Abstract
Metabolic acidosis is a relatively common pathological condition that is defined as a decrease in blood pH and bicarbonate concentration. The renal proximal convoluted tubule responds to this condition by increasing the extraction of plasma glutamine and activating ammoniagenesis and gluconeogenesis. The combined processes increase the excretion of acid and produce bicarbonate ions that are added to the blood to partially restore acid-base homeostasis. Only a few cytosolic proteins, such as phosphoenolpyruvate carboxykinase, have been determined to play a role in the renal response to metabolic acidosis. Therefore, further analysis was performed to better characterize the response of the cytosolic proteome. Proximal convoluted tubule cells were isolated from rat kidney cortex at various times after onset of acidosis and fractionated to separate the soluble cytosolic proteins from the remainder of the cellular components. The cytosolic proteins were analyzed using two-dimensional liquid chromatography and tandem mass spectrometry (MS/MS). Spectral counting along with average MS/MS total ion current were used to quantify temporal changes in relative protein abundance. In all, 461 proteins were confidently identified, of which 24 exhibited statistically significant changes in abundance. To validate these techniques, several of the observed abundance changes were confirmed by Western blotting. Data from the cytosolic fractions were then combined with previous proteomic data, and pathway analyses were performed to identify the primary pathways that are activated or inhibited in the proximal convoluted tubule during the onset of metabolic acidosis.
Keywords: metabolic acidosis, proximal convoluted tubule, cytosolic proteins, mass spectrometry, spectral counting
metabolic acidosis is a relatively common clinical condition that is characterized by a decrease in blood pH and bicarbonate concentration (17). Numerous conditions can produce acidosis including the excess production of ketone bodies (diabetic ketoacidosis), the buildup of lactic acid, kidney disease, or the excessive loss of bicarbonate ions that occurs with severe diarrhea (4). The kidney is a key contributor to the whole body response to restore acid-base balance (5). In the absence of acidosis, the kidneys extract very little of the circulating plasma glutamine (39). However, during acidosis the proximal convoluted tubule extracts more than one-third of the plasma glutamine in a single pass through the kidneys (6, 37). This glutamine is catabolized in the proximal convoluted tubule, forming two molecules of ammonia and two molecules of bicarbonate per glutamine (45). The resulting ammonium ions are primarily transported across the apical membrane by the apical Na+/H+ exchanger type 3 (NHE-3) and are eventually excreted in the urine, while the bicarbonate ions are returned to the blood via a basolateral Na+-3HCO3− cotransporter (6). The result is a net excretion of acids and an increase in blood bicarbonate, which partially restores the buffering capacity of the blood.
Although many of the primary responses of the proximal convoluted tubule to metabolic acidosis are well characterized, much is still unknown regarding what other changes occur in the proximal convoluted tubule and how such responses are regulated. To fully understand the entire cellular response to acidosis, a global approach must be employed to simultaneously monitor the many changes that occur in the cell. Nowik et al. (30) used a genome-wide expression profiling technique to monitor changes in the mouse renal transcriptome that occur during onset of acidosis. They identified nearly 13,000 transcripts in the whole kidney and found that over 4,000 mRNAs were differentially expressed during either acute or chronic acidosis compared with controls. However, changes in the transcriptome do not necessarily produce corresponding changes in protein expression or activity (12). By contrast, mass spectrometry (MS)-based proteomics offers the advantage of being able to monitor the levels of many different proteins simultaneously and to characterize functional differences that occur in response to perturbation.
Previous work has used proteomics techniques to characterize the proteome of the rat proximal convoluted tubule, as well as its response to metabolic acidosis (7, 11, 44). However, the complexity of the proximal convoluted tubule proteome made it difficult to analyze the response of the entire cell simultaneously. Thus previous studies used cellular fractionation to simplify the samples before MS analysis. This approach allowed for more in-depth analysis of the proteome, but left several fractions unanalyzed. To date, the apical membrane (44) and mitochondrial (11) proteomes have been analyzed, but no work has focused on the cytosolic proteins. It is well established that much of the response of the proximal convoluted tubule to metabolic acidosis involves processes such as gluconeogenesis and mRNA stabilization that occur primarily in the cytosol (6). Thus, to address this knowledge gap, the current study characterized the proteome of cytosolic fractions from proximal convoluted tubule cells isolated from both acidotic and control rats.
MATERIALS AND METHODS
Animals.
All samples were prepared from male Sprague-Dawley rats between 6 and 12 wk of age that were purchased from Charles River Laboratories (Kingston, NY). Rats were provided with free access to rodent chow (Harlan-Tekland, Madison, WI) throughout the duration of the study. Control rats were provided with normal tap water, while experimental rats were made acidotic by providing 0.28 M NH4Cl as their sole source of drinking water for a period of 1, 3, or 7 days (hereafter referred to as acutely acidotic, acidotic, or chronically acidotic, respectively). Fluid consumption was checked daily to ensure that all of the acidotic rats consumed a volume of NH4Cl at least as great as the water consumed by the control rats. In addition, acutely acidotic rats were stomach loaded with 20 mmol/kg of body weight of NH4Cl at the start of treatment. Previous work established that this protocol produced a pronounced metabolic acidosis (pH = 7.11, [HCO3−] = 8.8 mM) in the acutely acidotic and acidotic animals that is partially compensated (pH = 7.34, [HCO3−] = 15.6 mM) in the chronically acidotic rats (44). Three control rats and three biological replicates per acidotic time point were used, for a total of 12 animals. All animals were randomly assigned to their respective treatment groups. The Institutional Animal Care and Use Committee at Colorado State University approved all of these procedures.
Isolation of proximal convoluted tubules and soluble cytosolic fractions.
The procedure for the isolation of rat renal proximal convoluted tubules has been described previously (11, 43). In brief, the renal cortex was dissected from freshly extracted tissue and submitted to collagenase digestion in a buffer containing 1 mM PMSF to inhibit protease activity. Proximal convoluted tubules were then separated by Percoll density gradient centrifugation and homogenized in 0.3 M sucrose containing 1 mM PMSF, 1 mM EDTA, and 12 mM HEPES, pH 7.4. The resulting homogenate was centrifuged twice at 700 g for 10 min to remove nuclei and cell debris, and then again for 10 min at 7,000 g to pellet a crude mitochondrial fraction. Finally, the samples were centrifuged at 100,000 g for 20 min at 4°C to pellet the remaining organelles. The supernatants from this spin contained the soluble cytosolic fractions that were used for further analyses. All samples were stored at −80°C to limit protein degradation.
Western blot analyses.
Samples containing 8 μg of protein (unless otherwise noted) were separated by 10 or 15% SDS-PAGE, if the target protein was larger or smaller than 40 kDa, respectively. Separated proteins were then transferred to a polyvinylidene fluoride (PVDF) membrane and incubated for a minimum of 2 h in Pierce Protein-Free Blocking Buffer (Thermo Scientific, Rockford, IL). The blocking buffer was removed, and membranes were incubated overnight with primary antibody diluted in blocking buffer. Primary antibodies specific to cathepsin B (EMD Millipore, Billerica, MA), aconitase 2 (Aviva Systems Biology, San Diego, CA), binding immunoglobulin protein (BiP; Cell Signaling Technology, Danvers, MA), and cAMP response element-binding protein 1 (CREB-1; Santa Cruz Biotechnolgy, Santa Cruz, CA) were used to assess the purity of the cytosolic fractions. These antibodies react with soluble proteins that are localized specifically to lysosomes, mitochondria, endoplasmic reticulum, and nuclei, respectively. Either 680 Dylight-conjugated goat anti-rabbit or 800 Dylight-conjugated goat anti-mouse was used as a secondary antibody. Membranes were imaged using an Odyssey Infrared Imager (LI-COR Biosciences, Lincoln, NE), and bands were quantified using the accompanying Image Studio software.
Sample preparation for MS analysis.
Cytosolic fractions were dialyzed into 50 mM NH4HCO3, and their concentrations were determined using the bicinchoninic acid assay (38). Aliquots containing 50 μg of the control, 1-day acutely acidotic, and 7-day chronically acidotic samples were precipitated with acetone and then solubilized in 8 M urea by bath sonication for 5 min. ProteaseMAX surfactant trypsin enhancer (Promega, Madison, WI) was added to a final concentration of 0.2%. Disulfide bonds were then reduced with dithiothrietol and alkalyated with iodoacetamide. Proteins were digested for 3 h at 37°C using Trypsin Gold mass-spectrometry grade protease (Promega). The digestion was halted by the addition of 0.5% trifluoroacetic acid, and samples were dried in a vacuum evaporator. The resulting peptides were purified using a C18 TopTip (Glygen, Columbia, MD) and dried again using a vacuum evaporator. Dried peptides were resuspended in 3% acetonitrile/0.1% formic acid immediately before MS analysis.
MS analysis.
Sample fractionation was performed using an online two-dimensional liquid chromatography (2D-LC) system. A 10-μg aliquot of digested peptides was injected directly onto a strong-cation exchange (SCX) column (ZORBAX BioSCX Series II, 800 μm × 50 mm, 3.5-μm column, Agilent Technologies, Santa Clara, CA). Elution from the SCX column was performed by the injection of 20-μl salt bumps containing 30, 40, 50, 60, 90, and 500 mM NaCl. Peptides eluted from the SCX column were further separated on a subsequent C18 reverse phase column (1200 Nano HPLC, Zorbax C18, 5 μm, 75-μm ID × 150-mm column, Agilent Technologies) using a 60-min linear gradient from 25 to 55% of a 90% acetonitrile/0.1% formic acid solution. Flow rates were maintained at 300 nl/min throughout the gradient.
