Abstract
Crambe crambe is a marine sponge that produces high concentrations of the pharmacologically significant pentacyclic guanidine alkaloids (PGAs), Crambescines and Crambescidines. Although bio-mimetic chemical synthesis of PGAs suggests involvement of microorganisms in their biosynthesis, there are conflicting reports on whether bacteria are associated with this sponge or not. Using 16S rRNA gene pyrosequencing we show that the associated bacterial community of C. crambe is dominated by a single bacterial species affiliated to the Betaproteobacteria. Microscopy analysis of sponge tissue sections using a specific probe and in situ hybridization confirmed its dominance in the sponge mesohyl and a single microbial morphology was observed by transmission electron microscopy. If confirmed the presence of a simple bacteria community in C. crambe makes this association a very pertinent model to study sponge-bacteria interactions and should allow further research into the possible implication of bacteria in PGA biosynthesis.
Marine sponges (phylum Porifera) host a diverse array of associated microorganisms including unicellular algae, Cyanobacteria, heterotrophic bacteria and Archaea1,2,3,4,5. Although these associations are ubiquitous, the degree of association with microorganisms varies among host species. In bacteriosponges6 or high microbial abundance (HMA) sponges2 bacteria can constitute up to 40% of their biomass and generally present a relatively high diversity while low microbial abundance (LMA) sponges harbor much smaller bacterial communities with a lower bacterial diversity3,4,7,8,9. LMA sponges are usually dominated by one or two phyla, usually belonging to the Proteobacteria or the Cyanobacteria8,9 whereas HMA sponges can harbor more than 8 phyla7,9. Many of the bacteria associated with sponges fall into sponge-specific clusters that have been recovered from several different sponge species but not from the surrounding seawater5,10. However, an in depth pyrosequencing study on bacterial diversity in 32 sponges from 8 locations revealed that the majority of bacterial OTUs (operational taxonomic units) were specific to a given sponge4.
Sponges are also an important source of bioactive marine secondary metabolites making these organisms a target for research on compounds of pharmaceutical interest. The fact that many compounds found in sponges are complex polyketides or non-ribosomally synthesized peptides, whose biosynthesis is mostly associated with microorganisms suggests a bacterial origin for many sponge secondary metabolites11,12,13. However, this has only been unequivocally proven in a few cases. For example, a Salinospora strain isolated from the marine sponge Pseudoceratina clavata has been identified as a source of rifamycin antibiotics produced via polyketide biosynthesis14.
In addition to polyketides and non-ribosomally synthesized peptides, other chemical classes such as pentacyclic guanidine alkaloids (PGAs) that exhibit a wide range of activities15,16 have been extracted from the tissues of marine sponges. PGAs are mostly present in sponges of the Order Poeciloscleridae [e.g. Crambe sp.17 Monanchora sp.18 Batzella sp.19 Hemimycale sp.15] but also reported in Halichondrida sponges [e.g. Ptilocaulis sp.20]. This relatively wide phylogenetic distribution, as well as a hypothesized -albeit controversial (Olivier Thomas, personal communication), biosynthetic pathway involving a polyketide-like precursor21,22, points to a possible involvement of microorganisms in PGA biosynthesis. Remarkably, very few studies have been conducted to examine the microbial community associated with sponges known to produce these alkaloids, mostly using electron microscopy and focusing on the sponge Crambe crambe.
Crambe crambe (Schmidt, 1862) (Poecilosclerida) is a red incrusting marine sponge present in the Mediterranean Sea and reported to produce diverse PGAs, namely crambescidins 800, 816, 830, 844, as well as isocrambescidin 80017,22. The presence of bacteria associated with this sponge is somewhat controversial. Based on scanning electron microscopy C. crambe has a surface devoid of epibionts23. In addition Uriz and colleagues reported that cells and mesohyl of this sponge were free of microsymbionts (bacteria or cyanobacteria) when examined by transmission electron microscopy24. This observation contrasted to that of Sará25 who reported sporadic occurrence of cyanobacteria in specimens from Italian shores. Although several thousand sponge-derived 16S rRNA gene sequences are now available5, the few sequences originating from C. crambe are from unpublished studies. Two such partial sequences clustered together within a Betaproteobacteria cluster5. However, since in that study the sequences were added to a tree in a second parsimony step, their phylogenetic relationship to the other sponge-derived sequences is not clear.
In order to address these contrasting reports, we examined the diversity of 16S rRNA genes amplified from extracts from the sponge C. crambe, using 454-tag pyrosequencing and sequencing of 16S rRNA gene clone libraries for greater phylogenetic resolution. To demonstrate the presence of bacteria in C. crambe, the sponge tissue of a different specimen was examined using transmission electron microscopy (TEM), and the dominant bacterium was localized using a specific oligonucleotide probe combined with catalyzed reported deposition fluorescence hybridization (CARD-FISH).
Results
Bacterial community diversity analyses
16S rRNA gene community analysis of Crambe crambe
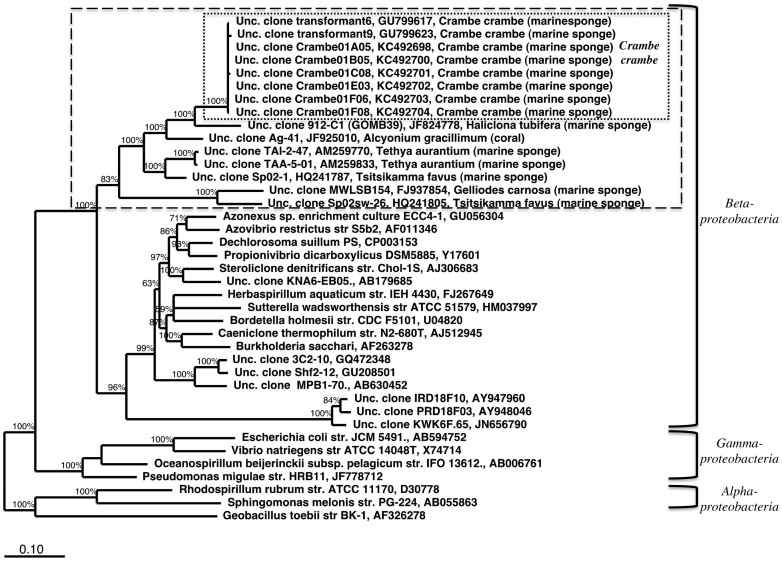
A total of 43 16S rRNA gene clones from C. crambe tissues were sequenced. After sequence quality and chimera checks a relatively high number of clones (11) were assigned as chimeras (26% of clones). The 32 remaining clones generated 12 OTUs (> 97% identity), which fell into 4 bacterial classes: Flavobacteriia, Gammaproteobacteria, Alphaproteobacteria, Betaproteobacteria and a few clones were assigned as chloroplasts. 17 sequences grouped into 9 OTUs were assigned as planktonic bacteria (> 98% identity). Phylogenetic analysis of the dominant non-chimeric and non planktonic OTU represented by 13 clones placed these sequences in a clade most closely related to the Betaproteobacteria, but separate from the known described betaproteobacterial orders (Figure 1; dashed line), and recently reported in a study of sponge-associated bacteria of the Mediterranean sponge Tethya aurantium26.
