Abstract
Desmosomes are cell adhesion junctions required for the normal development and maintenance of mammalian tissues and organs such as the skin, skin appendages and the heart. The goal of the present study was to investigate how desmocollins (DSC), transmembrane components of desmosomes, are regulated at the transcriptional level. We hypothesized that differential expression of the Dsc2 and Dsc3 genes is a prerequisite for normal development of skin appendages. We demonstrate that plakoglobin (Pg) in conjunction with Lef-1 differentially regulates the proximal promoters of these two genes. Specifically, we found that Lef-1 acts as a switch activating Dsc2 and repressing Dsc3 in the presence of Pg. Interestingly, we also determined that NFκB pathway components, down-stream effectors of the Eda/EDAR signaling cascade, can activate Dsc2 expression. We hypothesize that Lef-1 and Eda/EDAR/NFκB signaling contribute to a shift in Dsc isoform expression from Dsc3 to Dsc2 in placode keratinocytes. It is tempting to speculate that this shift is required for invasive growth of placode keratinocytes into the dermis, a crucial step in skin appendage formation.
INTRODUCTION
Desmosomes are cell adhesion complexes that are assembled at the plasma membrane where they serve as membrane anchors for intermediate filament (IF) proteins (Cheng et al., 2005; Cheng and Koch, 2004; Dubash and Green, 2011). The importance of this cell junction for organ stability and function is demonstrated by the severe acquired and inherited diseases of the skin, skin appendages (e.g. hair) and the heart that result from impaired desmosome function [e.g. (Amagai and Stanley, 2012; Petrof et al., 2012; Swope et al., 2013)]. These observations in human patients are further supported by the severe skin and heart phenotypes of mice with mutations in genes encoding desmosomal components [e.g. (Chen et al., 2008; Ganeshan et al., 2010; Koch et al., 1998; Koch et al., 1997; Li et al., 2012; Li et al., 2011)]. Desmosomes also contribute to cell sorting, morphogenetic cell movement and the formation of the proper histo-architecture during embryonic development (Cheng and Koch, 2004).
Desmosomes contain transmembrane adhesion molecules (desmosomal cadherins; Desmogleins, DSG; Desmocollins, DSC) and associated plaque proteins [reviewed in (Cheng et al., 2005; Dubash and Green, 2011)]. The main plaque proteins belong either to the armadillo family of structural and signaling proteins (plakoglobin, Pg; plakophilins, Pkp) or the plakin family (desmoplakin, Dp). The mouse genome encodes three Dsc and six Dsg genes. All DSG and DSC proteins are synthesized in the epidermis of the skin and in skin appendages: for example, Dsc2 and Dsc3 are present in the basal and immediate suprabasal layers while Dsc1 is mainly restricted to the granular layer of mouse epidermis [references in (Chen et al., 2008)]. It is thought that the specific complement of DSG and DSC proteins affects the adhesive and potentially the signaling properties of desmosomes [e.g. (Muller et al., 2008; Schmidt and Koch, 2007)]. However, little is known about the gene regulatory mechanisms that control the expression of these desmosomal genes [e.g. (Oshiro et al., 2005; Oshiro et al., 2003; Smith et al., 2004)]. In this study, we focused on the regulation of the Dsc2 and Dsc3 genes; their co-expression in the deep layers of the mouse epidermis suggests that they may play a role in keratinocyte differentiation and skin appendage formation [e.g. (Cheng and Koch, 2004; Chidgey et al., 1997)]. Further, we questioned whether signaling cascades known to play important roles during skin appendage formation affect the expression of Dsc2 and Dsc3, respectively.
Both the WNT and the EDA/EDAR/NFκB pathways have been shown to play critical roles in hair follicle (HF) formation (Schmidt-Ullrich et al., 2001; Zhang et al., 2009). The canonical Wnt cascade ultimately affects target gene expression via beta-catenin/TCF/Lef complexes. Plakoglobin (Pg), which is sequence-related to beta-catenin, has also been shown to act as a transcription factor, both in conjunction and independent of TCF/Lef factors [e.g. (Galichet et al., 2012; Maeda et al., 2004; Shtutman et al., 2002; Simcha et al., 1998; Teuliere et al., 2004; Williamson et al., 2006; Zhurinsky et al., 2000)]. Similar to beta-catenin, Pg does not possess a DNA binding domain, i.e. requires co-factors (such as TCF/Lef factors) to bind to promoter sequences.
It is well established that Pg, independent of beta-catenin signaling, is a key regulator of various cellular processes such as migration (Franzen et al., 2012; Rieger-Christ et al., 2005; Yin et al., 2005), proliferation (Li et al., 2012), apoptosis (Dusek et al., 2007) and gene expression (see above).
Here we demonstrate that Pg, in conjunction with Lef-1, can control Dsc2 and Dsc3 gene expression: In the presence of Lef-1, Pg activates Dsc2 and represses Dsc3 expression. Further, we show that NFκB proteins, the down-stream effectors of EDA/EDAR signaling (Schmidt-Ullrich et al., 2001), activate Dsc2 as well. Both the EDA/EDAR/NFκB and the TCF/Lef signaling play crucial roles in the early steps of HF formation. Our results predict that these two pathways shift expression from Dsc3 to Dsc2 in placode keratinocytes invading the dermis, a process that might be required for effective keratinocyte migration and thus appendage formation. To our knowledge, these results provide previously unreported evidence that the catenin/TCF/Lef and NFκB signaling cascades control Dsc gene expression in keratinocytes.
RESULTS
Desmocollin gene regulation in skin appendage formation
It is thought that changing the molecular composition of desmosomes is one mechanism used by keratinocytes to adapt their cell adhesion system to their specific environment. In the early stages of skin appendage formation, for example, desmocollins (Dsc) are down-regulated in placode keratinocytes invading the dermis (Nanba et al., 2000; Nanba et al., 2001). However, little is known regarding gene regulatory mechanisms that control Dsc gene expression in skin. To begin to identify gene regulatory pathways controlling Dsc gene expression, we cloned and sequenced the proximal promoters of mouse Dsc2 and Dsc3. These two Dsc genes are expressed in the basal layers of the interfollicular epidermis, the cellular compartment which maintains the epidermis and which forms hair follicles (HF).
DNA sequence analysis revealed the presence of putative TCF/Lef (Figure 1a-b) and NFκB (see below) target sites in the Dsc2 and Dsc3 promoters. Considering that the Wnt pathway, which affects gene expression via catenin/TCF/Lef transcription factors, and the NFκB pathways have been shown to play major roles in HF formation, we decided to focus on the role of these two signaling cascades in Dsc gene regulation.
