Abstract
Insulin release from pancreatic β-cells plays a critical role in blood glucose homeostasis, and β-cell dysfunction leads to the development of diabetes mellitus. In cases of monogenic type 1 diabetes mellitus (T1DM) that involve mutations in the insulin gene, we hypothesized that misfolding of insulin could result in endoplasmic reticulum (ER) stress, oxidant production, and mitochondrial damage. To address this, we used the Akita+/Ins2 T1DM model in which misfolding of the insulin 2 gene leads to ER stress-mediated β-cell death and thapsigargin to induce ER stress in two different β-cell lines and in intact mouse islets. Using transformed pancreatic β-cell lines generated from wild-type Ins2+/+ (WT) and Akita+/Ins2 mice, we evaluated cellular bioenergetics, oxidative stress, mitochondrial protein levels, and autophagic flux to determine whether changes in these processes contribute to β-cell dysfunction. In addition, we induced ER stress pharmacologically using thapsigargin in WT β-cells, INS-1 cells, and intact mouse islets to examine the effects of ER stress on mitochondrial function. Our data reveal that Akita+/Ins2-derived β-cells have increased mitochondrial dysfunction, oxidant production, mtDNA damage, and alterations in mitochondrial protein levels that are not corrected by autophagy. Together, these findings suggest that deterioration in mitochondrial function due to an oxidative environment and ER stress contributes to β-cell dysfunction and could contribute to T1DM in which mutations in insulin occur.
Keywords: diabetes mellitus, β-cell, mitochondrial respiration, endoplasmic reticulum stress, mitochondrial quality control
insulin release from pancreatic β-cells is largely dependent on mitochondrial and endoplasmic reticulum (ER) function and plays a critical role in maintaining blood glucose homeostasis. The synthesis of insulin in β-cells is an energy-requiring process, with as much as 50% of the total protein of these cells committed to generation of this single protein when stimulated (51). Insulin requires posttranslational processing before it is secreted, and it has been shown that mutations that lead to misfolding may cause neonatal diabetes (11, 51). Once considered rare, monogenic mutations in insulin are becoming recognized as causes of neonatal diabetes mellitus. They can also be causative factors for type 1b diabetes or maturity-onset diabetes of the young and in some cases early onset type 2 diabetes mellitus (T2DM) (27, 50). Mutations in the insulin gene lead to defective processing and accumulation of proinsulin in the ER, inducing ER stress. In addition, the consequent dysregulation of blood glucose homeostasis can initiate serious diabetic complications such as cardiovascular disease, neuropathy, and nephropathy (8). Thus, prevention or treatment of β-cell injury and diabetes mellitus onset/progression continues to be a challenge, particularly for the group of patients with mutations in the insulin gene.
It has been established that there is a potential link between the ER and the mitochondria, and this has been suggested to contribute to β-cell dysfunction in both type 1 diabetes mellitus (T1DM) and T2DM pathogenesis (3, 16, 43, 52, 58). One proposed sequence of events is that ER stress leads to disruption of Ca2+ flow to the mitochondria, causing mitochondrial dysfunction and triggering a series of cyclic events, such as oxidative stress, that culminates in induction of cell death (30). To counter the cell death processes and development of T1DM, prosurvival mechanisms such as autophagy can be initiated in β-cells (6). Autophagy is a multistep process that targets damaged proteins and organelles for degradation and efficiently regulates organelle turnover within the cell (57). The targeting of phagophores to dysfunctional mitochondria is dependent on mitochondrial quality and ubiquitinated proteins (17, 25). A decrease in mitochondrial quality can be identified by increased mitochondrial ROS production, mitochondrial fission, decreased membrane potential, mtDNA damage, and suppressed bioenergetic function (20). How mitochondrial morphology plays an important role in β-cell dysfunction is still unclear (52). In addition, how autophagy responds to metabolic stress and impacts bioenergetic function and cellular redox status is not well understood (26, 28, 47). This is important to understand since lack of degradation of damaged mitochondria can induce oxidative stress and cell death (20, 38).
As a means to identify mechanisms that contribute to T1DM and associated complications, several experimental models have been developed (4, 34, 49, 59). In particular, there are two animal models representative of the syndromes mentioned above known as the Munich Ins2C95S mutant mouse (19) and the AkitaIns2+/− mouse (60). The Akita mouse model contains a spontaneous dominant mutation in the insulin 2 gene allele in which tyrosine is replaced by cysteine at residue 96, which has also been reported in humans (1, 15, 46). This mutation causes misfolding and accumulation of proinsulin in the ER. The resulting ER stress has been proposed to lead to β-cell death, hyperglycemia, T1DM, and diabetic nephropathy (7, 30, 60). A progressive loss of β-cell viability is a characteristic feature of the progression of diabetes and may be largely influenced by ER stress. It has been shown that the ER stress response is induced in human islets from T1DM subjects (36), which if prolonged may also contribute to disease progression (16). Importantly, the link between ER stress and mitochondrial function in β-cells is not fully understood.
In this study, we used transformed pancreatic β-cell lines generated from wild-type Ins2+/+ (WT) and Akita+/Ins2 mice to investigate the role of mitochondrial bioenergetics, quality control, and autophagy in a spontaneous ER stress model of diabetes mellitus. Recently, we described that the Akita+/Ins2−-derived β-cells exhibit ER stress and that they undergo cell death via induction of the mitochondrial intrinsic apoptosis pathway (30). Since mitochondria play a critical role in β-cell function, we hypothesized that ER stress due to mutated insulin would cause mitochondrial damage, oxidative stress, and accumulation of damaged mitochondria in Akita+/Ins2-derived β-cells. In the present study, we show that the Akita+/Ins2-derived β-cells have impaired metabolic function, oxidative stress, mitochondrial DNA and protein damage, and fragmented mitochondria. In addition, p62, the ubiquitin-binding protein essential for targeting and transporting damaged mitochondria to autophagosomes, and Parkin, the protein that targets damaged mitochondria and is essential for mitophagy, were decreased. We have also tested the impact of ER stress on bioenergetic function in β-cells, INS-1 cells, and intact islets using thapsigargin. Our data suggest that ER stress induces mitochondrial dysfunction and that defects in the autophagic pathway contribute to β-cell dysfunction in humans with mutations in the insulin gene.
MATERIALS AND METHODS
Reagents and antibodies.
The following reagents were obtained from Sigma-Aldrich (St. Louis, MO): oligomycin, FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone), antimycin A, TMRM (tetramethylrhodamine methyl ester perchlorate), acetyl-coenzyme A sodium salt, DTNB [5,5′-dithiobis(2-nitrobenzoic acid)], oxaloacetatic acid, chloroquine, thapsigargin, d-glucose, Triton X-100, reduced glutathione, oxidized glutathione, and trypan blue solution. MitoSOX Red, BODIPY-FL-N-(2-aminoethyl)maleimide (Bodipy-NEM), and MitoTracker Deep Red FM were purchased from Invitrogen (Eugene, OR). DTPA (diethylenetriamine pentaacetic acid) was obtained from Fisher Scientific (Pittsburgh, PA).
Antibodies were purchased from a number of sources as follows: complex I-39 kDa, complex II-70 kDa, complex III core I-53 kDa, complex IV subunit I, voltage-dependent anion channel (VDAC), and ATP synthase-α and -β (MitoSciences, Eugene, OR); light chain 3 (LC3), p62 (Sigma-Aldrich St. Louis, MO); Parkin (Santa Cruz Biotechnology Santa Cruz, CA); manganese superoxide dismutase (MnSOD; Enzo Life Sciences, Ann Harbor, MI); Mfn1 (Abcam, Cambridge, MA); Drp1 (BD Biosciences, San Jose, CA); and citrate synthase (Epitomics, Burlingame, CA).
Cell culture and intact mouse islets.
WT and AkitaIns2+/− insulinoma pancreatic β-cell lines were generated from Akita mice or isogenic WT C57BL/6 littermates, as described previously (39, 41, 54). Both cell types were grown in 25 mM glucose Dulbecco's modified Eagle's medium (DMEM; 4.5 g/l d-glucose, 4 mM l-glutamine, and phenol red; Invitrogen, Eugene, OR) containing 15% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 1× penicillin-streptomycin (Invitrogen), and 150 μM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO). The INS-1 insulinoma cell line was maintained as described previously (31). All cell lines were maintained in a 37°C incubator under an atmosphere of (5% CO2-95% air) dioxide.
Intact mouse islets.
Male C57/BL6 mice were bred and maintained according to the Institutional Animal Care and Use Committee policies at the University of Alabama at Birmingham, which approved our experiments. Mice (4–5 wk of age) were euthanized with CO2 or cervical dislocation prior to the islets being isolated, as described previously (29). In brief, the pancreas was removed, distended, and processed through digestion and a series of washes using RPMI 1640 to isolate intact islets. The remaining islets were then selected under a microscope, counted, and incubated overnight in complete RPMI 1640 medium under an atmosphere of 5% CO2 and 95% air at 37°C. Typical islet yields were 150 per mouse.
Mitochondrial bioenergetics.
To assess cellular and intact islet bioenergetics, the Seahorse XF-24 extracellular flux analyzer from Seahorse Biosciences (North Billerica, MA) was utilized. WT and Akita+/Ins2-derived β-cells or INS-1 cells were seeded at 60,000 cells/well in Seahorse XF-24 specialized cell culture plates. The following day, medium was replaced with unbuffered DMEM for 1 h at 37°C (without CO2) prior to basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) being assessed simultaneously in the Seahorse XF-24 analyzer. Twenty-five minutes later, the effects of the mitochondrial inhibitors oligomycin (1 μg/ml), FCCP (2 μM), and antimycin A (10 μM) were measured. Both the OCR and ECAR were normalized to total protein in each well using the Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, CA). The cellular bioenergetic parameters ATP-linked respiration, proton leak respiration, maximal OCR, reserve capacity, and nonmitochondrial respiration were calculated as described previously (14). Briefly, the ATP-linked respiration was derived from the difference between basal OCR and respiration following complex V inhibition (oligomycin). The difference in respiration between antimycin A and oligomycin indicated the amount of oxygen consumed that is due to proton leak. Maximal OCR was determined by subtracting FCCP-induced respiration from the OCR after the addition of antimycin A. Finally, the reserve capacity was calculated as the difference between maximal respiration (OCR after FCCP) and basal OCR. In separate experiments, cells were treated with thapsigargin (1 μM), an ER stress inducer, for 6 h prior to assessing cellular bioenergetics with thapsigargin in the medium.
Islets (50/well) isolated from C57/BL6 mice and treated with vehicle (DMSO) alone or thapsigargin (2 μM) were plated in XF24 islet capture microplates containing 3 mM DMEM glucose using an established protocol (56). Following plating, islets were allowed to equilibrate for 1 h at 37°C (without CO2) prior to assessing basal OCR. After basal OCR was measured for 50 min, d-glucose (20 mM) was injected into the cellular medium to determine the islets' glucose response over time, and oligomycin (5 μM) was injected subsequently to inhibit OCR. Finally, the OCR was normalized to total protein in each well using the Bio-Rad DC protein assay (Bio-Rad Laboratories).
