Abstract
In this proof-of-concept study, we describe the use of the main red beet pigment betanin for the quantification of calcium dipicolinate in bacterial spores, including Bacillus anthracis. In the presence of europium(III) ions, betanin is converted to a water-soluble, non-luminescent orange 1∶1 complex with a stability constant of 1.4×105 L mol–1. The addition of calcium dipicolinate, largely found in bacterial spores, changes the color of the aqueous solution of [Eu(Bn)+] from orange to magenta. The limit of detection (LOD) of calcium dipicolinate is around 2.0×10–6 mol L–1 and the LOD determined for both spores, B. cereus and B. anthracis, is (1.1±0.3)×106 spores mL–1. This simple, green, fast and low cost colorimetric assay was selective for calcium dipicolinate when compared to several analogous compounds. The importance of this work relies on the potential use of betalains, raw natural pigments, as colorimetric sensors for biological applications.
Introduction
Bacteria of genus Bacillus can assume a dormant and resistant spore form (i.e., endospore) in order to survive harsh environmental conditions. Although some bacterial spores contribute to human activities, e.g., B. thurigiensis is used as a pesticide, other species, such as B. anthracis, B. cereus, pose a serious risk to human health [1]. Bacillus anthracis has emerged as a bioterrorism agent because of its high stability and virulence [2], [3] and became a public safety concern again after the anthrax attack in 2001 [4]. The infection usually takes place through skin contact with infected animals or animal products, but can also occur by inhalation or ingestion of spores [5]. Another example, Bacillus cereus, has negative economic impacts because this pathogenic bacterium can grow on food [6]. Consequently, there is a need for early detection of Bacillus species to ensure human health and safety.
Calcium dipicolinate (CaDPA) is the major component (up to 15% dry weight) of the bacterial spore core [7]. It stabilizes the bacterial DNA, contributes to the overall chemical and heat resistance of the spore and is released during germination [8], [9]. Endospores also release CaDPA upon thermal treatment or in the presence of reactivation agents that induce bacterial germination, such as inosine and L-alanine [10]. Although there are several methods for the detection of B. anthracis [11], [12], the simple and fast detection of dipicolinic acid (DPA) using luminescent lanthanide has been promising [9], [13], [14], [15].
Betanin (Bn) is the non-toxic, water-soluble pigment responsible for the deep red–magenta color of the red beet (λ = 536 nm, h° = 336, ε = 6.5×104 L mol–1 cm–1) [16], [17]. Bn is a food colorant (additive E-162) with a high antioxidant capacity [18], [19], [20], which has been used in several different applications such as dyes in solar cells [21], [22] or as a starting material in the semi-synthesis of fluorescent probes for the live-imaging of Plasmodium-infected red blood cells [23]. In this proof-of-concept study, betanin is used as a ligand in a new EuIII complex, which is sensitive to CaDPA but not to analogous compounds. In order to demonstrate the applicability of the method in detecting CaDPA, we used two representative Bacillus species (B. anthracis and B. cereus spores) as test samples.
Material and Methods
Complexation Studies
Determination of equilibrium constants
The stoichiometry of the EuIII/Bn complex was determined using the molar-ratio method (Yoe and Jones’ method) [24]. The concentration of a solution of Bn in MOPS buffer pH = 7.5 (10 mmol L–1) was kept constant (5.57 µmol L–1) and a variable amount of EuIII (0.3 to 22 equiv) was added. Experiments were carried out independently at 25±1°C in quartz cuvettes (o.p. 10 mm) with a final volume of 2 mL. The mole ratio of the metal ion to Bn was plotted versus absorbance at 536 nm and tangents were drawn. The perpendicular line located at the intersection of the tangents was drawn to the mole ratio axis showing the EuIII/Bn ratio.
Stability constants were determined using a simple metal–ligand complexation model [25] considering a 1∶1 stoichiometry (Eq. (1)):
| (1) |
where, L: ligand; M: metal; C: complex; a and b are the stoichiometric factors; [L]0 and [M]0: initial total concentration of the ligand and the metal, respectively; [L], [M] and [C]: equilibrium concentration of the ligand, the metal and the complex, respectively.
For the determination of K by UV/Vis spectrometry, it is necessary to determine the [C]. In case the metal does not absorb at the wavelength λ, [C] can be determined using Eq. 2. The mathematical derivation of this method and additional experimental details are presented in the File S1.
| (2) |
where, Aλ obs is the observed absorbance at a given wavelength, ελ L and ελ C are the molar absorption coefficient of the ligand and complex at the same wavelength, respectively.
The data obtained with the titration experiment were used for the determination of the lanthanide–betalain stoichiometry as well as for the determination of [C] using Eq. (2). The stability constant was determined by averaging the values obtained using Eq. (1) for each concentration of lanthanide (Table S1).
Calibration curve and limit of detection
In a 96 well microplate, solutions of [Eu(Bn)]+ were prepared by adding different amounts of a 1 mmol L–1 solution of EuCl3 in MOPS buffer to a solution of Bn in water. Next, a defined volume of a solution of CaDPA (1 mmol L–1) was added and the volume in each well was adjusted to 200 µL using MOPS buffer pH = 7.5. Final concentrations are as follows: [Bn] = 5.6 µmol L–1, [EuIII] = 5.6 to 33.4 µmol L–1, [CaDPA] varied from 0.6 up to 120 µmol L–1 depending on the [EuIII]. The absorbance at 536 nm was registered either in the absence or in the presence of variable amounts of CaDPA and used to calculate the variation in the response (ΔAbs536 nm = ΔAbs536 nm CaDPA – ΔAbs536 nm Control). The calibration curve was constructed by plotting the concentration of CaDPA in mol L–1 (y) versus the ΔAbs536 nm (x). The data points in the linear interval of the sigmoidal curve were submitted to linear regression analysis.