Figure 1. Evolutionary relationship of the dominant C. crambe bacterium to its closest relatives inferred from almost full-length 16S rRNA gene sequences, using Bayesian phylogenetic reconstruction.
Sequences obtained from the sponge C. crambe are shown in the smallest box. The accession numbers of the C. crambe sequences obtained in this study are pre-fixed by KC. A cluster of sequences associated with marine sponges and a coral is shown within the dashed lines. Percentage confidences for the Bayesian analysis are indicated at each node.
454- 16S rRNA Pyrosequencing analyses
From an initial 5973 raw bacterial 16S rRNA tag sequences obtained from C. crambe, 4421 sequences remained after denoising by AmpliconNoise. Subsequent stringent chimera detection removed a significant number of chimeric sequences, many of which were singletons, which would have otherwise significantly overinflated the total bacterial diversity (213 chimeric sequences representing 70 OTUs). Rarefaction analysis of non-chimeric sequences shows a low level of diversity with rarefaction curve reaching a plateau at about 115 OTUs (Figure S1). This low diversity was also reflected by a Chao1 value of 107.7 and a small total number of OTUs recovered (86 OTUs). Furthermore, among these OTUs, a single OTU, very similar (> 99% identity) in the V1-V3 region to the dominant clone in the 16S rRNA library, represented 74% of the total sequences (3271 sequences). Among the remaining 85 OTUs, 42 (representing 22% of total sequences) were classified as planktonic. Therefore, considering only non-planktonic bacteria, likely transient in the sponge, this dominant betaproteobacterium accounted for a remarkable 95% of the total sequences.
Catalyzed reporter deposition fluorescence in situ hybridization analyses (CARD-FISH)
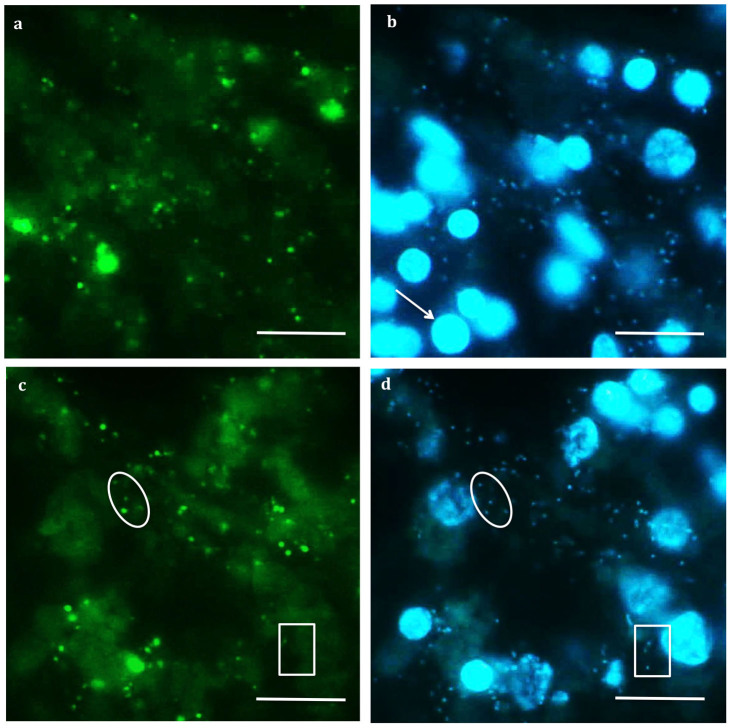
After optimization, CARD-FISH with the general bacterial probe confirmed the presence of bacteria in the sponge tissue. Initial hybridization experiments were performed with the general bacterial probe EUB338-I and the negative control probe NON338 according to a previously published protocol for worm symbionts27 except that a lysozyme permeabilization step was necessary to obtain a good fluorescent signal. Strong signals were observed with the EUB338-I probe (Figure 2a) demonstrating the presence of sparsely distributed bacteria within the sponge tissue corresponding to smaller cells labeled with DAPI (Figure 2b).
Figure 2. Detection of bacteria in the mesohyl of Crambe crambe by CARD-FISH on sponge-tissue sections.
Tissue section hybridized with the EUB 338 probe targeting the domain Bacteria, (a) and the corresponding DAPI-stained image, (b). Tissue section hybridized with the BET467 probe targeting the specific Betaproteobacteria, (c) and the corresponding DAPI-stained image, (d). Arrow: sponge cell nucleus. Oval: cells labeled with both DAPI and the BET467 probe. Rectangle: some cells labeled with DAPI do not hybridize with the BET467 probe.
The DAPI staining showed two types of cells with some variation in size (Figures 2b–d). Based on an overlap between DAPI staining and the bacterial probe hybridization, we identified the smaller cells as bacteria and the larger spherical cells (Figure 2b; arrow) as the nuclei of sponge cells. Observation of different tissue sections showed the same results indicating that bacteria were sparsely distributed in all tissues of the sponge. Negative controls including the anti-sense probe NON338 and a no-probe control yielded no signals indicating the stringency of the hybridization and a lack of endogenous peroxidases in the sponge tissue.
The same tissue sections were also stained with DAPI and hybridized with the specific BET 467 probe and its helper probes (sequences shown in Table 1). Strong signals were observed with this specific probe (Figure 2c). Probe BET467 hybridized to the overwhelming majority of the small cells stained with DAPI and identified as bacteria (Figure 2c–d; circles) demonstrating a dominant presence in tissues of C. crambe of the betaproteobacterial clade found in the clone library and pyrosequencing. Not all the bacteria present (EUB 338-I) in the sponge mesohyl were hybridized with the specific BET467 probe (Figure 2c–d; square) in accordance with the clone library and 454-pyrosequencing results that indicated that other bacteria were also associated with the sponge. Tissue sections were also hybridized with probe BET42a targeting the Betaproteobacteria class (along with a competitor unlabeled probe targeting Gammaproteobacteria). Surprisingly no signal was detected. Finally, no signal was detected in hybridizations with the ARCH915 probe suggesting the absence of Archaea in C. crambe. Even though FISH targeting eukaryotes was not performed, only sporadically red/orange fluorescence was observed under green excitation, thus it is unlikely that photosynthetic eukaryotes or cyanobacteria are important members of the microbial community associated with C. crambe.