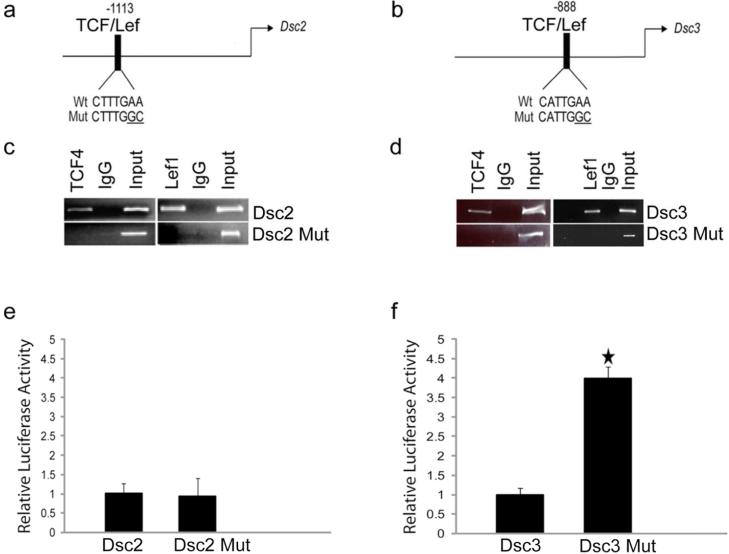
Figure 1. Identification and functional characterization of TCF/Lef factor-binding sites in the Dsc2 and Dsc3 promoters.
(a-b). Schematic representation of putative transcription factor binding sites in the proximal Dsc promoters. The arrows indicate translation start sites (ATG; A is defined as position +1). The DNA sequences of wild type (Wt) and mutant (Mut) transcription factor binding sites are shown. (c-d) ChIP assays demonstrating TCF/Lef binding to the predicted target sites in the Dsc promoters. Note that the point mutations introduced into the TCF/Lef target sequences (Dsc2 Mut, Dsc3 Mut) abrogate binding of the transcription factors. Input, chromatin used for immunoprecipitation; IgG, IP with unspecific IgG. (e-f) Reporter assays in mouse keratinocytes. Note that inactivation of the TCF/Lef binding sites in the Dsc3 construct increases reporter activity significantly. Error bars, standard deviation. Star indicates a statistically significant result (p-value < 0.05).
TCF/Lef regulation of the Dsc2 and Dsc3 promoters
We cloned 2kb of the genomic DNA sequences immediately upstream of the Dsc2 and Dsc3 translation start codons (Fig. 1a-b). These DNA fragments were cloned into a promoter-less Luciferase reporter vector designed to assess transcriptional activity of promoter fragments (reporter assays). Both the Dsc2 and Dsc3 construct showed reporter activity in primary mouse keratinocytes (data not shown). Next, we introduced point mutations abrogating DNA binding of TCF/Lef factors to their target sites into both promoters (Figure 1a-b). The wild type, but not the mutant TCF/Lef sites, showed binding of Lef-1 and TCF4 as demonstrated by chromatin immunoprecipitation (ChIP) assays (Figure 1c-d). TCF-3 behaved identical to TCF4 in all assays performed in this study. Consequently, only the Lef-1 and TCF4 data are shown. All ChIP assays and biochemical experiments were done in canine MDCK cells, while reporter assays and gene expression studies were done in primary mouse keratinocytes. MDCK cells express Dsc2 and Dsc3 (data not shown). Due to differences in the promoter DNA sequences of dog and mouse and the use of species-specific antibodies, we were able to investigate protein-DNA and protein-protein interactions in MDCK cells without interference of endogenous canine genes and proteins.
Next, we assessed whether loss of functional TCF/Lef sites affected reporter activity in primary keratinocytes. As shown in Figure 1, Dsc2 promoter activity was unchanged when wild type (Wt) and mutant (Mut) reporter constructs lacking a functional TCF/Lef site were compared (Figure 1e). Dsc3 promoter activity was significantly increased in the mutant construct, indicating that this site is repressive (Figure 1f).
Beta-catenin expression has no effect on the proximal Dsc2 and Dsc3 promoters
TCF/Lef sites are the primary target sites of beta-catenin/TCF/Lef transcription factor complexes, the principal effectors of the Wnt signaling pathway. ChIP assays demonstrated that beta-catenin can bind to the Dsc3 but not the Dsc2 promoter (Supplemental Figure, these experiments were done in MDCK cells expressing TCF4 as a co-factor, data not shown). In order to determine whether beta-catenin can influence Dsc promoter activity, we transfected the Dsc reporter constructs with different combinations of TCF/Lef factors into keratinocytes. We did not observe a statistically significant effect of these beta-catenin/TCF/Lef factors on Dsc2 or Dsc3 reporter activity (Supplemental Figure). A slight repression of Dsc3 reporter activity by beta-catenin and Lef-1 was often observed but did not reach statistical significance.
Plakoglobin (Pg) is a key regulator of Dsc2 and Dsc3 promoter activity
Pg has been shown to regulate gene expression in keratinocytes and other epithelial cell types (see references in the INTRODUCTION section). To determine whether Pg can affect Dsc promoter activity, we first assessed the ability of this protein to bind the Dsc2 and Dsc3 promoter fragments. As shown in Figure 2a-b, Pg binds to the Dsc2 promoter but not the Dsc3 promoter in the presence of Lef-1. Reporter assays demonstrated that Pg can affect both promoters, although its effects are dependent on Lef-1 (Figure 2c-d); in the presence of Lef-1, Pg activates the Dsc2 promoter, while Dsc3 promoter activation occurs in the absence of ectopic TCF/Lef expression. These results suggest that Lef-1 can act as a switch that shifts activity from the Dsc3 to the Dsc2 promoter in the presence of Pg.
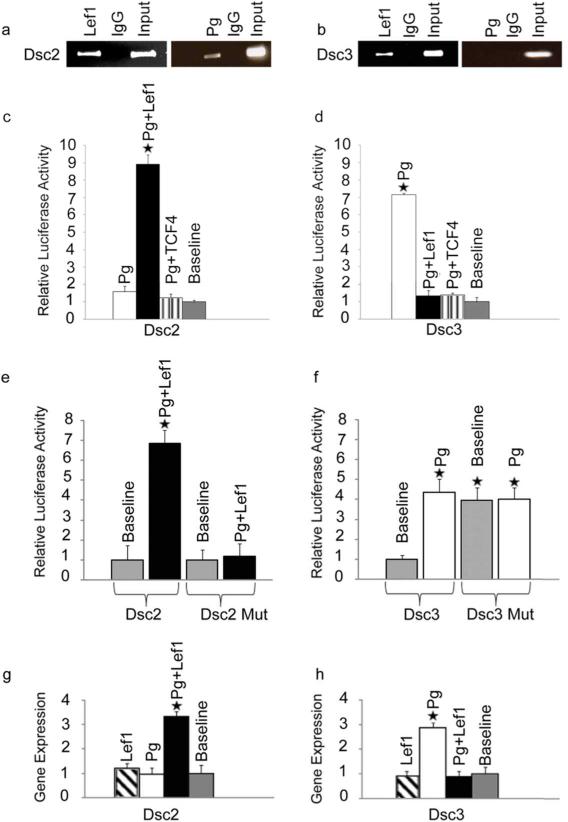
Figure 2. Effects of Pg on Dsc reporter activities.
