Abstract
The SGT1 protein is essential for R protein-mediated and PAMPs-triggered resistance in many plant species. Here we reported the isolation and characterization of the Hv-SGT1 gene from Haynaldia villosa (2n = 14, VV). Analysis of the subcellular location of Hv-SGT1 by transient expression of a fusion to GFP indicated its presence in the cytoplasm and nucleus. Levels of Hv-SGT1 transcripts were increased by inoculation with either the biotrophic pathogen Blumeria graminis DC. f. Sp. tritici (Bgt) or the hemi-biotrophic pathogen Fusarium graminearum (Fg). Levels of Hv-SGT1 showed substantial increase following treatment with H2O2 and methyl jasmonate (MeJA), only slightly induced following exposure to ethephon or abscisic acid, but not changed following exposure to salicylic acid. The demonstration that silencing of Hv-SGT1 substantially reduced resistance to Bgt indicated that Hv-SGT1 was an essential component of disease resistance in H . villosa . The over-expression of Hv-SGT1 in Yangmai 158 enhanced resistance to powdery mildew, and this correlated with increased levels of whole-cell reactive oxygen intermediates at the sites of penetration by the pathogens. Compared with wild-type plants, the expression levels of genes related to the H2O2 and JA signaling pathways were lower in the Hv-SGT1 silenced plants and higher in the Hv-SGT1 over-expressing plants. Therefore, the involvement of Hv-SGT1 in H2O2 production correlates with the hypersensitive response and jasmonic acid signaling. Our novel demonstration that wheat with over-expressed Hv-SGT1 showed enhanced resistance to both powdery mildew and FHB suggests that it could served as a transgenic genetic resource in wheat breeding for multiple disease resistance.
Introduction
Plants activate various regulated defense mechanisms in response to a broad range of pathogen invasions. Pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI) and effector-triggered immunity (ETI) are the two main classes of plant innate immune responses [1,2]. Several recent studies have indicated that plant SGT1 (suppressor of the G2 allele of skp1) was a critical signaling component required for both PTI and ETI mediated host cell death in several plant species against various plant pathogens, including fungi, bacteria and viruses [3–8].
The SGT1 protein was first identified as a suppressor of skp1–4, which was involved in kinetochore assembly in yeast [9]. Hereafter, conserved SGT1 homologues were found in many eukaryotes, and were shown to participate in various aspects of plant biology, including plant defense responses and development. All SGT1 proteins had three distinct regions: a tetratricopeptide repeat (TPR) domain, a CS (present in CHP and SGT1 proteins) domain, and an SGS (SGT1-specific sequence) domain. The CS domain interacted with different protein complexes to affect a range of biological processes [10].
In plants, SGT1 positively regulates many plant disease-resistance (R)-gene-mediated race-specific disease resistances [5,11]. Mutation or silencing of SGT1 will compromised disease resistance mediated by plant nucleotide-binding domain and leucine-rich repeat-containing (NLR) type R genes, such as MLA, N, Bs2, Bs4, Rx, RPS4, Prf, Mi, I2, R3a, and Lr21 or by non-NLR-type sensors such as Cf 4,9, or RPW8 [12]. Over-expression of SGT1 will sometimes enhance plant disease resistance. Over-expression of NbSGT1 in Nicotiana benthamiana accelerated the development of hypersensitive response (HR) during R-gene-mediated disease resistance [8]. Over-expression of OsSGT1 in rice significantly increased basal resistance to a virulent bacterial blight Xanthomonas oryzae pv. oryzae PXO99 and four virulent blast fungal Magnaporthe oryzae races [13]. Expression of SGT1 was tightly related to R-gene expression, the HR, and the activation of some R proteins [14,15]. Additionally, SGT1 is also required in some cases of non-host plant resistance [16].
During disease resistance, SGT1 appears to interact with molecular chaperones (RAR1, HSP70 or HSP90) to mediate target protein binding to initiate a specific signaling cascade that confers resistance. For example, the SGT1-RAR1-HSP90 complex was essential for Lr21-triggered wheat disease resistance to leaf rust [17]. The SGT1 protein interacted physically with SKP1, a part of the SCF E3 ubiquitin ligase, suggesting that its functions in the proteasome-mediated protein degradation pathway [16]. Both the NbSGT1 and NbSKP1 genes played important roles in the N-mediated resistance response to TMV [6].
Fungal pathogens can be classified as biotrophs, necrotrophs and hemi-biotrophs. Two globally important diseases that affect common wheat, powdery mildew and Fusarium head blight (FHB) (or scab), are caused by the biotrophic Blumeria graminis DC. f. sp. tritici (Bgt) and the hemi-biotrophic Fusarium graminearum (Fg) respectively. However, the mechanisms responsible for the resistance to biotrophs and necrotrophic pathogens are quite different. Active cell death (also known as the HR) in host-pathogen interaction sites is an effective resistance approach adopted by plant hosts to restrict the development of biotrophs. However, virulent necrotrophic pathogens must be able to damage or kill host tissues (necrosis) to support their successful infection and survival. Accordingly, it was proposed that it is difficult to co-regulate resistances to both the biotrophic and necrotrophic pathogens. Recent studies showed that SGT1 promotes both the HR and resistance to biotrophic pathogens, while suppressing resistance to necrotrophic pathogens. Silencing of SGT1 expression reduced necrosis, which in turn compromised resistance to the barley biothoph Blumeria graminis f. sp. Hordei (Bgh) [18] and the wheat biotroph Pucciniastriiformis f. sp. tritici (Pst) [17], while enhancing the resistance of tobacco to the necrotrophic pathogen Botrytis cinerea [19]. The resistance mechanisms used by hemi-biotrophs, were even more complicated. Cuzick et al. [20] reported that a lack of SGT1b reduced host-cell death and promoted the pathogenesis of the hemi-biotrophic fungal pathogen Fusarium culmorum . However, the potential role of SGT1 in powdery mildew resistance and FHB resistance in wheat was not known yet.
Haynaldia villosa , a diploid wild member of the genus Triticeae of the Poaceae, displays substantial broad-spectrum resistance to Bgt, and the disease resistance gene Pm21 that is responsible for this resistance has been transferred to common wheat by chromosome engineering [21]. Microarray analysis conducted in our laboratory on H . villosa plants that were either inoculated with Bgt or treated as controls were used to identify key genes in the response network [22] that mediated resistance to Bgt resistance. This included a key member of the Pm21 locus, the Stpk-V gene [23]. Besides Stpk-V, homologues of SGT1, RAR1, and HSP90 were also found to be induced in Bgt inoculated H . villosa . Here, we focused on the function of SGT1 in mediating the resistance of wheat towards fungal pathogens. A full-length H . villosa SGT1 gene was cloned and characterized in detail. Over-expression of Hv-SGT1 in common wheat and silencing of Hv-SGT1 in H . villosa revealed its pivotal roles in resistance to both the biotrophic pathogen Bgt and hemi-biotrophic pathogen Fg. It was shown that the different responses to attacks by these different pathogens are associated with activation or depression of the H2O2- and JA-mediated defense pathways.
Results
Identification of the Hv-SGT1
The Genechip microarray data collected from H . villosa (Cao et al. 2006a) plants inoculated with Bgt enabled us to design degenerate primer pairs to clone a full-length cDNA of Hv-SGT1 from H . villosa using RT-PCR and RACE (rapid-amplification of cDNA ends). The 1,429-bp cDNA fragment, which included the complete 1,122-bp open reading frame (ORF) that encoded a protein of 376 amino acid residues, was assigned an Accession No. JX885369 in GenBank.
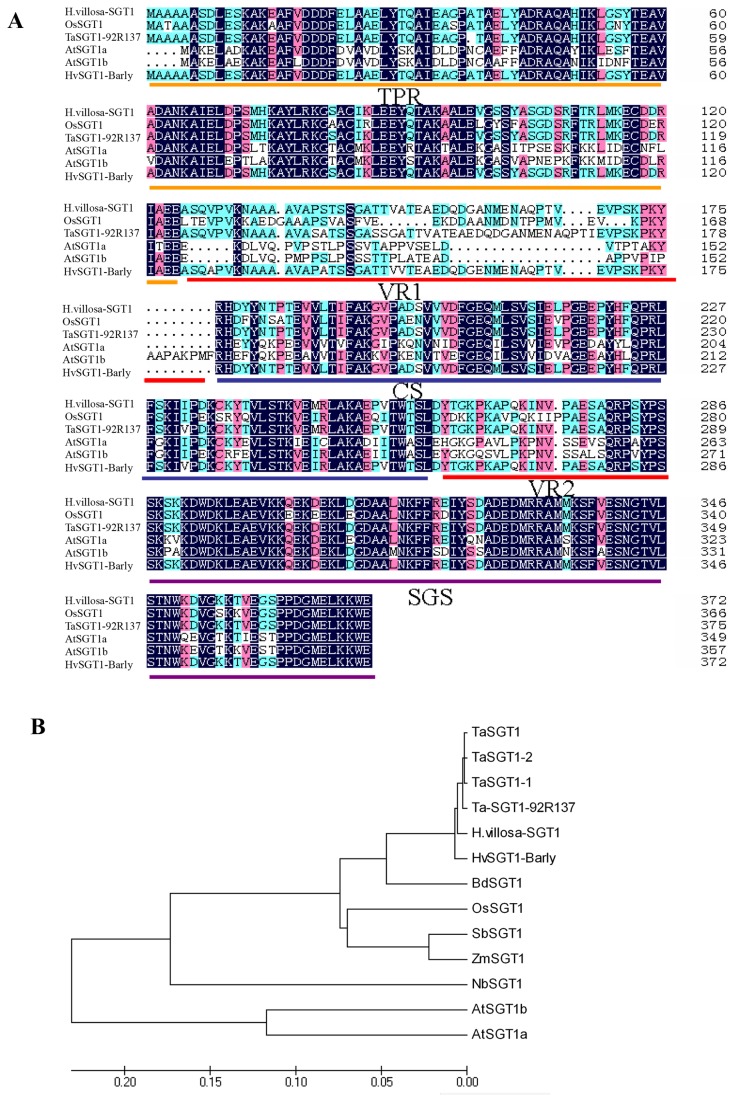
The predicted Hv-SGT1 protein contained five domains: a tetratricopeptide repeat domain (TPR), a P23 region with two variable domains (VR1 and VR2), a CS motif (present in the metazoan CHORD and SGT1 proteins), and an SGS (SGT1-specific) motif. Sequence alignment of SGT1 proteins from different species showed that Hv-SGT1 had the conserved domains present in other SGT1 proteins (Figure 1A). Phylogenetic analysis showed that Hv-SGT1 displayed different degree of similarity to other plant SGT1s, with the highest homology (98%) to barley SGT1. The SGT1 cloned from the common wheat line 92R137 (designated as Ta-SGT1-92R137) shared 95% s amino acid similarity with Hv-SGT1 (Figure 1B).
Figure 1. Analysis of primary structure and conserved domains of SGT1 proteins.