Mitochondrial membrane potential and superoxide generation.
The inner mitochondrial membrane potential was assessed using TMRM dye. Briefly, cells (80,000 cells/well) were seeded in a Costar 96-well black clear-bottom plate (Fisher Scientific, Pittsburgh, PA). The following day, cells were incubated for 30 min with 100 nM TMRM dye in culture medium alone or in medium containing FCCP (10 μM) or oligomycin (6 μg/ml). Next, the cellular medium containing dyes and inhibitors was removed, and trypan blue was added to the cells for 10 min to quench extracellular TMRM fluorescence. Subsequently, trypan blue was aspirated, and cells were washed once with medium. Fresh medium was added to the cells, and TMRM intracellular fluorescence was measured using a FLUOstar Omega Microplate Reader (BMG Labtech, Ortenberg, Germany) with excitation/emission filters (560/595 nm).
To detect basal mitochondrial superoxide generation in WT and Akita+/Ins2-derived β-cells, MitoSOX Red (Invitrogen), a cationic dye that fluoresces red when oxidized by mitochondrial superoxide, was utilized. Briefly, cells (50,000 cells/well) were plated in glass chamber slides (Nunc, Rochester, NY). The following day, cells were treated in the dark with MitoSOX Red (300 nM for 15 min). Subsequently, cells were washed with cellular medium and imaged using a Leica SP1 UV confocal laser scanning microscope (×100) with excitation/emission (510/580 nm) filters. All images were captured with equal exposure times and quantified using Simple PCI software (Compix, Cranberry Township, PA).
MnSOD activity.
Enzymatic activity of MnSOD was determined by the cytochrome c reduction method in the presence of 1 mM KCN to inhibit Cu/Zn-SOD activity, as described previously (37). Briefly, cells were harvested in PBS containing DTPA (10 μM), 0.1% Triton X-100, and protease inhibitors. The protein content of cleared lysates was measured by the Bradford protein determination method. Sample amounts were titrated to a reaction mixture consisting of 50 mM phosphate buffer, pH 7.8, containing 0.2 mM EDTA, 10 μM cytochrome c, 50 μM xanthine, and 1 mM KCN until the rate of xanthine-xanthine oxidase-generated superoxide production was reduced by approximately one-half. The amount of xanthine-xanthine oxidase required to achieve a rate of reduction of cytochrome c of 0.025 absorbance U/min was predetermined prior to beginning the assay.
Western blotting.
Protein extracts from WT and Akita+/Ins2-derived β-cells that were untreated or treated with chloroquine (40 μM for 5 h), a lysosomotropic agent that inhibits autophagic flux, or thapsigargin (1 μM for 2 h), an ER stress inducer, were separated on SDS-PAGE (10 or 15% gels) and transferred to polyvinylidene fluoride (PVDF) or nitrocellulose membranes. Subsequently, membranes were blocked in TBST (Tris-buffered saline with 0.05% Tween 20) containing 5% nonfat dry milk powder for 1 h and probed with primary antibodies overnight at 4°C. The following day, blots were washed three times with TBST and incubated with appropriate secondary antibodies for 1 h at room temperature (RT). Membranes were then washed with TBST three times prior to developing with SuperSignal West Dura chemiluminescent substrate (Thermo Scientific, Rockford, IL). Equal protein loading was established (10 or 20 μg) using the Lowry DC protein assay and verified by staining the membrane with Ponceau S or Amido Black. In all cases, the variation in protein loading was determined to be <10%, and no further correction was applied. Comparison of proteins was performed within the same gel to avoid variability in exposure and development conditions. Relative levels of protein expression were quantified using densitometry from the AlphaView SA software (Protein Simple, Santa Clara, CA).
Glutathione assays.
The total glutathione level was determined in cell lysates using the Tietze recycling assay (53). In brief, cells were lysed in 0.1% Triton X-100 in PBS buffer, pH 7.4, containing 10 μM DTPA. Total glutathione was determined on the basis of the reduction of DTNB at 412 nm, using an extinction coefficient of 13,000 M−1/cm−1. Subsequently, all values were normalized to cellular protein.
For GSH and GSSG measurements by mass spectrometry, cells were scraped in 10 mM Tris, pH 7.4, containing 10 mM N-ethylmaleimide (NEM) and allowed to incubate on ice for 30 min prior to the protein being precipitated with two volumes of cold methanol. Precipitated proteins were removed by centrifugation at 16,500 g for 15 min. Cleared supernatants (5 μl) were then injected onto the mass spectrometer. GSH-NEM standards were prepared by reacting 1 mM GSH with 10 mM NEM for 30 min at 37°C. To determine the efficiency of the alkylation procedure, the concentration of unreacted GSH was measured using the reduction of DTNB, and the reaction was shown to be 100% (data not shown). Calibration curves ranging from 0 to 0.05 nmol (injected on the column) of GSH-NEM and GSSG were established in conjunction with the samples. The linearities of the GSH-NEM and GSSG curves were r2 = 0.9861 and r2 = 0.9982, respectively. To separate the NEM-GSH from GSSG and excess unreacted NEM, a gradient elution was used. The elution profile was as follows: initial state 100% mobile phase A; 0- to 5-min linear 0–100% gradient of mobile phase B, followed by a reequilibration with 100% mobile A from 5.1 to 10 min. Mobile phase A consisted of isopropanol-acetonitrile-formic acid (50:50:0.5 vol/vol), respectively, and mobile phase B consisted of 0.5% formic acid. The column was a normal phase 2.1 × 20 mm ZIC-HILIC SeQuant (3.5 micron particle size). Positive ion electrospray ionization was employed for analysis using an AB/Sciex API-4000 Triple Quadrupole mass spectrometer equipped with a Shimadzu Prominence HPLC system. Selected ion monitoring in the positive mode was conducted to simultaneously monitor ions at m/z 433 and 613 that corresponded to the protonated molecular ions of GSH-NEM and GSSG, respectively.
Protein thiol oxidation.
To detect protein thiols in WT and Akita+/Ins2-derived β-cells, Bodipy-NEM, a fluorophore-labeled alkylating agent that reacts specifically with thiol groups in biological samples, was used (22). In brief, cells were treated with 100 μM Bodipy-NEM for 15 min, lysed, and separated using 10% SDS-PAGE with nonreducing conditions. To visualize the thiol redox state, in-gel fluorescence imaging of the BODIPY signal using a Typhoon imager (GE Healthcare Biosciences, Pittsburgh, PA) was implemented. ImageQuantTL analysis software (GE Healthcare Biosciences) was used to analyze the fluorescent signal intensity for each lane. In addition, the concentration of Bodipy in each sample was quantified using a Bodipy-GAPDH standard curve, as described previously (22).
Protein S-glutathiolation.
Cells (3 × 106) were suspended in 100 μl of 1 mM ethyl ester GSH-biotin (Invitrogen, Grand Island, NY) in serum-free medium and then incubated overnight at 37°C. Cell suspensions were centrifuged, and the resulting cell pellets were washed with fresh serum-free medium (500 μl). Cellular extracts were prepared in lysis buffer containing 20 mM Tris, pH 7.4, 1% Triton x-100, 10 μM DTPA, 25 mM NEM, and protease inhibitors. To enrich for S-glutathiolated proteins, cleared lysates were mixed with neutravidin beads and incubated overnight at 4°C. Beads were then washed and the modified proteins eluted and probed by Western blotting (21, 62). To identify mitochondrial proteins that were S-glutathiolated, affinity-purified material (10 μl) was loaded onto 4–18% gradient gels and then probed with an antibody specific to VDAC.
Mitochondrial copy number.
Mitochondrial copy number was determined using real-time quantitative PCR. Real-time PCR using SYBR Green (Life Technologies, Carlsbad, CA) was performed using 50 ng of total DNA as template. The primer sequences used for mtDNA were mtF (5′- CCCCAGCCATAACACAGTATCAAAC-3′) and mtR (5′-GCCCAAAGAATCAGAACAGATGC-3′) in an ABI 7500 system (Applied Biosystems, Grand Island, NY). Real-time PCR conditions were as follows: 94°C for 2 min, followed by 40 cycles of denaturation at 94°C for 15 s, annealing and extension at 60°C for 1 min. mtDNA copy number was normalized to amplification of an 18S nuclear amplicon using the following primer set: ms18s forward, aaacggctaccacatccaag; ms18s reverse, 5′-caattacagggcctcgaaag-3′.
Mitochondrial DNA damage.
To assess mitochondrial DNA (mtDNA) damage, total DNA was isolated from WT and Akita+/Ins2-derived β-cells using a QIAamp DNA Mini Kit according to the manufacturer's instructions (Qiagen, Valencia, CA). The 16-kb mtDNA was amplified by PCR using 50 ng of total DNA and the primer set M13597/M13361 (forward: cccagctactaccatcattcaagtag; reverse: 5′-gagagattttatgggtgtaatgcggtg-3′), as described previously (55). To normalize mtDNA copy number, a short 80-bp segment of mtDNA product was amplified using primer set M13281/M13361 (forward: 5′gcaaatccatattcatccttctcaac; reverse: gagagattttatgggtgtaatgcggtg-3′). Quantitation of the average lesion frequency in the 16-kp mtDNA was performed assuming a random distribution of lesions in accordance with the Poisson equation (55).
Citrate synthase activity.
Citrate synthase catalyzes the first reaction of the TCA cycle and was evaluated in WT and Akita+/Ins2-derived β-cells, as described previously (45). In brief, citrate synthase activity in cell lysates was measured spectrophotometrically at 412 nm based on the reaction between oxaloacetate, acetyl-CoA, and DTNB. Subsequently, cellular protein was determined and used to normalize citrate synthase activity.
Mitochondrial morphology.
To assess mitochondrial morphology, cells were grown in Lab-Tek chambered coverglass systems (Nunc, Rochester, NY). The following day, cells were washed with DMEM medium and incubated with 100 nM MitoTracker Deep Red FM (Invitrogen) for 15 min at 37°C. Subsequently, cells were washed with medium and imaged using a Carl Zeiss LSM 700 inverted confocal microscope (×40 oil; NA 1.4, Zoom ×3). Optical Z stacks were obtained, and thereafter the Zen software (Carl Zeiss) was utilized to assess the number of fragmented mitochondria per total mitochondrial area. Mitochondrial fragments that were 0.5 μm or smaller in diameter were considered for quantitation and counted in each optical slice of a z-stack after thresholding to account only for mitochondrial pixels. Subsequently, the total mitochondrial area per optical slice was determined by adding all of the mitochondrial area in the z-stacks. Finally, the number of fragmented mitochondria was normalized by total mitochondrial area and expressed as the number of mitochondrial fragments (≤0.5 μm diameter) in 100 μm2.
Immunocytochemistry.