The limit of detection (LOD) was determined using a procedure based on a linear model in different zones of the sigmoid response function; namely, the maximum and minimum slope (at low analyte activity) of the response function is fitted to a linear function by a statistical lack-of-fit test. Thus, the LOD is defined as the activity of the analyte that corresponds to the intersection between the two straight lines at the higher and lower slope (signal-to-noise ratio of 3) [26].
Microbiological Methods
Bacillus cereus sample preparation
Spore suspensions of Bacillus cereus ATCC 10987 were prepared by growing a uniform lawn of spores on solid PGSM [27]. After 7 d of growth (3 d at 37°C and the remainder at 21°C), plates were examined daily for spore formation by removing a small amount of spores and examining with phase microscopy (Olympus AX-70, Olympus America Inc, Center Valley, PA). Once 95% spore formation was noted, spores were suspended from the agar surface by pouring 2.0 mL of sterile water and gently scraping the agar surface with a clean, sterile glass cell spreader. Spore preparations were then washed 5 times in sterile water and then stored in water suspension at 4°C.
Suspensions of Green Fluorescent Protein (GFP)-labeled bacillus anthracis
Bacillus anthracis Sterne pAFp8gfp were prepared by growing a uniform lawn of spores on a modified Schaeffer media [28]. After 5 d of growth at 32°C, plates were examined daily for spore formation by removing a small colony of growth and examining with phase microscopy. Once 95% spore formation was noted, spores were harvested from the agar surface by pouring 2.0 mL of sterile water and gently scraping the agar surface with a clean, sterile glass cell spreader. Spore suspensions were washed 7 times with water by centrifugation (1,500×g, 2 min) and stored at 4°C as a water suspension.
Spore cleaning
Water suspensions of B. cereus or B. anthracis (200 µL) were washed 3 times with water by centrifugation (10,000×g, 2 min) and suspended in 40% v/v ethanol to eliminate any potential presence of CaDPA from germinated spores in the stock suspension. In addition, the samples were heated for 25 min at 65°C to inactivate any vegetative cells or germinated spores in the sample. The sample was named stock suspension and used to prepare a 12-fold diluted working suspension in water.
Determination of spore concentration
B. cereus and B. anthracis working suspensions were quantified by serial dilution in phosphate buffered saline pH = 7.4 containing 0.4% v/v Tween 80 (PBST), plated onto LB agar using the drop plate method (5 drops of 10 µL per dilution) and cultured for about 16 h at 30°C. Colony forming units (CFU) were enumerated and used to calculate the spore concentration in the working suspension [27].
Release of CaDPA from spores
Working suspensions (100 µL) of B. cereus ((1.3±0.2)×109 spores mL–1) or B. anthracis ((1.4±0.3)×108 spores mL–1 or (3.5±0.8)×108)) were autoclaved (20 min, 121°C) followed by sonication for 30 min. The samples were centrifuged (10,000×g, 2 min) and the supernatant containing CaDPA was transferred to another microfuge tube and used in the measurements.
Curve Fitting and Statistical Analysis
All values are expressed as mean ± standard deviation (sd) of three completely independent replicates, except when indicated. Statistical data analysis was performed by one-way analysis of variance (ANOVA) and the level of statistical significance was taken to be p<0.05. Curve fitting and statistical calculations were carried out using the software Origin (version 8.5; OriginLab: Northampton, MA, USA, 2011).
Linear regression analysis was carried out by the method of least squares. Sigmoidal curves, with a lower boundary near the background response and an upper asymptote near the maximum response, were fitter using the 4-parameter logistic model (Eq. (3)) [29].
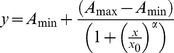 |
(3) |
Where, y is the response, Amax is the response at infinite analyte concentration, Amin is the response at zero analyte concentration, x is the analyte concentration, x0 is the inflection point on the curve (IC50), and α is a slope factor.