(a-b). ChIP assays demonstrating direct binding of Pg to the Dsc2 promoter in the presence of Lef-1. Note that Pg does not bind the Dsc3 promoter in the presence of Lef-1 (b) or TCF4 (data not shown). Input, chromatin used for immunoprecipitation; IgG, IP with unspecific IgG. (c-f) Reporter assays in keratinocytes. (c) Co-expression of Pg and Lef-1 dramatically increases Dsc2 reporter activity while all other combinations of Pg and TCF/Lef factors have no effect. (d) The Dsc3 promoter is activated in the presence of Pg. This activation is reversed in cells co-expressing Pg and TCF4 or Lef-1. (e) Dsc2 promoter activation by Pg and Lef-1 is dependent on the presence of a functional TCF/Lef binding site. (f) Pg over-expression and loss of a functional TCF/Lef binding site increase Dsc3 reporter activity to a similar extend. Note that expression is normalized to the baseline expression of the WT promoter (set to 1). (g-h) Endogenous Dsc gene expression in keratinocytes transfected with different combinations of Pg and TCF/Lef factors. Error bars, standard deviation. Stars indicate statistically significant results (p-value < 0.05).
Neither Lef-1, TCF3 nor TCF-4 had any effect on the Dsc2 or Dsc3 promoter in single transfection experiments (data not shown). These results are expected given that keratinocytes express endogenous TCF/Lef factors but show very low cytoplasmic and nuclear Pg levels (Williamson et al., 2006), conditions that favor low Dsc2 and Dsc3 reporter activity.
As shown in Figure 2e, Dsc2 promoter activation by Pg and Lef-1 was dependent on the presence of a functional TCF/Lef binding site. In the case of the Dsc3 promoter, we found no additive or synergistic effects of Pg and the TCF/Lef mutation (Figure 2f). These findings are consistent with the hypothesis that Pg asserts its activating effect on Dsc2 via the TCF/Lef site, while this was not the case for the Dsc3 promoter.
We next assessed the effects of Pg and Lef-1 co-expression on the endogenous Dsc2 and Dsc3 gene activity in primary keratinocytes by quantitative RT-PCR. As shown in Figures 2g-h, both genes responded to ectopic Pg/Lef expression as predicted by our reporter assays confirming the validity of our conclusions for the regulation of these genes in keratinocytes.
In order to gain further insights in the mechanisms by which Pg and Lef-1 control Dsc gene expression, we assessed whether Pg can bind to Lef-1. As shown in Figure 3a, Lef-1 and Pg can form a complex as shown by co-immunoprecipitation experiments using Lef-1 antibodies. Most interestingly, ChIP competition experiments suggested that the interaction of Pg and Lef-1 interferes with the ability of Lef-1 to bind to the Dsc3 promoter (Figure 3b). Considering that TCF/Lef-1 complexes can act as transcriptional repressors [e.g. (Hoverter and Waterman, 2008)], these results raise the possibility that Pg could activate Dsc3 expression by interfering with the binding of a TCF/Lef repressor complex to the Dsc3 promoter. Further support for this hypothesis is provided by Western blot experiments (Figure 3c-d) demonstrating that ectopically expressed Pg can interfere with the nuclear accumulation of Lef-1 and thus potentially suppress the formation of a repressor complex at the Dsc3 promoter. On the other hand, Lef-1 is required to shuttle Pg into the cell nucleus, where it activates the Dsc2 promoter as shown in Figure 3d-e. The data summarized above demonstrate that Lef-1 and Pg localize to the nucleus, which is predicted to lead to an activation of the Dsc2 gene and a suppression of the Dsc3 gene.
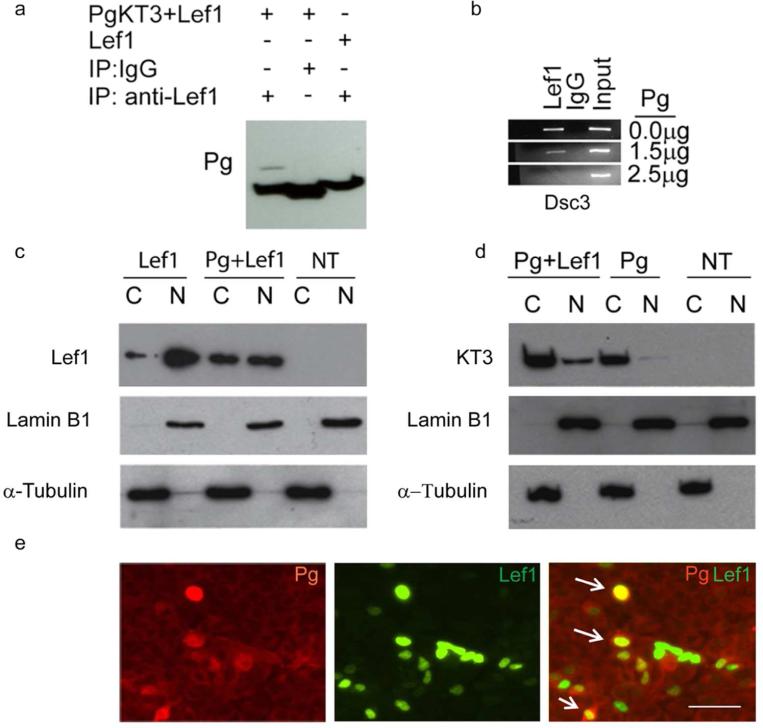
Figure 3. Pg co-localizes in the nucleus with Lef1 and disrupts TCF/Lef transcription factor binding to the Dsc3 promoter.