WT and Akita+/Ins2-derived β-cells were plated on glass coverslips in 12-well plates. The following day, cells were washed with cold PBS, fixed for 15 min with 4% paraformaldehyde in PBS (Affymetrix, Santa Clara, CA), washed gently with PBS, and permeabilized with 0.15% Triton X-100 in PBS for 15 min at RT. Cells were then washed with PBS and blocked with 1% bovine serum albumin (Sigma-Aldrich) plus 1% donkey serum host (Santa Cruz Biotechnology) in PBS for 30 min at RT. Subsequently, cells were incubated with the rabbit polyclonal LC3 antibody (Sigma-Aldrich) for 1 h at RT. Cells were then washed with 0.1% BSA in PBS for 10 min at RT and then incubated with a goat anti-rabbit IgG Alexa 488 secondary antibody (1:1,000; Invitrogen) for 30 min in the dark at RT. Finally, cells were washed gently with PBS for 15 min, coverslipped with mounting medium, and stored in the dark at 4°C. Fluorescence staining was evaluated using a Leica DM6000 epifluorescence microscope (×100). All images were captured with identical exposure settings and quantified using Simple PCI software (Compix, Cranberry Township, PA).
mRNA analyses.
RNA was isolated from cells using TRIzol (Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. RNA (0.5–2 μg) was used to make cDNA using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories) according to the manufacturer's protocol. Quantitative real-time PCR was performed with SYBR Green Mastermix (Life Technologies, Carlsbad, CA) with the following conditions: 95°C for 5 min, 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s × 40. Real-time quantitative RT-PCR results were normalized against an internal control (β-actin). Forward and reverse primer sequences for genes analyzed are as follows: Parkin (forward, gagcttccgaatcacctgac; reverse, ccctccagatgcatttgttt), P62 (forward, cgagtggctgtgctgttc; reverse, tgtcagctcctcatcactgg), and β-actin (forward, gacggccaggtcatcactat; reverse, aaggaaggctggaaaagagc).
Statistics.
All results are expressed as means ± SE (n = 3–5/group). Statistical analysis was performed using one-way ANOVA and Tukey's post hoc test. All values were considered statistically significant when P < 0.05.
RESULTS
Akita+/Ins2-derived β-cells have lower rates of oxidative phosphorylation.
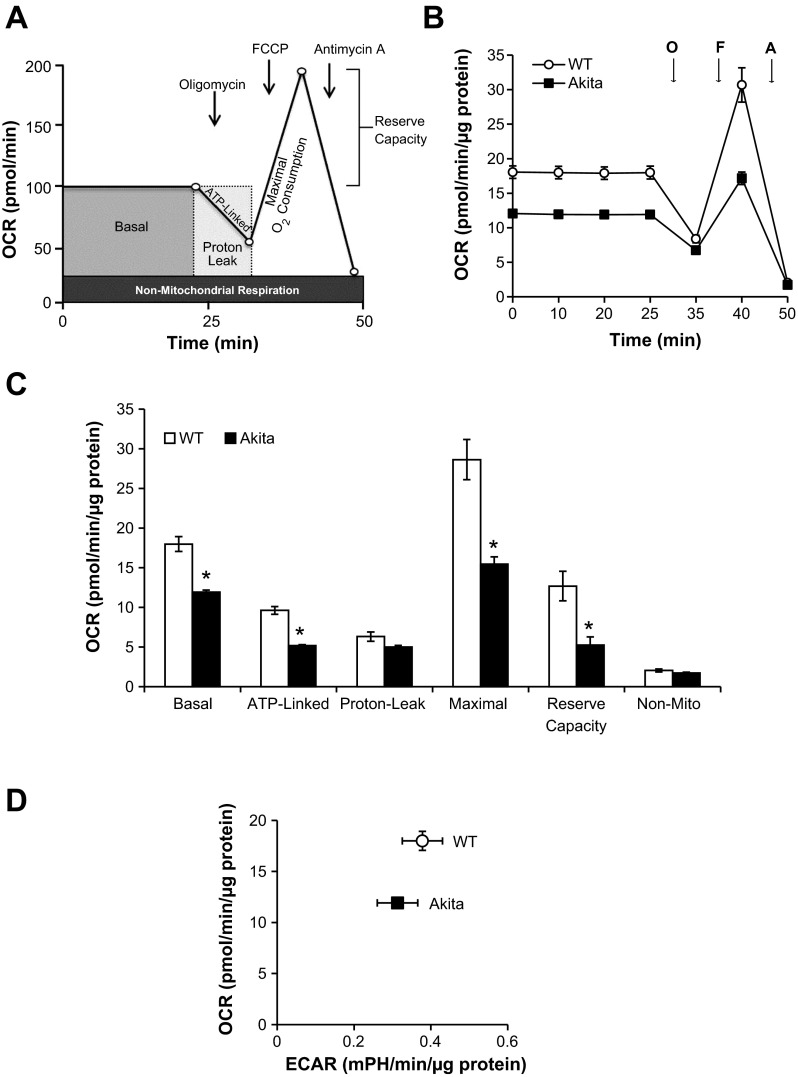
Mitochondrial bioenergetics in WT and Akita+/Ins2-derived β-cells were assessed using the Seahorse XF-24 analyzer (Fig. 1A). As shown in Fig. 1, basal OCR was measured for 25 min and was decreased in Akita+/Ins2-derived β-cells compared with WT β-cells. Subsequently, oligomycin, an ATP synthase inhibitor, was injected and as expected caused a rapid decline in ATP-linked respiration in both WT and Akita+/Ins2-derived β-cells. Next, FCCP, a mitochondrial uncoupler, was injected and stimulated maximal respiration in both cell types (Fig. 1B). Finally, injection of antimycin A inhibited mitochondrial respiration to an equal extent in both cell types. From this profile, several bioenergetic parameters were determined (Fig. 1C). Basal respiration, ATP-linked respiration, and maximal respiration were all significantly lower in Akita+/Ins2-derived β-cells compared with WT β-cells (Fig. 1C). The reserve capacity, which is an estimate of the potential bioenergetic capacity of the cell available to meet increased ATP demand or to combat oxidative stress (13), was also significantly lower in Akita+/Ins2-derived β-cells. Under the same conditions, the ECAR, which is an index of glycolysis, was found to be similar between WT and Akita+/Ins2-derived β-cells and is shown plotted against the basal OCR rates in Fig. 1D.
Fig. 1.
Oxidative phosphorylation in wild-type Ins2+/+ (WT) and Akita+/Ins2-derived β-cells. A: schematic of the mitochondrial function assay used to measure oxygen consumption rate (OCR) over time. B: WT and Akita+/Ins2-derived β-cells were seeded at 60,000 cells/well in 24-well Seahorse plates. The following day, cell culture medium was replaced with low-buffered DMEM for 1 h at 37°C prior to measuring basal OCR and the effects of the mitochondrial inhibitors oligomycin (O), FCCP (F), and antimycin A (A) on OCR. C: the individual bioenergetic parameters were calculated based on the measurements taken in B. Results are means ± SE; n = 4–5/group. *P < 0.05 compared with WT β-cells. D: the OCR and extracellular acidification rate (ECAR) of WT and Akita+/Ins2-derived β-cells were plotted against each other according to the final basal measurement shown in B.
ER stress induces bioenergetic defects in WT β-cells, INS-1 cells, and C57 mouse islets.
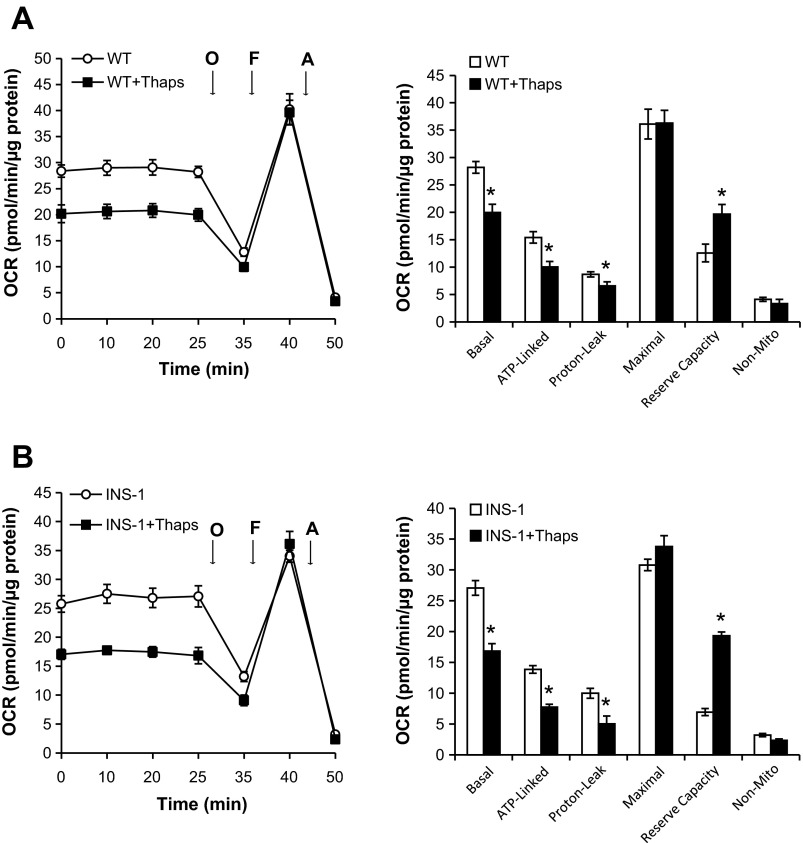
To assess the effect of ER stress on mitochondrial bioenergetics, WT β-cells and INS-1 cells were treated with thapsigargin for 6 h prior to the mitochondrial bioenergetic profile being assessed. As shown in Fig. 2A, thapsigargin pretreatment significantly decreased basal respiration, ATP-linked respiration, and proton leak in WT β-cells (Fig. 2A). However, thapsigargin had no effect on maximal respiration and increased reserve capacity significantly (Fig. 2A). As shown in Fig. 2B, thapsigargin also elicited similar effects in INS-1 cells.
Fig. 2.
Effect of endoplasmic reticulum (ER) stress on mitochondrial bioenergetics. WT β-cells (A) or INS-1 cells (B) were seeded at 60,000 cells/well in 24-well Seahorse plates. The following day, cells were treated with 1 μM thapsigargin (Thaps), an ER stress inducer, for 6 h prior to the medium being replaced with low-buffered DMEM and with or without 1 μM Thaps for 1 h at 37°C prior to basal OCR and the effects of the mitochondrial inhibitors O, F, and A being measured. Results are means ± SE; n = 4–5/group. *P < 0.05 compared with WT β-cells or INS-1 cells.