Results and Discussion
Complexation of Bn and EuIII
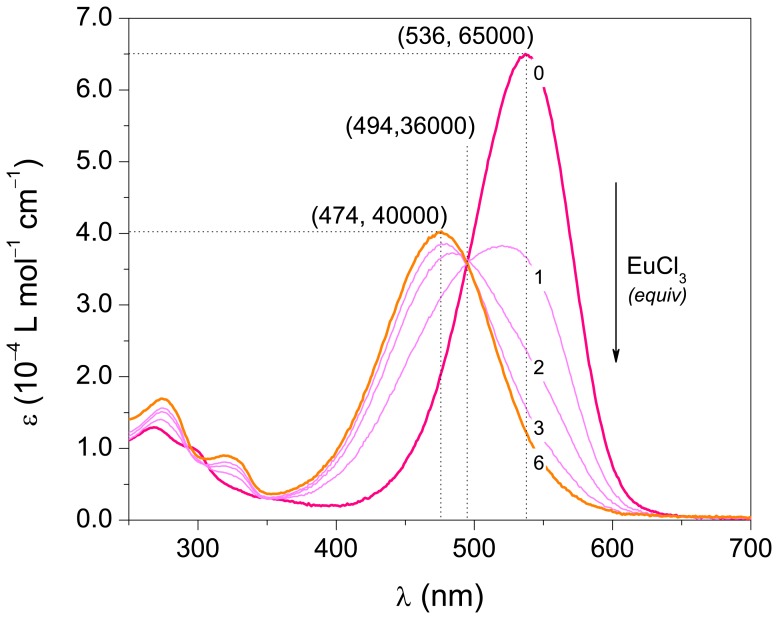
Most transition metals have been found to catalyze the decomposition of betanin (Bn) in aqueous solution. However, Bn has been described to form complexes with transition metals such as CuI, CuII and HgII in near-neutral aqueous media [30]. Interestingly, complexation of Bn and EuIII occurs spontaneously in MOPS buffer pH = 7.5, resulting in the bright orange and relatively stable [Eu(Bn)]+ complex (λ = 474 nm, h° = 53, ε474 nm = 4.0×104 L mol–1 cm–1, k obs = 2.7 10–4 min–1 in MOPS buffer pH = 7.5 at 25°C, Figure 1 and Figure S1).The EuIII–betanin stoichiometry was determined to be 1∶1 by using the molar-ratio method (Figure S2). Contrary to our expectations, the complex is not fluorescent in water probably due to water coordination to EuIII, which favors the non-radiative deactivation of the excited lanthanide [31]. The stability constant of the [Eu(Bn)]+ complex was determined to be (1.4±0.6)×105 L mol–1 (Table S1) using Eq. (1) and is similar to that reported for the complexation of CaII and DPA (log K = 5.4, K = 2.51×105 L mol–1) [32].
Figure 1. Absorptivity profile for the formation of [Eu(Bn)]+ by the addition of EuCl3 to a solution of Bn in MOPS buffer pH = 7.5.
The numbers in the curves indicate the concentration of EuCl3 in equivalents of Bn.
Despite our efforts, we were unable to obtain a single crystal of the [Eu(Bn)]+ complex suitable for crystallographic structure determination. However, insights into the structure of the complex were obtained by Raman spectroscopy associated with theoretical calculations. Figure S3 depicts the resonance Raman spectra of Bn and [Eu(Bn)]+ excited at 514.5 nm and 476.5 nm, respectively, as well as the theoretical Raman spectra of Bn (computational details and Cartesian coordinates are in File S1). The four intense Raman signals at 1610, 1518, 1506 and 1394 cm–1 in the spectrum of Bn can be assigned to ν(C = N)+ν(C = C)+φ8a, φ19a+ν(C = N), ν(C = C)pyr+ν(C = N), and ν(C = N), respectively. The observed resonance Raman enhancement profile is in accordance to HOMO-LUMO DFT calculations for the dianionic form of Bn (pK a1 = 3.4; pK a2 = 8.5, [33]) that show a charge transfer from the aromatic ring to the dihydropyridyl moiety [21], [34]. Raman signals of [Eu(Bn)]+ at 1616, 1529, and 1436 cm–1 are shifted to higher wavenumbers compared to that of Bn probably because the electronic delocalization of the π-system decreases upon complexation, i.e., bond orders increase. Furthermore, the Raman band of Bn at 1155 cm–1 (vibrational mode involving the tetrahydropyridyl N–H bending, δ(NH)) is missing in the Raman spectra of the [Eu(Bn)]+ complex. These results are in agreement with the blue shift (60 nm, 2440 cm–1) in the maximum absorption wavelength upon the [Eu(Bn)]+ complex formation and allow us to infer that EuIII complexation is likely to occur through the 2,6-dicarboxyl-1,2,3,4-tetrahydropyridine moiety.