Peptides eluted from the reverse phase column were injected directly into a linear ion trap mass spectrometer (LTQ, Thermo Scientific). Spectra were collected using a mass window of 200–2,000 m/z. A dynamic exclusion limit was used that allowed for a maximum of two MS/MS spectra for a given parent ion in 30 s, followed by a 90-s exclusion of that peptide. Compound lists of the resulting spectra were compiled using Xcaliber 2.2 software (Thermo Scientific) with an intensity threshold of 5,000 and 1 scan/group. Ion chromatograms were monitored throughout data collection to ensure proper function and consistency of the instrument. In addition, tryptic digests of bovine serum albumin were injected between each sample as a quality control.
Bioinformatics.
Tandem mass spectra (MS/MS) were searched against a UniProt-KB Rattus norvegicus reverse-concatenated database (downloaded December, 21 2011) using both Mascot (version 2.3.02, Matrix Science) and SEQUEST (version v.27, rev. 11, Sorcerer, Sage-N Research) search algorithms. The reverse database was searched to calculate the false discovery rates (FDR) (8). Search parameters were as follows: average mass, 2.5-Da peptide mass tolerance, 1.0-Da fragment ion mass tolerance, complete tryptic digestion with a maximum of two missed cleavages per peptide, a static modification of cysteine carbamidomethylation, and variable modifications of methionine oxidation and lysine acetylation. Search results were compiled and validated using Scaffold 3 software (version 3.6.0, Proteome Software, Portland, OR).
Filters were applied such that only proteins with 99% minimum protein probability, 95% minimum peptide probability, and at least two unique peptides were included in the data set. Protein and peptide probabilities were calculated using the Peptide and Protein Prophet algorithms included in the Scaffold 3 software (22, 28). Scaffold 3 software was also used to assign peptides shared between proteins to satisfy the laws of parsimony. Spectra from proteins identified by only two assigned unique peptides were manually validated using the following criteria: presence of at least three sequential y and b ions in the same spectrum, or at least five consecutive y or b ions with at least 5% relative intensity, as well as a lack of prominent unassigned peaks.
Relative protein abundance determination.
The data were first normalized using the method embedded in the Scaffold 3 software. In brief, a scaling factor was created by dividing the total number of spectra identified in a sample by the mean of spectra identified across all samples in the analysis. The spectral counts (SpC) and average total ion current values of the MS/MS spectra (MS2 TIC) values for each of the proteins in the sample were then divided by this scaling factor. Spectral counting uses the sum of the MS/MS spectra assigned to each protein as a measure of abundance (33). However, a major limitation to SpC analysis is the inaccuracy associated with quantification of low-abundance proteins (32). Therefore, this analysis was only applied to those proteins which had >10 SpC in at least one treatment when all of the biological replicates for that treatment were combined. By contrast, average MS2 TIC analysis uses the average ion intensity of all peptides for a particular protein to estimate abundance. The latter method provides improved sensitivity for the detection of relative changes in low-abundance proteins, but is less reliable for high-abundance proteins due to saturation of the acquired spectra (46). Thus the two methods are complementary, and their combined application increases the identification of significant changes (10). Therefore, both SpC and MS2 TIC values were used to calculate fold-changes in abundance, and statistical tests were applied to determine which of the observed changes were significant.
Statistical analysis.
One-way ANOVA was performed on the normalized values using DanteR (41), an R-based proteomics software, to determine which proteins showed significant abundance changes in samples from acidotic rats compared with those from controls. The ANOVA model used by DanteR was developed specifically for proteomics studies and has been shown to accurately detect changes in protein abundance (31). Fold-changes for the SpC data were calculated by first adding a pseudocount of one to all the quantitative values (to remove zeros), and then the mean value for each treatment was divided by the mean control value. MS2 TIC fold-changes were calculated in the same manner; with the exception that a pseudocount value of 506 (the lowest detected MS2 TIC value in the data set) was used instead of 1. The calculated fold-differences were considered to be significant if they had a P value <0.05 and a fold-change >1.5 compared with the control samples.
Validation of proteomics data.
A subset of the proteins which were found to be differentially expressed by the MS analysis was validated by Western blotting. Antibodies specific to glutathione S-transferase P1 (GSTP1), phenylalanine hydroxylase (PAH), and Na-K-ATPase α1 (AT1A1) were purchased from Novus Biologicals (Littleton, CO), while aconitase 1 (ACO1), glycerol kinase (GK), and d-dopachrome decarboxylase (DDT) antibodies were from Abcam (Cambridge, MA). Retinoid-inducible serine carboxypeptidases (RISC) as well as prostaglandin E-synthase 3 (PTGES3) antibodies were obtained from Acris (San Diego, CA), and a cytosolic phosphoenolpyruvate carboxykinase (PEPCK) antibody was obtained from Abgent (San Diego, CA). An antibody to GAPDH (GeneTex, Irvine, CA) was used as a loading control for all blots. A total of 8 μg of protein was loaded for each of the control, acutely acidotic (1-day), acidotic (3-day), and chronically acidotic (7-day) biological replicates (n = 3), with the exception of the AT1A1 and PAH blots where 12 μg was loaded. All blots were run in duplicate, and the data were combined into one analysis.
Renal hypertrophy.
Rats that had either undergone a sham procedure or a uninephrectomy (UNX) of the left kidney were obtained from Charles River Laboratories. They were analyzed to determine whether the changes in protein abundance observed in the acidotic samples were due to the acidosis or the associated renal hypertrophy (24). The removal of one kidney produces a greater hypertrophy in the remaining kidney than what occurs in both kidneys during acidosis (36). Kidneys dissected from the uninephrectomized rats were 36% larger on average than kidneys from the sham-operated animals. Rats were euthanized 21 days postoperation to achieve maximum renal hypertrophy, and soluble cytosolic protein samples were prepared from isolated proximal convoluted tubules. Western blotting was performed to determine whether renal hypertrophy altered the levels of PEPCK, GSTP1, ACO1, PAH, AT1A1, or RISC levels.
Pathway analyses.
A variety of tools were used to categorize the cellular pathways and molecular functions of the proteins that were differentially regulated during acidosis. Gene ontology (GO) terms are a collection of descriptors that are standardized to allow for clear annotation of genes and proteins across all fields of research. Scaffold 3 software has built-in tools that allow for easy extraction of GO annotations for proteins that are annotated in the UniProt-KB database. This tool was used to assess the cellular location and function of proteins identified in the cytosolic protein data set. For some analyses, the acquired knowledge of the authors was used to group the narrowly specified GO terms into broader classifications. In addition, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a freely available, web-based software that detects and maps known and predicted protein associations (40). STRING uses a variety of protein annotation databases including the GO database, the Kyoto Encyclopedia of Genes and Genomes (KEGG), as well as other methods to map protein interactions. This software was used to visualize interactions among proteins that were differentially regulated during acidosis. This was useful in determining what cellular processes and pathways are affected in the proximal convoluted tubule during the onset of acidosis. In addition, STRING allows users to download the raw annotation data that the software uses to map associations. This output was also used to elucidate the pathways that respond to acidosis in the proximal convoluted tubule.
RESULTS
Isolation of soluble cytosolic proteins.
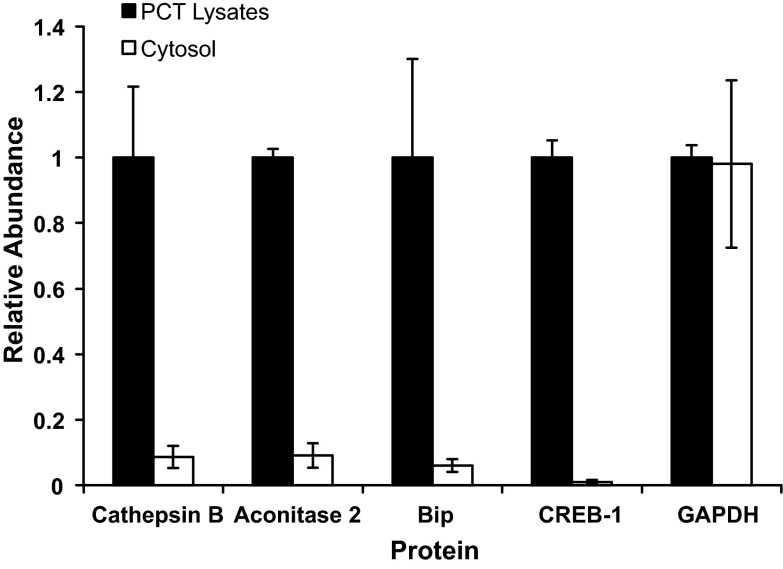
Proximal convoluted tubules were isolated from the rat renal cortex by Percoll gradient centrifugation (43). Previous work established that this method yields proximal convoluted tubules that are ∼95% pure (7, 43). Soluble cytosolic fractions were obtained by a differential centrifugation protocol that concluded with a 20-min 100,000-g spin. This step should pellet the remaining organelles, leaving only the soluble cytosolic proteins. Western blot analysis was used to determine the purity of the resulting cytosolic fractions. Primary antibodies specific to cathepsin B, aconitase 2, BiP, and CREB-1 were used to quantify the relative abundance of the respective proteins in the whole cell lysates and the cytosolic fractions. These proteins are soluble proteins that are contained specifically in the lysosomes, mitochondria, endoplasmic reticulum, and nucleus of intact cells, respectively. Thus their depletion in the cytosolic fractions is a measure of the disruption of the various organelles and the purity of the cytosolic fraction. Western blot analyses demonstrated that >90% of the four proteins were depleted in the cytosolic fractions compared with the crude homogenates (Fig. 1). By contrast, >95% of the initial GAPDH was recovered in the cytosolic fraction. Thus these data strongly suggest that the differential centrifugation procedure successfully enriched the soluble cytosolic proteins without an appreciable release of soluble proteins from various organelles. Analysis of the acidotic samples showed similar enrichment of the cytosolic fractions (data not shown).