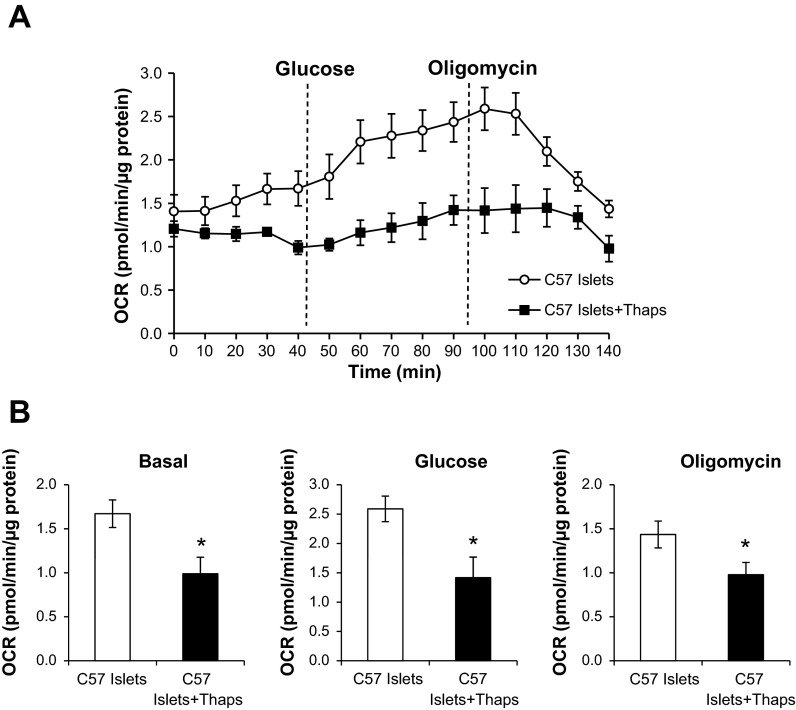
These findings were further confirmed in intact mouse islets treated with thapsigargin (24 h) prior to the islet glucose response being measured, using an established protocol (56) (Fig. 3). As shown in Fig. 3A, the basal OCR of mouse islets was measured for 45 min before glucose was injected to stimulate increased oxygen consumption. Approximately 50 min later, oligomycin, an ATP synthase inhibitor, was injected and caused a rapid decline in respiration. Islets treated with thapsigargin had a significantly lower basal OCR compared with control islets (Fig. 3, A and B). In addition, thapsigargin prevented a rapid increase in OCR following glucose stimulation and delayed the effects of oligomycin inhibition on respiration (Fig. 3, A and B). These data confirm that ER stress negatively affects cellular bioenergetics in both cell lines and isolated islets.
Fig. 3.
Glucose response of mouse islets exposed to ER stress. A: islets from C57/BL6 (C57) mice were isolated and plated 50 islets/well in 24-well Seahorse islet capture microplates. Following plating, islets were allowed to equilibrate for 1 h at 37°C prior to basal OCR being measured over time. Subsequently, glucose (20 mM) was injected into the medium to measure the glucose response, followed by oligomycin injection to inhibit respiration. B: the individual bioenergetic parameters were determined based on the measurements taken in A. Results are means ± SE; n = 4–5/group. *P < 0.05 compared with C57 islet controls.
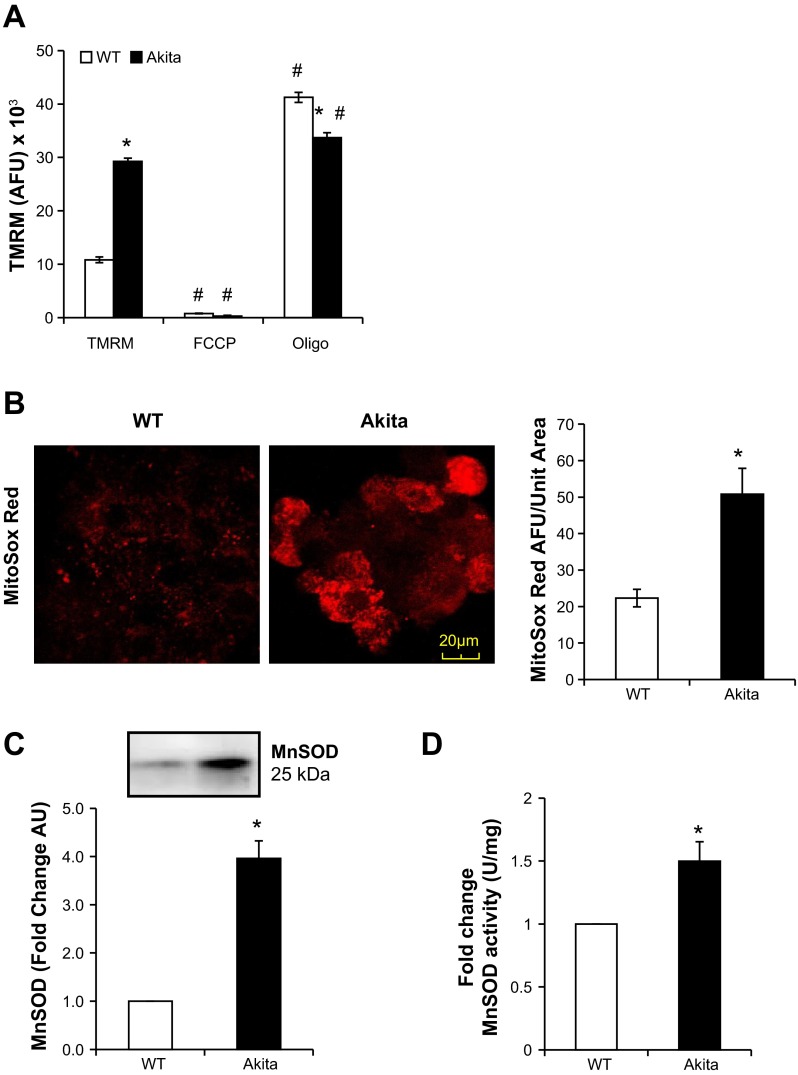
Akita+/Ins2-derived β-cells have increased mitochondrial membrane potential, mitochondrial superoxide, and MnSOD activity.
As shown in Fig. 4A, Akita+/Ins2-derived β-cells had a significantly higher basal mitochondrial membrane potential compared with WT cells (Fig. 4A). As expected, FCCP decreased the TMRM signal significantly in both WT and Akita+/Ins2-derived β-cells. However, oligomycin induced a further increase in membrane potential in the WT β-cells but to a much lesser extent in the Akita+/Ins2-derived β-cells. A higher mitochondrial membrane potential is frequently associated with increased superoxide production from the respiratory chain, which we assessed using MitoSOX Red fluorescence. As shown in Fig. 4B, the red fluorescence, reflecting increased mitochondrial superoxide generation, was increased significantly in Akita+/Ins2-derived β-cells. In addition, MnSOD protein and activity levels were increased significantly in the Akita+/Ins2-derived β-cells (Fig. 4, C and D). These data suggest that although MnSOD is increased in the Akita+/Ins2-derived β-cells, it is not sufficient to mitigate superoxide generation.
Fig. 4.
Mitochondrial membrane potential, superoxide generation, and manganese superoxide dismutase (MnSOD) in WT and Akita+/Ins2-derived β-cells. A: to detect mitochondrial membrane potential, WT and Akita+/Ins2−-derived β-cells were treated with 100 nM tetramethylrhodamine methyl ester perchlorate (TMRM) for 30 min. FCCP and oligomycin controls were included to depolarize and hyperpolarize mitochondrial membrane potential, respectively. Results are expressed as mean arbitrary fluorescence units (AFU) ± SE; n = 3/group. B: WT and Akita+/Ins2-derived β-cells were treated with 300 nM MitoSOX Red for 15 min and imaged using confocal microscopy (×100) to detect mitochondrial superoxide levels. The bar graph shows the arbitrary levels of mitochondrial superoxide detected in these cells per unit area. Results are means ± SE; n = 3/group. C: cell lysates from WT and Akita+/Ins2-derived β-cells were resolved by SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes, and probed for MnSOD. Western blot loading was controlled for by protein staining, and band intensities were quantified using AlphaView SA software. Results are expressed as mean fold change arbitrary units (AU) over WT β-cells ± SE; n = 3/group. D: MnSOD activity was determined in cell lysates using the cytochrome c reduction method. Results are expressed as mean fold change activity normalized to total protein over WT β-cells ± SE; n = 3/group. *P < 0.05 compared with WT β-cells; #P < 0.05 compared with respective controls.
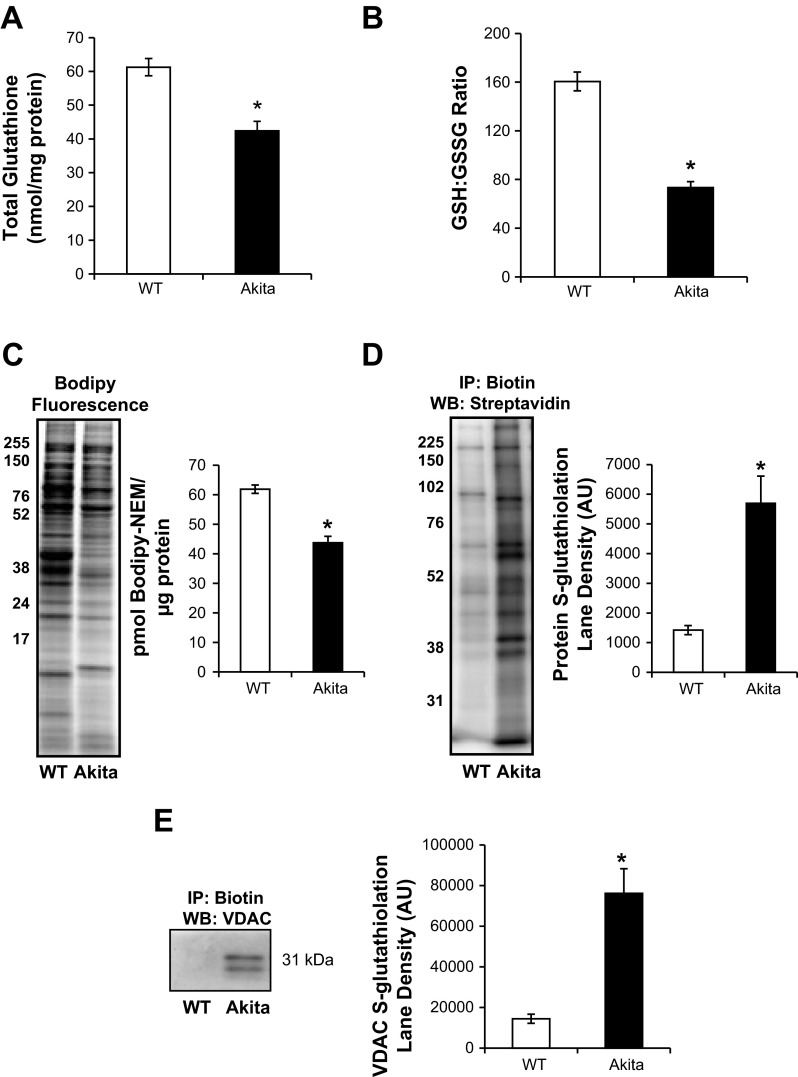
We found that the total glutathione pool was significantly lower in the Akita+/Ins2-derived β-cells compared with the WT β-cells (Fig. 5A). Since decreased glutathione levels would suggest changes in the ratio of reduced (GSH) to oxidized (GSSG) glutathione, we examined this using mass spectrometry and found the GSH/GSSG ratio to be significantly lower in Akita+/Ins2-derived β-cells (Fig. 5B). Given that glutathione oxidation is frequently associated with increased protein thiol oxidation, we evaluated the extent of oxidation using Bodipy-NEM, which introduces a fluorescent tag on reduced protein thiols. As shown in Fig. 5C, the overall Bodipy-NEM fluorescence intensity in the Akita+/Ins2-derived β-cell lane was ∼20% lower than the WT β-cells. An increased oxidation state of the GSH/GSSG redox couple could lead to S-glutathiolation, which would account for the decreased protein thiol content (23), and this was tested by incubating the cells with biotinylated GSH-ester and measuring protein S-glutathiolation. As shown in Fig. 5D, thiol modification of proteins was increased approximately fivefold in the Akita+/Ins2-derived β-cells. Next, using the biotin to affinity purify the S-glutathiolated proteins, we blotted for VDAC, a mitochondrial outer membrane protein, to determine whether it was modified. As shown in Fig. 5E, no detectable S-glutathiolated VDAC was detected in the WT cells, whereas the Akita+/Ins2-derived β-cells showed extensive modification. These findings suggest that there is increased oxidative stress and protein thiol oxidation in Akita+/Ins2-derived β-cells.