Determination of CaDPA
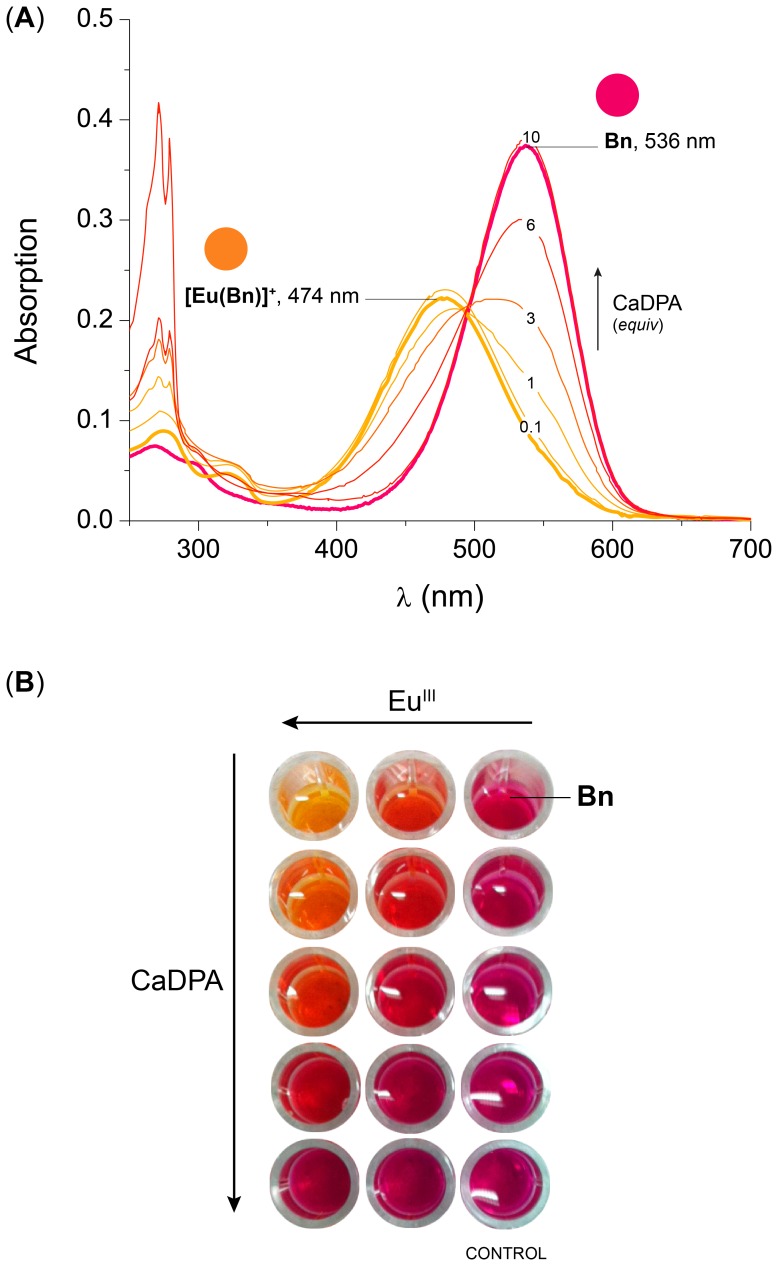
CaDPA competes with Bn for EuIII ions. The stability constants reported for complexes of type [Eu(DPA)n]3−2n (K1 = 6.92×108 (n = 1); K2 = 1.38×107 (n = 2); K3 = 3.23×105 (n = 3) [35] are higher than the stability constant determined for [Eu(Bn)]+ (i.e., K 1 = 1.4×105). Therefore, the reaction of CaDPA with the [Eu(Bn)]+ complex releases Bn due to the formation of [Eu(DPA)n]3−2n complexes, changing the color of the solution from orange to red-magenta (Figure 2).
Figure 2. Effect of the addition of CaDPA on an aqueous solution of [Eu(Bn)]+.
(A) Absorption profile for the formation of Bn by the addition of CaDPA to a solution of [Eu(Bn)]+; The numbers in the curves indicate the concentration of CaDPA in equivalents of Bn. (B) Picture of a microplate containing Bn and increasing amounts of EuCl3 and CaDPA, the background was removed for clarity. [Bn] = 5.8 µmol L–1, [EuCl3] = 34.8 µmol L–1 (6 equiv) in MOPS buffer pH = 7.5.
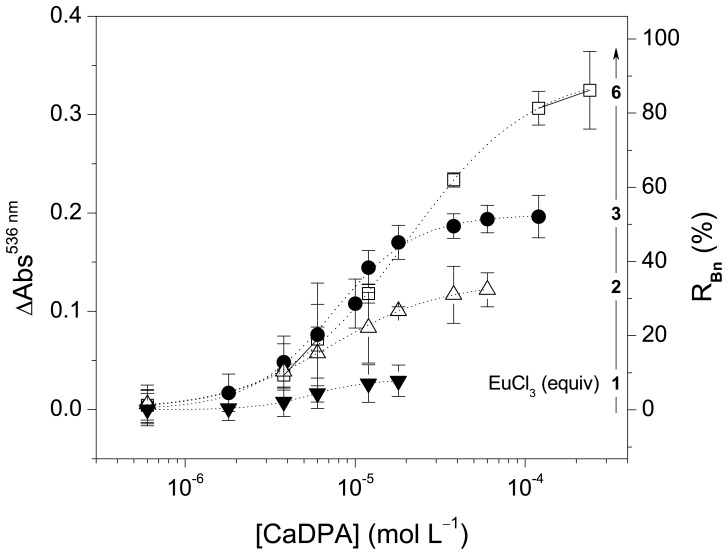
The effect of the amount of EuIII (1, 2, 3 or 6 equiv) on the quantification of CaDPA was investigated. All curves depicted in Figure 3 show the sigmoidal profile characteristic of ligand binding assays [29]. The limit of detection (LOD) in this experimental condition is around 2×10–6 mol L–1, independent of the concentration of EuIII. However, the recovery of Bn (RBn), i.e., the ratio between the initial absorbance at 536 nm before addition of EuIII and CaDPA and that after the addition of mole excess of CaDPA to the [Eu(Bn)]+ complex, increases with the amount of EuIII (Figure 3). This is probably related to the decrease in the concentration of unbound Bn in equilibrium caused by the increase in the concentration of EuIII. Furthermore, the variation in absorbance increases with the increase of the concentration of EuIII; consequently, the signal-to-noise ratio is improved using large mole excess of the lanthanide.
Figure 3. Effect of the concentration of EuIII on the quantification of CaDPA.
The variation of the absorbance at 536 nm and the percent recovery of Bn are plotted against the concentration of CaDPA (log scale). Lines indicate the fitting of data using Eq. (3) with parameter Amin set to zero; [Bn] = 5.8 µmol L–1 in MOPS buffer. LOD are: 6 equiv, (2.8±1.5)×10–6 mol L–1; 3 equiv, (2.2±1.1)×10–6 mol L–1; 2 equiv, (1.4±0.9)×10–6 mol L–1 and 1 equiv, (1.7±0.8)×10–6 mol L–1.