Fig. 1.
Western blot analysis of organelle-specific proteins in the initial lysates of isolated proximal convoluted tubules (PCT) and soluble cytosolic fractions. Cathepsin B, aconitase 2, binding immunoglobulin protein (BiP), and cAMP response element binding protein 1 (CREB-1) are soluble proteins that are contained specifically in lysosomes, mitochondria, endoplasmic reticulum, and nuclei, respectively. Levels of GAPDH were also quantified to assess the recovery of a cytosolic protein.
MS analysis of soluble cytosolic fractions.
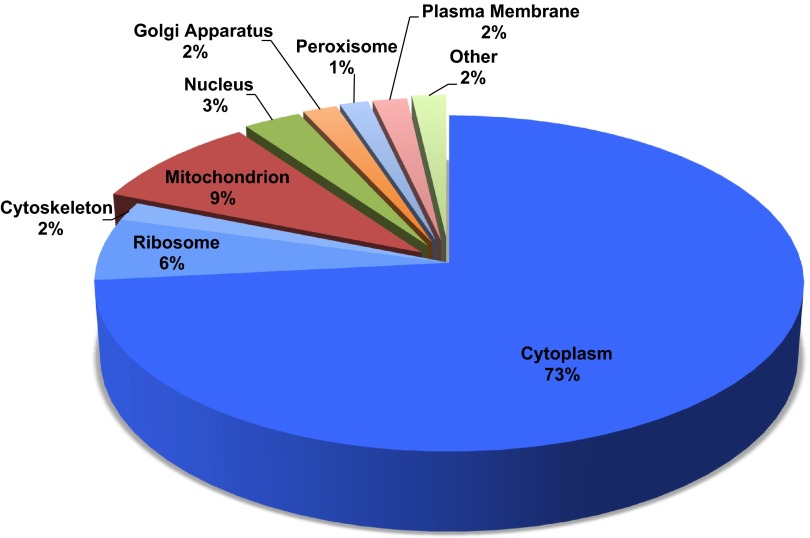
Cytosolic fractions were obtained from three biological replicates each of nonacidotic (control), acutely acidotic (1-day), or chronically acidotic (7-day) rats. Aliquots of these samples were digested with trypsin, and the resulting peptides were analyzed using 2D-LC coupled to MS/MS (2D-LC-MS/MS). Raw MS data were searched against a reverse concatenated R. norvegicus database using both MASCOT and SEQUEST protein search engines. In total, 91,044 spectra were identified, which led to the identification of 461 proteins in all of the analyzed samples. The data set had a 0.1% peptide and a 2.2% protein false discovery rate. The number of peptides and proteins identified in each sample was similar across all samples (data not shown). A list of 25 proteins with the highest number of spectral counts is provided in Table 1. A complete list of the identified proteins is available in a searchable online format at http://helixweb.nih.gov/ESBL/Database/PCT/ (18). Scaffold 3 software was used to extract GO terms for the identified proteins. Cellular location GO terms were available for 348 of the 461 proteins. Of those with annotations, 73% were cytosolic and another 8% were ribosomal or cytoskeletal, which would also be enriched in the fractionation procedure (Fig. 2).
Table 1.
List of 25 proteins identified during LC/MS/MS with the highest number of spectral counts
| Mean Spectral Counts ± SE |
|||||||
|---|---|---|---|---|---|---|---|
| Protein Name | UniProt Entry Name | UniProt Accession | Gene Name | Molecular Mass | Control | 1 day | 7 day |
| Hydroxyacid oxidase 2 | HAOX2_RAT | Q07523 | Hao2 | 39 kDa | 735.0 ± 141.6 | 1,001.0 ± 178.2 | 778.0 ± 87.2 |
| α-Enolase | ENOA_RAT | P04764 | Eno1 | 47 kDa | 562.7 ± 184.8 | 671.3 ± 151.2 | 466.7 ± 62.1 |
| Fructose-bisphosphate aldolase | Q66HT1_RAT | Q66HT1 | Aldob | 40 kDa | 457.0 ± 65.3 | 446.3 ± 47.5 | 559.7 ± 26.2 |
| Uncharacterized protein | D4AD25_RAT | D4AD25 | Cltc | 192 kDa | 309.3 ± 77.8 | 491.3 ± 59.5 | 389.3 ± 56.3 |
| Fructose-1,6-bisphosphatase 1 | F16P1_RAT | P19112 | Fbp1 | 40 kDa | 376.3 ± 90.6 | 417.3 ± 51.2 | 360.0 ± 23.0 |
| Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | PCKGC_RAT | P07379 | Pck1 | 69 kDa | 107.3 ± 23.1 | 644.3 ± 13.6 | 344.3 ± 55.8 |
| Actin, cytoplasmic 2 | ACTG_RAT | P63259 | Actg1 | 42 kDa | 303.0 ± 41.8 | 300.7 ± 41.3 | 243.3 ± 22.9 |
| Tubulin β-4B chain | TBB4B_RAT | Q6P9T8 | Tubb4b | 50 kDa | 271.3 ± 40.7 | 303.3 ± 16.2 | 230.7 ± 3.8 |
| Glyceraldehyde-3-phosphate dehydrogenase | D3ZGY4_RAT | D3ZGY4 | Gapdh-ps2 | 36 kDa | 276.7 ± 16.0 | 273.3 ± 29.9 | 240.3 ± 43.0 |
| Tubulin β-5 chain | TBB5_RAT | P69897 | Tubb5 | 50 kDa | 250.7 ± 40.1 | 267.0 ± 32.1 | 223.7 ± 17.8 |
| Major urinary protein | MUP_RAT | P02761 | N/A | 21 kDa | 328.0 ± 152.5 | 99.7 ± 27.9 | 249.7 ± 136.9 |
| Tubulin α-1C chain | TBA1C_RAT | Q6AYZ1 | Tuba1c | 50 kDa | 189.0 ± 64.0 | 254.3 ± 55.1 | 211.0 ± 39.1 |
| Tubulin α-4A chain | TBA4A_RAT | Q5XIF6 | Tuba4a | 50 kDa | 181.7 ± 60.7 | 237.3 ± 51.8 | 195.7 ± 43.0 |
| Glutathione S-transferase α-1 | GSTA1_RAT | P00502 | Gsta1 | 26 kDa | 200.0 ± 29.1 | 213.0 ± 8.0 | 201.7 ± 42.2 |
| Uncharacterized protein | D3ZXY4_RAT | D3ZXY4 | Aldh8a1 | 53 kDa | 202.0 ± 26.9 | 211.0 ± 18.0 | 184.7 ± 12.8 |
| Tubulin β-2A chain | TBB2A_RAT | P85108 | Tubb2a | 50 kDa | 247.0 ± 38.6 | 187.0 ± 93.6 | 153.3 ± 76.7 |
| Glutathione S-transferase α-3 | GSTA3_RAT | P04904 | Gsta3 | 25 kDa | 157.0 ± 52.5 | 216.7 ± 52.1 | 197.3 ± 73.7 |
| Malate dehydrogenase, cytoplasmic | MDHC_RAT | O88989 | Mdh1 | 36 kDa | 159.3 ± 63.8 | 225.7 ± 60.7 | 147.7 ± 29.6 |
| Catalase | CATA_RAT | P04762 | Cat | 60 kDa | 129.7 ± 20.2 | 195.0 ± 31.5 | 163.3 ± 20.3 |
| Argininosuccinate synthase | ASSY_RAT | P09034 | Ass1 | 46 kDa | 155.0 ± 36.8 | 181.0 ± 32.3 | 147.7 ± 15.6 |
| Alcohol dehydrogenase [NADP+] | AK1A1_RAT | P51635 | Akr1a1 | 37 kDa | 179.7 ± 74.7 | 132.3 ± 24.5 | 115.7 ± 42.1 |
| Transketolase, isoform CRA_a | G3V826_RAT | G3V826 | Tkt | 71 kDa | 104.3 ± 12.8 | 119.7 ± 6.1 | 118.0 ± 36.2 |
| Heat shock protein HSP 90-β | HS90B_RAT | P34058 | Hsp90ab1 | 83 kDa | 90.0 ± 18.8 | 136.0 ± 18.4 | 109.3 ± 19.4 |
| Aflatoxin B1 aldehyde reductase member 3 | ARK73_RAT | P38918 | Akr7a3 | 37 kDa | 117.0 ± 22.6 | 98.3 ± 36.1 | 107.7 ± 13.0 |
| Isocitrate dehydrogenase [NADP] cytoplasmic | IDHC_RAT | P41562 | Idh1 | 47 kDa | 103.0 ± 49.7 | 101.7 ± 17.7 | 63.7 ± 8.7 |
LC/MS/MS, liquid chromatography tandem mass spectrometry.
Fig. 2.
Gene ontology (GO) cellular location terms for the cytosolic proteins identified in isolated proximal convoluted tubules. Only proteins that were annotated with a cellular location (348 of 471 identified) are represented here. Sections colored blue contain proteins that should be enriched to be retained in the cytosolic fraction. Proteins were grouped into the “other” category if there were fewer than 3 proteins sharing the same cellular location annotation.
