most substantial overlap: E(ui,uj) ≤E(ui,rj)
A pathway then continues for the pairwise interactions between j and a residue k that satisfies these conditions. This process is continued until no more van der Waals overlaps are introduced. The CONTACT algorithm can calculate pathways using either side chain atoms only, or both main-chain and side chain atoms. The parameter Tstress can be varied (Supplementary Fig. 5). Although these alternative conformations need not be kinetically or thermodynamically mutually exclusive, the overlap of van der Waals radii indicates a likely coupling between the relative populations of conformations at each site.
Calculation of Contact Networks for CYPA and ecDHFR
Pathways identified by CONTACT are included in the network analysis of CYPA, WT ecDHFR and N23PP/S148A ecDHFR in the main text at the worst 30th percentile of van der Waal’s overlap, corresponding to 13%, 14%, and 14% overlaps respectively. We furthermore required that all clashes in a pathway were reduced to 10% overlap or less. We build a network graph where nodes are residues with edges indicating contacts identified by CONTACT. In analyzing the properties of these connected networks, we considered only subgraphs with more than 3 nodes and compute the edge weight as the number of distinct pathways between nodes.
Pathways and contact networks calculated from 35 pairs of cryogenic and room temperature data sets
We used qFit to rebuild 35 pairs of protein models with available high-resolution (2.0 Å or better) X-ray diffraction data collected at room and cryogenic temperatures (Supplementary Table 3). The majority were crystallized in nearly identical or very similar conditions. The datasets have Rmerge values indicating no unusual radiation damage6. When no Rfree set was deposited or could be extracted, we chose a test set using the standard parameters in phenix.refine. Structures were rebuilt using qFit and then refined as described above.
To examine the effect of severity of van der Waals overlap on pathway discovery the parameter Tstress was incremented in 5% steps starting at the worst 5th percentile of van der Waals overlaps. The search for additional pathways was terminated when its number exceeded 1,000,000. The effect of considering a higher percentile is that generally more van der Waals overlaps are identified, but that these are also less likely to be relieved below the threshold to continue the pathway. The number of all-atom pathways and their lengths diverge substantially between room and cryogenic temperatures, reaching a peak at the 30th–35th percentile overlap (Supplementary Fig. 5). For side chain only pathways the largest differences in the number of pathways between room and cryogenic temperature were observed at the 15th and 25th percentiles, but no significant difference was found for their lengths (Supplementary Fig. 2).
DHFR Chemical Shift Perturbations
Backbone amide assignments for the E:NADP+:FOL42 and E:FOL43 complexes of 13C,15N-labeled or 13C,15N,2H-labeled WT and G121V ecDHFR were made using standard triple resonance methods43. Weighted average of chemical shift differences were calculated using the formula:
ecDHFR X-ray crystallography
WT (E:NADP+:FOL) and N23PP/S148A (E:NADP+:FOL) were purified and concentrated to 30mg/mL32. Both WT and N23PP/S148A crystals were obtained by the hanging drop method by mixing protein 1:1 with well solution (100mM HEPES pH 7.5, 21% PEG8000, 200mM MgCl2). Data were collected at BL 8.3.1 of the Advanced Light Source. For room temperature data collection, the crystal is mounted in using loop and covered with RT tubing (MITEGEN) with 15uL of a 3:1 mixture of mother-liquor to water in the tip. The cryojet is set to 273 K and moved such that it lies just outside the capillary tube. The beam is heavily attenuated during test shots to establish a data collection strategy. During data collection, the attenuation is reduced by removing all but the Aluminum foil and the radiation dose is spread across the large (600uMx200uMx100uM) needle-shaped crystals by translation after each shot using a custom TCL script that interfaces with the standard BLUEICE interface. There are no unusual indications of radiation damage or problems with mosaic spread due to the translation: unit cell parameters and scale factors are stable during data processing, and the overall Rsym is low. The cryogenic dataset was collected after the addition of 10% PEG400 to the mother-liquor. Datasets were processed using XIA244 using the flag -3dii for XDS45 pipeline. Molecular replacement was performed using 1RX2 using Phaser46 through the Phenix GUI41.