(a) Co-Immunoprecipitation (Co-IP) assays demonstrating an interaction between Pg and Lef-1. (b) ChIP assays demonstrating that increasing amounts of Pg (measured in g plasmid transfected) interfere with the binding of Lef-1 to the Dsc3 promoter. Input, chromatin used for immunoprecipitation; IgG, IP with unspecific IgG. (c, d) Western blot analysis of MDCK cells transfected with Pg and Lef-1 (transfection constructs shown on top; NT, not transfected). The nuclear (N) and cytoplasmic (C) distribution of the proteins is shown. Antibodies used to detect Lef-1 and the KT3-tagged plakoglobin construct are shown on the left sides of the blots. Our Lef-1 antibody does not detect endogenous Lef-1 expression in MDCK cells. Lamin B1 (nuclear fraction) and α-tubulin (cytoplasmic fraction) antibodies were used as controls. (e) Immunofluorescence microscopy of MDCK cells transfected with Pg and Lef-1. The antibodies used for staining are indicated. Note the nuclear co-localization of Pg and Lef-1 in several cells (arrows). Bar, 50 μm
Topology of Lef-1 and Dsc3 expression in developing hair follicles is mutually exclusive
Our results thus far suggest that Lef-1 can act as a switch between Dsc2 and Dsc3 expression and that the presence of Lef-1 in keratinocytes can suppress the Dsc3 gene. We therefore compared the distribution of DSC3 and Lef-1 in transgenic mice expressing a nuclear LacZ reporter under the control of the Dsc3 promoter (Dsc3-LacZ mice). These animals were designed to identify cells and tissues that express low levels of Dsc3, or cells in which antigen masking prevented protein detection via antibodies. In all developmental stages and tissues examined thus far, DSC3 antibody staining and LacZ transgene activity perfectly overlapped (data not shown).
We conducted whole-mount β-galactosidase staining experiments using Dsc3-LacZ embryos (Figure 4a). At E15.5, β-galactosidase activity was prominent in whisker pads (vibrissae follicles), in mammary gland buds and in developing hair follicles over the entire body surface (Fig. 4a, and data not shown). Strikingly, the overall DSC3-LacZ expression pattern appeared similar to the staining patterns observed in transgenic embryos of the same age that expressed a TCF/Lef-regulated promoter driving a LacZ reporter [Figure 4b; Bat-Gal, (Maretto et al., 2003)]. However, a histochemical analysis revealed that the expression patterns of Wnt pathway components (Lef-1; TCF-3/-4; data not shown) and the DSC3-LacZ transgene did not overlap at the cellular level. In fact, Wnt activity (Lef-1 expression) and Dsc3 expression were mutually exclusive (Figure 4c-d). We observed that LacZ-positive (Dsc3-expressing) cells were found in the suprabasal layer on top of the newly forming hair follicles. Lef-1, on the other hand, was observed in the leading edge of keratinocytes growing down into the dermis and throughout the basal cell layer. These findings were confirmed by staining E16.5 wild type mouse epidermis with Lef-1 and DSC3 antibodies (Figure 4e-h). Strong Lef-1 antibody staining correlated with reduced or absent staining for DSC3. Unfortunately, we were not able to assess the distributions of DSC2, since antibodies that recognize mouse DSC2 are not available. Pg antibodies stained cell-cell borders in placode keratinocytes (data not shown). Nuclear Pg staining was not observed, possibly due to epitope masking or low nuclear Pg levels.
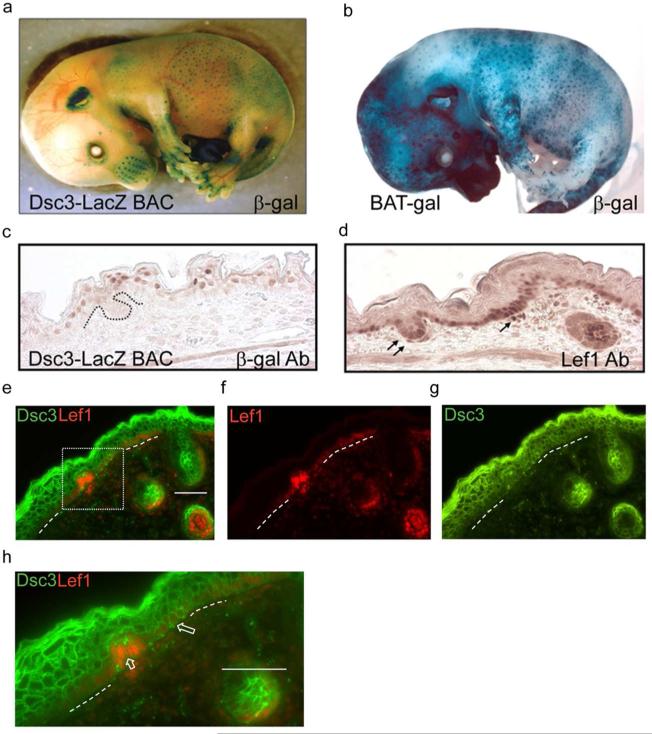
Figure 4. Wnt activity and Dsc3 expression during mouse appendage development.