(A) Sequence alignment analysis and the predicted conserved domains of SGT1 proteins. GenBank accession numbers: Rice (AAF18438), Wheat (EF546432.1), Arabidopsis (AF439975, AF439976), Barley (AF439974). The black (100%), pink (80%), and blue (60%) boxes represent levels of amino acid identity or similarity. The conserved domains were underlined. (B) The phylogenetic analysis of the amino acid sequence of SGT1 genes from different species. GenBank accession numbers: OsSGT1 (AAF18438), TaSGT1 (ABQ23992.1), TaSGT1-1 (ABO18602.1), TaSGT1-2 (ABO18603.1), HHvSGT1-Barley (AF439974), BdSGT1 (XP_003569394.1), SbSGT1 (EES01101.1), Zm-SGT1 (ACG34278.1), NbSGT1 (AAW82048.1), AtSGT1a (AF439975) and AtSGT1b (AF439976). The tree was generated by ClustalX1.83 analysis with the corrected full-length Hv-SGT1 protein sequences using Neighbor-Joining method (MEGA4.0 software). The bar beneath the dendrogram represents a distance of 0.05 change per amino acid.
The chromosomal location of Hv-SGT1 was determined following PCR-based amplification using DNA from the wheat- H . villosa addition lines as template. A 402-bp product was found in H . villosa , a T . durum – H . villosa amphiploid, and a wheat- H . villosa addition line DA3V, but not in Chinese spring and the remaining addition lines tested. This indicated that Hv-SGT1 was located on chromosome 3V (Figure 2).
Figure 2. Chromosomal location of Hv-SGT1 determined using the wheat–H. villosa addition lines as the templates.
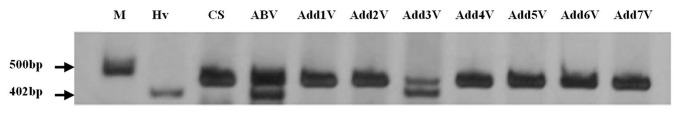
The arrow shows the specific amplicon from H . villosa .. M: DNA marker DL2000. HV:H. villosa, CS: wheat cv. Chinese spring, ABV: T . durum - H . villosa amphiploid, 2n=42, genome AABBVV, Add1V to Add7V: T. aestivum- H . villosa addition line, each contains one pair of chromosomes of H . villosa from 1V to 7V in the common wheat background, 2n=44, genome AABBDD add 1V1V~7V7V.
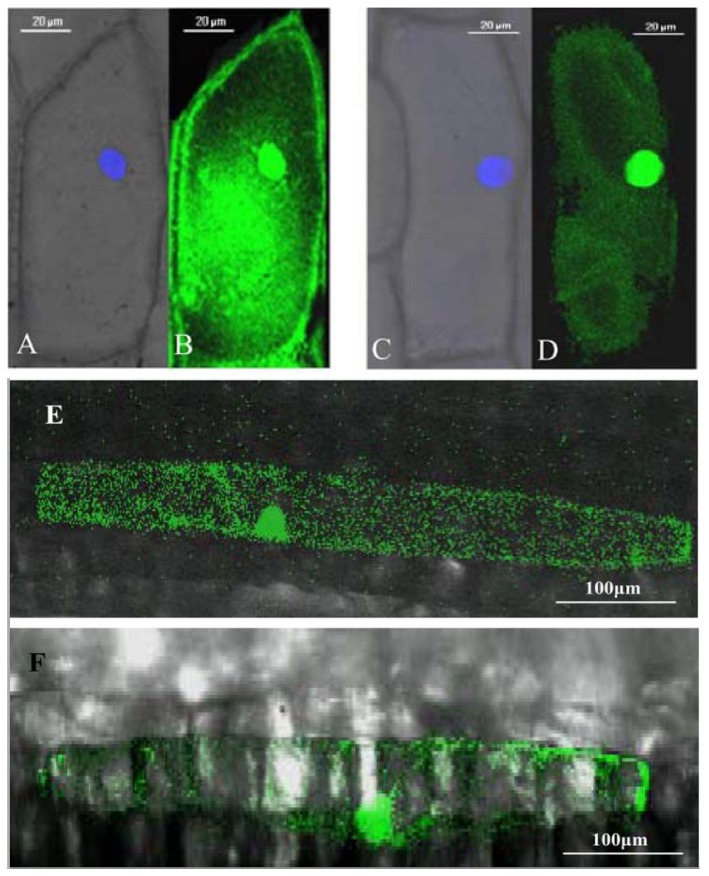
A construct that placed an in-frame fusion of the Hv-SGT1 coding sequence with the coding region for green fluorescent protein (Hv-SGT1::GFP) under the control of the cauliflower mosaic virus (CaMV) 35S promoter was transiently expressed in living onion epidermal cells. As shown in Figure 3, confocal microscopic examination revealed that cells that transiently expressing unconjugated GFP (control) exhibited a diffuse distribution of green fluorescence throughout the cell (Figure 3A-B). By contrast, the signal generated by cells that transiently expressing Hv-SGT1::GFP was confined to the cytoplasm and nucleus (DAPI stained) of onion epidermal cells (Figure 3C-D), suggesting that Hv-SGT1 is restricted to the cytoplasmic and nuclear compartments. Similar differences in the subcellular locations of the two constructs were evident following transient expression of GFP and the Hv-SGT1::GFP fusion in leaf epidermal cells of H . villosa (Figure 3E and 3F).
Figure 3. Subcellular localization of Hv-SGT1-GFP fusion protein by transient expression via biolistic bombardment.
(A–B) Detected GFP under blue emitting light (nucleus stainning with DAPI) and visible light (merged), and green emitting light of the onion epidermis cell and (E) in the merged figure of the H . villosa epidermis cell, indicating that GFP was sub-celluarly located to the whole cell. (C–D) Detected Hv-SGT1-GFP under blue emitting light (nucleus stainning with DAPI) and visible light (merged), and green emitting light of the onion epidermis cell and (F) in the merged figure of the H . villosa epidermis cell, indicating that the Hv-SGT1 was subcelluarly located to the nuclei and cytoplasm.
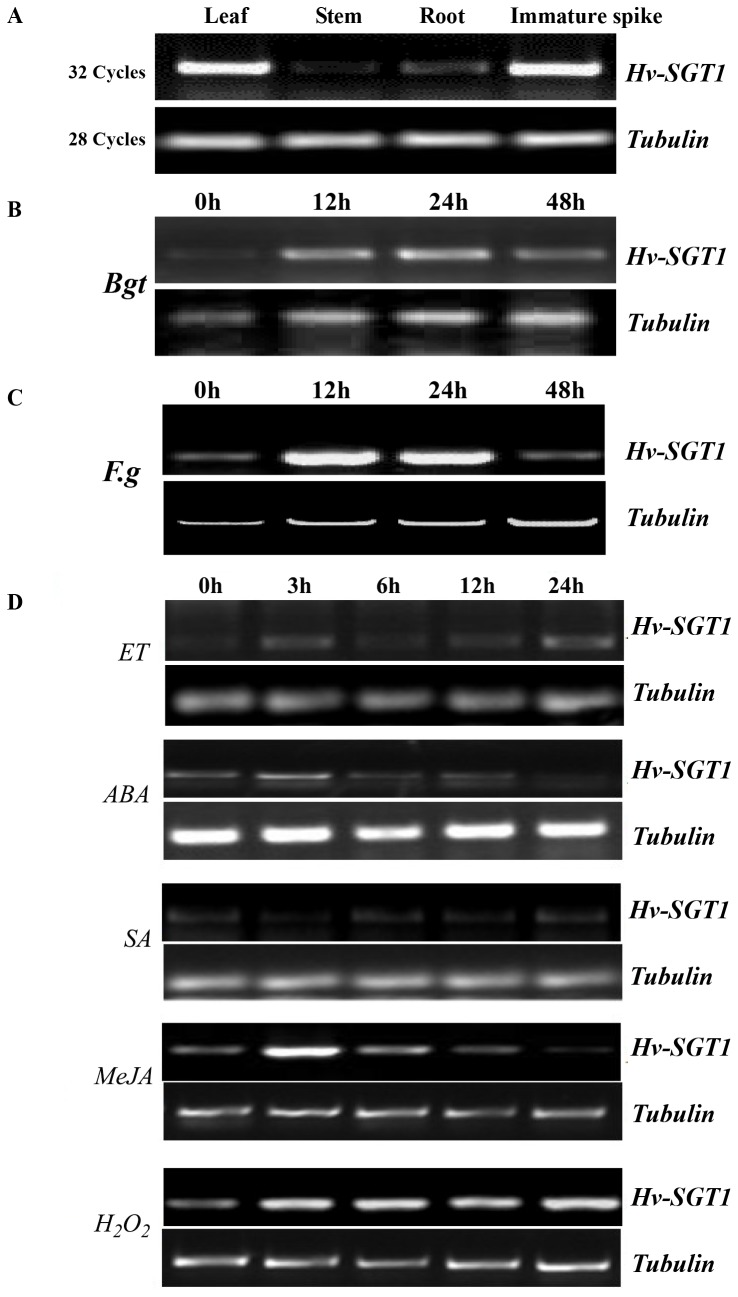
Expression profiling of Hv-SGT1 using different tissues of H . villosa showed that the highest expression was observed in leaves and immature spikes, with lower levels of expression observed in stems and roots (Figure 4A). Expression of the Hv-SGT1 gene was rapidly induced during infection of the leaves of seedlings of Bgt-resistant H . villosa , with levels of transcripts peaking 12 h after infection (Figure 4B). Similar inducible expression patterns were also observed in the immature spike of H . villosa after infection with Fg (Figure 4C). Treatment of the leaves of H . villosa seedlings with different phytohormones revealed that Hv-SGT1 transcripts were strongly induced by H2O2 and methyl jasmonate (MeJA), slightly induced by ethepon (ET) and abscisic acid (ABA), but not by salicylic acid (SA) (Figure 4D). The inducible expression of Hv-SGT1 following pathogen infection and treatment with phytohormones suggested its involvement in disease resistance reactions and defense pathways.
Figure 4. Responses of SGT1 to treatments of biotic and abiotic stresses, as well as phytohormone applications.
Expression patterns of SGT1 (A) in different organs of H . villosa , (B) in H . villosa leaves after Bgt inoculation, (C) in H . villosa immature spikes after Fg inoculation, and (D) in H . villosa leaves fter treatments with ET, ABA, SA, MeJA, and H2O2.
Knockdown of Hv-SGT1 by barley stripe mosaic virus–induced gene silencing in H. villosa enhances susceptibility of the host to Bgt
Barley stripe mosaic virus induced gene silencing (BSMV-VIGS) has been used to characterize gene function in wheat and H . villosa [24]. To confirm the role of Hv-SGT1 in the defense response of H . villosa to Bgt infection, we performed VIGS of Hv-SGT1 in H . villosa .