The linear section of the sigmoidal curve is crucial for quantitative measurements and for calibration [36]. The intersections of the slope at the point of inflection with the asymptote define the dynamic concentration range for which the assay delivers reliable quantitative data (Figure 4, solid line). The use of 3-fold stoichiometric excess of EuIII in relation to Bn results in a variation in the absorption at 536 nm of 0.2 and a dynamic concentration range between 2 and 25 µmol L–1 of CaDPA. As a proof-of-concept, this condition was defined as default for the quantification of CaDPA because the minimum amount of lanthanide necessary to provide reliable results is used. In this condition, the following calibration curve: [CaDPA] (mol L–1) = (9.4±0.5)×10–5 ΔAbs536 nm (Adj-R2 = 0.983, N = 5, Figure S4) was constructed for the determination of the concentration of CaDPA from the variation in the absorbance at 536 nm, if appropriate sample dilution is chosen. The LOD in this experimental condition is (2.2±1.1)×10–6 mol L–1, which is lower than the reported infectious dosage of spores (6×10–5 mol L–1 of CaDPA) [9], [37] and comparable to the LOD of more complex methods, such as surface-enhanced RAMAN spectroscopy on silver-coated silicon nanowire arrays (4×10–6 mol L–1) [37] and ratiometric fluorescent detection (0.2×10–6 mol L–1) [38].
Figure 4. CaDPA concentration (log scale) plotted against the variation in the absorbance at 536 nm (ΔAbs536 nm) and the normalized response (N Bn).
The diagonal straight-line shows when the plotted parameters are linearly correlated. Curved lines are the confidence bands at the 95% level. Error bars represent the sd of triplicates. [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv).
Finally, the selectivity of the method was tested by varying the concentration of the following ligands: CaDPA, acetic acid, benzoic acid, phthalic acids, picolinic acids, nicotinic acids, isonicotinic acid and inorganic phosphate (Figure S5). No false positive was produced for any aromatic acid as well as for acetic acid, indicating the high selectivity of the method for DPA. Phosphate was included in the study because it has been reported to produce false positives and false negatives in assays based on lanthanide complexes, impairing the analysis of complex samples [13], [14], [39], [40]. As most lanthanide complexes, [Eu(Bn)]+ is sensitive to the presence of phosphate [39].
Quantification of Bacterial Spores
Spores of B. anthracis and B. cereus were quantified using the plate count method (see Methods) and subjected to thermal treatment (autoclave) to induce the release of CaDPA. The concentration of CaDPA was determined using the colorimetric method and correlated to the concentration of spores (spores mL–1) (Figure 5). The uncertainty observed in the final result might be partially due to the plate count method because one single colony is not necessarily generated from a single spore [41]. However, the contribution of this to the overall uncertainty would be small. Other sources might come from sample preparation, such as extraction efficiency of CaDPA from spores.
Figure 5. Quantification of bacteria of the genus Bacillus by the amount of CaDPA released upon thermal treatment.
Vertical and horizontal error bars indicate uncertainties in spore counting and CaDPA quantification (sd, N = 3). The concentration of spores is in the log scale for clarity. [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv) in MOPS buffer pH = 7.5.
Control experiments indicate that CaDPA can only be detected in spore suspensions subjected to thermal treatment (Figure S6). Furthermore, after monitoring the germination, we assume that most CaDPA is released from the spores upon thermal treatment (data not shown). The LOD determined for both B. cereus and B. anthracis is (1.1±0.3)×106 spores mL–1. From the average slope of the linear section of the sigmoidal curves (Figure S7), we estimate that each spore has (1.6±0.5)×10–15 moles of CaDPA, i.e., (9.6±3.0)×108 molecules of CaDPA per spore. Although this result is in reasonable agreement with the values reported using some other methods, including SERS and fluorescence detection, e.g., Clostridium sporogenes (3.7×108 DPA molecules spore–1) [42] and Bacillus spp. spores (1.7×108 to 6.3×108 DPA molecules spore–1) [43], [44], [45], alternative methods requiring more sophisticate apparatus and data analysis may detect lower amounts of CaDPA in bacterial spores [13], [15], [46]. However, compared to the direct spectrophotometric detection of CaDPA, our method requires less sample preparation and is not susceptible to interferents absorbing in the middle UV region [47], [48].
The main limitation of the current methodology lies on the fact that the sensitivity depends on the molar absorptivity of Bn, implying that the detection limit of the present method cannot be easily enhanced. However, this approach introduces both the renewable natural pigment betanin as a biocompatible ligand for EuIII and the [Eu(Bn)]+ complex as a green, low cost and fast compound for the detection of CaDPA and bacterial spores, as well as opens the perspective of exploring other betalains as a platform for the development of sensors.
Conclusion
In this proof-of-concept study, we have shown that the orange complex formed between betanin, the main beetroot pigment, and EuIII is sensitive to the presence of CaDPA, but not to other structurally similar pyridinic, aromatic, and acid ligands. The [Eu(Bn)]+ complex can be applied for the qualitative (on/off) and quantitative detection of CaDPA with a LOD of (2.2±1.1)×10–6 mol L–1. A colorimetric assay using 3 equiv of EuIII in MOPS buffer was used to detect representative Bacillus species (B. anthracis or B.cereus spores) submitted to thermal treatment (autoclave). This low cost and ease of use approach indicates the potential use of betalains as sensors for biological applications.
Supporting Information
Absorption spectra of [Eu(Bn)]+ in MOPS buffer pH = 7.5 acquired over 5 d and decomposition kinetics monitored at 480 nm, N = 1.