Table 1. Details of the specific horse-radish peroxidase (HRP) labeled oligonucleotide probes used in this study, together with the unlabeled competitor and helper (H-) probes.
| Probe | Sequence (5′–3′) | Specificity | E.coli target position | Label | Reference |
|---|---|---|---|---|---|
| EUB338-I | GCTGCCTCCCGTAGGAGT | Eubacteria | 16S rRNA 338–355 | HRP | 62 |
| NON-EUB338-I | ACTCCTACGGGAGGCAGC | Eubacteria Negative control | 16S rRNA 338–355 | HRP | 63 |
| BET42a | GCCTTCCCACTTCGTTT | Betaproteobacteria | 23S rRNA 1027–1043 | HRP | 64 |
| BET42a competitor | GCCTTCCCACATCGTTT | Gammaproteobacteria | 23S rRNA 1027–1043 | – | 64 |
| ARCH915 | GTGCTCCCCCGCCAATTCCT | Archaea | 16S rRNA 915–934 | HRP | 65 |
| BET467 | CGTCATGGACGCGGCCTG | C. crambe Betaproteobacteria | 16S rRNA 467–484 | HRP | This study |
| H-485 | GGCGCTTCTTCTCAGGGTAC | Helper | 16S rRNA 485–504 | – | This study |
| H-446 | TTCGCCCGCGTTTTTTCTTTC | Helper | 16S rRNA 446–466 | – | This study |
Although we did not use a confocal microscope, and thus did not accurately quantify bacterial cells, an analysis of 20 different 20 μm2 areas showed an average of 6 bacterial cells/sponge cell (5 Betaproteobacteria/sponge cell), resulting roughly in 3000 bacterial cells (2500 Betaproteobacteria cells) per cm2 of thin section. Since these micrographs were not randomly visualized, these numbers are likely upper estimates.
Transmission electron microscopy of thin sections of Crambe crambe tissue
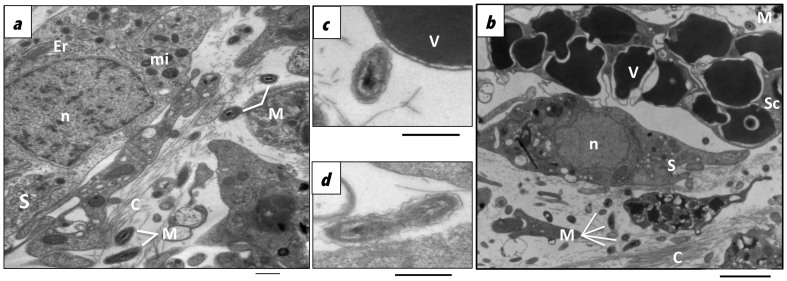
Ultra thin sections were prepared from one specimen of C. crambe and inspected by transmission electron microscopy to observe the localization and the morphotypes of bacteria associated with this sponge. The results revealed a low density of morphologically uniform intercellular bacteria scattered in the mesohyl compartment of C. crambe. Bacteria were found embedded in the collagen matrix (Figure 3a–b) in the mesohyl, surrounding sponge cells remarkably including spherulous cells (Figure 3b–c). The rod-shaped bacteria observed of approximately 0.25 μm width and 0.5 μm length present dense chromatin condensation and an undulating surface. Bacteria in a potential state of cell division were also found in the mesohyl of the sponge, suggesting that the betaproteobacterium may be actively growing in the sponge tissue (Figure 3d). Ultra thin sections from the 10 subsamples cut in different orientations were observed under the transmission electron microscope and similar results were found.
Figure 3. Transmission electron microscopy of thin sections of C. crambe tissues.
Microorganisms in the mesohyl of C. crambe embedded in the collagen matrix of the sponge, (a). Proximity between microorganisms and spherulous cells, (b–c). Bacteria in division, (d). C: collagen; Er: endoplasmic reticulum; mi: mitochondrion; M: Microorganisms; n: nucleus; S:sponge cell; Sc: Spherulous cell whose vesicles (V) contain an electron dense material.
Discussion
To date, all investigations of microorganisms associated with sponges in the genus Crambe, were performed by electron microscopy and microbial cultivation. Early studies1,25, reported low densities and diversity of microorganisms associated with these sponges. Later studies suggested that in fact those sponges were virtually devoid of epibionts and mesohyl-associated bacteria and this absence was suggested to be linked to the high toxicity associated of extracts of the sponge24.
Our results demonstrate, through a combination of molecular techniques and microscopic studies, that bacteria can be associated with the tissues of the PGA containing sponge C. crambe, and that in our samples these bacteria overwhelmingly belong to a specific clade affiliated to the Betaproteobacteria (95% of the total non planktonic sequences generated with 454 pyrosequencing) in agreement with a recent report that specific bacteria (particularly within the phylum Proteobacteria) were found as dominant members of microbial communities associated with LMA sponges8. We believe that confirmation of a single dominant OTU was possible because we used a stringent denoising and chimera detection pipelines, as well as our bioinformatic approach to “filter” out primarily planktonic organisms. This dominant bacterial OTU was affiliated to a specific uncultured betaproteobacterial clade, which was not identified amongst the planktonic bacteria compiled from the literature, and a large 16S rRNA cloning effort29. Attempts to cultivate this microorganism in rich and diluted R2A broth, as well as in media targeting ammonium-oxidizing bacteria were unsuccessful (data not presented).
The phylogenetic analyses of full-length 16S rRNA genes demonstrated that the clade including the C. crambe Betaproteobacteria branches separately from the clade containing the remainder of previously described and cultured Betaproteobacteria (dashed box in Figure 1). This clade contains exclusively sequences of bacteria associated with marine invertebrates including sequences of clones in an unpublished clone library of bacteria retrieved from C. crambe (D. Sipkema personal communication) that was sampled from a different site in the Mediterranean Sea, clones retrieved from the sponge Haliclona tubifera and clones from another LMA sponge Tethya aurantium, also from the Mediterranean Sea26. It is important to remark that in the available Silva 108 taxonomy this clade was assigned as Nitrosomonadales (thus in the main Betaproteobacteria branch), but our refined phylogenetic placement supports the basal placement of the clade within the Betaproteobacteria as reported by Thiel and collaborators26. This clade placement could be 1) real or 2) an artifact caused by a higher evolutionary rate known to occur in symbiotic microorganisms30,31. Case 1) above is supported by the observation that the probe Bet42a for the class Betaproteobacteria (targeting the 23S rRNA) did not yield a signal in CARD-FISH experiments targeting the C. crambe symbiont, indicating that this clade is distant from the remainder of the Betaproteobacteria. Metagenomic analyses in progress should allow us to clarify the real phylogenetic placement of this clade by the analysis of additional conserved genes.