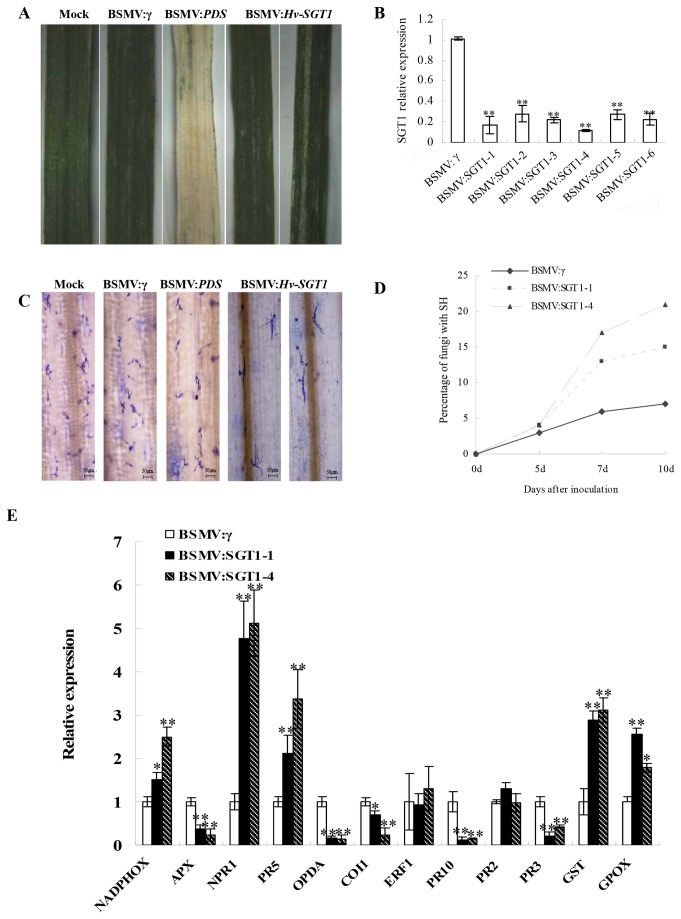
First, checking the efficiency of the VIGS system revealed that mild chlorotic mosaic symptoms were observed on the fourth leaves inoculated with BSMV at 10 days after inoculation. Photobleaching was observed on leaves infected with BSMV:PDS at 15 days, whereas no obvious changes were observed for mock leaves inoculated with 1×GKP buffer (Figure 5A). The level of expression of Hv-SGT1 was checked by qRT-PCR and found to decrease dramatically in H . villosa plants inoculated with BSMV:Hv-SGT1 compared with the Bgt inoculation plants preciously infected with BSMV: γ (Figure 5B).
Figure 5. Functional analysis of the Hv-SGT1 gene by BSMV-induced gene silencing.
(A) Following infection of H . villosa with the virus, chlorotic mosaic symptoms were observed on the 4th leaves 10 days after inoculation (dpi) with BSMV:γ or BSMV:Hv-SGT1. Photobleaching was observed on leaves infected with BSMV:PDS 15 dpi, whereas no detectable phenotype was observed in the mock-treated plants inoculated with 1×GKP buffer. Representative photographs were taken 15 days after virus inoculation. (B) The Hv-SGT1 gene was efficiently silenced in the BSMV: Hv-SGT1 inoculated leaves compared to the control inoculated with BSMV:γ, as indicated using qRT-PCR analysis. (C) More infected Bgt could developed into secondary hyphae in the BSMV: Hv-SGT1 inoculated leaves compared to the BSMV:γ infected leaves 10 days after inoculation with Bgt. (D) The rate of formation of secondary hyphae (SH) was higher in the Hv-SGT1 silenced leaves inoculated with BSMV:Hv-SGT1 compared with that in BSMV:γ-infected leaves at 5, 7, and 10 days after inoculation with Bgt. (E) Levels of NADPHOX, APX, NPR1, PR5, OPDA, COI1, ERF1, PR10, PR2, PR3, GSTand GPOXin Hv-SGT1 silenced leaves. * p < 0.05, ** p < 0.01 compared to the BSMV:γ-infected leaves.
The VIGS data indicated that 10 days after inoculation with Bgt, only the primary germ tubes (pp) and appressorium penetration peg (app) could be detected for most geminated spores (Figure 5C), and fewer than 5% of all spores could produce secondary hyphae (SH) in H . villosa plants that either received mock inoculation or inoculation with BSMV:γ. However, for those BSMV:Hv-SGT1 treated leaves, 15–20% of the spores could produce SH and even conidial chains (Figure 5D). The evidence that the resistance of H . villosa to Bgt was compromised when Hv-SGT1 was silenced underscores the notion that Hv-SGT1 is essential for the resistance of H . villosa to powdery mildew.
Given that Hv-SGT1 expression was phytohormone-dependent and related to disease resistance, we next examined the expression of a set of genes associated with the H2O2, SA, and JA pathways in the Hv-SGT1-silenced plants. Whereas levels of the transcript that encoded NADPH oxidase (NADPHOX, a H2O2-producing gene product) were elevated in the Hv-SGT1-silenced plants, levels of the transcript that encoded APX (a H2O2-scavenging gene product) were lower in Hv-SGT1-silenced plants than in control plants, while the transcript levels of both GST and GPOX (a ROS-scavenging gene product) were higher, revealing close link between SGT1 and the activity of antioxidative protection system. Transcript levels of NPR1 and PR5, which mediate the SA-related defense pathway [25], were significantly higher in Hv-SGT1-silenced plants than those in the control. However, the association of the transcription level of PR2 (marker gene of the SA pathway) with SGT1 was not observed. The transcript levels of PR3 (marker gene of the ET pathway) were significantly lower in Hv-SGT1-silenced plants than that in the control. Investigation of transcripts rrelated to the JA-mediated defense signaling (OPDA, COI1, ERF1, and PR10) [26,27], revealed a substantial reduction in levels of OPDA, COI1, and PR10 transcripts in Hv-SGT1-silenced plants, but no change in expression of ERF1 (Figure 5E). These results implied that Hv-SGT was intimately related to phytohormone-dependent defense pathways.
Over-expression of Hv-SGT1 enhances the resistance of common wheat to the biotrophic pathogen Bgt
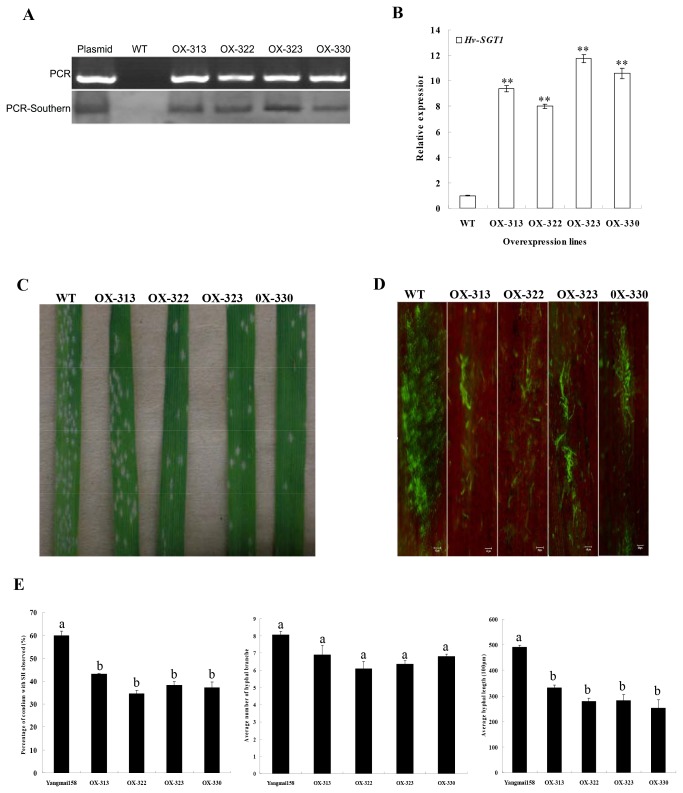
Although it has been well documented that SGT1 regulated defense responses triggered by various pathogens, less research had investigated its effectiveness to both biothophic and hemi-biothophic pathogens. Transgenic Yangmai 158 plants that over-express Hv-SGT1 under the regulation of the CAMV35S promoter were obtained by using the particle bombardment-mediated transformation approach. Overall, 52 independent T0-generation transgenic plants were identified after analysis of 432 regenerants using PCR amplification. Four of these lines (OX-313, OX-322, OX-323, OX-330) were characterized by further Southern blot of the PCR products (Figure 6A). qRT-PCR of the identified four lines indicated that expression of Hv-SGT1 in the transgenic plants was significantly increased up to 7.5-fold compared with that in the non-transformed Yangmai 158 (Figure 6B). The progenitors derived from these four positive transgenic plants were analyzed to assess their resistances to powdery mildew.
Figure 6. Characterization of the transgenic wheat of Hv-SGT1.
(A) PCR and PCR-Southern blot of four transgenic lines that over-expressing Hv-SGT1 (OX), and the non-transformed control Yangmai 158. The plasmid pBI220.6-Hv-SGT1 and Yangmai 158 were used as the positive and negative controls, respectively. (B) qRT-PCR of the expression of Hv-SGT1 in the four transgenic lines and Yangmai 158. ** p < 0.01 compared with the control. (C) Reduced disease symptoms in transgenic plants. Seedling resistance of transgenic line or wild-type plants was assessed following in vitro infection with the native pathogen population (Sumai 3). (D) Microscopic observation of Bgt hyphae spreading after DioC6 staining of the transgenic plants and Yangmai 158. (E) Quantitative comparisons of the percentage of infection sites with secondary hyphae (SH), the average number of hyphal branches and average hyphal length emerging on the leaves of infection sites. Means (± SE) were calculated using the measurements from five seedlings, and at least 30 infection sites for each seedling. Significance was determined according to paired sample t-test method (b indicates P < 0.05).
Compared with the wild type (WT) control Yangmai 158, transgenic T2 plants that over-express Hv-SGT1 (a total of 151 individuals were derived from the four T0 plants mentioned above) showed enhanced resistance both in the seedling and adult stages (Table 1, Figure 6C), and the development of Bgt was observed under the microscope. The area of Bgt spreading at the infection sites was much smaller in the transgenic plants than that in Yangmai 158 (Figure 6D). It was proposed that the increased resistance of the transgenic plants was related to the effective suppression of the hyphal development after successful penetration. The percentage of infection sites with secondary hyphae (SH), the average number of hyphal branches and average hyphal length at the infection site were investigated by histological observations at 48h after Bgt inoculation. A significantly (P < 0.05) lower percentage of infection sites producing SH (37.1%-43.1%) and shorter hyphal length (25,700-34,600 µm) at infection sites were observed in transgenic plants compared with corresponding values of 59.9% and 48,900 µm in Yangmai158. However, there was no significant difference in number of hyphal branches between control and transgenic plants (Figure 6E).
Table 1. Bgt responses of Hv-SGT1 transgenic plants and Yangmai158.
| Lines | No. of T2 positive plants | Disease Index of T2 generation |
Significance of ANOVA* (p<0.05) | Response a | |
|---|---|---|---|---|---|
| Seedling | Adult | ||||
| OX-313 | 36 | 52.47+18.52 | 47.84+11.52 | B | MR |
| OX-322 | 38 | 61.70+24.33 | 54.09+16.78 | B | MS |
| OX-323 | 47 | 54.61+21.71 | 47.52+14.60 | B | MR |
| OX-330 | 30 | 62.22+22.90 | 52.22+14.92 | B | MS |
| Yangmai 158 | 50 | 91.33+7.54 | 92.22+6.44 | A | HS |
a Means different responses to Bgt. HS: highly sensitive; MS: moderately sensitive; MR: moderately resistant. * Means with the same letter are not significantly different.