Among the 10 most abundant proteins identified in the cytosolic fraction of the proximal convoluted tubule are three enzymes that catalyze reversible reactions of glycolysis and the two cytosolic enzymes that are unique to gluconeogenesis, PEPCK and fructose 1,6-bisphosphatase. This finding is consistent with the high capacity of the proximal convoluted tubule to synthesize glucose. Interestingly, hexokinase and phosphofructokinase-1 were the only enzymes of glycolysis that were not identified in the complete analysis. This finding suggests that the proximal convoluted tubule has limited ability to phosphorylate and catabolize the glucose that is absorbed from the glomerular filtrate. By contrast, the more distal segments of nephron consume significant amounts of glucose (15). As a result, arterial-venous differences cannot be used to estimate the synthesis of glucose by the proximal convoluted tubule.
Relative quantitation of protein abundance.
A combination of SpC and MS2 TIC was used to estimate the relative abundance of proteins in the proximal convoluted tubules following different treatments. SpC uses the number of identified MS/MS spectra assigned to a particular protein as a measure of abundance (29, 49), whereas average MS2 TIC is based on the average ion intensity of the MS/MS spectra for each peptide assigned to a given protein (2). Before statistical analysis, the data were normalized as described in materials and methods. Relative fold-changes were calculated by dividing the average SpC or MS2 TIC value for a protein in a particular treatment by the corresponding value for the control samples, and P values were calculated by ANOVA. A protein was considered to be significantly altered if the comparison produced a fold-change of >1.5 or <0.67 and had a P value <0.05.
Of the 461 identified proteins, 24 were found to be differentially expressed in the cytosolic fractions in at least 1 time point compared with control tubules. Of those, 10 were significant only in the acutely acidotic samples (Table 2), 9 in only the chronically acidotic samples (Table 3), and 5 in both conditions. Furthermore, SpC detected more differentially regulated proteins at both time points, with 10 proteins being identified in the acutely acidotic samples and 9 in the chronic acidotic. Among all of the enzymes required for gluconeogenesis, only PEPCK was significantly increased during acute and chronic acidosis, suggesting that it catalyzes a rate-determining step in renal glucose synthesis. Interestingly, only RISC met the cut-offs for significance by both methods in the acutely acidotic samples. However, SpC indicated a significant increase, whereas MS2 TIC identified a slight decrease. Western blot analysis was performed to resolve this discrepancy.
Table 2.
Differentially regulated cytosolic proteins during acute metabolic acidosis
| Protein Name | Accession Number | Entry Name | Gene Name | P Value (SpC/TIC)* | Fold-Change (SpC/TIC)* | Detection Method |
|---|---|---|---|---|---|---|
| Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | P07379 | PCKGC_RAT | Pck1 | 0.001 | 4.94 | SpC |
| Sodium/potassium-transporting ATPase subunit α-1 | P06685 | AT1A1_RAT | Atp1a1 | 0.003 | 0.10 | SpC |
| Prostaglandin E synthase 3 | P83868 | TEBP_RAT | Ptges3 | 0.005 | 3.73 | SpC |
| Low-density lipoprotein receptor-related protein 2 | P98158 | LRP2_RAT | Lrp2 | 0.008 | 0.33 | SpC |
| Phenylalanine-4-hydroxylase | P04176 | PH4H_RAT | Pah | 0.012 | 0.49 | SpC |
| Aconitase 1 | D4ACL3 | D4ACL3_RAT | Aco1 | 0.013 | 2.27 | SpC |
| RCG32401, isoform CRA_a | D3ZPA9 | D3ZPA9_RAT | Ctsa | 0.014 | 4.60 | SpC |
| WD repeat-containing protein 1 | Q5RKI0 | WDR1_RAT | Wdr1 | 0.024 | 3.53 | SpC |
| 6-Phosphogluconolactonase | P85971 | 6PGL_RAT | Pgls | 0.025 | 0.33 | SpC |
| Ubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast) (Predicted), isoform CRA_a | D3ZNQ6 | D3ZNQ6_RAT | Ube2m | 0.034 | 2.49 | SpC |
| Retinoid-inducible serine carboxypeptidase | Q920A6 | RISC_RAT | Scpep1 | 0.011/0.049 | 1.88/0.42 | Both |
| Peroxisomal 2,4-dienoyl-CoA reductase | Q9Z2M4 | DECR2_RAT | Decr2 | 0.009 | 0.17 | TIC |
| Pyruvate carboxylase, mitochondrial | P52873 | PYC_RAT | Pc | 0.010 | 2.56 | TIC |
| 3-Hydroxyisobutyrate dehydrogenase, mitochondrial | P29266 | 3HIDH_RAT | Hibadh | 0.014 | 5.25 | TIC |
| 40S ribosomal protein S3 | P62909 | RS3_RAT | Rps3 | 0.027 | 2.35 | TIC |
Table 3.
Differentially regulated cytosolic proteins during chronic metabolic acidosis
| Protein Name | Accession Number | Entry Name | Gene Name | P Value | Fold-Change | Detection Method |
|---|---|---|---|---|---|---|
| Sodium/potassium-transporting ATPase subunit α-1 | P06685 | AT1A1_RAT | Atp1a1 | 0.003 | 0.14 | SpC |
| Low-density lipoprotein receptor-related protein 2 | P98158 | LRP2_RAT | Lrp2 | 0.010 | 0.36 | SpC |
| Phosphoenolpyruvate carboxykinase, cytosolic | P07379 | PCKGC_RAT | Pck1 | 0.012 | 3.37 | SpC |
| Triosephosphate isomerase | P48500 | TPIS_RAT | Tpi1 | 0.020 | 0.54 | SpC |
| Glutathione S-transferase P | P04906 | GSTP1_RAT | Gstp1 | 0.026 | 3.15 | SpC |
| Transgelin-2 | Q5XFX0 | TAGL2_RAT | Tagln2 | 0.027 | 0.46 | SpC |
| Uncharacterized protein | D3ZPL5 | D3ZPL5_RAT | RGD1562953 | 0.041 | 3.07 | SpC |
| N-acylglucosamine 2-epimerase | F1LQ78 | F1LQ78_RAT | Renbp | 0.047 | 0.23 | SpC |
| Acyl-coenzyme A thioesterase 1 | O88267 | ACOT1_RAT | Acot1 | 0.047 | 0.48 | SpC |
| d-Dopachrome decarboxylase | P80254 | DOPD_RAT | Ddt | 0.031 | 5.47 | TIC |
| Aromatic l-amino acid decarboxylase | P14173 | DDC_RAT | Ddc | 0.016 | 0.28 | TIC |
| Prostaglandin E synthase 3 | P83868 | TEBP_RAT | Ptges3 | 0.039 | 15.86 | TIC |
| Ketohexokinase | Q02974 | KHK_RAT | Khk | 0.040 | 2.09 | TIC |
| Pyruvate carboxylase, mitochondrial | P52873 | PYC_RAT | Pc | 0.045 | 2.05 | TIC |
Validation of MS data.
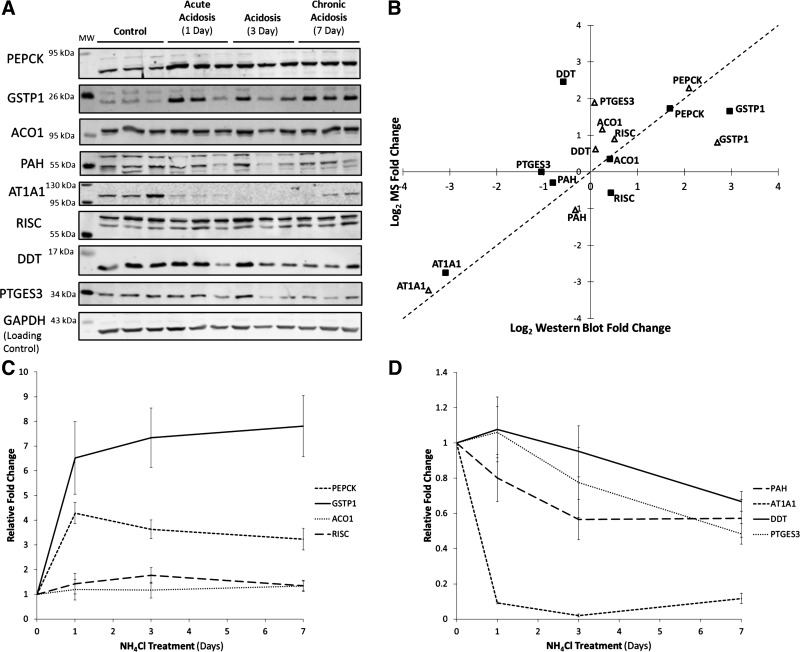
Western blotting was also used to validate a subset of the differentially expressed proteins. Primary antibodies were obtained that are specific to cytosolic PEPCK, glutathione S-transferase P (GSTP1), aconitase 1 (ACO1), phenylalanine-4-hydroxylase (PAH), sodium/potassium-transporting ATPase subunit alpha-1 (AT1A1), RISC, d-dopachrome decarboxylase (DDT), and prostaglandin E synthase 3 (PTGES3). Triplicate biological replicates from each time point, including the 3-day acidotic samples, were separated on a single polyacrylamide gel and probed with a primary antibody and an antibody specific for GAPDH, which was used as a loading control. The blots were probed with fluorescently labeled secondary antibodies and imaged using an infrared imager. Each analysis was run in duplicate and representative blots are shown in Fig. 3A. Fluorescence intensities were quantified and used as a measure of relative protein abundance. For most proteins, the Western blot data correlated well with the MS data (Fig. 3B). For the RISC protein, the Western blot analysis confirmed the results obtained with spectral counting. This result illustrates the importance of independent validation of MS data. Time courses showing the levels of each protein during the progression of acidosis are shown in Fig. 3, C and D.