Interestingly, several attempts to amplify a large fragment of the rRNA operon as previously described by Suzuki and coworkers32 failed, despite the use of several combinations of forward primers targeting the 16S rRNA gene and reverse primers targeting the 23S rRNA, strongly suggesting that the 16S rRNA gene is unlinked to the 23S rRNA gene in the C. crambe associated bacterium (data not shown). Both unlinked 16S-23S rRNA genes and large insertions in the rRNA operon are common in endosymbiotic bacteria and obligate intracellular parasites33,34, thus supporting Case 2) above and if proven would suggest a stronger and perhaps obligatory symbiosis between this betaproteobacterium and C. crambe. If this were the case, it would be interesting to determine how the dominant C. crambe associated bacterium is acquired. This betaproteobacterium could be part of the “rare biosphere” which would be filtered by sponge and whose growth is favored in the sponge tissue. Alternatively there could be a vertical transfer of the bacteria via their larvae since bacteria were observed in the mesohyl, the surface and in the posterior region of C. crambe larvae28. This form of transmission is common in sponges and insects when the bacteria are obligate symbionts35,36. Interestingly, the reproductive stages of LMA sponges were generally reported as bacteria-free37. Regardless of their origin, the maintenance of the desired symbionts and exclusion of other bacteria by the host sponge may be mediated through the mechanism of antibiosis38. This mechanism is based on the ability of few bacteria to adapt to the presence of antibiotics produced by the host. C. crambe is known to produce a wide variety of toxic compounds with high antimicrobial activity23, which could help explain the selection of a single symbiont.
To confirm the localization of bacteria within the tissues of C. crambe, two complementary microscopy techniques were employed. The TEM pictures revealed a low density of morphologically uniform intercellular bacteria scattered in the mesohyl of C. crambe, already noted by Vacelet and Donadey in 1977 for Crambe sp.1 and in agreement with previous observations reporting that microorganisms associated with LMA sponges, are sparse and free living in the sponge mesohyl1. Interestingly the dominant bacterial morphotype was also observed surrounding the spherulous cells (Figure 3) that are specialized cells that seem to be responsible for the storage of secondary metabolites24,39,40. The proximity between bacteria and these specialized cells could indicate their possible resistance to the highly-toxic compounds isolated from C. crambe, and further support the symbiosis between these organisms. The observation of several bacteria in the process of cell division suggests that these bacteria are active. In the LMA sponge Polymastia sp., the dominant Alphaproteobacteria phylotypes were represented in 16S rRNA clone libraries constructed from both DNA and RNA also suggesting that the bacteria in these LMA sponges were active7.
The application of a specific oligonucleotide probe in conjunction with CARD-FISH allowed us to microscopically demonstrate the presence of the dominant betaproteobacterium within the C. crambe tissues, which to our knowledge has not been shown for other single associations between Proteobacteria and LMA sponges. The majority of the bacteria (Figure 2d) were also labeled with the specific probe (Figure 2c) in agreement with the dominance of this bacterial OTU in the 16S rRNA gene clone library and in the pyrosequencing data.
In our study, two sponge specimens collected one year apart were used, and using alternative techniques showed agreeing results. Sequencing of 16S rRNA genes in the 2011 sample and CARD-FISH with the 2012 sample, show the dominance of the same specific bacterium in these two specimens suggesting that the association between the betaproteobacterium and C. crambe is stable, although sampling through an annual cycle with replicate sponge specimens should be done to confirm this. Such a study was carried out on the LMA sponge Tethya stolonifera. In that sponge, the dominant bacterial OTU was also affiliated to a sponge-specific Betaproteobacteria cluster and the community remained very stable over a two-year period9.
A comprehensive study of sponge symbionts revealed 26 different bacterial phyla with most of the diversity occurring within the Proteobacteria4. The bacterial sequences retrieved from tropical sponges were more closely related to each other than from the sub-tropical ones, indicating that bacterial community composition could be influenced more by temperature than their localization in the same water mass4. This latitudinal separation of bacterial communities was also shown for five LMA sponges where the temperate sponge Raspailia topsenti was dominated by Betaproteobacteria whereas the tropical LMA sponges were dominated by Alphaproteobacteria, Gammaproteobacteria or Cyanobacteria8. Betaproteobacteria sequences also represented the dominant OTU in the temperate LMA sponges Tethya stolonifera9, and in Scopolina sp. and Tedania anhelans41. These independent studies and our own indicate that Betaproteobacteria-LMA sponge associations in temperate waters may be a global feature but that within this clade, each sponge appears to harbor a phylogenetically distinct betaproteobacterial OTU.
Our study adds further weight to others, which have shown the low diversity within LMA sponges, making them simpler models to study the roles and molecular basis of these symbioses. From a practical standpoint in contrast to HMA sponges which harbor dense and diverse microbial communities2,6, metagenomic or single-cell genomic techniques42 would be simpler to conduct with simpler models such as LMA sponges harboring a single dominant OTU, as would be techniques such as transcriptomics and proteomics to study the expression of specific bacterial genes. Furthermore, a possible role of bacteria in the synthesis of secondary metabolites extracted from highly active sponges would be easier to evaluate in LMA sponges. In the specific case of C. crambe, and its secondary metabolites, earlier results reporting the lack of associated microflora23,24, as well as the known high concentrations of guanidine alkaloids (about 1% of dry mass) strongly supported the idea that the sponge cells were solely responsible for the biosynthesis of these highly bioactive metabolites. However, biomimetic studies indicate that crambescidine-like molecules are produced via a condensation of guanidine to a polyketide-like molecule21, and since polyketides are primarily produced by microorganisms, this biosynthesis model suggests a microbial implication in PGA biosynthesis. Our report of the dominance of a single OTU in the tissues of C. crambe, means that the implication of these bacteria in the biosynthesis of PGAs cannot be completely ruled out.