Fig. 5.
Total glutathione and free protein thiol levels in WT and Akita+/Ins2-derived β-cells. A: total glutathione content in WT and Akita+/Ins2-derived β-cells was determined and normalized to total protein, as described in materials and methods. B: GSH (reduced glutathione)/GSSG (oxidized glutathione) ratio was determined using mass spectrometry, as described in materials and methods. C: representative in-gel fluorescence image and quantification of the BODIPY signal in WT and Akita+/Ins2-derived β-cells treated with 100 μM BODIPY-FL-N-(2-aminoethyl)maleimide (Bodipy-NEM) for 15 min (lighter fluorescence indicates increased oxidized protein thiols; darker fluorescence represents reduced protein thiols). D: WT and Akita+/Ins2-derived β-cells were treated with biotinylated glutathione ethyl ester prior to affinity purification of S-glutathiolated proteins using streptavidin beads. Affinity-purified proteins were separated using SDS-PAGE and probed for total glutathiolated proteins using streptavidin-horseradish peroxidase. E: affinity-purified S-glutathiolated proteins were probed with an antibody specific to voltage-dependent anion channel (VDAC). Results are means ± SE; n = 3/group, *P < 0.05 compared with WT β-cells. IP, immunoprecipitation; WB, Western blot.
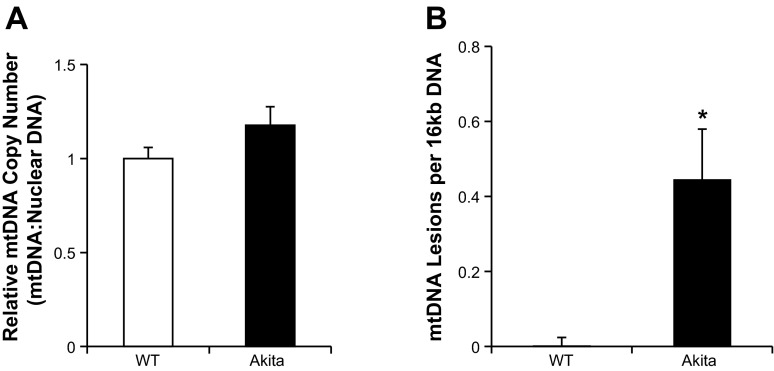
MtDNA is highly susceptible to oxidation, and therefore, we hypothesized that it would also be modified in Akita+/Ins2-derived β-cells compared with WT β-cells. As shown in Fig. 6A, real-time quantitative PCR detected no significant difference in mtDNA copy number between the two cell types. When we assessed mtDNA damage, as expected, the short product (80 bp) was not significantly different between the two cell types (Fig. 6B). However, the long product (16 kb) was decreased significantly in Akita+/Ins2-derived β-cells, which is consistent with increased mtDNA damage (Fig. 6B).
Fig. 6.
Mitochondrial DNA (mtDNA) copy number and damage in WT and Akita+/Ins2-derived β-cells. A: mtDNA copy number was quantified from WT and Akita+/Ins2-derived β-cells total DNA using real-time quantitative PCR. mtDNA copy numbers were normalized to nuclear DNA copy numbers (18S) and represented as the fold change compared with WT β-cells. B: mitochondrial lesion frequency was calculated using the ratio of the long PCR (16 kb) product normalized to the short PCR (80 bp) based on the assumption that the lesion is distributed randomly in the 16-kp mtDNA according to the Poisson equation mentioned in materials and methods. Results are mean ± SE; n = 3/group. *P < 0.05 compared with WT β-cells.
Akita+/Ins2-derived β-cells have altered mitochondrial protein composition.
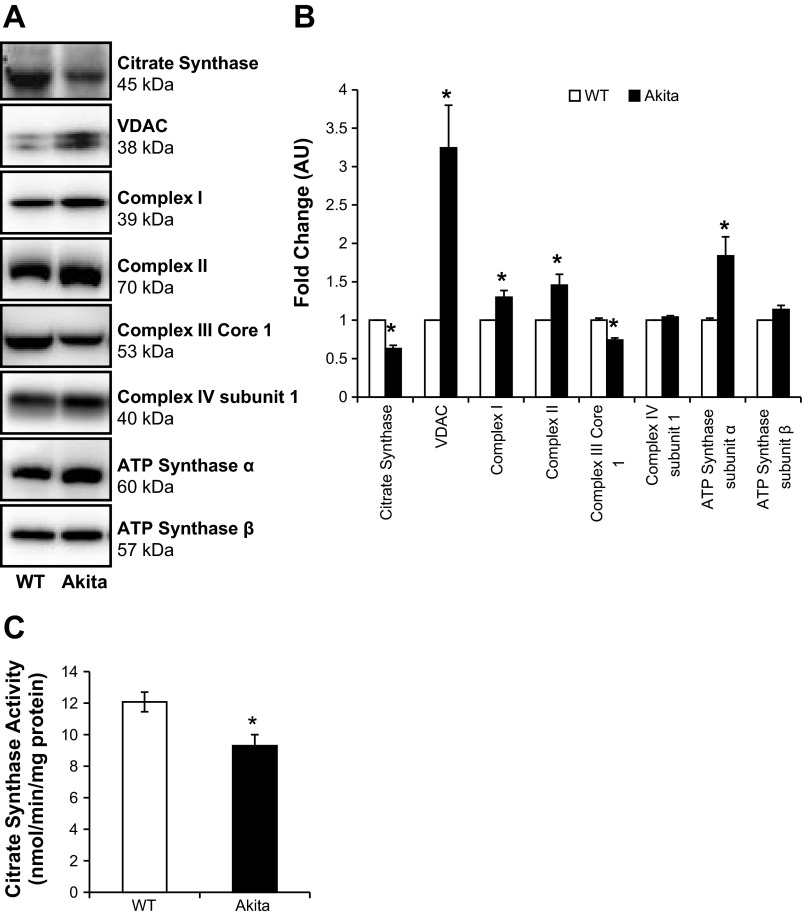
Since the total mitochondrial number, assessed by mtDNA, appears not to have influenced the bioenergetic changes described above, these changes could arise due to differential regulation of components of the mitochondrial respiratory chain (42). To assess this, we performed both activity assays and Western blot analyses of proteins in different mitochondrial compartments. Both the activity and levels of the matrix enzyme citrate synthase were significantly lower in Akita+/Ins2-derived β-cells compared with WT β-cells (Fig. 7, A–C). The outer membrane mitochondrial protein VDAC is essential for substrate transport and permeability transition and was found to be approximately threefold higher in Akita+/Ins2-derived β-cells compared with WT β-cells (Fig. 7, A and B). Next, we assessed selected mitochondrial respiratory complexes (Fig. 7, A and B) and found that complex I (39 kDa) and II (70 kDa) protein levels were increased in Akita+/Ins2-derived β-cells. However, complex III core 1 protein was significantly decreased in Akita+/Ins2-derived β-cells compared with WT β-cells. In contrast, complex IV subunit I protein levels were expressed equally between the two groups. Furthermore, ATP synthase α-levels were increased in Akita+/Ins2-derived β-cells, whereas ATP synthase-β expression was similar to WT β-cell levels (Fig. 7, A and B). Together, these data indicate significant changes in mitochondrial composition in the Akita+/Ins2-derived β-cells compared with WT β-cells.
Fig. 7.
Mitochondrial protein composition in WT and Akita+/Ins2-derived β-cells. A: cell lysates from WT and Akita+/Ins2-derived β-cells were prepared and resolved on SDS-PAGE gels, transferred to PVDF membranes, and subjected to immunoblot analysis for the following antibodies: citrate synthase, VDAC, complex I (39-kDa subunit), complex II (70-kDa subunit), complex III core protein 1, complex IV subunit I, and ATP synthase α- and β-subunits. B: Western blot loading was controlled for by protein staining, and band intensities were quantified using AlphaView SA software. Results are expressed as mean fold change AU over WT β-cells ± SE; n = 3/group. C: citrate synthase activity was determined and normalized to cellular protein, as described in materials and methods. Results are means ± SE; n = 3/group. *P < 0.05 compared with WT β-cells.
Mitochondrial morphology is altered in Akita+/Ins2-derived β-cells.
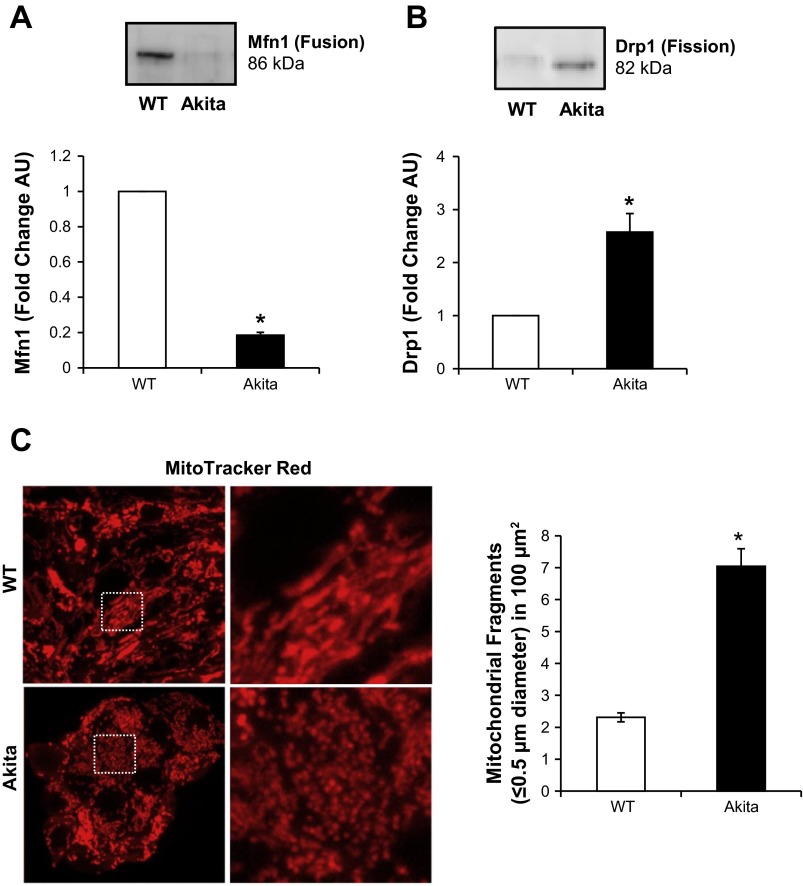
Mitochondrial morphology is established by equilibrium between fusion and fission, which are important cellular processes that control mitochondrial dynamics and turnover (33). Using specific antibodies, we determined the levels of the key fusion protein mitofusin 1 (Mfn1) and the key fission protein dynamin-related protein 1 (Drp1) in the WT and Akita+/Ins2-derived β-cells. As shown in Fig. 8A, Mfn1 protein expression was significantly lower in Akita+/Ins2-derived β-cells. In contrast, Drp1 was approximately threefold higher in Akita+/Ins2-derived cells than in WT β-cells (Fig. 8B). These results suggested that mitochondrial integrity is compromised in Akita+/Ins2-derived β-cells. To assess this, we visualized mitochondrial morphology using the fluorescent dye MitoTracker Red. As illustrated in Fig. 8C, WT β-cell mitochondria were larger and had longer tubular structures and filamentous networks. However, the Akita+/Ins2-derived β-cell mitochondria were smaller, fragmented, and punctate, which is consistent with a higher level of mitochondrial fission. In addition, Akita+/Ins2-derived β-cells had a significant increase in the number of fragmented mitochondria per mitochondrial area compared with the WT β-cells (Fig. 8C).