(a). Whole mount in-situ staining for beta-galactosidase (β-gal) activity of a transgenic mouse (Dsc3-LacZ BAC, E15.5) expressing β-gal under the control of the Dsc3 promoter. Note that whisker pads, hair follicles and mammary glands (not shown) are strongly stained. (b) Whole mount β-gal staining of a BAT-gal transgenic mouse (Maretto et al., 2003) at E15.5. Note the similar staining patterns of the transgenic mice shown in a. and b. (c.) Immunohistochemistry staining of a skin section from a Dsc3-LacZ BAC mouse at E15.5 with (c) β-gal antibodies and (d) Lef-1 antibodies. The LacZ transgene contains a nuclear localization signal. Note that β-gal and Lef-1 expression appears to be mutually exclusive. Arrows point to Lef-1 expressing cells in a placode and a dermal papilla of a forming hair follicle. (e-g) Immunofluorescence staining of a developing hair follicle in wild type mouse skin at E16.5 with antibodies against Lef-1 and DSC3. The area demarcated by the dotted box is shown at higher magnification in panel (h). (h) The large arrow points to DSC3 positive keratinocytes in the basal cell layer. The short arrow points to DSC3-negative and Lef-1-positive cells in a forming hair placode. Dotted lines in (e-h), basement membrane area._Bar, 50μm
NFκB regulation of the Dsc2 gene
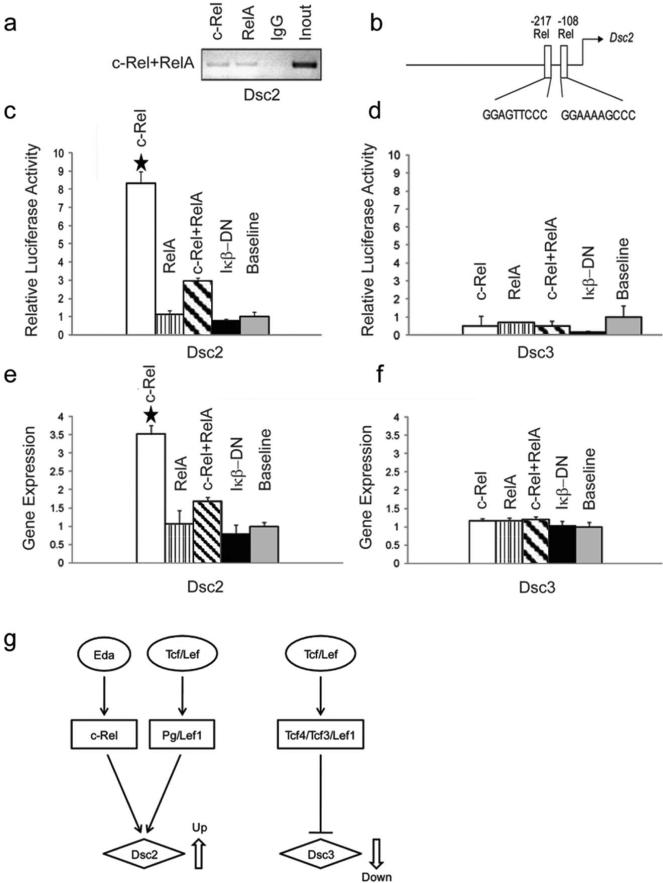
Our initial analysis of the Dsc promoters indicated the presence of putative NFκB (Rel) binding sites in both Dsc promoters. We thus conducted ChIP assays and reporter assays to determine the effect of Rel factors and a dominant-negative construct blocking NFκB signaling (IκB-DN) on Dsc promoter activity. The results shown in Figure 5a-c demonstrate that c-Rel specifically binds to and activates the proximal Dsc2 promoter whereas we did not observe any effect of the NFκB factors on the proximal Dsc3 promoter (Figure 5d). We then transfected primary keratinocytes with the NFκB factors and assessed endogenous Dsc gene expression (Figure 5e-f). We confirmed the specific activation of the endogenous Dsc2 gene by ectopic expression of c-Rel.
Figure 5. Effects of NFκB transcription factors on the proximal Dsc2 and Dsc3 promoters.
(a) ChIP assays demonstrating that both c-Rel and Rel A bind to a Dsc2 promoter fragment that contains two predicted Rel binding sites. Input, chromatin used for immunoprecipitation; IgG, IP with unspecific IgG. (b) Schematic representation of putative NFκB transcription factor binding sites in the proximal Dsc2 promoter. The arrows indicate translation start sites (ATG; A is defined as position +1). (c) c-Rel dramatically increases the reporter activity of the Dsc2 construct while Rel A did not. The IκB_DN construct encodes a dominant-negative inhibitor of the NFκB pathway, i.e. this construct can block endogenous NFκB activity. The promoter activities in the absence of ectopically expressed transcription factors are defined as relative expression level 1. (d) The Dsc3 reporter construct does not show any significant changes in activity in response to NFκB transcription factors. (e-f) Endogenous Dsc gene expression in mouse keratinocytes. (e) c-Rel ectopic expression significantly increases endogenous Dsc2 expression. (f) None of the NFκB transcription factors affects endogenous Dsc3 expression. (g) Simplified model of signaling pathways active in hair follicle placodes and their proposed effects on Dsc2 and Dsc3 gene expression. Note that open arrows symbolize up- and down-regulation of gene expression, respectively. Error bars, standard deviation. Stars indicate statistically significant results (p-value < 0.05).
DISCUSSION
It has been shown that deregulated expression or loss of desmosomal cadherins, including Dsc, can lead to abnormal differentiation of the epidermis and defects in hair follicles (HF) [e.g. (Chidgey et al., 2001; Elias et al., 2001; Hardman et al., 2005; Merrittet al., 2002)], suggesting that tight control of Dsc gene expression is required for normal epidermal development and homeostasis. In the present study, we focused on the regulation of Dsc2 and Dsc3, the two desmocollins expressed in the deep epidermis, the compartment that maintains the skin and which plays a crucial role in skin appendage development.
Little is known regarding gene regulatory pathways that control the expression of Dsc genes in processes such as epidermal differentiation and hair follicle (HF) formation. Epidermal appendage formation requires extensive remodeling of cell adhesion systems, including desmosomes (Kurzen et al., 1998). The first step in the development of these appendages is placode formation. Keratinocytes in these structures segregate from the surrounding epidermis and then begin to invade the dermis. Nanba and colleagues have demonstrated that DSCs are down-regulated both in the hair and mammary gland placodes (Nanba et al., 2000; Nanba et al., 2001). However, the antigen specificity of the antibody used by these authors was not determined, i.e. it was not known which of the DSC proteins was actually down-regulated. Given the results presented in the present study, it is likely that the main desmosomal cadherin isoform down-regulated in this process is Dsc3.
The expression of classical cadherins is also switched during hair follicle formation. E-cadherin is down-regulated in HF placodes while P-cadherin is upregulated. Further, it was shown that forced expression of E-cadherin inhibits HF formation (Jamora et al., 2003), demonstrating that control of cadherin isoform expression is crucial for HF formation.
Given our results, it is tempting to speculate that an analogous switch occurs from Dsc3 (TCF/Lef factors-mediated repression) to Dsc2 (Pg/Lef-1-mediated induction) in placode keratinocytes (Figure 5g). Unfortunately, due to a lack of appropriate antibodies, we currently do not have the tools required to assess DSC2 expression in mouse epidermis. Nevertheless, expression studies in human embryonic epidermis indicated increased DSC2 and reduced DSC3 expression in bulbous hair pegs, suggesting that the above postulated switch from Dsc3 to Dsc2 expression is likely to occur in mammalian skin (Kurzen et al., 1998).
Pg appears to activate both the Dsc2 and the Dsc3 promoter. Previous studies have suggested that Pg can signal by changing the levels of signaling active beta-catenin in cells. Our experiments failed to demonstrate a role of beta-catenin in regulating the two Dsc genes, suggesting that the signaling we observed is a specific function of Pg.
The Dsc2 promoter requires Lef-1 for activation and this regulation is dependent on the TCF/Lef binding site in the Dsc2 promoter. Interestingly, this effect is specific for Lef-1 since TCF-3 and TCF-4 do not appear to be able to substitute for Lef-1. It is noteworthy that Lef-1 expression facilitates nuclear accumulation of Pg, a prerequisite for activation of the Dsc2 gene.