Methods
Sample collection
The Crambe crambe specimen (one sponge individual growing in a rocky substrate) for 16S rRNA gene analysis were collected in the Bay of Banyuls-sur-Mer (Western Mediterranean, France; 42° 28.828′ N −3° 08′666′ E) at a depth of approximately 10 meters on 5 January 2011. For CARD-FISH and TEM, one sponge individual (meaning a sponge associated to both shells of the bivalve Arca noae) was collected at the same location on 4 January 2012. For both collections samples were placed in plastic bags underwater to avoid contact with air and immediately transported to the laboratory. The sponge specimens were rinsed 3 times in calcium magnesium free - artificial seawater (CMF-ASW) to remove exogenous particles and loosely attached bacteria present in the sponge's aquiferous channels. Sponge tissue was processed 30 minutes after sampling to avoid possible modifications of the microbial community associated with the sponge during processing. The washed specimen collected in 2011 was frozen at −80°C for subsequent DNA extraction whereas the 2012 specimen was fixed immediately for CARD-FISH and TEM analysis (see below).
DNA extraction
Cleaned sponge fragments (2011 sample) were pooled (35 g total) and homogenized in two volumes of CMF-ASW using a Waring blender with 3 cycles of: 15 s at 22000 rpm, 30 s pause, 15 s again at 22000 rpm, with a 60 s pause in between each cycle. The homogenate was centrifuged for 5 min at 3000 rpm to pellet larger particles and sponge cells. The remaining sponge cells and bacteria in the supernatant were pelleted by centrifuging for 30 min at 8000 rpm. The pellet was washed twice by resuspending in 35 ml of CMF-ASW and centrifuging at 8000 rpm for 30 min. The final pellet was resuspended in 35 ml of CMF-ASW, filtered though GF/A glass fiber filters (1.6 μm nominal retention size) to remove remaining sponge cells and centrifuged once more at 8000 rpm for 30 min. This pellet was frozen in 35% glycerol and stored at −80°C until further analysis.
To extract DNA, cells present in the pellet were disrupted by bead beating (3 pulses of 30 s at 25 Hz) using a model MM301 cell disruptor (Retsch, Germany) and 200 μm low-protein binding zirconium beads (OPS Diagnostics, Bridgewater, NJ, USA). The lysate was then gently mixed with an equal volume of guanidinium thiocyanate buffer43. Nucleic acids were recovered using a standard phenol and chloroform extraction, and washed and concentrated in TE buffer using Centricon 30 microconcentrators (Amicon, Danvers MA).
16S rRNA gene library analysis
Extracted DNA was used as template for 16S rRNA gene amplification using modified universal primers targeting Bacteria, 27Fmod (5′ AGRGTTTGATCMTGGCTCAG 3′) and 1492Rmod (5′ TACGGYTACCTTGTTAYGACTT 3′) as previously described44. Amplification was performed in a 40 μL total-reaction volume with 1X Platinum Taq DNA polymerase buffer, 500 nM of each primer, 0.8 mM dNTPs, 2 mM MgCl2, 2 μg/ml BSA, 0.025 U/μl Platinum Taq DNA polymerase, and 200 ng of extracted DNA. PCR cycling was as follows: 94°C for 3 min, 27 cycles of 94°C for 1 min, 50°C for 1 min 72°C for 2 min followed by 72°C for 10 min.
PCR products were run in a modified TAE agarose gel and purified with Ultrafree DNA columns (Millipore, Billerica, MA) using the manufacturer's protocol. Purified PCR products were cloned using the TOPO TA Cloning Kit for Sequencing using One Shot TOP 10 Electrocompetent Cells (Life Technologies) according to manufacturers instructions and plasmid purification was performed with the Montage Plasmid Miniprep 96 Kit (Millipore). Plasmid inserts were sequenced using the dideoxy-terminator reaction using the 907 reverse primer [5′ CCG TCA ATT CCT TTG AGT TT 3′]45 and the BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies). Cycle sequencing products were cleaned using the Agencourt CleanSeq Dye-Terminator Removal kit (Beckman Coulter, Brea, CA) and run in an AB3130xl Genetic Analyzer (Life Technologies).
Partial, raw sequences were assembled using 20 nucleotides minimum overlap and 99% maximum mismatches and visually edited using the gap4 program of the Staden package46. Edited contigs were exported in fasta format including the number of reads per contig as weights, and chimeric sequences were eliminated by using a stringent chimera checking pipeline consisting of uchime47, Bellerophon48 and by querying the sequences against a curated version of the Silva database SSURef_111_SILVA_ NR_98_26_07 (http://www.arb-silva.de/) using blastn (see Supplementary Information for detailed parameters used). Non-chimeric sequences resulting from this pipeline were filtered for clones of planktonic origin by blastn queries against a local database including 1493 OTUs from 37, 860 16S rRNA PCR library sequences from the Global Ocean Sampling project plus 8,471 marine bacterial 16S rRNA sequences obtained from the ribosomal database project dataset, and graciously provided by S. Yooseph.
The 16S rRNA sequences of the clones used in this study have been deposited in the GenBank database under accession numbers KC492587-KC492611 and KC492698-KC492704.
Phylogenetic analysis
Clone sequences classified as non-chimeric and non-planktonic were fully sequenced using primers 907R and 785F (5′ GGATTAGATACCCTGGTAGT 3′), individually assembled using gap4, and exported. These sequences were aligned using the mothur49 and imported into the SSURef_111_SILVA_18_07_12 database of the Silva project50 using the ARB software51. Phylogenetic trees were constructed by neighbor-joining using the phylip package V3.652 and by Bayesian analysis using Mr Bayes v3.2.1×64 53. For both methods, the aligned clone sequences and reference sequences from the Betaproteobacteria, Gammaproteobacteria and Alphaproteobacteria were filtered using an 1112 bp majority filter.
454-pyrosequencing analyses
The PCR products used to construct the clone library were used as template for 454 pyrosequencing by the Research and Testing Laboratory (Lubbock, TX) according to a standard multiple protocol of this sequencing facility and described by Dowd and coworkers54. Briefly a PCR step (30 cycles) with a HotStart HiFidelity Polymerase and primers 28F (5′ TTTGATCNTGGCTCAG 3′) and 519R (5′ GTNTTACNGCGGCKGCTG 3′) was performed in order to amplify the hypervariable V1-V3 region of the 16S rRNA gene. Tag-encoded FLX amplicons were sequenced on the Roche 454 FLX sequencer using Titanium (Roche) reagents. Multiplex raw sff files were analyzed using a hybrid analysis pipeline. In brief, denoising was done by AmpliconNoise V1.2555 implemented in Qiime V1.556 and rather than using chimera analysis by Perseus, de novo chimera detection and removal was done by using the uchime47 module of Usearch V5.057 followed by the same analyses as for the clone library (details of parameters and commands in Supplementary Information). Non-chimeric were unweighted and grouped into OTUs using uclust with 97% sequence identity. OTU representatives were selected based on abundance. OTUs of planktonic origin were identified by blastn searches as above.