Histological examination revealed that lines that over-expressed Hv-SGT1 showed a higher frequency of Bgt-induced whole-cell oxidative burst. Staining with 3, 3′-diaminobenzidine (DAB), which allows detection of H2O2, did not uncover genotype-specific differences at an early stage of the interaction (12 h post inoculation [hpi], data not shown). At 24 hpi, the fungus had penetrated epidermal host cells in up to 60% of interaction sites in wild-type (WT) plants as well as in the transgenic lines OX-323 and OX-330. Detection of DAB polymerization showed a similar whole-cell accumulation ratio of H2O2 in both OX-323 and OX-330 (Figure 7A and 7B). In contrast, only a few DAB-stained cells (Figure 7D) was detected in WT Yangmai 158, although appressorial germ tube (AGT) penetration sites and primary germ tube (PGT) penetration sites with oxidative burst points were observed (Figure 7C). More ROIs accumulated in most pathogen-inoculated transgenic leaves (11.4% in OX-323, 9.7% in OX-330) than in the inoculated Yangmai 158 leaves (4.2%), although there was no significant difference in the frequencies with which oxidative burst points formed at Bgt interaction sites (Figure 7E) in leaves from WT, OX-323, or OX-330 plants (Figure 7F). These revealed that the pathways responsible for the oxidative burst were activated for the host defense response though regulation of Hv-SGT1.
Figure 7. Hydrogen peroxide accumulation in leaves of Hv-SGT1 over-expressing plants and the WT Yangmai 158.
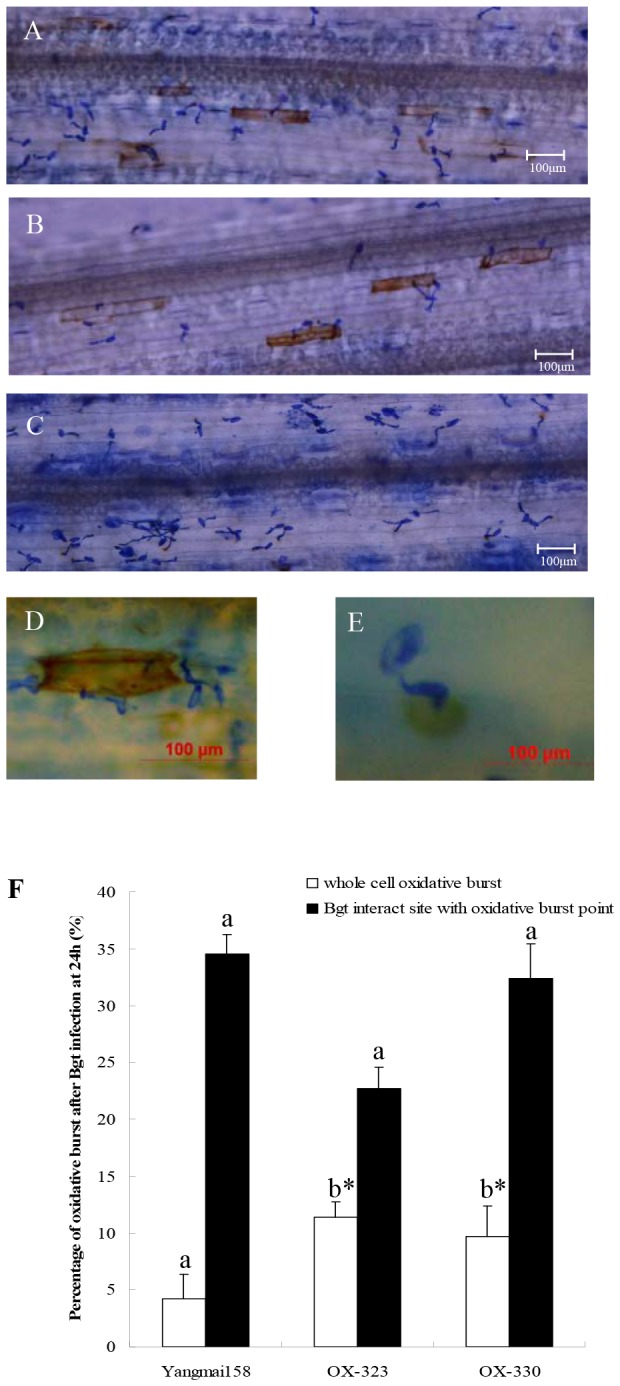
Hydrogen peroxide accumulated in Yangmai 158 (A), and Hv-SGT1 over-expressing lines OX-323 (B) and OX-330 (C). (D) Whole-cell ROI accumulation. (E) Oxidative burst at the Bgt interaction site. (F) Comparison of the percentage of cells with H2O2 accumulation throughout the entire cell or only around the infection sites in wild-type Yangmai 158 and the transgenic plants (* means p < 0.05).
Plant NADPH oxidasetive (NADPH-OX) enzyme was called Rboh (respiratory burst oxidase homologue), rapidly increases or decreases reactive oxygen species (ROS) through self-activation or inactivation during plant responses to biotic stresses. Two Hv-SGT1 transgenic lines OX-323, OX-330, and the Yangmai158 (WT) control, were analyzed for NADPH-OX activities using fully expanded leaves. NADPH-OX activities in the two transgenic lines were enhanced by 173.9% and 10.2% before Bgt inoculation (0 hai), but were decreased at 24 hai, respectively, in contrast to WT plants (Figure 8). These results suggested that varied NADPH-OX activities in the transgenic lines were likely associated with expression of Hv-SGT1.
Figure 8. NADPH-OX activities in transgenic and WT plants.
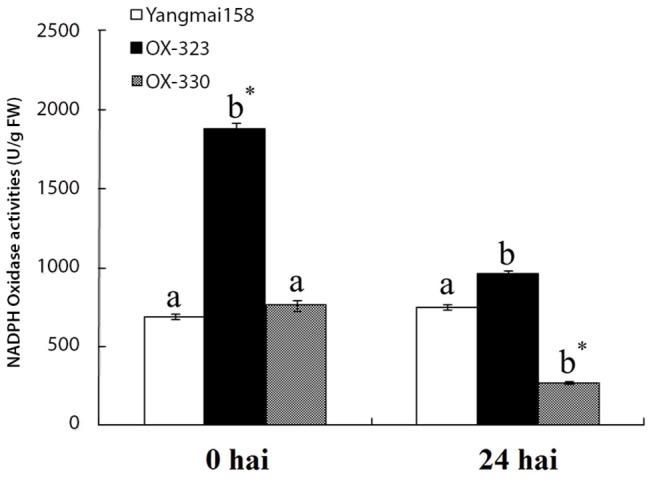
Each point represents the mean of three replicates. Bars indicate ±SE, Mean values followed by different letters are significantly different from each other (b* indicates P < 0.01 and b indicates P < 0.05).
Over-expression of Hv-SGT1 contributes to the resistance of common wheat to the hemi-biotrophic pathogen Fg
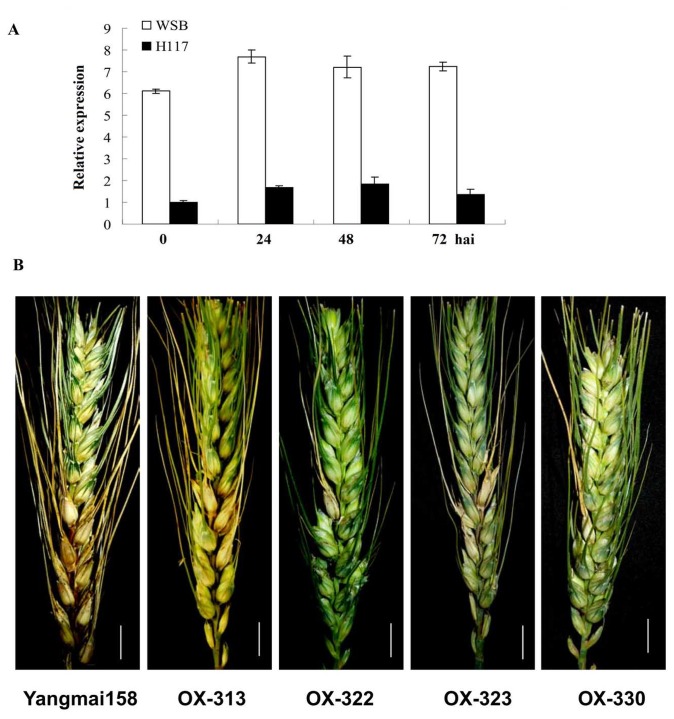
Unlike the inducible expression pattern of Hv-SGT1 in spikes of H . villosa after inoculation with Fg, Ta-SGT1 was constitutively expressed in both the FHB-resistant Wangshuibai (WSB) cultivar and its susceptible mutant NAUH117 (Figure 9A), and no obvious change was detected after inoculation with Fg. Nonetheless, an approximately 6-fold lower expression level of Ta-SGT1 transcript was detected in the susceptible mutant NAUH117 than that in resistant WSB. To determine whether higher expression levels of SGT1 was required for the resistance to Fg, we further investigated the over-expression plants transformed to express Hv-SGT1 under the regulation of the CAMV35S promoter for resistance to infection by the hemi-biotrophic Fg.
Figure 9. Relative expression levels of Ta-SGT1 in spikes of scab-resistant wheat variety Wangshuibai (WSB) and its susceptible mutant NAUH117 at different times after inoculation with Fg.
(A) Ta-SGT1 was constitutively expressed in both WSB and NAUH117, and no obvious change was detected after inoculation with Fg. Nonetheless, an approximately 6-fold lower expression level of Ta-SGT1 transcript was detected in the susceptible mutant NAUH117 than that in resistant WSB. (B) The FHB symptom of spikes of Yangmai158 and transgenic plants at 21 days after Fusarium inoculation, scale bar represents 1cm.
To evaluate resistance to FHB, transgenic T1 (164 individuals derived from the four positive T0 plants during 2010–2011) and T2 (151 individuals derived from the four positive T0 plants during 2011–2012) plants were grown under both greenhouse and field conditions, respectively. For both T1 and T2 plants, their average percentages of diseased spikelets were significant reduced compared with those of Yangmai 158 (Figure 9B, Table 2). This indicated that over-expression of Hv-SGT1 also contributed to the enhanced resistance of the transgenic plants to wheat FHB.
Table 2. Average rate of scab diseased spikes of the T1 and T2 transgenic plants.
| Lines | No. of positive plants | Average rate of scab diseased spikes |
Significance of ANOVA* (p<0.01) | |
|---|---|---|---|---|
| T1 generation | T2 generation | |||
| OX-313 | 20 | 6.7±2.6 | 6.0±1.6 | B |
| OX-322 | 20 | 4.5±2.4 | 5.8±0.1 | B |
| OX-323 | 20 | 5.8±2.3 | 5.9±1.3 | B |
| OX-330 | 20 | 7.4±1.1 | 6.3±1.2 | B |
| Yangmai 158 a | 5 | 8.4±2.7 | 15.8±5.1 | A |
| Sumai 3 b | 5 | 2.5±0.1 | 4.5±0.5 | B |
| Mianyang 85-45 c | 5 | 20.6±5.5 | 24.0±3.2 | A |
a Yangmai 158 is the parental control; b Sumai 3 and c Mianyang 85-45 are the disease resistant and susceptible controls, respectively. * Means with the same letter are not significantly different.