(TIF)
Absorption of solutions of Bn and EuIII (536 nm) at a fixed [Bn] = 5.75×10–6 mol L–1.
(TIF)
Spectroscopic data on the [Eu(Bn)]+ complex. (A) Experimental Raman spectra (Ir), second derivative of Raman Intensities relative to wavenumber (d2Ir/dRS 2) and theoretical intensities determined the B3LYP/6-31+G(d)/SDM level and corrected by a factor of 0.98 (Ir DFT) and optimized structure of Bn; (B) Experimental Raman spectra (Ir), second derivative of Raman Intensities relative to wavenumber (d2Ir/dRS 2) of [Eu(Bn)]+ and non-optimized illustration of a possible structure. [Bn] = 1×10–4 mol L–1, [EuCl3] = 3.6×10–3 mol L–1.
(TIF)
Calibration curve for the determination of the [CaDPA] from the variation in absorbance at 536 nm. The y-axis is in the log scale to show the sigmoidal profile of the curve and the linear fitting of the data. Curved lines are the confidence bands at the 95% level. Error bars represent the sd of triplicates. [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv). [CaDPA] = (9.4±0.5)×10–5 ΔAbs536 nm (Adj-R2 = 0.983, N = 5).
(TIF)
Effect of the addition of carboxylic acids and phosphate to the [Eu(Bn)]+ complex monitored by the change in the absorption maxima of Bn. [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv), [analyte] = 69.6 µmol L–1 (12 equiv) in MOPS buffer pH = 7.5.
(TIF)
Control experiments for the determination of CaDPA in samples containing spores of B. cereus not submitted to thermal treatment. [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv) in MOPS buffer pH = 7.5, N = 1.
(TIF)
Quantification of bacteria of the genus Bacillus by the amount of CaDPA released upon thermal treatment for the determination of the amount of CaDPA per spore. The red dotted line is the linear data fitting and the green lines are the confidence bands at 95% confidence level. Vertical and horizontal error bars indicate uncertainties in spore counting and CaDPA quantification (sd, N = 3). [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv) in MOPS buffer pH = 7.5.
(TIF)
Stability constants determined using Eqs. (1) and (2) and the corresponding concentration of EuIII.
(DOCX)
Supplementary methods.
(DOCX)
Acknowledgments
E.L.B thanks CNPq for a productivity fellowship. Disclosure: All opinions expressed in this paper are the authors’ and do not necessarily reflect the policies and views of NIST or affiliated venues. Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Funding Statement
Funding was provided by the São Paulo Research Foundation (FAPESP, 07/00684-6 and 11/23036-5). LCPG was supported by a FAPESP fellowship (07/59407-1) and the Coordination for the Improvement of Higher Education Personnel (CAPES, PNPD 427-10/2009) fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vilas-Boas GT, Peruca APS, Arantes OMN (2007) Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can J Microbiol 53: 673–687. [DOI] [PubMed] [Google Scholar]
- 2. Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. (1999) Anthrax as a biological weapon: Medical and public health management. JAMA, J Am Med Assoc 281: 1735–1745. [DOI] [PubMed] [Google Scholar]
- 3. Inglesby TV, O’Toole T, Henderson DA, Bartlett JG, Ascher MS, et al. (2002) Anthrax as a biological weapon, 2002: Updated recommendations for management. JAMA, J Am Med Assoc 287: 2236–2252. [DOI] [PubMed] [Google Scholar]
- 4. Sanderson WT, Stoddard RR, Echt AS, Piacitelli CA, Kim D, et al. (2004) Bacillus anthracis contamination and inhalational anthrax in a mail processing and distribution center. J Appl Microbiol 96: 1048–1056. [DOI] [PubMed] [Google Scholar]
- 5. Dragon DC, Rennie RP (1995) The ecology of anthrax spores: tough but not invincible. Can Vet J 36: 295–301. [PMC free article] [PubMed] [Google Scholar]
- 6. Salter SJ (2011) You cannot B. cereus. Nat Rev Microbiol 9: 83–83. [DOI] [PubMed] [Google Scholar]
- 7. Gould GW (1977) Recent Advances in the Understanding of Resistance and Dormancy in Bacterial Spores. J Appl Microbiol 42: 297–309. [DOI] [PubMed] [Google Scholar]
- 8. Setlow B, Wahome PG, Setlow P (2008) Release of Small Molecules during Germination of Spores of Bacillus Species. J Bacteriol 190: 4759–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oh WK, Jeong YS, Song J, Jang J (2011) Fluorescent europium-modified polymer nanoparticles for rapid and sensitive anthrax sensors. Biosensors Bioeletron 29: 172–177. [DOI] [PubMed] [Google Scholar]
- 10. Setlow P (2003) Spore germination. Curr Opin Microbiol 6: 550–556. [DOI] [PubMed] [Google Scholar]
- 11. Rao SS, Mohan KVK, Atreya CD (2010) Detection technologies for Bacillus anthracis: Prospects and challenges. J Microbiol Methods 82: 1–10. [DOI] [PubMed] [Google Scholar]