Pyrosequencing sequence reads representing the 86 OTUs have been deposited in the GenBank database under accession numbers KC492612-KC492697.
Oligonucleotide probe design
The specific oligonucleotide probe BET467 targeting the prevalent betaproteobacterial sequences in the clone library was designed using the probe design tool of the ARB software package51. The specificity of the probe sequence was confirmed by the probe_match tool of ARB, the probe match tool of Ribosomal Database Project (http://rdp.cme.msu.edu/) and blastn searches. Only 3 non-target sequences in the probe_match tool of ARB database had less than 3 mismatches with this specific probe. As the 16S rRNA target region was potentially inaccessible58, two unlabeled helper oligonucleotides H-485 and H-446 were also designed. Oligonucleotide sequences of probes used, labeling and details are shown in Table 1. All probes were synthesized by ThermoFisher Scientific (Germany).
Catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH)
A summary of all the steps performed for CARD-FISH is given in Table 2. To determine the distribution of the bacteria in the sponge, 10 tissue samples of C. crambe, from different anatomic locations of two sponges specimens (about 1 cm3) were fixed as in steps 1–3 (Table 2), embedded in paraffin (at different orientations), cut into 10 μm thick sections with a microtome (MICROM HM340E; Thermo Scientific) and mounted on polysilane-coated slides (steps 4 and 559). Subsequent sample treatment and CARD-FISH was carried out according to a previously described protocol27 with some modifications. After drying, slides were deparaffined (steps 6 and 7). A step to inactivate endogenous peroxidases was performed, followed by a permeabilization step where a solution of Proteinase K (0.5 μg/ml) was added to tissue sections. A lysozyme permeabilization step was not included in the protocol from Blazejak and coworkers, but since this step has been shown to improve significantly CARD-FISH signals60 our tissue sections were subject to lysozyme permeabilization steps [10 mg ml−1 lysozyme (Fluka) in 0.05 M EDTA, 0.1 M Tris-HCl (pH8.0) (step 12–15)].
Table 2. Summary of CARD-FISH protocol optimized for the detection of bacteria in marine sponge tissue sections. RT, room temperature.
| Stage | Step N° | Description |
|---|---|---|
| Fixation | 1 | Incubate sponge tissue in 4% buffered paraformaldehyde (4°C, 4 h) |
| 2 | Wash 3 times in same phosphate buffer (RT) | |
| 3 | Dehydrate in 70% ethanol (4°C, 24 h) | |
| Tissue Sectioning | 4 | Embed tissue samples in paraffin |
| 5 | Cut 10 μm thick sections with a microtome onto polysilane-coated microscope slides, leave to dry 3 h at 35°C | |
| Deparaffinization | 6 | Incubate slides twice in xylene (RT, 10 min each) |
| 7 | Rehydrate in an ethanol series (95%, 80%, 70%; 10 min each) | |
| Inactivation of endogenous peroxidases | 8 | Incubate in 0.2 M HCl (RT,12 min) |
| 9 | Incubate in 20 mM Tris.HCl (RT, 10 min) | |
| Permeabilization 1 | 10 | Cover tissue sections with proteinase K, incubate in humid chamber (37°C, 5 min) |
| 11 | Wash slides in 20 mM Tris.HCl (RT,10 min), air dry | |
| Permeabilization 2 | 12 | Dip slides in 0.1% low melting point agarose and air dry (10 min) |
| 13 | Cover tissue sections with lysozyme, incubate in humid chamber (37°C, 1 h) | |
| 14 | Wash slides in milliQ water (1 min) | |
| 15 | Dehydrate in 96% ethanol (1 min) and air dry | |
| Hybridization | 16 | Cover tissue sections with hybridization buffer containing probe at 1/20 dilution of working solution (1 probe per slide) |
| 17 | Insert slides into prepared humid chambers | |
| 18 | Incubate at 35°C for 3 h | |
| 19 | Wash slides in prewarmed wash buffer (37°C, 15 min) | |
| CARD | 20 | Incubate slides in 1X PBS (RT, 15 min) |
| 21 | Dab around tissue sections to remove excess buffer but do not let sections dry | |
| 22 | Incubate tissue sections with substrate mix (1/200 dilution of tyramide in amplification buffer) in humid chamber in the dark (37°C, 20 min) | |
| 23 | Wash slides in 1X PBS (RT, 15 min) | |
| 24 | Wash slides in milliQ (1 min) and air dry | |
| 25 | Counterstain with DAPI using mounting medium supplemented with DAPI |
RT, Room temperature.
Hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH7.5], 10% dextran sulfate, 0.02% sodium dodecyl sulfate [SDS], 55% formamide [Sigma] and 1% Blocking Reagent [Roche]) - probe mixtures were added to the tissue sections on slides, and incubated in hybridization chambers prepared with a 55% formamide-water mixture at 35°C for 3 h. Due to the absence of a cultured representative of the targeted betaproteobacterium, the optimum formamide concentration to be used for the specific probe BET467 was determined by carrying out hybridizations with the sponge tissue sections, varying formamide concentrations (50%, 55%, 60%), and selecting the highest formamide concentration before a reduction in fluorescence signal was observed. This was determined to be 55%. Hybridizations for the BET467 probe were carried out as above and also included the helper probes H-485 and H-446 at a concentration of 2.5 ng/μl. Following hybridization, the slides were prepared as in steps 20–25 (Table 2). Washing buffer contained 5 mM EDTA [pH 8], 20 mM Tris–HCl [pH 8], 0.01% [w/v] sodium dodecyl sulfate and 13 mM NaCl). The CARD substrate mix was prepared by mixing amplification buffer (10% [w/v] dextran sulfate, 2 M NaCl, 0.1% [w/v] blocking reagent, in 1X PBS [pH 7.6]) with a freshly prepared H2O2 solution (0.15% in 1X PBS) at a ratio of 100:1. Mounting medium contained Citifluor and Vectashield at a 4:1 ratio with DAPI at 0.5 μg/ml.
Slides were observed under a model BX61 microscope (Olympus) equipped with DAPI (U-MNU2) and FITC (U-N41001 HQIF) filter sets. Images analyses were performed using a model Olympus DP72 (Olympus) camera system and CellA (Olympus) imaging software. To obtain a rough estimate of bacterial cell numbers in the sponge tissue, 20 square zones of 20 μm2 were selected. In each of these zones, the Betaproteobacteria signals were counted from the specific probe signals, and total bacterial cells and sponge nuclei were counted from the corresponding DAPI image.