Fig. 8.
Mitochondrial dynamics and morphology in WT and Akita+/Ins2-derived β-cells. Cell lysates from WT and Akita+/Ins2-derived β-cells were prepared and resolved on SDS-PAGE gels, transferred to PVDF membranes, and subjected to immunoblot analysis for mitofusin 1 (Mfn1; mitochondrial fusion; A) and dynamin-related protein 1 (Drp1; mitochondrial fission; B). Western blot loading was controlled for by protein staining, and band intensities were quantified using AlphaView SA software. Results are expressed as mean fold change arbitrary units over WT β-cells ± SE; n = 3/group. C: representative maximum intensity projection of 3 consecutive optical z-slices acquired from WT and Akita+/Ins2-derived β-cells stained with MitoTracker Red. Quantification of mitochondrial fragments was performed as described in materials and methods. *P < 0.05 compared with WT β-cells.
Autophagy in Akita+/Ins2-derived β-cells.
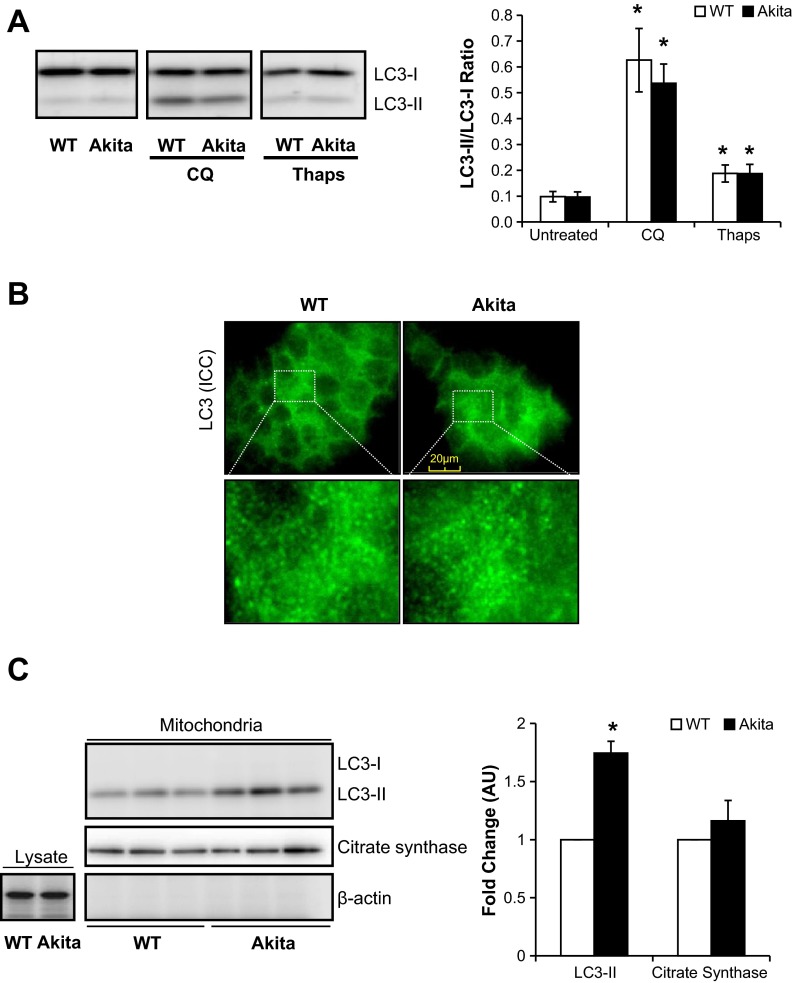
The microtubule-associated LC3 protein is a marker of autophagy and is comprised of two isoforms, LC3-I and LC3-II (28). Upon induction of autophagy, LC3-I is lipidated to form LC3-II. As shown in Fig. 9A, the ratio of LC3-II to LC3-I was not significantly different between WT and Akita+/Ins2-derived β-cells. To assess autophagic flux, we treated cells with chloroquine (CQ), a compound that prevents autophagy completion and degradation of LC3-II (28). CQ treatment had no effect on LC3-I in either cell type but increased LC3-II as expected, resulting in an increase in the LC3-II/LC3-I ratio, similarly in both cell types, indicating that Akita+/Ins2-derived β-cells exhibit normal autophagic flux (Fig. 9A). Furthermore, both cells exhibited similarly significant increases in the LC3-II/LC3-I ratio after being exposed to thapsigargin (Fig. 9A). Consistent with the data shown in Fig. 9A, there was no difference in LC3 between the two cell types, as evidenced by immunocytochemistry (Fig. 9B). We also examined LC3 levels in the mitochondria and found that LC3-II was increased significantly in the Akita+/Ins2-derived β-cells (Fig. 9C). In addition, the isolated mitochondria showed no detectable actin and were enriched in citrate synthase content, as expected (Fig. 9C).
Fig. 9.
Light chain 3 (LC3) protein levels in WT and Akita+/Ins2-derived β-cells. A: cell lysates from WT and Akita+/Ins2-derived β-cells ± chloroquine (CQ; 20 μM for 5 h) or Thaps (1 μM for 2 h) were resolved on SDS-PAGE and subjected to immunoblot analysis for microtubule-associated protein LC3. Western blot loading was controlled for by protein staining, and the band intensity was quantified using AlphaView SA software. The ratio of LC3-II to LC3-I results was expressed as mean AU ± SE; n = 3/group. *P < 0.05 compared with respective controls. B: cells were immunostained with LC3 antibody, and images were captured using immunofluorescence. C: protein levels of LC3, citrate synthase, and β-actin in whole cell lysates and/or isolated mitochondria. Western blot loading was controlled for by protein staining and LC3, and citrate synthase band intensities in isolated mitochondria were quantified using AlphaView SA software and are expressed as mean fold change AU over WT β-cells ± SE; n = 3/group. *P < 0.05 compared with WT β-cells. ICC, immunocytochemistry.
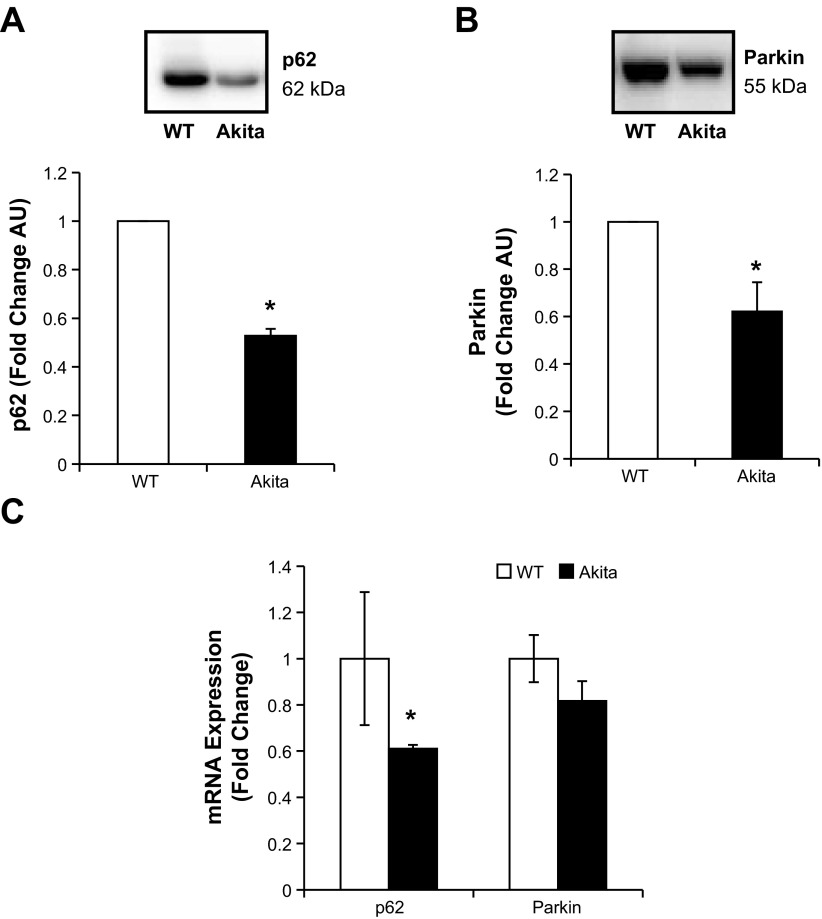
Although overall autophagic flux is unaltered, both p62 and Parkin, which are important in targeting damaged mitochondria for degradation, were decreased significantly in the Akita+/Ins2-derived β-cells compared with WT β-cells (Fig. 10, A and B). To determine whether this was due to transcription, we further examined p62 and Parkin mRNA levels. As shown in Fig. 10C, Akita+/Ins2-derived β-cells had a significant decrease in p62 mRNA levels; however, Parkin mRNA was not significantly different between the two cell types.
Fig. 10.
p62 and Parkin levels in WT and Akita+/Ins2-derived β-cells. A and B: cell lysates from WT and Akita+/Ins2-derived β-cells were resolved on SDS-PAGE, transferred, and probed for p62 (A) and Parkin (B). Western blot loading was controlled for by protein staining, and the band intensity was quantified using AlphaView SA software. Results are expressed as mean fold change AU over WT β-cells ± SE; n = 3/group. C: real-time RT-PCR analysis of p62 and Parkin mRNA levels in WT and Akita+/Ins2-derived β-cells. Results are expressed as fold change to WT β-cells ± SE; n = 3/group. *P < 0.05 compared with WT β-cells.