Fig. 3.
Validation of mass spectrometry (MS) data by Western blotting. A: representative Western blots for cytosolic PEPCK, glutathione S-transferase P (GSTP1), aconitase 1 (ACO1), phenylalanine-4-hydroxylase (PAH), sodium/potassium-transporting ATPase subunit α-1 (AT1A1), Retinoid-inducible serine carboxypeptidases (RISC), d-dopachrome decarboxylase (DDT), and prostaglandin E synthase 3 (PTGES3). B: comparison of fold changes for acute (△) and chronic (■) acidotic samples compared with controls, as determined by MS and Western blot analysis. The MS fold-changes shown are those calculated using the method (SpC/TIC) in which the protein was significantly altered. In the case of RISC, the fold-change calculated by SpC was used, as it had the lower P value. C and D: temporal changes in protein abundance during the progression of acidosis as determined by Western blotting.
Effects of renal hypertrophy.
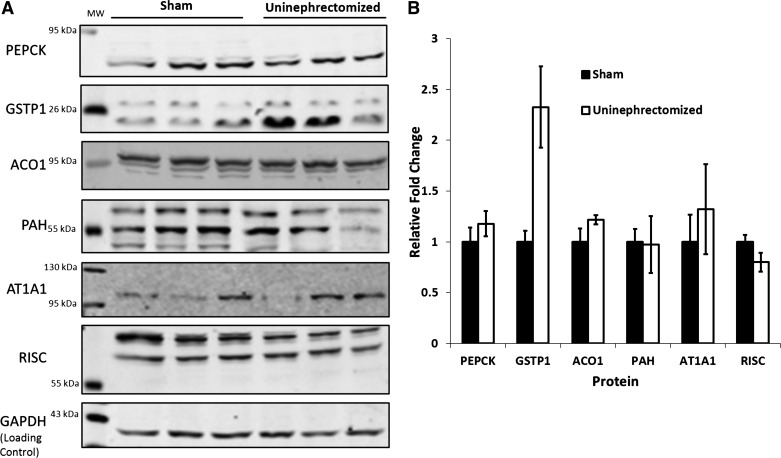
Metabolic acidosis causes hypertrophy of the proximal convoluted tubule (24, 34). To determine whether the observed changes in protein abundance were due to renal hypertrophy or the decrease in blood pH, rats were obtained that had either undergone a sham procedure or a uninephrectomy (UNX) where the left kidney was removed (n = 3/treatment). The removal of one kidney causes a renal hypertrophy that is greater than that which occurs during acidosis (36). After 21 days postoperation, the right kidneys were removed and soluble cytosolic fractions were prepared using the same method as used for the acidotic animals. The samples were then analyzed in duplicate by Western blotting to determine the relative levels of PEPCK, GSTP1, ACO1, PAH, AT1A1, and RISC. These proteins were chosen because both MS and Western blotting showed similar changes in abundance during the development of acidosis. A representative blot from each analysis is shown in Fig. 4A, along with quantification from the combined experiments (Fig. 4B). Overall, no difference was observed between treatments for any of the analyzed proteins, except for GSTP1. Levels of GSTP1 were about twofold higher in the UNX samples compared with controls. Although this increase is significant, it is unlikely that hypertrophy alone was responsible for the nearly eightfold increase in GSTP1 that was observed in chronically acidotic animals.
Fig. 4.
Effects of renal hypertrophy on expression of proteins that exhibit altered expression during metabolic acidosis. Cytosolic samples were prepared from proximal convoluted tubules isolated from the right kidneys of rats that had undergone a sham operation or a monolateral nephrectomy (left kidney removed). Protein levels were quantified by Western blot analysis. A: representative blots for each of the proteins analyzed (abbreviations are the same as in Fig. 3). B: quantification of the levels of each protein relative to the level of GAPDH, which was used as a loading control.
Detection of acetylated peptides.
Previous work has suggested that increased lysine acetylation of mitochondrial proteins may play a role in the response of the proximal convoluted tubule to metabolic acidosis (11, 48). Therefore, the MS data were searched for the variable modification of acetylated lysine. It is important to note that due to the small mass difference between lysine acetylation and trimethylation, the low-resolution ion trap mass spectrometer used in this study was not able to differentiate between the two modifications. Therefore, the sites identified as acetylated may actually be trimethylated. In total, 15 different acetylated/trimethylated peptides were identified across all of the analyzed samples, 9 of which were not previously identified (Table 4). No single acetylated peptide was detected more than five times in any treatment.
Table 4.
Acetylated proteins detected in the cytosolic fractions
| Number of Modified Spectra |
||||||||
|---|---|---|---|---|---|---|---|---|
| Protein Name | Accession Number | Control | Acute acidosis | Chronic acidosis | Peptide Sequence | Modified Site* | Site-Determining Ion | Novel |
| Heat shock protein HSP 90-β | P34058 | 1 | TKPIWTRNPDDITQEEYGEFYK | K286 | No | Yes | ||
| Sorbitol dehydrogenase | P27867 | 1 | 1 | 1 | AAPAKGENLSLVVHGPGDIR | K6 | No | No |
| Transitional endoplasmic reticulum ATPase | P46462 | 3 | ASGADSKGDDLSTAILK | K8 | Yes | No | ||
| 40S ribosomal protein S24 | D4A6H5 | 1 | QMVIDVLHPGKATVPK | K32 | Yes | Yes | ||
| Similar to RIKEN cDNA C630028N24 gene (Predicted), isoform CRA_b | D3ZUX1 | 4 | SSGDDQQSQAFTKPTFTEAQASALVESIFGFK | K14 | No | No | ||
| Uncharacterized protein | F1LNM3 | 1 | 1 | DPSKELAGLFEHK | K2224 | No | Yes | |
| 14-3-3 Protein ζ/Δa | P63102 | 1 | NLLSVAYKNVVGAR | K49 | Yes | No | ||
| Aldo-keto reductase family 1, member C1 | Q3MHS3 | 1 | GVVVLAKSFTEKR | K275 | Yes | No | ||
| Acyl-CoA-binding protein | P11030 | 1 | ENAMKTYVEKVEELK | K72 or K77 | No | No | ||
| Cytoplasmic dynein 1 heavy chain 1 | P38650 | 1 | VQSKVNLK | K1113 | Yes | Yes | ||
| Uncharacterized protein | D4A5F2 | 5 | 4 | 5 | KNILLEEKLNK | K2445 | Yes | Yes |
| Uncharacterized protein | D3Z8J0 | 1 | KLKAQMD | K199 | No | Yes | ||
| Uncharacterized protein | F1LP82 | 1 | 1 | MITIDGKQIK | K53 | Yes | Yes | |
| 26S Protease regulatory subunit 4 | P62193 | 1 | TLLAKAVANQTSATFLR | K237 | No | Yes | ||
| RCG21481, isoform CRA_b | D3ZLD5 | 1 | 1 | DKFMLQAKVSELK | K1308 | Yes | Yes | |
If no site-determining ions were detected, all possible modification sites are listed. Trypsin does not cleave after an acetylated lysine. Thus a single internal lysine must be the site of modification for peptides with a single internal lysine, and no site-determining ion.
Pathway analysis of available data characterizing the response of the proximal convoluted tubule to acidosis.
Previous studies have used MS-based proteomics to study the response of apical membrane (43) and mitochondrial (11) proteins of the proximal convoluted tubule to acidosis. These data were combined with the results of the current study to perform a pathway analysis of how the proximal convoluted tubule responds to metabolic acidosis. The previous study of isolated brush-border membrane vesicles (43) suggested that a significant fraction of the enzymes of glycolysis associates with the apical membrane during normal acid-base balance but dissociates during acidosis. In addition, the level of many of the more abundant mitochondrial proteins trapped in the isolated brush-border membrane vesicles increased significantly during acidosis. None of these enzymes were differentially expressed in the cytosolic or mitochondrial fractions. Therefore, it is likely that the changes in these proteins observed in the apical membrane vesicles were not representative of differences in cellular concentration. Thus these proteins were excluded from the remainder of the analyses. After exclusion of these proteins, a total of 98 proteins were found to be differentially regulated across all three data sets, of which 58 were upregulated and 40 were downregulated during either acute (1-day) or chronic (7-day) metabolic acidosis. Of these, 92 were annotated in the STRING database (http://www.string-db.org).