Transmission electron microscopy
TEM was performed according to Uriz and collaborators61 with minor modifications. Briefly, small samples of sponge tissue (3–4 mm3), were fixed overnight at 4°C in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.4 (osmolarity adjusted to 980 mOsM with sucrose), washed in the same buffer at room temperature, and post-fixed for 1 hour in 1% osmium tetroxide. The samples were dehydrated in an ethanol-graded series (70%; 95%; 100%) and embedded in Epon 812 (EMS). Ten sub samples of C. crambe tissue (two sponges specimens) were prepared and embedded with different orientation. Contrasted ultra thin sections (produced using an ultramicrotome (Ultracut R Leica)) were observed at 80 kV on a transmission electron microscope (Hitachi, H7500, Japan).
Author Contributions
Experiment concept and design: M.T.S., P.L. Laboratory work: J.C., N.J.W., M.L.E., L.I. Data analysis: M.T.S., J.C., N.J.W., M.L.E. Wrote the first draft of the manuscript: M.T.S., J.C., N.J.W. The manuscript was written and approved by all authors.
Supplementary Material
Supplementary Information
Acknowledgments
We thank the OOB diving team for collecting the sponge specimens and Shibu Yooseph (JCVI) for providing us with a database of planktonic rRNA genes associated with his previous publication. Molecular biology, and DNA sequencing analyses were performed using the Marine Biodiversity and Biotechnology platform (Bio2Mar) and microscopy analysis using the Cytometry and Imaging platform (http://www.obs-banyuls.fr/pci/index.php) both at OOB. We thank Olivier Thomas (Univ. Nice), Detmer Sipkema (Univ Wageningen), Maria-Jesús Uriz (CSIC Blanes), Nataly Bontemps and Bernard Banaigs (Univ. Perpignan) and Thierry Perez (Univ. Marseille) for suggestions to the experimental work and very fruitful discussions. We thank the UPMC doctoral school 392 (Diversity of Life) for its doctoral fellowship to Julie Croué.
References
- Vacelet J. & Donadey C. Electron microscope study of the association between some sponges and bacteria. J Exp Mar Biol Ecol 30, 301–314 (1977). [Google Scholar]
- Hentschel U. et al. Microbial diversity of marine sponges. Prog Mol Subcell Biol 37, 59–88 (2003). [DOI] [PubMed] [Google Scholar]
- Hentschel U., Usher K. M. & Taylor M. W. Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55, 167–177 (2006). [DOI] [PubMed] [Google Scholar]
- Schmitt S. et al. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6, 564–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister R. L., Deines P., Botté E. S., Webster N. S. & Taylor M. W. Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ Microbiol 14, 517–524 (2012). [DOI] [PubMed] [Google Scholar]
- Reiswig H. M. Partial Carbon and Energy Budgets of the Bacteriosponge Verohgia fistularis (Porifera: Demospongiae) in Barbados. Mar Ecol 2, 273–293 (1981). [Google Scholar]
- Kamke J., Taylor M. W. & Schmitt S. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. The ISME J 4, 498–508 (2010). [DOI] [PubMed] [Google Scholar]
- Giles E. C. et al. Bacterial community profiles in low microbial abundance sponges. FEMS Microbiol Ecol 83, 232–241 (2013). [DOI] [PubMed] [Google Scholar]
- Simister R., Taylor M. W., Rogers K. M., Schupp P. J. & Deines P. Temporal molecular and isotopic analysis of active bacterial communities in two New Zealand sponges. FEMS Microbiol Ecol 10.1111/1574-6941.12109 (2013). [DOI] [PubMed] [Google Scholar]
- Hentschel U. et al. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68, 4431–4440 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep 21, 519–538 (2004). [DOI] [PubMed] [Google Scholar]
- Faulkner D. J. Highlights of marine natural products chemistry (1972-1999). Nat Prod Rep 17, 1–6 (2000). [DOI] [PubMed] [Google Scholar]
- Schwarzer D. & Marahiel M. A. Multimodular biocatalysts for natural product assembly. Naturwissenschaften 88, 93–101 (2001). [DOI] [PubMed] [Google Scholar]
- Kim T. K. & Fuerst J. A. Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: culture-dependent and culture-independent approaches. Environ Microbiol 8, 1460–1470 (2006). [DOI] [PubMed] [Google Scholar]
- Kashman Y. et al. Ptilomycalin A: a novel polycyclic guanidine alkaloid of marine origin. J Am Chem Soc 111, 8925–8926 (1989). [Google Scholar]
- Jares-Erijman E. A., Ingrum A. L., Carney J. R., Rinehart K. L. & Sakai R. Polycyclic guanidine-containing compounds from the mediterranean sponge Crambe crambe: the structure of 13, 14, 15-isocrambescidin 800 and the absolute stereochemistry of the pentacyclic guanidine moieties of the crambescidins. J Org Chem 58, 4805–4808 (1993). [Google Scholar]
- Jares-Erijman E. A., Sakai R. & Rinehart K. L. Crambescidins: new antiviral and cytotoxic compounds from the sponge Crambe crambe. J Org Chem 56, 5712–5715 (1991). [Google Scholar]
- Tavares R. et al. Isolation of Crambescidin 800 from Monanchora arbuscula (Porifera). Biochem Syst Ecol 22, 645–646 (1994). [Google Scholar]
- Patil A. D. et al. Batzelladines F-I, novel alkaloids from the sponge Batzella sp.: Inducers of p56lck-CD4 dissociation. J Orga Chem 62, 1814–1819 (1997). [Google Scholar]
- Harbour G. C. et al. Ptilocaulin and isoptilocaulin, antimicrobial and cytotoxic cyclic guanidines from the Caribbean sponge Ptilocaulis aff. P. spiculifer. (Lamarck, 1814). J Am Chem Soc 103, 5604–5606 (1981). [Google Scholar]
- Snider B. B. & Shi Z. Biomimetic synthesis of the pentacyclic nucleus of ptilomycalin A. J Am Chem Soc 116, 549–557 (1994). [Google Scholar]
- Berlinck R. G. S. et al. Polycyclic guanidine alkaloids from the marine sponge Crambe crambe and Ca++ channel blocker activity of crambescidin 816. J Nat Prod 56, 1007–1015 (1993). [DOI] [PubMed] [Google Scholar]
- Becerro M. A., Lopez N. I., Turon X. & Uriz M. J. Antimicrobial activity and surface bacterial film in marine sponges. J Exp Mar Biol Ecol 179, 195–205 (1994). [Google Scholar]
- Uriz M. J., Becerro M. A., Tur J. M. & Turon X. Location of toxicity within the Mediterranean sponge Crambe crambe (Demospongiae: Poecilosclerida). Mar Biol 124, 583–590 (1996). [Google Scholar]
- Sarà M. Associazionia fra Poriferi e alghe in acque superficiali det litorale marino. Ricerca scient 36, 277–282 (1966). [Google Scholar]
- Thiel V., Neulinger S. C., Staufenberger T., Schmaljohann R. & Imhoff J. F. Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol Ecol 59, 47–63 (2007). [DOI] [PubMed] [Google Scholar]