DISCUSSION
Mitochondrial changes, ER stress, and oxidative stress have all been suggested to contribute to pancreatic β-cell dysfunction (10, 16, 24). In the present study, we examined the effects of endogenous ER stress induced by a mutation in the insulin B chain in pancreatic β-cell lines generated from AkitaIns2+/− mice and by inducing ER stress pharmacologically using thapsigargin in WT β-cells, INS-1 cells, and C57/BL6 mouse islets. This is important to understand because mutations in insulin have been shown to occur in rare cases of neonatal diabetes in humans (1, 15, 46). Spontaneous mutations of insulin result in progressive β-cell death and secondary complications of T1DM such as diabetic nephropathy (7, 9). Previous studies with β-cells generated from Akita+/Ins2 and WT mice have shown that Akita+/Ins2-derived β-cells exhibited decreased insulin release, increased ER stress, and increased apoptotic cell death through the intrinsic mitochondrial cell death pathway (30, 41). We hypothesized that ER stress due to mutated insulin would cause mitochondrial damage, oxidative stress, and accumulation of damaged mitochondria in Akita+/Ins2-derived β-cells and that the failure to maintain a healthy mitochondrial population may underlie the mechanisms leading to cytotoxicity. Consistent with our hypothesis, we found that Akita+/Ins2-derived β-cells have decreased mitochondrial function and increased mitochondrial ROS, mitochondrial fission, and mtDNA damage.
Increased intramitochondrial oxidative stress observed was associated with changes to the cellular bioenergetics that were evident by decreased ATP-dependent and maximal respiration. The lower levels of respiration could arise from a decrease in the number of mitochondria, but this explanation is unlikely since the total mtDNA copy numbers were similar between the Akita+/Ins2-derived β-cells and controls. Under the same conditions, glycolysis was also measured and found to be not significantly different between WT and Akita+/Ins2-derived β-cells, suggesting that the Akita+/Ins2-derived β-cells had overall lower energetic requirements than WT β-cells. In support of this finding, the resting mitochondrial membrane potential was higher in Akita+/Ins2-derived β-cells and was not further hyperpolarized following the addition of oligomycin. It is well established that an increased mitochondrial membrane potential contributes to higher rates of mitochondrial superoxide production (40), and this was also evident in the current study. Indeed, both MnSOD protein and activity levels were also increased, but the level of enzyme activity was only 1.4-fold compared with a fourfold increase in protein levels. It has been shown previously that modifications such as S-glutathiolation and nitration cause decreased MnSOD activity, and it is possible that these could be occurring and explain that the increase in enzyme activity is substantially lower than the increase in protein levels (35, 44).
Pancreatic β-cells are extremely susceptible to oxidative stress due to their low levels of antioxidant enzymes such as glutathione peroxidase 1 and Cu/Zn-superoxide dismutase 1 (32). Consistent with these findings, we found total glutathione to be lower in Akita+/Ins2-derived β-cells. We further evaluated the GSH/GSSG redox couple in the Akita+/Ins2-derived β-cells and found it to be more oxidized, consistent with an increased level of protein S-glutathiolation, including S-glutathiolation of the mitochondrial outer membrane protein VDAC. In addition, increased protein thiol oxidation was observed in the Akita+/Ins2-derived β-cells, which correlates with increased mitochondrial superoxide generation and the formation of downstream oxidants. In agreement with this conclusion, it has been shown that oxidative stress, tyrosine nitration, and mitochondrial swelling are induced in Akita+/Ins2-derived β-cells (61).
It is clearly evident that an oxidative environment can alter mtDNA and protein composition and function. Evaluation of both mtDNA and selective mitochondrial proteins showed evidence of increased mtDNA damage and a significant restructuring of the mitochondrial proteins. Citrate synthase protein levels and activity, a TCA cycle enzyme generally regarded as resistant to oxidative stress, was significantly lower, and VDAC levels were enhanced. The latter was particularly interesting because VDAC is essential for mitochondrial Ca2+ uptake from the ER and plays a key role in mitochondrially mediated apoptosis. The increase in VDAC observed in the Akita+/Ins2-derived β-cells suggests that VDAC may be contributing to Ca2+ overload and cell death. Indeed, VDAC protein levels have been shown to be increased in diabetic mouse coronary vascular endothelial cells and were suggested to cause increased mitochondrial superoxide due to Ca2+ overload within the mitochondria (48). This could be an additional mechanism for increased mitochondrial superoxide in the Akita+/Ins2-derived β-cells since it is well documented that disruption of electron transport in the mitochondria can lead to increased mitochondrial superoxide generation (40). Hence, we also evaluated proteins related to oxidative phosphorylation in these cells because mitochondrial protein composition is critical in regulating the respiratory chain. In the Akita+/Ins2-derived β-cells the distribution of several proteins was significantly different and may explain the changes observed in cellular bioenergetics. In particular, complex I and II subunits were increased in Akita+/Ins2-derived β-cells compared with WT β-cells, but complex III was significantly decreased, and complex IV subunit I levels were not significantly different between the two groups. Interestingly, ATP synthase α-subunit was twofold higher in Akita+/Ins2-derived β-cells compared with WT β-cells. However, the ATP synthase β-subunit was not changed, which suggests defective assembly of this mitochondrial complex. Consistent with our findings, both complex I and ATP synthase protein expression have been shown to be increased in islets isolated from T2DM subjects (2), suggesting that the composition of mitochondrial proteins in diabetes plays an important role in β-cell viability. In addition, the composition of mitochondrial proteins and their function is largely dependent on mitochondrial dynamics, where the balance between fusion and fission is essential for regulating mitochondrial morphology and maintaining a healthy population of mitochondria (12). In the Akita+/Ins2-derived β-cells, mitochondrial fission was elevated and resulted in smaller fragmented mitochondria.
Damaged mitochondria are typically programmed for removal by mitophagy, and it has been shown previously that inhibition of mitophagy results in the accumulation of damaged mitochondria that generate increased levels of reactive oxygen species (28). However, although Akita+/Ins2-derived β-cells exhibited more mitochondrial fragmentation, LC3 protein levels as well as autophagic flux are comparable in the Akita+/Ins2-derived β-cells and the WT β-cells. These data suggest that, although there are more damaged mitochondria in the Akita+/Ins2-derived β-cells, autophagic flux is not adequate to meet the demand for mitochondrial clearance. Intriguingly, higher levels of LC3-II were detected in the mitochondrial fraction, perhaps due to a redistribution of LC3-II to the mitochondria rather than the autophagosomes. Paradoxically, although there is evidence for deterioration in mitochondrial quality, perhaps due to a lowering in energetic demand in Akita+/Ins2-derived β-cells, the mitochondrial population appears to be able to maintain an energized state. A decrease in mitochondrial membrane potential is one of the characteristics of mitochondria targeted for mitophagy (20), and this is not occurring in Akita+/Ins2-derived cells. On the contrary, the mitochondrial membrane potential is higher in Akita+/Ins2-derived β-cells than in WT cells and is associated with increased superoxide. Notably, the levels of both p62 and Parkin, proteins vital for targeting and transporting mitochondria to the phagophore, are lower in Akita+/Ins2-derived β-cells. These lowered levels may result in an insufficient mitophagy activity, contributing to accumulation of damaged mitochondria in the Akita+/Ins2-derived β-cells. It is important to note that the decrease in p62 levels was due to a decrease in transcription, whereas the decrease in Parkin appears to be independent of transcriptional repression. The mechanisms for transcriptional repression of p62 in Akita+/Ins2-derived β-cells are not known, although we speculate that this may be mediated by the ROS-Nrf pathway. Furthermore, the increased mitochondrial ROS may contribute to the increased cytosolic oxidation of protein thiols and glutathione. Recently, it has been shown in two reports that stimulation of autophagy with rapamycin in Akita+/Ins2 mice and in two different β-cell lines attenuates stress and T1DM pathogenesis (5, 6), supporting the hypothesis that an increase in mitophagy may be required for maintaining mitochondrial quality in Akita+/Ins2-derived β-cells.
One of the limitations of the study is that transformed cell lines were used. However, our findings clearly demonstrate the ER stress induced by thapsigargin causes mitochondrial dysfunction in two β-cell lines and in normal intact islets, and a recent study has shown that thapsigargin can inhibit autophagy (18). Furthermore, our data suggest that the accumulation of damaged mitochondria due to ER stress and insufficient autophagy in combination with oxidative stress may contribute to poor pancreatic β-cell function. As a result, this may contribute to dysregulation of physiological mechanisms involved in insulin release and hyperglycemia and suggests that promotion of autophagy may preserve β-cells in individuals with T1DM mutations in the insulin gene.
GRANTS
We appreciate support from the following: T32-DK-007545 (T. Mitchell), DK-69455 (S. Ramanadham), NS-064090 and a Veterans Affairs Merit Award (J. Zhang), AA-13395 and DK-075865 (V. M. Darley-Usmar), and the O'Brien Center P30-DK-079337 (S. Barnes and V. M. Darley-Usmar). Funds for the mass spectrometer used in this study were provided by a grant from the University of Alabama at Birmingham Health Services Foundation General Endowment Fund.
DISCLOSURES
V. M. Darley-Usmar is a member of the Seahorse Biosciences Scientific Advisory Board.
AUTHOR CONTRIBUTIONS
T.M., K.M., X.L., S.B., A.K., S.R., and V.M.D.-U. contributed to the conception and design of the research; T.M., M.S.J., X.O., B.K.C., K.M., Y.G., and D.R.M. performed the experiments; T.M., M.S.J., X.O., B.K.C., K.M., Y.G., D.R.M., and V.M.D.-U. analyzed the data; T.M., M.S.J., B.K.C., K.M., X.L., D.R.M., S.B., J.Z., S.R., and V.M.D.-U. interpreted the results of the experiments; T.M., M.S.J., B.K.C., and K.M. prepared the figures; T.M. drafted the manuscript; T.M., M.S.J., K.M., S.B., J.Z., S.R., and V.M.D.-U. edited and revised the manuscript; T.M., M.S.J., X.O., B.K.C., K.M., X.L., Y.G., D.R.M., S.B., J.Z., A.K., S.R., and V.M.D.-U. approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank Dr. Asta Jurkuvenaite and Dr. Tomader Ali for technical assistance.