Hv-SGT1 over-expression regulates the expression of genes related to both the H2O2 and JA pathways
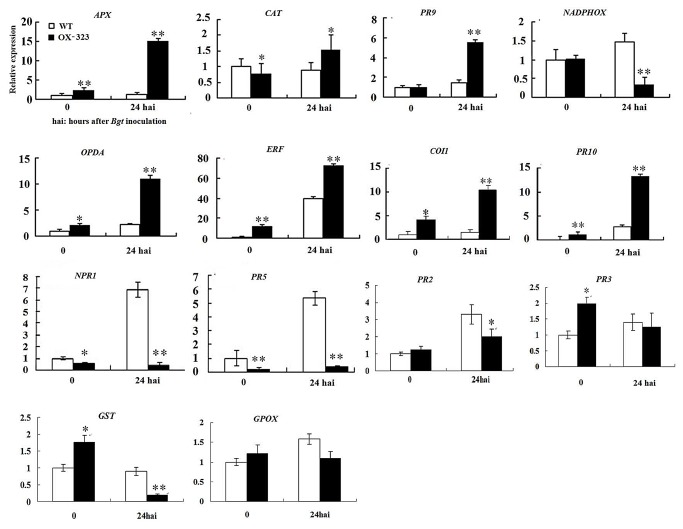
Given our demonstrations that both H2O2 and JA could induce the expression of Hv-SGT1, and that over-expression of Hv-SGT1 resulted in H2O2 accumulation in transgenic plants after inoculation with Bgt, we deduced that and H2O2 and JA might be linked to the expression of Hv-SGT1. The SA pathway is also closely related to plant disease resistance, and has been associated with HR induced by H2O2. We therefore analyzed the expression patterns of a subset of genes related to producing or scavenging H2O2, as well as genes related to SA- and JA-mediated signaling.
The H2O2-producing gene PR9 was significantly up-regulated in the transgenic plants after Bgt inoculation. Interestingly, another gene responsible for H2O2 production, NADPHOX, was down-regulated following inoculation with Bgt. The H2O2-scavenging gene, APX, was significantly up-regulated in the transgenic plants after inoculation with Bgt, and the CAT gene, which has a similar function, was only slightly up-regulated. Unlike enzymatic antioxidative system genes, GST (marker gene of non-enzymatic antioxidative system) was significantly down-regulated in the transgenic plants after inoculation with Bgt, but the GPOX gene, which has a similar function, was not regulated by SGT1 (Figure 10). After Bgt inoculation, the expression levels of genes related to the JA-signaling pathway, including those involved in JA synthesis (OPDA), JA signal transduction (COI1 and ERF), and response to the JA signal (PR-10) were substantially higher (Figure 10) in OX-323 transgenic plants than in WT plats after inoculation with Bgt. However, expression of marker genes of the SA pathway, such as NPR1, PR5 andPR2, was higher in the transgenic plants than in the WT plants, but down-regulated after inoculation with Bgt (Figure 10). Meanwhile, we found expression of PR3 (marker gene of ET pathway) was hardly changed in the transgenic plants both before and after inoculation with Bgt. We presumed that the over-expression of Hv-SGT1 might regulate the balance between the production and scavenging of H2O2, stimulate JA signaling, and suppress the SA defense pathway, but was not directly involved in the ET defense pathway.
Figure 10. Real-time qPCR analysis of the expression patterns of ten genes related to pathogenesis pathway in the wild-type Yangmai 158 and Hv-SGT1-over-expressing transgenic plants (OX-323) before inoculation of Bgt and 24 h post inoculation with Bgt.
* p < 0.05, ** p < 0.01 compared with the wild type. Over-expression Hv-SGT1 regulates the expression of genes related to the H2O2, SA and JA pathways.
Discussion
This study characterized the induction of the Hv-SGT1 gene by both biotrophic and hemi-biotrophic pathogens. Homology and cluster analyses indicated that Hv-SGT1 belongs to the SGT protein family found in eukaryotes, and that the homologies of SGTs from various organisms are relatively high. The main similarities between SGT proteins lie in the conserved TPR domain and SGS motifs, although the considerable diversity in the CS region, which was required for its binding to HSP90 [28], suggested that the functions of Hv-SGT1 may differ from other SGTs. At the subcellular level, Arabidopsis SGT1b fused to Cerulean localized to the cytosol but could be seen in nuclei of 25% of 55 transformed cells examined. This suggested the movement of SGT1b between the cytosol and nucleus [29]. Moreover, a fusion of rice OsSGT1 to GFP localized to both the cytoplasm and nuclei of onion epidermal cells [13], which was consistent with our localization of Hv-SGT1-GFP fusion protein. This suggested that the rice and Arabidopsis SGT1 proteins might have similar functions associated with plant disease resistance.
Microarray analysis identified Hv-SGT1 as a candidate gene that regulates broad-spectrum resistance to Bgt in H . villosa . However, it is challenging to generate α Bgt susceptible mutant or SGT1 knockout mutant of H . villosa , and although immature embryo culture and regeneration system for transformation were established for wheat, these approaches were not yet available for H . villosa . Therefore, the goal of this research was to demonstrate stable constitutive expression of Hv-SGT1 in Yangmai 158, a wheat variety with moderate susceptibility to both powdery mildew and scab.
As an ubiquitylation pathway component, SGT1 plays important roles in the plant defense. Unver et al. [30] found that mRNA levels of SGT1 was up-regulated following NbUbc2 silencing. SGT1 positively regulated the disease resistance via interaction with the UBC2 or ubiquitylation in Nicotiana benthamiana . Similar results were also obtained in TaUbc2-silenced wheat leaves [31]. Our results were consistent with the above results, showing that HvSGT1 was also a positive regulator in the plant hypersensitive response, although we haven’t found its interaction substrate or UBC in this paper.
The interplay among complex signaling networks, including various pathways regulated by phytohormones such as salicylic acid (SA), jasmonic acid (JA), ethylene and ABA, dramatically influences plants’ disease resistances. By precisely regulation of the accumulation and perception of phytohormones, plants counteract disease stresses. In recent years, the biosynthesis, signal transduction, and physiological functions of phytohormones in various plant species have been better understood. The roles of SGT1 in plant disease resistance, especially its link with phytohormone pathways have been intensively studied. Meldau et al. [15] found that SGT1 was required for herbivory-induced SA and JA homeostasis and normal MeJA-induced transcriptional responses, and was important for the biosynthesis of secondary metabolites defense responses against phytophagous insects in N . attenuate . Our results further confirmed the involvement of SGT1 in different phytohormone signaling (H2O2, JA and SA) both in H . villosa and wheat. Further characterization of the involvement of SGT1 in defense responses mediated by the phytohormone pathways will help us to unravel the roles of Hv-SGT1 in powdery mildew resistance of wheat.
Several previous studies have shown that SGT1 is required for cell death during incompatible interactions associated with HR [3,8,32,33]. The accumulation of H2O2 accumulation is believed to be involved in HR during plant-pathogen interactions. A previous study demonstrated that a serine/threonine kinase gene Stpk-V, which is a key member of the broad-spectrum resistance locus, can regulate H2O2 accumulation in the wheat-Bgt interaction. In this study, we demonstrated differential expression of both H2O2-producing and H2O2-scavenging genes in the SGT1 transgenic wheat and the parental line from which these were derived. We observed a far more obvious accumulation of H2O2 after inoculation of Bgt in the transgenic plants compared with the WT line from which they were derived. This indicated that the increased H2O2 level was correlated with the over-expression of SGT1. The accumulation of H2O2 and the subsequent cell death usually resulted in the disease resistance caused by the biothophic pathogens [34]. Over-expression of NbSGT1 accelerates cell death associated with resistance to a non-adapted pathogen [8], although over-expression of SGT1 in rice did not enhance disease-associated cell death [13]. It was proposed that different SGT1-pathogen interactions and the complexity of the cell death pathways determine whether cell death can be induced by the over-expression of SGT1. But expression levels of NPR1 and PR5, which could be accounted for the component of SA pathway activity, were decreased after Bgt inoculation when the cellular H2O2 content was increased. We supposed that SGT1 positively regulated the H2O2 pathway, meanwhile negatively regulated the SA pathway. Moreover, H2O2 might not directly transfer the SA signal to the downstream response genes in the SGT1 transgenic plants.
In Arabidopsis , the CWP (cell wall protein fraction)-induced defense system appeared to be regulated by JA-mediated and SGT1-dependent signaling pathways [35]. What role does the JA pathway play in Hv-SGT1-mediated disease resistance in wheat? The enzyme 12-oxophytodienoate reductase (OPDA) plays a key role in the biosynthesis of JA [36]. Arabidopsis SGT1b is also required for the activities of the SCFCOI1 -mediated JA response [37,38]. The COI1 protein contains an F-box motif and associates physically with AtCUL1, AtRbx1, and the Skp1-like proteins to assemble SCFCOI1 ubiquitin-ligase complexes for ubiquitination and subsequent degradation by the 26S proteasome [39,40]. The JA-responsive gene that encodes ethylene response factor (ERF) plays a crucial role in the cross-talk between JA and ET signal transduction [26,41]. The PR10 gene have been implicated in the signaling pathway that responds to JA, and is critical to effective functioning of the JA defense pathway [42]. In our research, expression of genes involved in JA signaling and response pathways was enhanced in Hv-SGT1 transgenic plants and in Hv-SGT1-silenced plants. Expression of NPR1, PR2 and PR5, which are markers of SA pathway activity, was either reduced in Hv-SGT1-overexpressing plants after Bgt inoculation or enhanced in Hv-SGT1-silenced plants. Differently, expression level of PR3 was unobviously changed in the transgenic plants both before and after inoculation with Bgt. This suggests that the transactivation of PR genes in SA pathway was independent of SGT1, and SGT1 was not directly involved in the ET defense pathway to regulate the PR3 expression. We propose that Hv-SGT1 activates resistance mechanisms through JA-dependent defense pathways, but suppresses the activities of SA-dependent defense pathways, and does not affect the ET-dependent defense pathways.
EI Oirdi & Bouarab [19] demonstrated that silencing SGT1 in N . benthamiana compromises the hypersensitive response induced by Botrytis cinera . They hypothesized that B . cinera promotes NbSGT1 expression to exploit the antagonistic effects between SA and JA. Higher basal concentrations of SA were also found in NaCOI1-silenced N . attenuata , suggesting that impaired JA signaling increases SA concentrations [43]. It is not enough to detect the transcription level of pathway-related genes only. Whether SGT1 also mediates the homeostasis of pathogen-elicited SA and JA levels by measurement changes of the SA and JA in both SGT1-silenced and SGT1-overexpressing plants, deserves further study.