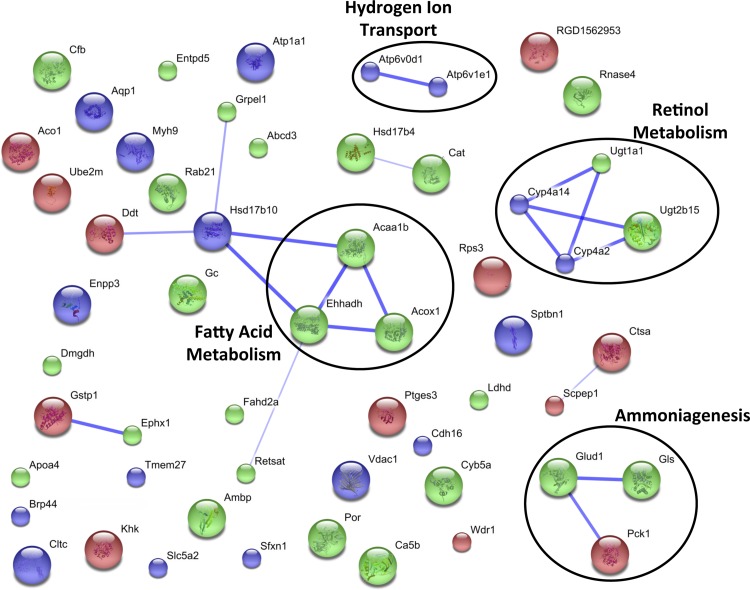
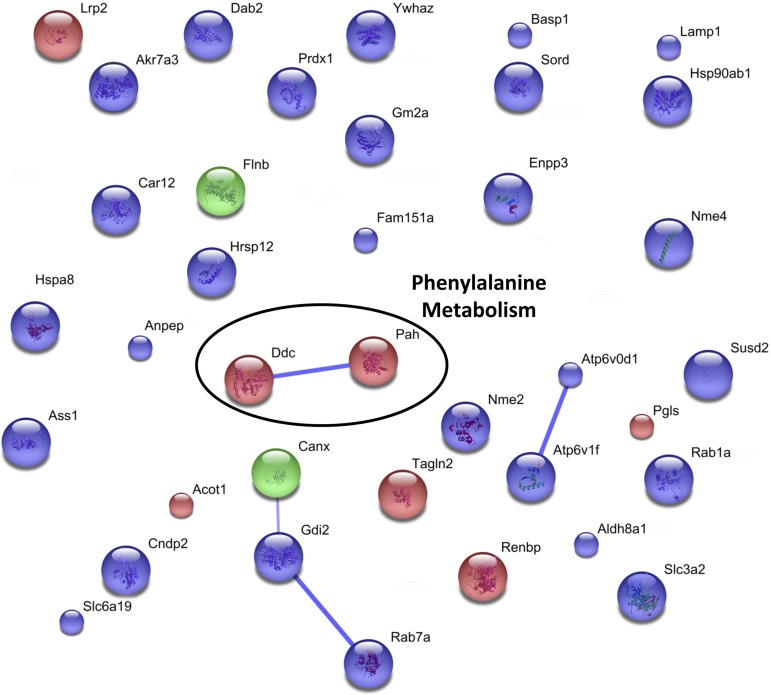
To elucidate which cellular pathways were affected by acidosis, the altered proteins were grouped into the general categories of increasing or decreasing, regardless of the time point at which the change was observed. The gene names for all of the proteins in each of these groups were separately submitted to the web-based STRING protein interaction network tool (40). All of the settings in the software were kept at their default parameters, with the exception that text mining-based associations were omitted. The resulting interaction maps for the increasing and decreasing proteins are shown in Figs. 5 and 6, respectively. In these figures, each individual protein is color coded to illustrate whether it was identified in the analysis of the brush-border membrane (blue), mitochondrial (green), or cytosolic (red) fraction, and the predicted interactions are represented by blue lines. The thickness of the line is proportional to the amount of evidence suggesting the association. Clusters of interconnected proteins are often representative of a particular cellular process or pathway. In the map of increasing proteins (Fig. 5), several such clusters are evident, including ammoniagenesis, retinol metabolism, hydrogen ion transport, and fatty acid metabolism. In the analysis of the decreasing proteins (Fig. 6), the only identifiable cluster contains proteins involved in phenylalanine metabolism.
Fig. 5.
STRING pathway analysis of proteins that are increased in the proximal convoluted tubule during acidosis. MS data collected from the cytosolic, mitochondrial (11), and apical membrane (44) studies were included for this analysis. Each individual protein is color coded to illustrate whether they were identified in the analysis of the brush border membrane (blue), mitochondrial (green) or cytosolic (red) fraction. Thicker lines are indicative of higher evidence of association. All proteins are labeled by their gene name. The identified groupings of interactive proteins participate in ammoniagenesis, hydrogen ion transport, retinol metabolism and fatty acid metabolism.
Fig. 6.
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) pathway analysis of proteins that are decreased in the proximal convoluted tubule during acidosis. MS data collected from the cytosolic, mitochondrial (11), and apical membrane (44) studies were included for this analysis. Each individual protein is color coded to illustrate whether it is identified in the analysis of the brush-border membrane (blue), mitochondrial (green), or cytosolic (red) fraction. Individual proteins are labeled by gene name, and thicker lines represent more evidence of association. This analysis indicated that renal metabolism of phenylalanine may be decreased during metabolic acidosis.
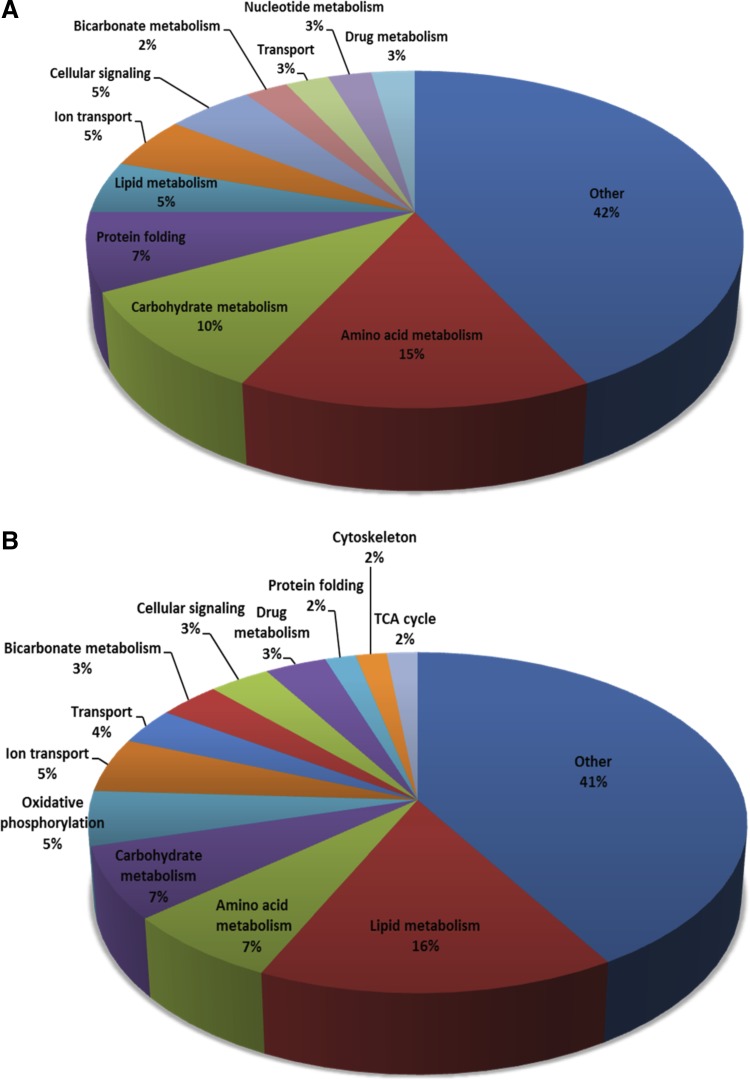
In addition to the STRING analysis, each protein in the combined list was manually annotated with a single cellular process or function. Manual assignment of such functions was used because proteins are often annotated with multiple GO terms or KEGG pathways. With the manual assignments, related categories were combined to group the proteins into broad categories. Pie charts showing the percentage of the increasing proteins belonging to each of the broad cellular processes are presented in Fig. 7A, where the decreasing proteins are shown in Fig. 7B. Many of the proteins found to be increasing were related to amino acid metabolism (15%), carbohydrate metabolism (10%), lipid metabolism (5%), and ion transport (5%). For the downregulated proteins, many were involved in lipid (15%), carbohydrate (12%), and amino acid metabolism (7%), and oxidative phosphorylation (5%). Other annotated processes include transport, protein folding, and cellular signaling.
Fig. 7.
Cellular functions of the proteins that are regulated in the proximal convoluted at any stage of metabolic acidosis. Data from cytosolic, mitochondrial (11), and apical membrane (44) fractions were combined, and functions were manually assigned for all proteins. A: proteins identified by MS analysis as being upregulated during metabolic acidosis. B: proteins that are downregulated in acidosis.
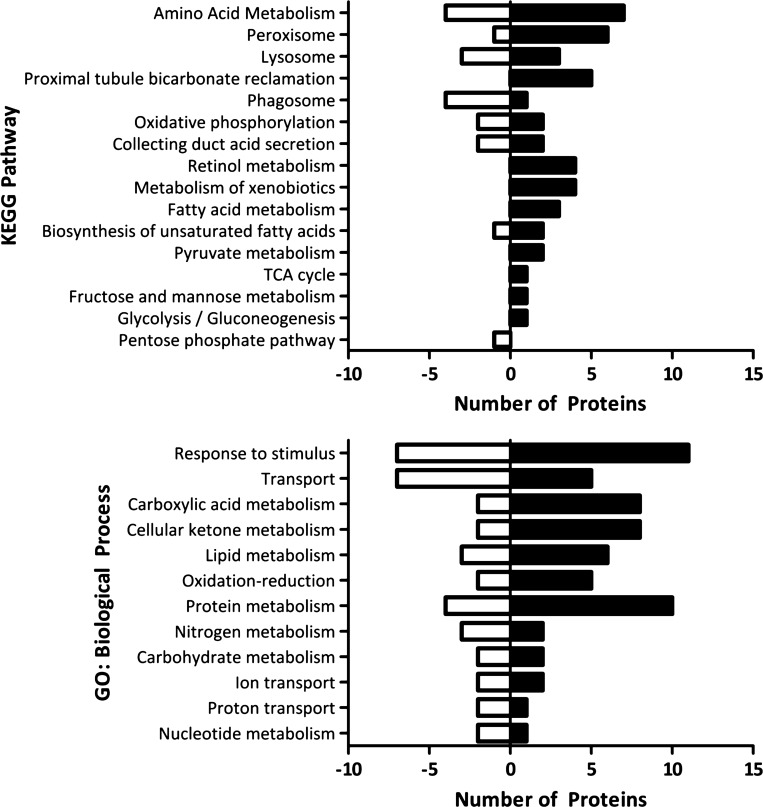
Finally, KEGG annotations and GO term biological processes were compiled from STRING for all of the differentially regulated proteins. A subset of the resulting annotations was compared to visualize the number of proteins that were found to be increasing or decreasing for each annotation (Fig. 8). Pathways and processes that were annotated with a number of upregulated proteins, but few downregulated included amino acid metabolism, proximal convoluted tubule bicarbonate reclamation, TCA cycle, fatty acid metabolism, retinol metabolism, metabolism of xenobiotics, and ketone metabolism. Interestingly, relatively few pathways contained primarily downregulated proteins.