Pg activated the Dsc3 promoter without direct binding. Interestingly, ectopic expression of TCF/Lef factors completely blocked Pg-mediated activation of the Dsc3 promoter. This suggests that Pg interferes with the activity of a repressor complex-containing TCF/Lef factors that inhibits the Dsc3 promoter. A likely mechanism that could explain this observation is that binding of Pg to TCF/Lef proteins causes a depletion of the TCF/Lef pool; thus preventing these factors from forming an inhibitory complex on the Dsc3 promoter.
Previous experiments in mouse models suggested that Pg is not required for the formation of hair follicles (Li et al., 2012; Teuliere et al., 2004). These results suggest the existence of alternative mechanisms that can regulate a switch of Dsc isoforms, specifically the activation of Dsc2. NFκB signaling might be such an alternative mechanism. Our experiments revealed a role of c-Rel, a NFκB protein, in activating the Dsc2 gene. NFκB proteins are downstream effectors of the EDA/EDAR/NFκB pathway, which interact with the Wnt pathway, a process crucial for HF formation during embryogenesis (Zhang et al., 2009). The observation that both catenin/TCF/Lef and NFκB signaling are required for pelage hair development, and our in-vitro data demonstrating differential regulation of Dsc genes by these two pathways, suggest the possibility that both signaling cascades partially function via modulating desmosomal cell adhesion.
Based on the observation summarized above, we suggest the following mechanisms for the regulation of Dsc2 and Dsc3 gene expression during the early stages of skin appendage formation: Wnt activation in the early placode leads to an increase accumulation of Lef1 in placodes. Lef-1, potentially in conjunction with other co-repressors (Arce et al., 2009; Hoverter and Waterman, 2008), binds to the Dsc3 promoter and inhibits expression, thus leading to a reduction in the desmosomal adhesion receptors present at the plasma membrane. In turn, this could lead to a transient increase of cytoplasmic Pg, which is usually bound to the carboxy-terminal domain of DSC3. Given the abundance of Lef-1 in placode keratinocytes, Pg would then form a transcription complex with Lef-1 and activate the Dsc2 promoter. This chain of events would lead to a shift from DSC3 to DSC2 as the main desmocollin synthesized in placode keratinocytes, which would be consistent with the proposed distribution of these two proteins in skin placodes [our data and (Kurzen et al., 1998)]. Activation of the EDA/EDAR pathway, via its NFκB effectors, would then further facilitate the shift from DSC3 to DSC2. It is possible that this change from DSC3 to DSC2 is better suited to support invasively keratinocytes growth. In this context, it is noteworthy that we have previously shown in a mouse model that invasive growth of skin squamous cell carcinoma (SCC) is associated with a specific loss of Dsc3 expression in tumor cells (Chen et al., 2011). Similar results have also been reported in other types of cancer, such as breast cancer (Klus et al., 2001).
Further experiments will be required to unequivocally prove this model. It is tempting to speculate that changes in desmosomal cell adhesion might affect the signaling pool of plakoglobin and thus, in a feedback loop, control expression of genes that encode central desmosomal components. This represents a previously unreported and exciting concept that can now be tested in-vivo.
MATERIALS AND METHODS
Animal protocols
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Colorado Denver (UC Denver).
Generation of Dsc promoter constructs and luciferase reporter assays
2kb of the promoter sequences immediately upstream of the Dsc2 and Dsc3 translation start codons were cloned into pGL3-basic vector (Promega, Madison, WI), which contains a promoter-less luciferase reporter cassette. TCF/Lef-1 mutations were generated by PCR using primers DSC2-5F/5R and DSC3-2F/2R (Supplemental Table). The following expression vectors were used: CMV-βgal (Dennis Roop, UC Denver); pcDNA1.1p65 and pcDNA-Iκβ-DN (Rune Toftgård, Karolinska Institute, Sweden); pcmv4c-Rel (Warner Greene, University of California, San Francisco) (Doerre et al., 1993); pCS2 ΔNPβ-catenin (Pamela Cowin, New York University) (Imbert et al., 2001); pRcCMV plakoglobin expression vector tagged with a KT3 epitope (Ansgar Smith ,University of Marburg, Germany); pCS2-hLef-1 (Rolf Kemler, Max-Planck Institute, Freiburg, Germany) (Huber et al., 1996); and pGLOWMYC-hTcf-4 (Hans Clevers, University Hospital, Utrecht, The Netherlands) (Korinek et al., 1997). Plasmids were transiently transfected into the canine kidney epithelial cell line MDCK (ATCC CCL-34) and mouse keratinocytes (MPEK-BL6, Cellntec, Bern, Switzeland), using the Nucleofector Technology (Lonza, Walkersville, MD). Reporter activities were measured with the “Chemiluminescent Reporter Gene Assay System” (Applied Biosystems, Bedford, MA), using a ‘Glomax Multi Detection System (Promega, Madison, WI.). The results shown represent average expression levels from three independent experiments, each performed in triplicate (error bars in all figures: standard deviations, p-values<0.05 were considered statistically significant).
Chromatin immunoprecipitation (ChIP) experiments
ChIP assays were done essentially as described (Koster et al., 2007). The following antibodies were used: Pg (Cell Signaling, Boston, MA), β-catenin (Santa Cruz Biotechnology, Santa Cruz, CA), Lef1 (Millipore) and TCF4 (Millipore). The following primers (Supplemental Table) were used for QRT-PCR: Dsc2-4F/4R (Rel-binding site in Dsc2 promoter), DSC2-6F/6R (TCF/Lef binding site in Dsc2 promoter) and DSC3-3F/3R (TCF/Lef binding site in Dsc3 promoter). The data shown are based on three independent experiments.
Generation of Dsc3-LacZ transgenic mice
The BAC vector RP23-290M4 (BACPAC Resource, Children's Hospital Oakland Research Institute) which contains the entire mouse Dsc3 gene, including 29kb of 5’ upstream and 90kb of 3’ downstream sequences, was used to generate the Dsc3-LacZ transgene. A promoter-less LacZ cassette (NLS-LacZ-PA cassette; provided by Dr. Ming-Jer Tsai, Baylor College of Medicine) was inserted into exon 1 of the Dsc3 gene immediately downstream of the start codon. The LacZ cassette contains a nuclear localization sequence, i.e. transgenic mice can be identified by the detection of β-galactosidase activity in nuclei.
Immunostaining, β-galactosidase staining, Western blotting and Co-IP
All experiments were done following standard protocols. The following antibodies were used: Lef1 (Cell Signaling), KT3 antibody (Ansgar Schmidt, University of Marburg); normal IgG (Millipore, Billerica, MA), Lamin B1 (Santa Cruz; Biotechnology, Santa Cruz, CA.), α-tubulin (Sigma, Saint-Louis, MO), DSC3 (Cheng et al., 2004), plakoglobin (Firtzgerald, North Acton, MA), β-galactosidase (a gift from Dennis Roop, UC Denver); HRP conjugated and biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA) Alexa Fluor coupled secondary antibodies (Invitrogen, Grand Island, NY). Antibody binding was detected and quantified as described (Koch et al., 2000).