There is compelling evidence that SGT1 contributes to the resistance to powdery mildew mediated by race-specific R genes in barley [18]. In the present study, both the VIGS and gene transformation experiments proved that SGT1 was tightly correlated with the powdery mildew resistance in H . villosa , which showed substantial broad-spectrum resistance to powdery mildew. The increased cell death due to the over-expression of the SGT1 and the activated JA pathway might both contribute to the improved resistance. Our results indicated that SGT1 was also related to the observed broad-spectrum resistance, but the role of SGT1 in broad-spectrum disease resistance was still poorly understood. Further analysis of the function of SGT1 will enhance our understanding of the roles of SGT1 in the interaction of H . villosa with Bgt.
There are differences in the resistance mechanisms and patterns of fungal development seen in biothophic and necrotrophic pathogens, and Fg is a hemi-biotrophic pathogen with both biotrophic and necrotrophic features. The earliest stages of Fg infection are characterized by biothophic features, although the pathogen proceeds to the necrotrophic phase after successful infection. The SA pathway is critical for scab resistance at the early stage of infection, when Fg attacks the host as a biothroph, although the JA pathway plays a predominant role at later stage when the pathogen develops necrotrophic tendencies [44]. The SA pathway is usually correlated with HR and cell death, which effectively restricts infection by the biothophic pathogen. A study conducted using tobacco indicated that SGT1 is responsible to cell death, and SGT1 was a positive regulator of the JA pathway [15]. In this study, the over-expression of SGT1 in wheat variety Yangmai 158 can increase its resistance to Fg, which may be the result of the increased cell death caused by over-expression of SGT1 at the site of primary infection. The activated JA pathway in the transgenic wheat may contribute to the resistance to the Fg during a later stage of infection.
Previous studies have showed that SGT1 plays an important role in the resistance to necrotrophic pathogens. For example, SGT1 of Nicotiana benthamiana contributed symptom development and disease susceptibility after infection by the necrotrophic fungus Botrytis cinerea [19]. However, our research has implied an important role for SGT1 in this hemi-biotrophic pathogen. Although our preliminary results showed that the transgenic plants showed increased resistance to the Fg, further experiments (including gene silencing) should be performed using a resistant variety, such as Sumai 3 or Wangshuibai, to clarify the involvement of SGT1 in resistance to Fg.
Some studies have demonstrated an essential role for SGT1 in basal resistance to phytopathogens. For example, over-expression of OsSGT1 from rice significantly increases basal resistance to the virulent bacterial blight pathogen Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99 and to several blast fungal M. grisea races [13]. Many studies also implied a role for SGT1 in R-gene-mediated resistance. Although SGT1 is associated with resistance to both Bgt and Fg in wheat, it remains unknown whether it functions in the PAMP-triggered basal resistance stage or in the effector-triggered resistance stage.
Owing to the different (sometimes even contradictory) mechanisms of resistance to biotrophic and necrotrophic pathogens, the co-regulation of the resistance to both classes of pathogens seems to be complicated. The co-regulation of resistances to the bithophic Bgt and the hemi-biotrophic Fg demonstrated in the present study indicates the feasibility of improving the resistance to different types of pathogens if the universal regulatory factors are explored and used properly. Our results provide further evidence for the universal role of the SGT1 gene in various plant defense responses. Over-expression of Hv-SGT1 conferred significantly increased resistance to wheat powdery mildew and scab, providing an effective strategy for developing wheat germplasm with improved resistance to the pathogens responsible for these diseases.
Materials and Methods
Plant materials
Haynaldia villosa (2n=14, VV), T . durum - H . villosa amphiploid (AABBVV), DA1V-DA7V (T. aestivum- H . villosa addition line, each contains one pair of chromosomes of H . villosa from 1V to 7V in the common wheat background), powdery mildew susceptible wheat varieties Chinese spring and Yangmai 158, powdery mildew resistant wheat variety T. aestivum–H. villosa T6VS/6AL translocation line 92R137, FHB resistant wheat variety Wangshuibai to and its susceptible mutant NAUH117 were all developed or preserved by Cytogenetic Institute, Nanjing Agricultural University (CINAU).
Fungal and chemical treatments
Mixed races of Bgt were maintained on susceptible variety Sumai 3 seedlings in a spore-proof greenhouse under 14 h light/10 h dark (24/18°C, 70% humidity) regime. Wheat seedlings at the two-leaf stage were inoculated with Bgt or treated with exogenous hormone or signal molecules, including 5 mM salicylic acid (SA), 100 μΜ methyl jasmonate (MeJA), 200 μΜ ethepon (ET), 5 μΜ abscisic acid (ABA), and 7 mM hydrogen peroxide (H2O2). All chemicals were administered as a 0.05% Tween-20 solution, with 0.05% Tween 20 used as a mock treatment, and all of the treatments followed Cao et al. [22]. Leaves were harvested at different time points after treatments for gene expression analysis. The young spikes of Wangshuibai and its susceptible mutant NAUH117 were inoculated by Fg using the single-floret inoculation method [45].
Primers designing
All primers used for RACE (Rapid Amplification of the cDNA ends, RACE), RT-PCR, qRT-PCR and plasmid constructions are listed in Table 3.
Table 3. Sequences of the gene-specific primer pairs used in the study.
| Primers | Sequence 5' to 3' |
|---|---|
| RT-PCR Primers from wheat EST for SGT1-1 | SGT1-1-F: CGCCATGTTCACTGACGA; SGT1-1-R: GATCCATGGTCGGAGCAA |
| RACE nest Primers | 5’-SGT1-S1: AATCTCCTCAACCTCCAGCA; 3’-SGT1-A1: TTGTTATCAGGGCCTGTTCC |
| RT-PCR Primers for Ta-SGT1 | SGT1-RT-F: ACTGAGGCTGTAGCTGATG; SGT1-RT-R: CATATCTTCATCAGCATCAC |
| RT-PCR Primers for Hv-SGT1 | Hv-SGT1-RT-F: TCGGATCTGGAGAGCAAG; Hv-SGT1-RT-R: TCCTGGTCCTCAGCTT |
| RT-PCR Primers for Tubulin | Tubulin-F AGAACACTGTTGTAAGGCTCAAC; Tubulin-R: GAGCTTTACTGCCTCGAACATGG |
| Chromosomal Location | SGT1-F4: AGGCTGTAGCTGATGCCA; SGT1-R5: TCTTCATATGCATCTGGTGACT |
| Sub-cellular localization Primers | Hv-SGT1-SalⅠ-F: CGG T C G A CATGGCCGCCGCCGCCG; Hv-SGT1-NcoⅠ-R: CGC C A T G G AATACTCCCACTTCTTG |
| VIGS Primers | Hv-SGT1-VIGS-F: GCTG C T A G CAGCTGATGCCAACAAAG; Hv-SGT1-VIGS-R: GCTG C T A G CATACGATCACACTCCT |
| Over-expression Primers | Hv-SGT1-BamHI-F: CGCG G A T C CTCGACGCAGACATGG; Hv-SGT1-KpnI-R: CG G T A C CTCATTAATACTCCCAC |
| Identification Primers for Hv-SGT1 | CaMV35S-F: AGTTCATTTCATTTGGAGAGAACAC; Hv-SGT1-R4: CAGCTGGAACACCCTTAGC |
| qRT-PCR Primers for Hv-SGT1 | Hv-SGT1-F: ATACTCCCACTTCTTG; Hv-SGT1-R: AGCTGATGCCAACAAAG |
| qRT-PCR Primers for Ta-PR9 (EU264058.1) | F: AGGCTGTGGTTTTTGTGCTT; R: GCTTAAAGCTGAGGCTGCAT |
| qRT-PCR Primers for Ta-NADPHOX (AY561153.1) | F: ATGCTCCAGTCCCTCAACCAT; R: TTCTCCTTGTGGAACTCGAATTT |
| qRT-PCR Primers for Ta-CAT (HM989895.1) | F: TGCCTGTGTTTTTTATCCGAGA; R: CTGCTGATTAAGGTGTAGGTGTT |
| qRT-PCR Primers for Ta-APX (EF555121.1) | F: GGTTTGAGTGACCAGGACATTG; R: GCATCCTCATCCGCAGCAT |
| qRT-PCR Primers for Ta-GST (AJ441055) | F: GGAGCACAAGAGCCCCGAGC; R: CGGGTTGTAGGTGTGCGCGT |
| qRT-PCR Primers for Ta-GPOX (AJ010455.1) | F: AACTACCCGCTCTGCTCCT; R: GCCTTGGTCCTTGTACTTCG |
| qRT-PCR Primers for Ta-NPR1 (AX049430.1) | F: CTGTCCGACTTTGTGAGCATA; R: CCCGCTGTCATTCTTCAGGTTG |
| qRT-PCR Primers for Ta-PR5 (AF384146.1) | F: CAAGCAGTGGTATCAACGCAGAG; R: GTGAAGCCACAGTTGTTCTTGAT |
| qRT-PCR Primers for Ta-PR2 (DQ090946) | F: GCGTGAAGGTGGTGATTT; R: GTGCCCGTTACACTTGGAT |
| qRT-PCR Primers for Ta-PR3 (CK207575) | F: ACCTCCTTGGCGTCAGCT; R: TCGCACCATTATTCCCTT |
| qRT-PCR Primers for Ta-OPDA (JQ409278.1) | F: CCATAAACGCCATCAAAGCAGG; R: TGCATCGGGTTCGAGTCATAGG |
| qRT-PCR Primers for Ta-ERF (GU452719.1) | F: CACCTTGACCTCCTCCTCTTCGC; R: TTGTTCCCTTTGGACGCCAGG |
| qRT-PCR Primers for Ta-PR10 (CV778999) | F: ACGGAGCGGATGTGGAAG; R: GCCACCTGCGACTTGAGC |
| qRT-PCR Primers for Ta-COI1 (HM447645.1) | F: AAGGAGTTGCTGCTTTAGTGAACG; R: TCAGAGTGGGTCGCTTTTACTTG |
Isolation of the cDNA of SGT1 from H. villosa and the translocation line 92R137
Twenty-six wheat EST sequences with high homology to barley SGT1 were found and aligned for the design of specific primers. First-strand cDNA was reversely transcribed using 2 µg of total RNA extracted from either the powdery mildew–resistant line 92R137 or H . villosa 24 h after inoculation. A 600-bp fragment was obtained by RT-PCR using the specific primers SGT1-1-F and SGT1-1-R (Table 3). The 3ʹ-RACE was conducted using the 3ʹ-RACE System for Rapid Amplification of cDNA Ends kit (Invitrogen, USA), and the 5ʹ-Race was conducted using the 5ʹ-Full RACE Core Set (Takara, Japan). The primers Hv-SGT1-5’RACE and Hv-SGT1-3’RACE (Table 3) were used for PCR-mediated amplification of two fragments, which were 1100 bp and 400 bp. These sequences were cloned, sequenced, and spliced in order to clone the full-length cDNA sequences of both Ta-SGT-92R137 and Hv-SGT1.
Sequence analysis and phylogenetic tree construction
The putative function of the cloned genes were analyzed using the BLAST software (http://www.ncbi.nlm.nih.gov/blast/), the ORF was predicted by the ORF Finder software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), the multiple sequence aligment was conducted by ClustalX 1.83 program, and the phylogenetic tree was constructed using the MEGA 4.0 software.