Fig. 8.
Pathways and biological processes that are up- or downregulated in the proximal convoluted tubule during metabolic acidosis. Black bars to the right of the x-axis represent the number of proteins involved in the pathway or process that were upregulated during acidosis, whereas white bars to the left represent the number of downregulated proteins. Data from cytosolic, mitochondrial (11), and apical membrane (44) fractions were concatenated before analysis.
DISCUSSION
It is well established that the onset of acidosis triggers the uptake and catabolism of plasma glutamine in the proximal convoluted tubule and that this process leads to increased gluconeogenesis (6). The increased conversion of glutamine to glucose results in the net production of bicarbonate ions that are transported into the blood to partially restore acid-base balance (6). Although glutamine uptake and catabolism are dependent upon apical and basolateral transporters and mitochondrial proteins, respectively, the process of gluconeogenesis occurs in the cytoplasm. The current study identified significant levels of all nine of the cytosolic proteins that participate in gluconeogenesis. In fact, 5 of the enzymes of gluconeogenesis were among the 10 most abundant proteins identified in the proximal convoluted tubule. However, of these proteins, only PEPCK exhibits a rapid and pronounced increase in protein abundance following the acute onset of acidosis. Thus this adaptation is likely to constitute a primary site of regulation of renal gluconeogenesis during acidosis. Previous studies have established that the initial increase in cytosolic PEPCK results primarily from an increased rate of transcription, while the sustained increase is also mediated by stabilization of PEPCK mRNA (16, 20, 27).
Although the role of gluconeogenesis in the response of the proximal convoluted tubule to acidosis has been well established, the effects of acidosis on other cytosolic processes have not been elucidated. This study presents the first comprehensive analysis of the proteome of the cytosolic fraction of the proximal convoluted tubule and its response to metabolic acidosis. These data, when combined with the results of other proteomics studies (11, 43), increase our knowledge of how this segment of the nephron responds to metabolic acidosis.
Previous microarray analysis of the whole mouse kidney transcriptome (30) demonstrated that the mRNAs, which encode N-acylglucosamine 2-epimerase, aromatic-l-amino-acid decarboxylase, and PAH, are significantly increased during the onset of acidosis. The current study established that the reported increases in the three mRNAs lead to similar changes in protein expression in the proximal convoluted tubule. In addition, the current LC/MS/MS analyses confirmed the increases in GSTP1 and pyruvate carboxylase that were previously identified by difference gel electrophoresis analysis of isolated rat renal proximal convoluted tubules (7). Surprisingly, peptides from the low-density lipoprotein receptor-related protein-2 (megalin) and AT1A1, which are very abundant integral proteins of the apical and basolateral membranes, respectively, were also identified in the cytosolic fractions. The two proteins are likely to be associated with small vesicles or endosomes that were not removed by the high-speed centrifugation. Interestingly, the 7- to 10-fold decrease observed for AT1A1 in the cytosolic fraction contrasts with the 1.5- to 2-fold increase previously observed in the analysis of isolated brush-border membrane vesicles (43). Thus it is possible that more of the AT1A1 protein is sequestered in cytoplasmic vesicles during normal acid-base balance and then moved to the plasma membrane during acidosis.
Although a number of the differentially regulated proteins observed in this study were previously described, 15 of the 24 responding proteins are novel. Transgelin-2, an actin-binding protein that plays a role in the regulation of angiogenesis (47), was found to decrease greater than twofold during chronic acidosis. Previous data suggest that transgelin-2 inhibits endothelial migration and tube formation in vivo (47). Hence the observed downregulation of this protein may be representative of increased angiogenesis. Although angiogenesis is necessary for proper maintenance of renal tissue, it has also been implicated in the progression of chronic kidney disease (23, 25). This information combined with the renal hypertrophy and increased glutamine extraction suggests that there may be an increase in renal angiogenesis occurring during chronic acidosis.
PTGES3 was another protein that previously had not been shown to be upregulated in acidosis. PTGES3 is a molecular chaperone that aides in activation of transcription by the disassembly of transcriptional regulatory complexes in response to hormone stimulation (9). Although this method of transcriptional regulation has not been previously associated with acidosis, it is possible that the greater than 3.5-fold increase in this protein contributes to some of the other changes in protein abundance that occur in the proximal convoluted tubule during metabolic acidosis.
The increase in cytosolic isoform of aconitase (ACO1) was another novel change. Cytoplasmic citrate can be converted to isocitrate by ACO1, which is then converted to α-ketoglutarate by the NADP+-dependent isocitrate dehydrogenase enzyme (42). This reaction reduces NADP+ to NADPH, which is an important cofactor in fatty acid synthesis. The upregulation of the ACO1 enzyme may be representative of the need to increase NADPH production. In addition, previous work has found that the mitochondrial isoform of this protein increases in both abundance and activity during acidosis (7, 26), which leads to an increased rate of isocitrate synthesis in the mitochondrial TCA cycle. Thus, as an alternative, the isocitrate produced by cytosolic ACO1 could be transported into the mitochondria via known isocitrate transporters (13) and enter the TCA cycle. Other cytosolic proteins, which exhibit novel changes during acidosis, function in actin disassembly, carbohydrate metabolism, immune response, protein translation, and lipid metabolism, but the potential roles of these proteins in the response to acidosis is uncertain.
Previous work has established that mRNA stabilization is responsible for the differential expression observed for several proteins in the proximal convoluted tubule during metabolic acidosis (21, 27, 35). These proteins include PEPCK, glutaminase, and glutamate dehydrogenase, all of which contain one or two 8-nucleotide, AU-rich elements in the 3′-untranslated region that function as pH-responsive elements (pHREs) (27, 35). Interestingly, 6 of the 14 cytosolic proteins found in our study to be increased in acidosis contain at least one AU-rich sequence with 85% sequence homology to a pHRE, suggesting these proteins may be increased due to mRNA stabilization. Further investigation is necessary to determine whether this mode of regulation actually occurs in these proteins.
In recent years, the role of lysine acetylation in the regulation of nonhistone proteins has become more apparent (1, 14, 48). Previous work also suggested that lysine acetylation of mitochondrial proteins in the proximal convoluted tubule may play a role in acid-base homeostasis (11). Therefore, we sought to determine which cytosolic proteins of the proximal convoluted tubule undergo lysine acetylation. Due to the limited mass accuracy and mass range limitations of the linear ion trap mass spectrometer, it was not feasible to distinguish between lysine acetylation and trimethylation, which have nearly identical masses. However, most trimethyl lysine modifications are found in the nucleus (1, 19). Interestingly, only 15 unique peptides with modified lysines were identified (Table 1), each with far fewer spectra than observed in a similar analysis of acetylation of mitochondrial proteins in the proximal convoluted tubule (11). Overall, the low number of total acetylated peptides identified in the MS analysis suggests that lysine acetylation may play a minor role in the regulation of cytosolic proteins in the proximal convoluted tubule.
Using just the cytosolic protein data, it is difficult to determine which cellular pathways are affected during metabolic acidosis. However, when the data are combined with previous proteomic characterization of apical membrane (44) and mitochondrial (11) proteins, several trends appear. STRING analysis revealed several clusters that are indicative of activated cellular processes (Figs. 5 and 6). STRING analysis identified ammoniagenesis, hydrogen ion transport, and retinol and fatty acid metabolism as cellular processes that increased during acidosis (Fig. 5). The increase in hydrogen ion transport is expected during acidosis, due to the increased rate of hydrogen ion secretion into the tubular lumen. Interestingly, renal disease associated with diabetes causes a dysregulation of retinol metabolism in the renal cortex (19). Thus the observed changes in the pathway of retinol metabolism may be indicative of a cellular stress that occurs in the kidneys during acidosis. Alternatively, it is well established that retinol (vitamin A) plays an important role in epithelium maintenance (3). Therefore, it is also possible that the observed changes in retinol metabolism are associated with the maintenance of the renal epithelium. Further investigation is necessary to verify either hypothesis. The observed increases in proteins involved in fatty acid synthesis and elongation correlate with the predicted increase in cytosolic NADPH production. Together, the observed changes suggest that lipid synthesis is increased during acidosis to support the membrane expansion and remodeling, which are necessitated by the associated hypertrophy.