Quantitative Real Time RT-PCR
Real time RT-PCR was performed using a LightCycler 480 from Roche (Indianapolis, Indiana) following manufacturer's recommendations. Dsc2 and Dsc3 cDNA were amplified with “assay-on-demand probes” Mm00516355_m1 and Mm00492270_m1, respectively from Applied Biosystems (Bedford, MA). A GAPDH probe set from Applied Biosystems was used as an internal control. The data shown are based on three independent experiments
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Charlene O'Shea for expert technical assistance, Maranke Koster and Jason Dinella for critical reading of the manuscript, and Sarah Millar (University of Pennsylvania) for providing Bat-Gal mice with permission from Stefano Piccolo (University of Padova, Italy). Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number 5RO1AR053892. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- DSG
Desmoglein
- DSC
Desmocollin
- HF
Hair Follicle
- Pg
Plakoglobin
- ChIP
Chromatin Immunoprecipitation
Footnotes
The authors state no conflict of interest.
REFERENCES
- Amagai M, Stanley JR. Desmoglein as a target in skin disease and beyond. J Invest Dermatol. 2012;132:776–84. doi: 10.1038/jid.2011.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce L, Pate KT, Waterman ML. Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC cancer. 2009;9:159. doi: 10.1186/1471-2407-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Den Z, Koch PJ. Loss of desmocollin 3 in mice leads to epidermal blistering. J Cell Sci. 2008;121:2844–9. doi: 10.1242/jcs.031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, O'Shea C, Fitzpatrick JE, Koster MI, Koch PJ. Loss of desmocollin 3 in skin tumor development and progression. Molecular carcinogenesis. 2011 doi: 10.1002/mc.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Den Z, Koch PJ. Desmosomal cell adhesion in mammalian development. Eur J Cell Biol. 2005;84:215–23. doi: 10.1016/j.ejcb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Cheng X, Koch PJ. In vivo function of desmosomes. J Dermatol. 2004;31:171–87. doi: 10.1111/j.1346-8138.2004.tb00654.x. [DOI] [PubMed] [Google Scholar]
- Cheng X, Mihindukulasuriya K, Den Z, Kowalczyk AP, Calkins CC, Ishiko A, et al. Assessment of splice variant-specific functions of desmocollin 1 in the skin. Mol Cell Biol. 2004;24:154–63. doi: 10.1128/MCB.24.1.154-163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey M, Brakebusch C, Gustafsson E, Cruchley A, Hail C, Kirk S, et al. Mice lacking desmocollin 1 show epidermal fragility accompanied by barrier defects and abnormal differentiation. J Cell Biol. 2001;155:821–32. doi: 10.1083/jcb.200105009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey MA, Yue KK, Gould S, Byrne C, Garrod DR. Changing pattern of desmocollin 3 expression accompanies epidermal organisation during skin development. DevDyn. 1997;210:315–27. doi: 10.1002/(SICI)1097-0177(199711)210:3<315::AID-AJA11>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Doerre S, Sista P, Sun SC, Ballard DW, Greene WC. The c-rel protooncogene product represses NF-kappa B p65-mediated transcriptional activation of the long terminal repeat of type 1 human immunodeficiency virus. Proc Natl Acad Sci U S A. 1993;90:1023–7. doi: 10.1073/pnas.90.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubash AD, Green KJ. Desmosomes. Current biology : CB. 2011;21:R529–31. doi: 10.1016/j.cub.2011.04.035. [DOI] [PubMed] [Google Scholar]
- Dusek RL, Godsel LM, Chen F, Strohecker AM, Getsios S, Harmon R, et al. Plakoglobin deficiency protects keratinocytes from apoptosis. J Invest Dermatol. 2007;127:792–801. doi: 10.1038/sj.jid.5700615. [DOI] [PubMed] [Google Scholar]
- Elias PM, Matsuyoshi N, Wu H, Lin C, Wang ZH, Brown BE, et al. Desmoglein isoform distribution affects stratum corneum structure and function. J Cell Biol. 2001;153:243–9. doi: 10.1083/jcb.153.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen CA, Todorovic V, Desai BV, Mirzoeva S, Yang XJ, Green KJ, et al. The desmosomal armadillo protein plakoglobin regulates prostate cancer cell adhesion and motility through vitronectin-dependent Src signaling. PloS one. 2012;7:e42132. doi: 10.1371/journal.pone.0042132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galichet A, Strauss C, Sayar BS, Howald D, Mueller EJ. Desmosomal cadherins crosstalk to the nucleus via a novel potent transcriptional regulator of the Wnt pathway: Plakoglobin (PG, JUP, γ-catenin). Journal Investigative Dermatology. 2012:S60. [Google Scholar]
- Ganeshan R, Chen J, Koch PJ. Mouse models for blistering skin disorders. Dermatol Res Pract. 2010;2010:584353. doi: 10.1155/2010/584353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman MJ, Liu K, Avilion AA, Merritt A, Brennan K, Garrod DR, et al. Desmosomal cadherin misexpression alters beta-catenin stability and epidermal differentiation. MolCell Biol. 2005;25:969–78. doi: 10.1128/MCB.25.3.969-978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoverter NP, Waterman ML. A Wnt-fall for gene regulation: repression. Science signaling. 2008;1:e43. doi: 10.1126/scisignal.139pe43. [DOI] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mechanisms of development. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- Imbert A, Eelkema R, Jordan S, Feiner H, Cowin P. Delta N89 beta-catenin induces precocious development, differentiation, and neoplasia in mammary gland. J Cell Biol. 2001;153:555–68. doi: 10.1083/jcb.153.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–22. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klus GT, Rokaeus N, Bittner ML, Chen Y, Korz DM, Sukumar S, et al. Down-regulation of the desmosomal cadherin desmocollin 3 in human breast cancer. IntJOncol. 2001;19:169–74. doi: 10.3892/ijo.19.1.169. [DOI] [PubMed] [Google Scholar]
- Koch PJ, de Viragh PA, Scharer E, Bundman D, Longley MA, Bickenbach J, et al. Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J Cell Biol. 2000;151:389–400. doi: 10.1083/jcb.151.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch PJ, Mahoney MG, Cotsarelis G, Rothenberger K, Lavker RM, Stanley JR. Desmoglein 3 anchors telogen hair in the follicle. J Cell Sci. 1998;111(Pt 17):2529–37. doi: 10.1242/jcs.111.17.2529. [DOI] [PubMed] [Google Scholar]