Supplementary Material
Acknowledgments
H.v.d.B. is supported by the National Institute of General Medical Sciences Protein Structure Initiative (U54GM094586) at the Joint Center for Structural Genomics and a SLAC National Accelerator Laboratory LDRD grant SLAC-LDRD-0014-13-2; G.B. is supported as a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2136-12); K.Y. is supported by the General Wang Yaowu Stanford graduate fellowship; P.E.W. is supported by the US National Institutes of Health (GM75995); J.S.F. is supported by the US National Institutes of Health Early Independence Award (DP5OD009180). We acknowledge: T. Alber, T. Kortemme, D. Keedy, D. Kern, D. Tawfik, and R. Woldeyes for helpful discussions; N. Echols for advice on different treatments of hydrogen atoms in Phenix; J. Holton, J. Tanamachi and G. Meigs at Advanced Light Source Beamline 8.3.1 for support with X-ray data collection; B. Duggan and D. Boehr for chemical shift data on G121V ecDHFR complexes; H. Axelrod and C. Trame for data collection and refinement of RT RBM39.
Footnotes
AUTHOR CONTRIBUTIONS
H.v.d.B., G.B., P.E.W. and J.S.F. designed and performed experiments, analyzed data and prepared the manuscript; G.B. and J.S.F. collected data; and H.v.d.B., K.Y. and J.S.F. developed analytical tools.
The authors declare no competing financial interests.
ACCESSION CODES
Protein Databank: 4KJL, 4KJK, 4KJJ
References
- 1.Fraser JS, Jackson CJ. Mining electron density for functionally relevant protein polysterism in crystal structures. Cell Mol Life Sci. 2011;68:1829–1841. doi: 10.1007/s00018-010-0611-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci USA. 2005;102:6679–85. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat Chem Biol. 2010;6:352–8. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser JS, et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–73. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140:246–56. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser JS, et al. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc Natl Acad Sci USA. 2011;108:16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitten ST, García-Moreno EB, Hilser VJ. Local conformational fluctuations can modulate the coupling between proton binding and global structural transitions in proteins. Proc Natl Acad Sci USA. 2005;102:4282–7. doi: 10.1073/pnas.0407499102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–11. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–82. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 10.Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Burling F, Brunger A. Thermal motion and conformational disorder in protein crystal structures: Comparison of multi-conformer and time-averaging models. Israel J Chem. 1994;34:165–175. [Google Scholar]
- 12.Levin EJ, Kondrashov DA, Wesenberg GE, Phillips GN., Jr Ensemble refinement of protein crystal structures. Structure. 2007;15:1040–1052. doi: 10.1016/j.str.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight JL, et al. Exploring structural variability in X-ray crystallographic models using protein local optimization by torsion-angle sampling. Acta Cryst. 2008;D64:383–96. doi: 10.1107/S090744490800070X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuriyan J, et al. Exploration of disorder in protein structures by X-ray restrained molecular dynamics. Proteins. 1991;10:340–58. doi: 10.1002/prot.340100407. [DOI] [PubMed] [Google Scholar]
- 15.Terwilliger TC, et al. Interpretation of ensembles created by multiple iterative rebuilding of macromolecular models. Acta Cryst. 2007;D63:597–610. doi: 10.1107/S0907444907009791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson MA, Brunger AT. The 1.0 A crystal structure of Ca(2+)-bound calmodulin: an analysis of disorder and implications for functionally relevant plasticity. Journal of molecular biology. 2000;301:1237–56. doi: 10.1006/jmbi.2000.4029. [DOI] [PubMed] [Google Scholar]