- 12. Irenge L, Gala JL (2012) Rapid detection methods for Bacillus anthracis in environmental samples: a review. Appl Microbiol Biotechnol 93: 1411–1422. [DOI] [PubMed] [Google Scholar]
- 13. Cable ML, Kirby JP, Sorasaenee K, Gray HB, Ponce A (2007) Bacterial Spore Detection by [Tb3+(macrocycle)(dipicolinate)] Luminescence. J Am Chem Soc 129: 1474–1475. [DOI] [PubMed] [Google Scholar]
- 14. Cable ML, Kirby JP, Levine DJ, Manary MJ, Gray HB, et al. (2009) Detection of Bacterial Spores with Lanthanide-Macrocycle Binary Complexes. J Am Chem Soc 131: 9562–9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee I, Oh WK, Jang J (2013) Screen-printed fluorescent sensors for rapid and sensitive anthrax biomarker detection. J Hazard Mater 252–253: 186–191. [DOI] [PubMed] [Google Scholar]
- 16. Gonçalves LCP, Trassi MAD, Lopes NB, Dörr FA, dos Santos MT, et al. (2012) A comparative study of the purification of betanin. Food Chem 131: 231–238. [Google Scholar]
- 17. Bartoloni FH, Gonçalves LCP, Rodrigues ACB, Dörr FA, Pinto E, et al. (2013) Photophysics and hydrolytic stability of betalains in aqueous trifluoroethanol. Monats Chemie 144: 567–571. [Google Scholar]
- 18. Ravichandran K, Ahmed AR, Knorr D, Smetanska I (2012) The effect of different processing methods on phenolic acid content and antioxidant activity of red beet. Food Res Intern 48: 16–20. [Google Scholar]
- 19. Wootton-Beard PC, Ryan L (2011) A beetroot juice shot is a significant and convenient source of bioaccessible antioxidants. J Funct Foods 3: 329–334. [Google Scholar]
- 20. Gonçalves LCP, Di Genova BM, Dörr FA, Pinto E, Bastos EL (2013) Effect of dielectric microwave heating on the color and antiradical capacity of betanin. J Food Eng 118: 49–55. [Google Scholar]
- 21. Sandquist C, McHale JL (2011) Improved efficiency of betanin-based dye-sensitized solar cells. J Photochem Photobiol, A 221: 90–97. [Google Scholar]
- 22. Zhang D, Lanier SM, Downing JA, Avent JL, Lum J, et al. (2008) Betalain pigments for dye-sensitized solar cells. J Photochem Photobiol, A 195: 72–80. [Google Scholar]
- 23. Gonçalves LCP, Tonelli RR, Bagnaresi P, Mortara RA, Ferreira AG, et al. (2013) A Nature-Inspired Betalainic Probe for Live-Cell Imaging of Plasmodium-Infected Erythrocytes. PLoS One 8: e53874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoe JH, Jones AL (1944) Colorimetric determination of iron with disodium-1,2-dihydroxybenzene-3,5-disulfonate. Ind Eng Chem Anal Ed 16: 111–115. [Google Scholar]
- 25. Hirose K (2001) A Practical Guide for the Determination of Binding Constants. J Inclusion Phenom Macrocyclic Chem 39: 193–209. [Google Scholar]
- 26. Fernandez-Ramos MD, Cuadros-Rodriguez L, Arroyo-Guerrero E, Capitan-Vallvey LF (2011) An IUPAC-based approach to estimate the detection limit in co-extraction-based optical sensors for anions with sigmoidal response calibration curves. Anal Bioanal Chem 401: 2881–2889. [DOI] [PubMed] [Google Scholar]
- 27. Downey AS, Da Silva SM, Olson ND, Filliben JJ, Morrow JB (2012) Impact of Processing Method on Recovery of Bacteria from Wipes Used in Biological Surface Sampling. Appl Environ Microbiol 78: 5872–5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Faille C, Lebret V, Gavini F, Maingonnat JF (1997) Injury and lethality of heat treatment of Bacillus cereus spores suspended in buffer and in poultry meat. J Food Prot 60: 544–547. [DOI] [PubMed] [Google Scholar]
- 29. Findlay J, Dillard R (2007) Appropriate calibration curve fitting in ligand binding assays. AAPS J 9: E260–E267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Attoe EL, Von Elbe JH (1984) Oxygen involvement in betanin degradation. Oxygen uptake and influence of metal ions. Z Lebensm-Unters Forsch 179: 232–236. [Google Scholar]
- 31. Kropp JL, Windsor MW (1967) Luminescence and energy transfer in solutions of rare earth complexes. II. Studies of the solvation shell in europium(III) and terbium(III) as a function of acetate concentration. J Phys Chem 71: 477–482. [Google Scholar]
- 32. Lewis JC (1967) Determination of dipicolinic acid in bacterial spores by ultraviolet spectrometry of the calcium chelate. Anal Biochem 19: 327–337. [DOI] [PubMed] [Google Scholar]
- 33. Nilsson T (1970) Studies into the pigments in beetroot (Beta vulgaris L. ssp.vulgaris var. rubra L.). Lantbrukshoeg Ann 36: 179–219. [Google Scholar]
- 34. Qin CY, Clark AE (2007) DFT characterization of the optical and redox properties of natural pigments relevant to dye-sensitized solar cells. Chem Phys Lett 438: 26–30. [Google Scholar]
- 35. Grenthe I (1961) Stability Relationships Among the Rare Earth Dipicolinates. J Am Chem Soc 83: 360–364. [Google Scholar]
- 36. Saleh L, Plieth C (2010) A coelenterazine-based luminescence assay to quantify high-molecular-weight superoxide anion scavenger activities. Nat Protocols 5: 1635–1641. [DOI] [PubMed] [Google Scholar]