The number of proteins found to decrease in response to acidosis is less than the number that increase. As a result, the STRING analysis of this group of proteins did not identify large clusters of proteins (Fig. 6). However, the proteomic characterizations of the cytosolic proteins of the proximal convoluted tubule, combined with previously reported analyses (11, 44), have identified several cellular pathways in the proximal convoluted tubule that may be activated during metabolic acidosis. Thus the combined analyses have generated a number of novel hypotheses that now require further research to assess their physiological significance and characterize the mechanisms responsible for the observed changes.
GRANTS
This research was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK-37124 and DK-75517 awarded to N. P. Curthoys.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: K.L.S., D.M.F., and N.P.C. performed experiments; K.L.S., D.M.F., and J.E.P. analyzed data; K.L.S., D.M.F., J.E.P., and N.P.C. interpreted results of experiments; K.L.S. prepared figures; K.L.S. drafted manuscript; K.L.S., D.M.F., J.E.P., and N.P.C. approved final version of manuscript; D.M.F., J.E.P., and N.P.C. edited and revised manuscript; N.P.C. provided conception and design of research.
ACKNOWLEDGMENTS
All mass spectrometry was performed in the Proteomics and Metabolomics Facility at Colorado State University (http://www.pmf.colostate.edu).
REFERENCES
- 1.Arif M, Selvi BR, Kundu TK. Lysine acetylation: the tale of a modification from transcription regulation to metabolism. Chembiochem 11: 1501–1504, 2010 [DOI] [PubMed] [Google Scholar]
- 2.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics 8: 994–999, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Blomhoff R, Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol 66: 606–630, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Charles J, Heilman R. Metabolic acidosis. Hospital Physician 41: 37–42, 2005 [Google Scholar]
- 5.Curthoys NP. Renal ammonium ion production and excretion. In: The Kidney: Physiology and Pathophysiology, 5th ed., edited by Alpern RJ, Moe OW, Caplan M. San Diego: Elsevier, 2012, chapt. 57, p. 1991, 2018 [Google Scholar]
- 6.Curthoys NP, Gstraunthaler G. Mechanism of increased renal gene expression during metabolic acidosis. Am J Physiol Renal Physiol 281: F381–F390, 2001 [DOI] [PubMed] [Google Scholar]
- 7.Curthoys NP, Taylor L, Hoffert JD, Knepper MA. Proteomic analysis of the adaptive response of rat renal proximal tubules to metabolic acidosis. Am J Physiol Renal Physiol 292: F140–F147, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol 604: 55–71, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296: 2232–2235, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Freund DM, Prenni JE. Improved detection of quantitative differences using a combination of spectral counting and MS/MS total ion current. J Proteome Res [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freund DM, Prenni JE, Curthoys NP. Response of the mitochondrial proteome of rat renal proximal convoluted tubules to chronic metabolic acidosis. Am J Physiol Renal Physiol 304: F145–F155, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen PZ, Brewer H, Weitz K, Camp DG, 2nd, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genetics 7: e1001393, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gnoni GV, Priore P, Geelen MJ, Siculella L. The mitochondrial citrate carrier: metabolic role and regulation of its activity and expression. IUBMB Life 61: 987–994, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Guarente L. The logic linking protein acetylation and metabolism. Cell Metab 14: 151–153, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Guder WG, Ross BD. Enzyme distribution along the nephron. Kidney Int 26: 101–111, 1984 [DOI] [PubMed] [Google Scholar]
- 16.Gummadi L, Taylor L, Curthoys NP. Concurrent binding and modifications of AUF1 and HuR mediate the pH-responsive stabilization of phosphoenolpyruvate carboxykinase mRNA in kidney cells. Am J Physiol Renal Physiol 303: F1545–F1554, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halperin ML. Metabolic aspects of metabolic acidosis. Clin Invest Med 16: 294–305, 1993 [PubMed] [Google Scholar]
- 18.Huling JC, Pisitkun T, Song JH, Yu MJ, Hoffert JD, Knepper MA. Gene expression databases for kidney epithelial cells. Am J Physiol Renal Physiol 302: F401–F407, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huq MD, Tsai NP, Khan SA, Wei LN. Lysine trimethylation of retinoic acid receptor-alpha: a novel means to regulate receptor function. Mol Cell Proteomics 6: 677–688, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Hwang JJ, Curthoys NP. Effect of acute alterations in acid-base balance on rat renal glutaminase and phosphoenolpyruvate carboxykinase gene expression. J Biol Chem 266: 9392–939, 1991 [PubMed] [Google Scholar]
- 21.Hwang JJ, Perera S, Shapiro RA, Curthoys NP. Mechanism of altered renal glutaminase gene expression in response to chronic acidosis. Biochemistry 30: 7522–7526, 1991 [DOI] [PubMed] [Google Scholar]
- 22.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Lerman LO, Chade AR. Angiogenesis in the kidney: a new therapeutic target? Curr Opin Nephrol Hypertens 18: 160–165, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotspeich WD. Renal hypertrophy in metabolic acidosis and its relation to ammonia excretion. Am J Physiol 208: 1135–1142, 1965 [DOI] [PubMed] [Google Scholar]
- 25.Maeshima Y, Makino H. Angiogenesis and chronic kidney disease. Fibrogenesis Tissue Repair 3: 13, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melnick JZ, Preisig PA, Moe OW, Srere P, Alpern RJ. Renal cortical mitochondrial aconitase is regulated in hypo- and hypercitraturia. Kidney Int 54: 160–165, 1998 [DOI] [PubMed] [Google Scholar]
- 27.Mufti J, Hajarnis S, Shepardson K, Gummadi L, Taylor L, Curthoys NP. Role of AUF1 and HuR in the pH-responsive stabilization of phosphoenolpyruvate carboxykinase mRNA in LLC-PK1-F+ cells. Am J Physiol Renal Physiol 301: F1066–F1077, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Nikolov M, Schmidt C, Urlaub H. Quantitative mass spectrometry-based proteomics: an overview. Methods Mol Biol 893: 85–100, 2012 [DOI] [PubMed] [Google Scholar]
- 30.Nowik M, Lecca MR, Velic A, Rehrauer H, Brändli AW, Wagner CA. Genome-wide gene expression profiling reveals renal genes regulated during metabolic acidosis. Physiol Genomics 32: 322–334, 2008 [DOI] [PubMed] [Google Scholar]
- 31.Oberg AL, Mahoney DW, Eckel-Passow JE, Malone CJ, Wolfinger RD, Hill EG, Cooper LT, Onuma OK, Spiro C, Therneau TM, Bergen HR., III Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J Proteome Res 7: 225–233, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 4: 1487–1502, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Paoletti AC, Washburn MP. Quantitation in proteomic experiments utilizing mass spectrometry. Biotechnol Genet Eng Rev 22: 1–19, 2006 [DOI] [PubMed] [Google Scholar]
- 34.Pfaller W, Seppi T, Ohno A, Giebisch G, Beck FX. Quantitative morphology of renal cortical structures during compensatory hypertrophy. Exp Nephrol 6: 308–319, 1998 [DOI] [PubMed] [Google Scholar]
- 35.Schroeder JM, Liu W, Curthoys NP. pH-responsive stabilization of glutamate dehydrogenase mRNA in LLC-PK1-F+ cells. Am J Physiol Renal Physiol 285: F258–F265, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Shechter P, Shi JD, Rabkin R. Renal tubular cell protein breakdown in uninephrectomized and ammonium chloride-loaded rats. J Am Soc Nephrol 5: 1201–1207, 1994 [DOI] [PubMed] [Google Scholar]
- 37.Silbernagl S. Tubular reabsorption of l-glutamine studied by free-flow micropuncture and microperfusion of rat kidney. Int J Biochem 12: 9–16, 1980 [DOI] [PubMed] [Google Scholar]
- 38.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85, 1985 [DOI] [PubMed] [Google Scholar]
- 39.Squires EJ, Hall DE, Brosnan JT. Arteriovenous differences for amino acids and lactate across kidneys of normal and acidotic rats. Biochem J 160: 125–128, 1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: 561–568, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taverner T, Karpievitch YV, Polpitiya AD, Brown JN, Dabney AR, Anderson GA, Smith RD. DanteR: an extensible R-based tool for quantitative analysis of -omics data. Bioinformatics 28: 2404–2406, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong WH, Rouault TA. Metabolic regulation of citrate and iron by aconitases: role of iron-sulfur cluster biogenesis. Biometals 20: 549–564, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Walmsley SJ, Broeckling C, Hess A, Prenni J, Curthoys NP. Proteomic analysis of brush-border membrane vesicles isolated from purified proximal convoluted tubules. Am J Physiol Renal Physiol 298: F1323–F1331, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walmsley SJ, Freund DM, Curthoys NP. Proteomic profiling of the effect of metabolic acidosis on the apical membrane of the proximal convoluted tubule. Am J Physiol Renal Physiol 302: F1465–F1477, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiner ID, Hamm LL. Molecular mechanisms of renal ammonia transport. Ann Rev Physiol 69: 317–340, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Q, Zhao Q, Liang Z, Qu Y, Zhang L, Zhang Y. NSI and NSMT: usages of MS/MS fragment ion intensity for sensitive differential proteome detection and accurate protein fold change calculation in relative label-free proteome quantification. Analyst 137: 3146–3153, 2012 [DOI] [PubMed] [Google Scholar]
- 47.Xiao Y, Li Y, Han J, Pan Y, Tie L, Li X. Transgelin 2 participates in lovastatin-induced anti-angiogenic effects in endothelial cells through a phosphorylated myosin light chain-related mechanism. PLoS ONE 7: e46510, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotech 2010: 1–7, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]