- Blazejak A., Erséus C., Amann R. & Dubilier N. Coexistence of bacterial sulfide oxidizers, sulfate reducers, and spirochetes in a gutless worm (Oligochaeta) from the Peru margin. Appl Environ Microbiol 71, 1553–1561 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriz M. J., Turon X. & Becerro M. A. Morphology and ultrastructure of the swimming larvae of Crambe crambe (Demospongiae, Poecilosclerida). Invert Biol 120, 295–307 (2001). [Google Scholar]
- Yooseph S. et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468, 60–66 (2010). [DOI] [PubMed] [Google Scholar]
- Peek A. S., Vrijenhoek R. C. & Gaut B. S. Accelerated evolutionary rate in sulfur-oxidizing endosymbiotic bacteria associated with the mode of symbiont transmission. Mol Biol Evol 15, 1514–1523 (1998). [DOI] [PubMed] [Google Scholar]
- Moran N. A. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr Opin Microbiol 6, 512–518 (2003). [DOI] [PubMed] [Google Scholar]
- Suzuki M. T., Beja O., Taylor L. T. & DeLong E. F. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ Microbiol 3, 323–331 (2001). [DOI] [PubMed] [Google Scholar]
- Bensaadi-Merchermek N., Salvado J. C., Cagnon C., Karama S. & Mouchès C. Characterization of the unlinked 16S rDNA and 23S-5S rRNA operon of Wolbachia pipientis, a prokaryotic parasite of insect gonads. Gene 165, 81–86 (1995). [DOI] [PubMed] [Google Scholar]
- Rurangirwa F. R., Brayton K. A., McGuire T. C., Knowles D. P. & Palmer G. H. Conservation of the unique rickettsial rRNA gene arrangement in Anaplasma. IJSEM 52, 1405–1409 (2002). [DOI] [PubMed] [Google Scholar]
- Usher K. M., Kuo J., Fromont J. & Sutton D. C. Vertical transmission of cyanobacterial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Hydrobiologia 461, 9–13 (2001). [Google Scholar]
- Sharp K. H., Eam B., Faulkner D. J. & Haygood M. G. Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl Environ Microbiol 73, 622–629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S., Angermeier H., Schiller R., Lindquist N. & Hentschel U. Molecular Microbial Diversity Survey of Sponge Reproductive Stages and Mechanistic Insights into Vertical Transmission of Microbial Symbionts. Appl Environ Microbiol 74, 7694–7708 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvin J., Ninawe A. S., Seghal Kiran G. & Lipton A. P. Sponge-microbial interactions: Ecological implications and bioprospecting avenues. Crit Rev Microbiol 36, 82–90 (2010). [DOI] [PubMed] [Google Scholar]
- Bretting H. & Königsmann K. Investigations on the lectin-producing cells in the sponge Axinella polypoides (Schmidt). Cell Tissue Res 201, 487–497 (1979). [DOI] [PubMed] [Google Scholar]
- Thompson J. E., Barrow K. D. & Faulkner D. J. Localization of two brominated metabolites, aerothionin and homoaerothionin, in spherulous cells of the marine sponge Aplysina fistularis ( = Verongia thiona). Acta Zool 64, 199–210 (1983). [Google Scholar]
- Fan L. et al. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. PNAS 109, E1878–E1887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl A. et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J 5, 61–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N. S., Wilson K. J., Blackall L. L. & Hill R. T. Phylogenetic Diversity of Bacteria Associated with the Marine Sponge Rhopaloeides odorabile. Appl Environ Microbiol 67, 434–444 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergin K. L. et al. Screening of a Fosmid Library of Marine Environmental Genomic DNA Fragments Reveals Four Clones Related to Members of the OrderPlanctomycetales. Appl Environ Microbiol 64, 3075–3078 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G. et al. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Kluwer Academic (ed Akkermans A., Van Elsas J., & De Bruijn F.) 3.4.4, 1–27 (Dordrecht, 1998). [Google Scholar]
- Bonfield J. K., Smith K. F. & Staden R. A new DNA sequence assembly program. Nucleic Acids Res 23, 4992–4999 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. & Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T., Faulkner G. & Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319 (2004). [DOI] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W. et al. ARB: a software environment for sequence data. Nucleic Acids Res 32, 1363–1371 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences., University of Washington, Seattle (2005).
- Ronquist F. et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst Biol 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S. E. et al. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3, e3326 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C., Lanzen A., Davenport R. J. & Turnbaugh P. J. Removing noise from pyrosequenced amplicons. BMC bioinformatics 12, 38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- Fuchs B. M., Glöckner F. O., Wulf J. & Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol 66, 3603–3607 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauzet S. et al. Cloning and retinal expression of melatonin receptors in the European sea bass, Dicentrarchus labrax. Gen Comp Endocrinol 157, 186–195 (2008). [DOI] [PubMed] [Google Scholar]
- Pernthaler A., Pernthaler J. & Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68, 3094–3101 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriz M. J., Turon X. & Becerro M. A. Silica deposition in Demosponges: spiculogenesis in Crambe crambe. Cell Tissue Res 301, 299–309 (2000). [DOI] [PubMed] [Google Scholar]
- Amann R. et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56, 1919–1925 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner G., Amann R. & Beisker W. Optimizing fluorescent in situ hybridization of suspended cells with rRNA-targeted oligonucleotide probes for the flow cytometric identification of microorganisms. Cytometry 14, 136–143 (1993). [DOI] [PubMed] [Google Scholar]
- Manz W., Amann R., Ludwig W. & Schleifer K. H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst and Appl Microbiol 15, 593–600 (1992). [Google Scholar]
- Stahl D. A. & Amann R. Development and application of nucleic acid probes. In: Nucleic acid techniques in bacterial systematics (eds Stackebrandt, E. & Goodfellow, M.) 205–248 (Wiley & Sons, 1991). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information