- 37. Zhang B, Wang H, Lu L, Ai K, Zhang G, et al. (2008) Large-Area Silver-Coated Silicon Nanowire Arrays for Molecular Sensing Using Surface-Enhanced Raman Spectroscopy. Adv Funct Mater 18: 2348–2355. [Google Scholar]
- 38. Yilmaz MD, Hsu SH, Reinhoudt DN, Velders AH, Huskens J (2010) Ratiometric Fluorescent Detection of an Anthrax Biomarker at Molecular Printboards. Angew Chemie Intern Ed 49: 5938–5941. [DOI] [PubMed] [Google Scholar]
- 39. Ammann AB, Kölle L, Brandl H (2011) Detection of Bacterial Endospores in Soil by Terbium Fluorescence. Intern J Microbiol 2011: ID435281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pellegrino PM, Fell NF, Rosen DL, Gillespie JB (1998) Bacterial endospore detection using terbium dipicolinate photoluminescence in the presence of chemical and biological materials. Anal Chem 70: 1755–1760. [DOI] [PubMed] [Google Scholar]
- 41. Rosen DL, Sharpless C, McGown LB (1997) Bacterial Spore Detection and Determination by Use of Terbium Dipicolinate Photoluminescence. Anal Chem 69: 1082–1085. [Google Scholar]
- 42. Yang WW, Ponce A (2009) Rapid endospore viability assay of Clostridium sporogenes spores. Intern J Microbiol 133: 213–216. [DOI] [PubMed] [Google Scholar]
- 43. Shafaat HS, Ponce A (2006) Applications of a Rapid Endospore Viability Assay for Monitoring UV Inactivation and Characterizing Arctic Ice Cores. Appl Environ Microbiol 72: 6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng HW, Chen YY, Lin XX, Huan SY, Wu HL, et al. (2011) Surface-enhanced Raman spectroscopic detection of Bacillus subtilis spores using gold nanoparticle based substrates. Anal Chim Acta 707: 155–163. [DOI] [PubMed] [Google Scholar]
- 45. Zhang X, Young MA, Lyandres O, Van Duyne RP (2005) Rapid Detection of an Anthrax Biomarker by Surface-Enhanced Raman Spectroscopy. J Am Chem Soc 127: 4484–4489. [DOI] [PubMed] [Google Scholar]
- 46. Hindle AA, Hall EAH (1999) Dipicolinic acid (DPA) assay revisited and appraised for spore detection. Analyst 124: 1599–1604. [DOI] [PubMed] [Google Scholar]
- 47. Perry JJ, Foster JW (1955) Studies on the biosynthesis of dipicolinic acid in spores of Bacillus cereus var. mycoides. J Bacteriol 69: 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murty GGK, Halvorson HO (1957) Effect of duration of heating, L-alanine and spore concentration on the oxidation of glucose by spores of Bacillus cereus var. terminalis. J Bacteriol 73: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Absorption spectra of [Eu(Bn)]+ in MOPS buffer pH = 7.5 acquired over 5 d and decomposition kinetics monitored at 480 nm, N = 1.
(TIF)
Absorption of solutions of Bn and EuIII (536 nm) at a fixed [Bn] = 5.75×10–6 mol L–1.
(TIF)
Spectroscopic data on the [Eu(Bn)]+ complex. (A) Experimental Raman spectra (Ir), second derivative of Raman Intensities relative to wavenumber (d2Ir/dRS 2) and theoretical intensities determined the B3LYP/6-31+G(d)/SDM level and corrected by a factor of 0.98 (Ir DFT) and optimized structure of Bn; (B) Experimental Raman spectra (Ir), second derivative of Raman Intensities relative to wavenumber (d2Ir/dRS 2) of [Eu(Bn)]+ and non-optimized illustration of a possible structure. [Bn] = 1×10–4 mol L–1, [EuCl3] = 3.6×10–3 mol L–1.
(TIF)
Calibration curve for the determination of the [CaDPA] from the variation in absorbance at 536 nm. The y-axis is in the log scale to show the sigmoidal profile of the curve and the linear fitting of the data. Curved lines are the confidence bands at the 95% level. Error bars represent the sd of triplicates. [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv). [CaDPA] = (9.4±0.5)×10–5 ΔAbs536 nm (Adj-R2 = 0.983, N = 5).
(TIF)
Effect of the addition of carboxylic acids and phosphate to the [Eu(Bn)]+ complex monitored by the change in the absorption maxima of Bn. [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv), [analyte] = 69.6 µmol L–1 (12 equiv) in MOPS buffer pH = 7.5.
(TIF)
Control experiments for the determination of CaDPA in samples containing spores of B. cereus not submitted to thermal treatment. [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv) in MOPS buffer pH = 7.5, N = 1.
(TIF)
Quantification of bacteria of the genus Bacillus by the amount of CaDPA released upon thermal treatment for the determination of the amount of CaDPA per spore. The red dotted line is the linear data fitting and the green lines are the confidence bands at 95% confidence level. Vertical and horizontal error bars indicate uncertainties in spore counting and CaDPA quantification (sd, N = 3). [Bn] = 5.8 µmol L–1, [EuCl3] = 17.4 µmol L–1 (3 equiv) in MOPS buffer pH = 7.5.
(TIF)
Stability constants determined using Eqs. (1) and (2) and the corresponding concentration of EuIII.
(DOCX)
Supplementary methods.
(DOCX)