Chromosome location of Hv-SGT1 gene
DNA samples from the H . villosa , T . durum - H . villosa amphiploid, DA1V-DA7V, and Chinese spring were used as templates for PCR analysis to determine the chromosomal location of the Hv-SGT1 in the H . villosa genome using the primer pairs Hv-SGT1–F4 and Hv-SGT1–R5 (Table 3). The PCR was performed in 25-µl reaction volumes including 1× PCR buffer, 2 mmol/l MgCl2, 0.15 mmol/l dNTPs, 20 ng of each primer, 2 µl template and 1U Taq DNA polymerase (Takara Bio, Japan). The conditions for thermal cycling involved incubation at 94°C for 3 min, followed by 33 cycles that each involved 94°C for 45 s, 50°C for 45 s, and 72°C for 1 min. The PCR products were separated in 8% non-denaturing polyacrylamide gels.
qRT-pcr and RT-PCR for gene expression analysis
Total RNA was isolated using Trizol Reagent (Invitrogen, USA) and quantified with a NanoDropTM 1000 spectrophotometer (Thermo, Fisher Scientific, USA). The first-strand cDNA was synthesized using 2 µg of total RNA by the AMV reverse transcriptase (Takara) following the manufacturer’s instruction. For expression pattern analysis, Hv-SGT1-RT-F and Hv-SGT1-RT-R (Table 3) specific primers for Hv-SGT1, SGT1-RT-F and SGT1-RT-R (Table 3) primers for Ta-SGT1-92R137, were used. To evaluate the effectiveness of the gene silencing by VIGS, the expression of the Hv-SGT1 was analyzed by qRT-PCR with a pair of primers (Hv-SGT1-qRT-F and Hv-SGT1-qRT-R, Table 3) specific to Hv-SGT1 using the tubulin as the internal control for normalization. The PCR reaction was performed in 25 µl of reaction mixture containing 1×SYB Premix Ex Taq (Takara), 0.2 µM of each primer, 1× Rox Reference Dye, and about 30 ng cDNA per sample using the ABI Prism 7500 system (Applied Biosystems, USA). The program used was as follows: 1 min at 95°C, followed by 40 cycles at 95°C for 10 s, 60°C for 20 s, and 72°C for 40s. Three independent biological replications were performed for each treatment. Dissociation curves were generated for each reaction to ensure specific amplification. Threshold values (CT) generated from the ABI PRISM 7500 Software Tool (Applied Biosystems) were used to quantify relative gene expression using the comparative 2–ΔΔCT method [46]. The expression of Hv-SGT1 and a set of H2O2, SA and JA pathogenesis-related genes in both transgenic plants and Hv-SGT1 silencing plants were analyzed by the qRT-PCR, using the tubulin gene (amplified with specific primers, Table 3) as the internal control. The PCR was performed in 25 µl of reaction mixture containing. The program used was as follows: 3 min at 94°C, followed by 27 cycles at 94°C for 30 s, 55-60°C for 30 s, and 72°C for 40s, and then retained at 10°C.
Sub-cellular localization of the Hv-SGT1 in epidermal cells of onion and H. villosa
Sal I and Nco I sites were added to the 5ʹ and 3ʹ ends of the full length open reading frame of Hv-SGT1, respectively, with the stop codon deleted by appropriate design of the Hv-SGT1- Nco I -R primer. The PCR product and the sGFP (s65t) vector were cut by Sal I and Nco I, and the fragments were ligated to produce the fusion gene expression vector p35S::Hv-SGT1-GFP::Nos3. After confirmation of the recombinant construct, it was delivered either to epidermal cells on the adaxial surfaces of layers peeled from an onion or to H . villosa leaves by particle bombardment, as described by Chen et al. [25]. The sub-cellular localization of the Hv-SGT1-GFP fusion protein was observed using a Leica confocal microscope (SP2) fitted with 20× numerical aperture plan apochromat objectives (the nucleus was stained with DAPI).
Functional analysis of Hv-SGT1 by VIGS
A pair of primers Hv-SGT1-VIGS-F/R with NheⅠ sites was introduced to the 5' and 3' end of the Hv-SGT1 partial sequence (202bp fragment flanking the stop codon) and then was reversely inserted into the RNAγ gammab strand of the BSMV. The methods of recombinant vector RNAγ gammab: Hv-SGT1 construction, plasmid linearization, in vitro transcription and virus infection were as described as Cao et al. [23]. The 5th full expanded young leave inoculated with BSMV were detached from the plant and mounted on the culture medium in a Petri dish. Fresh powdery mildew spores were brushed off onto the leaf surface. After fourteen hours incubation in dark, the Petri dish was placed in a chamber in which the temperature was set at 25°C and the photoperiod was 14/10 h light/dark. Five days later leaves were fixed, bleached and stained with Commassie blue for observation of fungal development in bright field under Olympus BX-60 Stereo-Fluoroscope. 10 leaves per sample were observed to evaluating the effect of silencing. The 5th leaves challenged with BSMV: Hv-SGT1 or not were also used to check expression level of Hv-SGT1 on the iCycler iQTM Multi-Color Real Time PCR Detection System (BioRad).
Development and molecular characterization of Hv-SGT1 transgenic wheat plants
The over expression vector pBI220-35S::Hv-SGT1::Nos3 was constructed by inserting Hv-SGT1 into the plant expression vector pBI220, using the forward primer (Hv-SGT1-BamH1-F) and the reverse primer (Hv-SGT1-Kpnl-R) for amplifying the ORF fragment. Another plant expression vector pAHC25 containing report gene β-glucuronidase gene (Gus) and the herbicide tolerance gene (Bar) driven by the ubiqutin (Ubi) promoter was used as the co-transformation vector. Putative transgenic wheat plants (Yangmai 158) were produced by particle bombardment of calli cultured from immature embryo and subsequently selected by biolaphos in callus maintain, differentiation and regeneration medium as described [47]. Fully developed plants were transferred into small pots in the greenhouse [20–25°C, 14/10 h (d/night) photoperiod].
Total genomic DNA was isolated from fresh leaves of transgenic plants and untransgenic control using a modified SDS-Na+ salts protocol [48]. PCR amplification use primers specific for the Hv-SGT1-R4 and the promoter CaMV35S-F (Table 3). PCR procedure was the same as described in chromosome location and the PCR products were separated in 0.8% agarose gel. The PCR products amplified from the PCR positive transgenic plants and Yangmai 158 were transferred to HybondTM-N+ membrane (Amersham Biosciences, UK) and southern blotting was conducted using the partial coding region of Hv-SGT1 and the promoter CaMV35S as the probe which was labeled with digoxigenin by PCR using specific primer CaMV35S-F and Hv-SGT1-R4 (Table 3). Southern blot hybridization was the same as described in the copy number of Hv-SGT1 detecting. qRT-PCR analysis of the expression levels of Hv-SGT1 in the transgenic lines and wild Yangmai158 was taken as the same as described in VIGS analysis.
Tests of Bgt resistance in plants that over-express Hv-SGT1
The characterized positive transgenic T2 plants (151 individuals derived from four positive T0 lines) were identified by PCR analysis at the seedling stage, and were grown under both greenhouse and field conditions. The WT line Yangmai 158 was used as a negative control. Transgenic T2 plants were planted in the greenhouse, with ten T2 seeds derived from each T1 accession planted in a row. The susceptible cultivar Suimai 3 were planted in every fifth row for Bgt propagating and spraying. To estimate the resistance of adult T2 plants following natural infection with the native population of the pathogen, the mildew was assessed on the upper two leaves as a visually estimated percentage of leaves covered with mildew. The Bgt infection types (ITs) were scored on a 0–9 scale [49] and disease indexes (DI) of the plants were calculated as described by Chen et al. [25]. The ANOVA (analysis of variance) was performed to determine the significance of differences between the transgenic plants and wild type. The Bgt hyphae were stained with DioC6 and photographed using an Olympus MVX10 microscope (Japan). Wheat leaves of four transgenic lines and Yangmai158 control infected with Bgt were sampled at 48 hpi and stained as described [24]. Bleached leaf segments were examined with an Olympus a BX-51 microscope (Olympus, Japan) for infection sites. The the average number of hyphal branches and average hyphal length were calculated by DP-BSW software, and the final data of each index was the mean of at least 30 infection sites for each of the five randomly selected leaf segments per line. Standard deviations and a paired sample t-test for statistical analysis were performed with SAS software.
Evaluation of FHB resistance in transgenic plants that over-express Hv-SGT1
Transgenic T1 (164 individuals derived from four positive T0 lines during 2010–2011) and T2 (151 individuals derived from four positive T0 lines during 2011–2012) were planted under greenhouse and field conditions, respectively. Yangmai 158 was used as the negative control, and Sumai 3 and Mianyang 85-45 were used as the disease-resistant and disease-susceptible controls, respectively. The resistance to wheat FHB was evaluated using single floret injection, and at least three heads per plant were inoculated. The percentages of diseased spikelets were calculated 20 days after initial inoculation. Thirty characterized positive transgenic plants were recorded for each line and the average percentages of diseased spikelets were calculated. Analysis of variance (ANOVA) was performed to determine the significance of differences between the transgenic and wild-type plants.
Histochemical detection of H2O2 using DAB
We detected H2O2 using the DAB (Bio Basic Inc., Shanghai, China) staining method described by Thordal-Christensen et al. [50] Eight hours before sampling, inoculated primary leaves of wheat were cut, and the cut ends were placed in a solution containing 1 mg/ml DAB dissolved in NaOH-acidified (pH 3.8) distilled water. Leaves were incubated for an additional 8 h to allow DAB uptake and reaction with H2O2 and peroxidase. The inoculated wheat leaves were then cut into 1.5-cm segments at the indicated time (12 or 24 h) and discolored in boiling 95% ethanol for 10 min before being cleared in saturated chloral hydrate. The cleared leaf segments were then stored in 50% glycerol. For microscopic observation, the treated leaf segments were mounted on glass slides in 50% glycerol, and examined (200×) using an Olympus microscope (Japan). Four leaf pieces from each transgenic line and wild-type plant were observed. Each point represents at least 100 infection sites on each leaf piece. Bars represent standard deviation. Standard deviations and a paired sample t-test for statistical analysis were performed using the SAS software. The NADPH oxidase activity in wheat leaf tissues was assayed as described [51].