- 17.Rader SD, Agard DA. Conformational substates in enzyme mechanism: The 120K structure of a-lytic protease at 1.5 A resolution. Protein Sci. 1997;6:1375–1386. doi: 10.1002/pro.5560060701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burnley BT, Afonine PV, Adams PD, Gros P. Modelling dynamics in protein crystal structures by ensemble refinement. eLife. 2012;1:1–29. doi: 10.7554/eLife.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van den Bedem H, Dhanik A, Latombe J-C, Deacon AM. Modeling discrete heterogeneity in X-ray diffraction data by fitting multi-conformers. Acta Cryst. 2009;D65:1107–1117. doi: 10.1107/S0907444909030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang PT, et al. Automated electron-density sampling reveals widespread conformational polymorphism in proteins. Protein Sci. 2010;19:1420–31. doi: 10.1002/pro.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindorff-Larsen K, Best RB, Depristo Ma, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–32. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 22.Serrano P, et al. Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites. Acta Cryst. 2010;F66:1393–405. doi: 10.1107/S1744309110033658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young Ma, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell. 2001;105:115–26. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 24.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009;138:774–86. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–21. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 26.Schlegel J, Armstrong GS, Redzic JS, Zhang F, Eisenmesser EZ. Characterizing and controlling the inherent dynamics of cyclophilin-A. Protein Sci. 2009;18:811–24. doi: 10.1002/pro.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–42. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 28.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 29.Boehr DD, et al. A Distal Mutation Perturbs Dynamic Amino Acid Networks in Dihydrofolate Reductase. Biochemistry. 2013 doi: 10.1021/bi400563c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rod TH, Radkiewicz JL, Brooks CL. Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc Natl Acad Sci USA. 2003;100:6980–5. doi: 10.1073/pnas.1230801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mauldin RV, Sapienza PJ, Petit CM, Lee AL. Structure and dynamics of the G121V dihydrofolate reductase mutant: lessons from a transition-state inhibitor complex. PloS ONE. 2012;7:e33252. doi: 10.1371/journal.pone.0033252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–8. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClendon CL, Friedland G, Mobley DL, Amirkhani H, Jacobson MP. Quantifying Correlations Between Allosteric Sites in Thermodynamic Ensembles. J Chem Theory Comput. 2009;5:2486–2502. doi: 10.1021/ct9001812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubay KH, Bothma JP, Geissler PL. Long-range intra-protein communication can be transmitted by correlated side-chain fluctuations alone. PLoS Comput Biol. 2011;7:e1002168. doi: 10.1371/journal.pcbi.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kidd Ba, Baker D, Thomas WE. Computation of conformational coupling in allosteric proteins. PLoS Comput Biol. 2009;5:e1000484. doi: 10.1371/journal.pcbi.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boutet S, et al. High-resolution protein structure determination by serial femtosecond crystallography. Science. 2012;337:362–4. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halle B. Biomolecular cryocrystallography: structural changes during flash-cooling. Proc Natl Acad Sci USA. 2004;101:4793–8. doi: 10.1073/pnas.0308315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhuravlev PI, Papoian G. a Protein functional landscapes, dynamics, allostery: a tortuous path towards a universal theoretical framework. Q Rev Biophys. 2010;43:295–332. doi: 10.1017/S0033583510000119. [DOI] [PubMed] [Google Scholar]
- 39.Khersonsky O, et al. Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution. J Mol Biol. 2011;407:391–412. doi: 10.1016/j.jmb.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dror RO, Dirks RM, Grossman JP, Xu H, Shaw DE. Biomolecular simulation: a computational microscope for molecular biology. Annu Rev Biophys. 2012;41:429–52. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Backbone dynamics in dihydrofolate reductase complexes: role of loop flexibility in the catalytic mechanism. Biochemistry. 2001;40:9846–59. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 43.Falzone CJ, et al. 1H, 15N and 13C resonance assignments, secondary structure, and the conformation of substrate in the binary folate complex of Escherichia coli dihydrofolate reductase. J Biomol NMR. 1994;4:349–366. doi: 10.1007/BF00179346. [DOI] [PubMed] [Google Scholar]
- 44.Winter G, Lobley CMC, Prince SM. Decision making in xia2. Acta Cryst. 2013;69:1260–73. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch WXDS. Acta Cryst. 2010;D66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.