REFERENCES
- 1.Ahamed A, Unnikrishnan AG, Pendsey SS, Nampoothiri S, Bhavani N, Praveen VP, Kumar H, Jayakumar RV, Nair V, Ellard S, Edghill EL. Permanent neonatal diabetes mellitus due to a C96Y heterozygous mutation in the insulin gene. A case report. JOP 9: 715–718, 2008 [PubMed] [Google Scholar]
- 2.Anello M, Lupi R, Spampinato D, Piro S, Masini M, Boggi U, Del Prato S, Rabuazzo AM, Purrello F, Marchetti P. Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia 48: 282–289, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Araki E, Oyadomari S, Mori M. Impact of endoplasmic reticulum stress pathway on pancreatic beta-cells and diabetes mellitus. Exp Biol Med (Maywood) 228: 1213–1217, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology 130: 167–178, 1992 [DOI] [PubMed] [Google Scholar]
- 5.Bachar-Wikstrom E, Wikstrom JD, Ariav Y, Tirosh B, Kaiser N, Cerasi E, Leibowitz G. Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes. Diabetes 62: 1227–1237, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartolome A, Guillen C, Benito M. Autophagy plays a protective role in endoplasmic reticulum stress-mediated pancreatic β cell death. Autophagy 8: 1757–1768, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breyer MD, Böttinger E, Brosius FC, 3rd, Coffman TM, Harris RC, Heilig CW, Sharma K AMDCC. Mouse models of diabetic nephropathy. J Am Soc Nephrol 16: 27–45, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes 54: 1615–1625, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Chacko BK, Reily C, Srivastava A, Johnson MS, Ye Y, Ulasova E, Agarwal A, Zinn KR, Murphy MP, Kalyanaraman B, Darley-Usmar V. Prevention of diabetic nephropathy in Ins2(+/)−(AkitaJ) mice by the mitochondria-targeted therapy MitoQ. Biochem J 432: 9–19, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cnop M, Welsh N, Jonas JC, Jörns A, Lenzen S, Eizirik DL. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes 54, Suppl 2: S97–S107, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Colombo C, Porzio O, Liu M, Massa O, Vasta M, Salardi S, Beccaria L, Monciotti C, Toni S, Pedersen O, Hansen T, Federici L, Pesavento R, Cadario F, Federici G, Ghirri P, Arvan P, Iafusco D, Barbetti F; Early Onset Diabetes Study Group of the Italian Society of Pediatric Endocrinology and Diabetes (SIEDP) Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. J Clin Invest 118: 2148–2156, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol 8: 870–879, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, Zou L, Chatham JC, Hill BG, Zhang J, Landar A, Darley-Usmar VM. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med 51: 1621–1635, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radic Biol Med 48: 905–914, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edghill EL, Flanagan SE, Patch AM, Boustred C, Parrish A, Shields B, Shepherd MH, Hussain K, Kapoor RR, Malecki M, MacDonald MJ, Støy J, Steiner DF, Philipson LH, Bell GI, Neonatal Diabetes International Collaborative Group. Hattersley AT, Ellard S. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes 57: 1034–1042, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev 29: 42–61, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Fujitani Y, Ueno T, Watada H. Autophagy in health and disease. 4. The role of pancreatic β-cell autophagy in health and diabetes. Am J Physiol Cell Physiol 299: C1–C6, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 42: 731–743, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbach N, Rathkolb B, Kemter E, Pichl L, Klaften M, de Angelis MH, Halban PA, Wolf E, Aigner B, Wanke R. Dominant-negative effects of a novel mutated Ins2 allele causes early-onset diabetes and severe beta-cell loss in Munich Ins2C95S mutant mice. Diabetes 56: 1268–1276, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Hill BG, Benavides GA, Lancaster JR, Jr, Ballinger S, Dell'italia L, Jianhua Z, Darley-Usmar VM. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem 393: 1485–1512, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill BG, Ramana KV, Cai J, Bhatnagar A, Srivastava SK. Measurement and identification of S-glutathiolated proteins. Methods Enzymol 473: 179–197, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill BG, Reily C, Oh JY, Johnson MS, Landar A. Methods for the determination and quantification of the reactive thiol proteome. Free Radic Biol Med 47: 675–683, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol 295: C849–C868, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajimoto Y, Kaneto H. Role of oxidative stress in pancreatic beta-cell dysfunction. Ann NY Acad Sci 1011: 168–176, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH. Ubiquitinated-protein aggregates form in pancreatic beta-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes 56: 930–939, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Kaushik S, Singh R, Cuervo AM. Autophagic pathways and metabolic stress. Diabetes Obes Metab 12, Suppl 2: 4–14, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupa T, Skupien J, Malecki MT. Monogenic models: what have the single gene disorders taught us? Curr Diab Rep 12: 659–666, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J 441: 523–540, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei X, Bone RN, Ali T, Wohltmann M, Gai Y, Goodwin KJ, Bohrer AE, Turk J, Ramanadham S. Genetic modulation of islet β-cell iPLA(2)β expression provides evidence for its impact on β-cell apoptosis and autophagy. Islets 5: 29–44, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei X, Zhang S, Barbour SE, Bohrer A, Ford EL, Koizumi A, Papa FR, Ramanadham S. Spontaneous development of endoplasmic reticulum stress that can lead to diabetes mellitus is associated with higher calcium-independent phospholipase A2 expression: a role for regulation by SREBP-1. J Biol Chem 285: 6693–6705, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei X, Zhang S, Bohrer A, Bao S, Song H, Ramanadham S. The group VIA calcium-independent phospholipase A2 participates in ER stress-induced INS-1 insulinoma cell apoptosis by promoting ceramide generation via hydrolysis of sphingomyelins by neutral sphingomyelinase. Biochemistry 46: 10170–10185, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei XG, Vatamaniuk MZ. Two tales of antioxidant enzymes on β cells and diabetes. Antioxid Redox Signal 14: 489–503, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab 17: 491–506, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Like AA, Rossini AA. Streptozotocin-induced pancreatic insulitis: new model of diabetes mellitus. Science 193: 415–417, 1976 [DOI] [PubMed] [Google Scholar]
- 35.MacMillan-Crow LA, Thompson JA. Tyrosine modifications and inactivation of active site manganese superoxide dismutase mutant (Y34F) by peroxynitrite. Arch Biochem Biophys 366: 82–88, 1999 [DOI] [PubMed] [Google Scholar]
- 36.Marhfour I, Lopez XM, Lefkaditis D, Salmon I, Allagnat F, Richardson SJ, Morgan NG, Eizirik DL. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia 55: 2417–2420, 2012 [DOI] [PubMed] [Google Scholar]
- 37.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem 244: 6049–6055, 1969 [PubMed] [Google Scholar]
- 38.Mitchell T, Chacko B, Ballinger SW, Bailey SM, Zhang J, Darley-Usmar V. Convergent mechanisms for dysregulation of mitochondrial quality control in metabolic disease: implications for mitochondrial therapeutics. Biochem Soc Trans 41: 127–133, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, Shibasaki Y, Oka Y, Yamamura K. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127: 126–132, 1990 [DOI] [PubMed] [Google Scholar]
- 40.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 417: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nozaki J, Kubota H, Yoshida H, Naitoh M, Goji J, Yoshinaga T, Mori K, Koizumi A, Nagata K. The endoplasmic reticulum stress response is stimulated through the continuous activation of transcription factors ATF6 and XBP1 in Ins2+/Akita pancreatic beta cells. Genes Cells 9: 261–270, 2004 [DOI] [PubMed] [Google Scholar]
- 42.Oliva CR, Nozell SE, Diers A, McClugage SG, Sarkaria JN, Markert JM, Darley-Usmar VM, Bailey SM, Gillespie GY, Landar A, Griguer CE. Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J Biol Chem 285: 39759–39767, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SY, Ye H, Steiner DF, Bell GI. Mutant proinsulin proteins associated with neonatal diabetes are retained in the endoplasmic reticulum and not efficiently secreted. Biochem Biophys Res Commun 391: 1449–1454, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patil NK, Saba H, MacMillan-Crow LA. Effect of S-nitrosoglutathione on renal mitochondrial function: a new mechanism for reversible regulation of manganese superoxide dismutase activity? Free Radic Biol Med 56: 54–63, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez J, Hill BG, Benavides GA, Dranka BP, Darley-Usmar VM. Role of cellular bioenergetics in smooth muscle cell proliferation induced by platelet-derived growth factor. Biochem J 428: 255–267, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polak M, Dechaume A, Cavé H, Nimri R, Crosnier H, Sulmont V, de Kerdanet M, Scharfmann R, Lebenthal Y, Froguel P, Vaxillaire M; French ND (Neonatal Diabetes) Study Group Heterozygous missense mutations in the insulin gene are linked to permanent diabetes appearing in the neonatal period or in early infancy: a report from the French ND (Neonatal Diabetes) Study Group. Diabetes 57: 1115–1119, 2008 [DOI] [PubMed] [Google Scholar]
- 47.Rabinowitz JD, White E. Autophagy and metabolism. Science 330: 1344–1348, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasaki K, Donthamsetty R, Heldak M, Cho YE, Scott BT, Makino A. VDAC: old protein with new roles in diabetes. Am J Physiol Cell Physiol 303: C1055–C1060, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solomon M, Sarvetnick N. The pathogenesis of diabetes in the NOD mouse. Adv Immunol 84: 239–264, 2004 [DOI] [PubMed] [Google Scholar]
- 50.Støy J, Edghill EL, Flanagan SE, Ye H, Paz VP, Pluzhnikov A, Below JE, Hayes MG, Cox NJ, Lipkind GM, Lipton RB, Greeley SA, Patch AM, Ellard S, Steiner DF, Hattersley AT, Philipson LH, Bell GI; Neonatal Diabetes International Collaborative Group Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci USA 104: 15040–15044, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Støy J, Steiner DF, Park SY, Ye H, Philipson LH, Bell GI. Clinical and molecular genetics of neonatal diabetes due to mutations in the insulin gene. Rev Endocr Metab Disord 11: 205–215, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Supale S, Li N, Brun T, Maechler P. Mitochondrial dysfunction in pancreatic β cells. Trends Endocrinol Metab 23: 477–487, 2012 [DOI] [PubMed] [Google Scholar]
- 53.Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem 27: 502–522, 1969 [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Takeuchi T, Tanaka S, Kubo SK, Kayo T, Lu D, Takata K, Koizumi A, Izumi T. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. J Clin Invest 103: 27–37, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westbrook DG, Anderson PG, Pinkerton KE, Ballinger SW. Perinatal tobacco smoke exposure increases vascular oxidative stress and mitochondrial damage in non-human primates. Cardiovasc Toxicol 10: 216–226, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wikstrom JD, Sereda SB, Stiles L, Elorza A, Allister EM, Neilson A, Ferrick DA, Wheeler MB, Shirihai OS. A novel high-throughput assay for islet respiration reveals uncoupling of rodent and human islets. PLoS One 7: e33023, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M, Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 1: 425–437, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 115: 2656–2664, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Y, Santamaria P. Lessons on autoimmune diabetes from animal models. Clin Sci (Lond) 110: 627–639, 2006 [DOI] [PubMed] [Google Scholar]
- 60.Yoshioka M, Kayo T, Ikeda T, Koizumi A. A novel locus, Mody4, distal to D7Mit189 on chromosome 7 determines early-onset NIDDM in nonobese C57BL/6 (Akita) mutant mice. Diabetes 46: 887–894, 1997 [DOI] [PubMed] [Google Scholar]
- 61.Yuan Q, Tang W, Zhang X, Hinson JA, Liu C, Osei K, Wang J. Proinsulin atypical maturation and disposal induces extensive defects in mouse Ins2+/Akita β-cells. PLoS One 7: e35098, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zmijewski JW, Banerjee S, Abraham E. S-glutathionylation of the Rpn2 regulatory subunit inhibits 26 S proteasomal function. J Biol Chem 284: 22213–22221, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]