- Koch PJ, Mahoney MG, Ishikawa H, Pulkkinen L, Uitto J, Shultz L, et al. Targeted disruption of the pemphigus vulgaris antigen (desmoglein 3) gene in mice causes loss of keratinocyte cell adhesion with a phenotype similar to pemphigus vulgaris. J Cell Biol. 1997;137:1091–102. doi: 10.1083/jcb.137.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, et al. p63 induces key target genes required for epidermal morphogenesis. P Natl Acad Sci USA. 2007;104:3255–60. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzen H, Moll I, Moll R, Schafer S, Simics E, Amagai M, et al. Compositionally different desmosomes in the various compartments of the human hair follicle. Differentiation. 1998;63:295–304. doi: 10.1046/j.1432-0436.1998.6350295.x. [DOI] [PubMed] [Google Scholar]
- Li D, Zhang W, Liu Y, Haneline LS, Shou W. Lack of plakoglobin in epidermis leads to keratoderma. J Biol Chem. 2012;287:10435–43. doi: 10.1074/jbc.M111.299669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Swope D, Raess N, Cheng L, Muller EJ, Radice GL. Cardiac tissue-restricted deletion of plakoglobin results in progressive cardiomyopathy and activation of {beta}-catenin signaling. Mol Cell Biol. 2011;31:1134–44. doi: 10.1128/MCB.01025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda O, Usami N, Kondo M, Takahashi M, Goto H, Shimokata K, et al. Plakoglobin (gamma-catenin) has TCF/LEF family-dependent transcriptional activity in beta-catenin-deficient cell line. Oncogene. 2004;23:964–72. doi: 10.1038/sj.onc.1207254. [DOI] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, et al. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. ProcNatlAcadSciUSA. 2003;100:3299–304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt AJ, Berika MY, Zhai W, Kirk SE, Ji B, Hardman MJ, et al. Suprabasal desmoglein 3 expression in the epidermis of transgenic mice results in hyperproliferation and abnormal differentiation. MolCell Biol. 2002;22:5846–58. doi: 10.1128/MCB.22.16.5846-5858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EJ, Williamson L, Kolly C, Suter MM. Outside-in signaling through integrins and cadherins: a central mechanism to control epidermal growth and differentiation? J Invest Dermatol. 2008;128:501–16. doi: 10.1038/sj.jid.5701248. [DOI] [PubMed] [Google Scholar]
- Nanba D, Hieda Y, Nakanishi Y. Remodeling of desmosomal and hemidesmosomal adhesion systems during early morphogenesis of mouse pelage hair follicles. J Invest Dermatol. 2000;114:171–7. doi: 10.1046/j.1523-1747.2000.00842.x. [DOI] [PubMed] [Google Scholar]
- Nanba D, Nakanishi Y, Hieda Y. Changes in adhesive properties of epithelial cells during early morphogenesis of the mammary gland. Development, growth & differentiation. 2001;43:535–44. doi: 10.1046/j.1440-169x.2001.00596.x. [DOI] [PubMed] [Google Scholar]
- Oshiro MM, Kim CJ, Wozniak RJ, Junk DJ, Munoz-Rodriguez JL, Burr JA, et al. Epigenetic silencing of DSC3 is a common event in human breast cancer. Breast Cancer Res. 2005;7:R669–R80. doi: 10.1186/bcr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro MM, Watts GS, Wozniak RJ, Junk DJ, Munoz-Rodriguez JL, Domann FE, et al. Mutant p53 and aberrant cytosine methylation cooperate to silence gene expression. Oncogene. 2003;22:3624–34. doi: 10.1038/sj.onc.1206545. [DOI] [PubMed] [Google Scholar]
- Petrof G, Mellerio JE, McGrath JA. Desmosomal genodermatoses. Br J Dermatol. 2012;166:36–45. doi: 10.1111/j.1365-2133.2011.10640.x. [DOI] [PubMed] [Google Scholar]
- Rieger-Christ KM, Ng L, Hanley RS, Durrani O, Ma H, Yee AS, et al. Restoration of plakoglobin expression in bladder carcinoma cell lines suppresses cell migration and tumorigenic potential. Br J Cancer. 2005;92:2153–9. doi: 10.1038/sj.bjc.6602651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Aebischer T, Hulsken J, Birchmeier W, Klemm U, Scheidereit C. Requirement of NF-kappaB/Rel for the development of hair follicles and other epidermal appendices. Development. 2001;128:3843–53. doi: 10.1242/dev.128.19.3843. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Koch PJ. Desmosomes: just cell adhesion or is there more? Cell Adh Migr. 2007;1:28–32. doi: 10.4161/cam.1.1.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Oren M, Levina E, Ben-Ze'ev A. PML is a target gene of beta-catenin and plakoglobin, and coactivates beta-catenin-mediated transcription. Cancer Res. 2002;62:5947–54. [PubMed] [Google Scholar]
- Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, et al. Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. J Cell Biol. 1998;141:1433–48. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Zhu K, Merritt A, Picton R, Youngs D, Garrod D, et al. Regulation of desmocollin gene expression in the epidermis: CCAAT/enhancer-binding proteins modulate early and late events in keratinocyte differentiation. The Biochemical journal. 2004;380:757–65. doi: 10.1042/BJ20040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swope D, Li J, Radice GL. Beyond cell adhesion: The role of armadillo proteins in the heart. Cellular signalling. 2013;25:93–100. doi: 10.1016/j.cellsig.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuliere J, Faraldo MM, Shtutman M, Birchmeier W, Huelsken J, Thiery JP, et al. beta-catenin-dependent and -independent effects of DeltaN-plakoglobin on epidermal growth and differentiation. Molecular and cellular biology. 2004;24:8649–61. doi: 10.1128/MCB.24.19.8649-8661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson L, Raess NA, Caldelari R, Zakher A, de Bruin A, Posthaus H, et al. Pemphigus vulgaris identifies plakoglobin as key suppressor of c-Myc in the skin. EMBO J. 2006;25:3298–309. doi: 10.1038/sj.emboj.7601224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T, Getsios S, Caldelari R, Kowalczyk AP, Muller EJ, Jones JC, et al. Plakoglobin suppresses keratinocyte motility through both cell-cell adhesion-dependent and -independent mechanisms. ProcNatlAcadSciUSA. 2005;102:5420–5. doi: 10.1073/pnas.0501676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, et al. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Developmental cell. 2009;17:49–61. doi: 10.1016/j.devcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze'ev A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Mol Cell Biol. 2000;20:4238–52. doi: 10.1128/mcb.20.12.4238-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.