Funding Statement
This research was supported by grants from the Important National Science & Technology Specific Projects of Transgenic Research (Grant No. 2011ZX08002-001, 2009ZX08002-001B); Nature Science Foundation of China (Grant No. 31101206, 31171540); the National University Doctoral Fund of China (Grant No. 20100097120036); and Jiangsu Natural Science Foundation (Grant No. BK2011636). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Hostmicrobe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814. doi:10.1016/j.cell.2006.02.008. PubMed: 16497589. [DOI] [PubMed] [Google Scholar]
- 2. Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329. doi:10.1038/nature05286. PubMed: 17108957. [DOI] [PubMed] [Google Scholar]
- 3. Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD et al. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080. doi:10.1126/science.1067747. PubMed: 11847308. [DOI] [PubMed] [Google Scholar]
- 4. Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K et al. (2002a) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076. doi:10.1126/science.1067554. PubMed: 11847307. [DOI] [PubMed] [Google Scholar]
- 5. Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noël L et al. (2006b) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25: 2007–2016. doi:10.1038/sj.emboj.7601084. PubMed: 16619029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y, Schiff M, Serino G, Deng X, Dinesh-Kumar SP (2002a) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N genemediated resistance response to tobacco mosaic virus. Plant Cell 14: 1483–1496. doi:10.1105/tpc.002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP (2004b) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chemi 279: 2101–2108. doi:10.1074/jbc.M310029200. PubMed: 14583611. [DOI] [PubMed] [Google Scholar]
- 8. Wang K, Uppalapati SR, Zhu X, Dinesh-Kumar SP, Mysore KS (2010) SGT1 positively regulates the process of plant cell death during both compatible and incompatible plant–pathogen interactions. Mol Plant Pathol 11: 597–611. PubMed: 20695999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33. doi:10.1016/S1097-2765(00)80184-7. PubMed: 10445024. [DOI] [PubMed] [Google Scholar]
- 10. Bansal PK, Abdulle R, Kitagawa K (2004) Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Mol Cell Biol 24: 8069–8079. doi:10.1128/MCB.24.18.8069-8079.2004. PubMed: 15340069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muskett P, Parker J (2003) Role of SGT1 in the regulation of plant R gene signalling. Microbes Infect 5: 969–976. doi:10.1016/S1286-4579(03)00183-7. PubMed: 12941389. [DOI] [PubMed] [Google Scholar]
- 12. Shirasu K (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60: 139–164. doi:10.1146/annurev.arplant.59.032607.092906. PubMed: 19014346. [DOI] [PubMed] [Google Scholar]
- 13. Wang Y, Gao M, Li Q, Wang L, Wang J et al. (2008): OsRAR1 and OsSGT1 physically interact and function in rice basal disease resistance. Mol Plant Microbe Int 21: 294–303. [DOI] [PubMed] [Google Scholar]
- 14. Bhaskar PB, Raasch JA, Kramer LC, Neumann P, Wielgus SM et al. (2008) Sgt1, but not Rar1, is essential for the RB-mediated broad-spectrum resistance to potato late blight. BMC Plant Biol 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meldau S, Baldwin IT, Wu J (2011) SGT1 regulates wounding- and herbivory-induced jasmonic acid accumulation and Nicotiana attenuata’s resistance to the specialist lepidopteran herbivore Manduca sexta . New Phytol 189: 1143–1156. doi:10.1111/j.1469-8137.2010.03558.x. PubMed: 21118264. [DOI] [PubMed] [Google Scholar]
- 16. Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P et al. (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci U S A 99: 10865–10869. doi:10.1073/pnas.152330599. PubMed: 12119413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138: 2165–2173. doi:10.1104/pp.105.061861. PubMed: 16024691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K et al. (2003) Recognition specificity and RAR1/SGT1 dependency in barley Mla disease resistance alleles to the powdery mildew fungus. Plant Cell 15: 732–744. doi:10.1105/tpc.009258. PubMed: 12615945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. El Oirdi M, Bouarab K (2007) Plant signalling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea . New Phytol 175: 131–139. doi:10.1111/j.1469-8137.2007.02086.x. PubMed: 17547673. [DOI] [PubMed] [Google Scholar]
- 20. Cuzick A, Maguire K, Hammond-Kosack KE (2009) Lack of the plant signalling component SGT1b enhances disease resistance to Fusarium culmorum in Arabidopsis buds and flowers. New Phytol 181: 901–912. doi:10.1111/j.1469-8137.2008.02712.x. PubMed: 19140951. [DOI] [PubMed] [Google Scholar]
- 21. Chen P, Qi L, Zhang S, Liu D (1995) Development and molecular cytogenetic analysis of wheat 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91: 1125–1128. [DOI] [PubMed] [Google Scholar]
- 22. Cao A, Wang X, Chen Y, Zou X, Chen P (2006a) A sequence-specific PCR marker linked with Pm21 distinguishes chromosomes 6AS, 6BS, 6DS of Triticum aestivum and 6VS of Haynaldia villosa . Plant Breeding 125: 201-205. doi:10.1111/j.1439-0523.2006.01222.x. [Google Scholar]
- 23. Cao A, Xing L, Wang X, Yang X, Wang W et al. (2011b) Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc Natl Acad Sci USA 108: 7727–7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X, Cao A, Yu C, Wang D, Wang X et al. (2010) Establishment of an effective virus induced gene silencing system with BSMV in Haynaldia villosa . Mol Biol Rep 37: 967–972. doi:10.1007/s11033-009-9766-1. PubMed: 19714483. [DOI] [PubMed] [Google Scholar]
- 25. Chen YP, Xing LP, Wu GJ, Wang HZ, Wang XE et al. (2007) Plastidial glutathione reductase from Haynaldia villosa is an enhancer of powdery mildew resistance in wheat (Triticum aestivum). Plant Cell Physiol 48: 1702–1712. doi:10.1093/pcp/pcm142. PubMed: 17947258. [DOI] [PubMed] [Google Scholar]
- 26. Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R (2003a) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178. doi:10.1105/tpc.007468. PubMed: 12509529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu Z, Gaudet D, Puchalski B, Despins T, Frick M et al. (2005) Inducers of resistance reduce common bunt infection in wheat seedlings while differentially regulating defense-gene expression. Physiol Mol Plant Pathol 67: 138–148. doi:10.1016/j.pmpp.2005.12.001. [Google Scholar]
- 28. Takahashi A, Casais C, Ichimura K, Shirasu K (2003) HSP90 interacts with RAR1 and SGT1, and is essential for RPS2-mediated resistance in Arabidopsis . Proc Natl Acad Sci U S A 100: 11777–11782. doi:10.1073/pnas.2033934100. PubMed: 14504384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noël LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S et al. (2007) Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19: 4061–4076. doi:10.1105/tpc.107.051896. PubMed: 18065690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Unver T, Turktas M, Budak H (2013) In planta evidence for the involvement of a ubiquitin conjugating enzyme (UBC E2 clade) in negative regulation of disease resistance. Plant Mol Biol Rep 31: 323–334. doi:10.1007/s11105-012-0502-y. [Google Scholar]
- 31. Unver T (2008) Detection and characterization of plant genes involved ın various biotic and abiotic stress conditions using DDRTPCR and isolation of interacting proteins. In: Biotechnology. Ankara: Middle East Technical University. [Google Scholar]
- 32. Wang K, Uppalapati SR, Zhu X, Dinesh-Kumar SP, Mysore KS (2010) SGT1 positively regulates the process of plant cell death during both compatible and incompatible plant-pathogen interactions. Mol Plant Pathol 11: 597–611. PubMed: 20695999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holt BF III, Belkhadir Y, Dangl JL (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science 309: 929–932. doi:10.1126/science.1109977. PubMed: 15976272. [DOI] [PubMed] [Google Scholar]
- 34. Li A, Zhang R, Pan L, Tang L, Zhao G et al. (2011) Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive ratty acid desaturase gene participating in powdery mildew resistance. PLOS ONE 12: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawamura Y, Takenaka S, Hase S, Kubota M, Ichinose Y, et al. (2009) Enhanced defense responses in Arabidopsis induced by the cell wall protein fractions from Pythium oligandrum require SGT1, RAR1, NPR1 and JAR1 . Plant Cell Physiol 50(5): 924–934. doi:10.1093/pcp/pcp044. PubMed: 19304739. [DOI] [PubMed] [Google Scholar]
- 36. Hill-Ambroz K, Webb CA, Matthews AR, Li W, Gill BS et al. (2006) Expression analysis and physical mapping of a cDNA library of Fusarium head blight infected wheat spikes. The Plant Genome (a Supplement to Crop Science) 46: S15–S26. [Google Scholar]
- 37. Gray WM, Muskett PR, Chuang HW, Parker JE (2003) Arabidopsis SGT1b is required for SCFTIR1-mediated auxin response. Plant Cell 15: 1310–1319. doi:10.1105/tpc.010884. PubMed: 12782725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lorenzo O, Solano R (2005b) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8: 532–540. doi:10.1016/j.pbi.2005.07.003. PubMed: 16039901. [DOI] [PubMed] [Google Scholar]
- 39. Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. doi:10.1126/science.280.5366.1091. PubMed: 9582125. [DOI] [PubMed] [Google Scholar]
- 40. Xu Y, Sun Y, Liang W, Huang H (2002) The Arabidopsis AS2 gene encoding a predicted leucine-zipper protein is required for the leaf polarity formation. Acta Bot Sin 44: 1194–1202. [Google Scholar]
- 41. Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink JPre’ M, Atallah M, Champion A, De VM, Pieterse CMJ, et al (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357. doi:10.1104/pp.108.117523. PubMed: 18467450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomma BP, Eggermont K, Tierens KF, Broekaert WF (1999) Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea . Plant Physiol 121: 1093–1102. doi:10.1104/pp.121.4.1093. PubMed: 10594097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kallenbach M, Alagna F, Baldwin IT, Bonaventure G (2010) Nicotiana attenuata SIPK, WIPK, NPR1, and fatty acid-amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway. Plant Physiol 152: 96–106. doi:10.1104/pp.109.149013. PubMed: 19897603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ding L, Xu H, Yi H, Yang L, Kong Z et al. (2011) Resistance to hemi-biotrophic F. Graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLOS ONE 6: e19008. doi:10.1371/journal.pone.0019008. PubMed: 21533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiao J, Jia X, Wang H, Zhao R, Fang Y et al. (2011) A fast-neutron induced chromosome fragment deletion of 3BS in wheat landrace Wangshuibai increased its susceptibility to Fusarium head blight. Chromosome Res 19: 225–234. doi:10.1007/s10577-011-9192-9. PubMed: 21331795. [DOI] [PubMed] [Google Scholar]
- 46. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2-ΔΔCt method. Methods 25: 402-408. doi:10.1006/meth.2001.1262. PubMed: 11846609. [DOI] [PubMed] [Google Scholar]
- 47. Xing L, Wang H, Jiang Z, Ni J, Cao A et al. (2008) Transformation of wheat thaumatin-like protein gene and diseases resistance analysis of the transgenic plants. Acta Agron Sin 34: 349–354. doi:10.1016/S1875-2780(08)60014-0. [Google Scholar]
- 48. Dellaporta SL, Wood J, Hicks JB (1983) A rapid method for DNA extraction from plant tissue. Plant Mol Bion Rep 1: 19–21. [Google Scholar]
- 49. Jakobson I, Peusha H, Timofejeva L, Järve K (2006) Adult plant and seedling resistance to powdery mildew in a Triticum aestivum×Triticum militinae hybrid line. Theor Appl Genet 112: 760–769. doi:10.1007/s00122-005-0181-2. PubMed: 16362813. [DOI] [PubMed] [Google Scholar]
- 50. Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194. doi:10.1046/j.1365-313X.1997.11061187.x. [Google Scholar]
- 51. Jing H, Zhang L, Li H, Shao K, Wu H et al. (2012) Exogenous phytic acid inhibits early growth of wheat seedlings (Triticum aestivum L.) by decreasing superoxide anion derived from NADPH oxidase. Aus J Crops Sci 8: 1221–1227. [Google Scholar]