Abstract
Between January 2012 and March 2012, the infection rates of porcine epidemic diarrhea virus (PEDV) increased substantially in vaccinated swine herds in many porcine farms in Gansu Province, China. The spike (S) glycoprotein is an important determinant for PEDV biological properties. To determine the distribution profile of PEDV outbreak strains, we sequenced the full-length S gene of five samples from two farms where animals exhibited severe diarrhea and high mortality rates. Five new PEDV variants were identified, and the molecular diversity, phylogenetic relationships, and antigenicity analysis of Gansu field samples with other PEDV reference strains were investigated. A series of insertions, deletions, and mutations in the S gene was found in five PEDV variants compared with classical and vaccine strains. These mutations may provide stronger pathogenicity and antigenicity to the new PEDV variants that influenced the effectiveness of the CV777-based vaccine. Our results suggest that these new PEDV variant strains in Gansu Province might be from South Korean or South China, and the effectiveness of the CV777-based vaccine needs to be evaluated.
Keywords: molecular characterization, spike glycoprotein gene, phylogenetic analysis, porcine epidemic diarrhea virus (PEDV)
1. Introduction
Porcine epidemic diarrhea virus (PEDV), a member of Coronaviridae, is an enveloped, single-stranded RNA genome with a 5' cap and a 3' polyadenylated tail. The size of its genome is approximately 28 Kb [1]. The genome comprises a 5' untranslated region (UTR); a 3' UTR; at least seven open reading frames (ORFs) that encode four structural proteins, namely, spike (S), envelope (E), membrane (M), and nucleocapsid (N); and three non-structural proteins, namely, replicases 1a and 1ab as well as ORF3 [2]. The PEDV S protein is a type I glycoprotein composed of 1,383 amino acids (aa). Similar to other coronavirus S proteins, the PEDV S protein is a glycoprotein peplomer (surface antigen) on the viral surface and contains four neutralizing epitopes (499–638, 748–755, 764–771, and 1,368–1,374 aa) [3,4,5]. The PEDV S protein has a pivotal function in regulating interactions with specific host cell receptor glycoproteins to mediate viral entry [6]. Thus, the S glycoprotein is a primary target for the development of vaccines against PEDV. The S glycoprotein is also the major envelope glycoprotein of the virion, which serves as an important viral component to understand the genetic relationships of different PEDV strains and the epidemiological status of PEDV in the field [2,7,8,9].
Porcine epidemic diarrhea (PED), caused by PEDV, is an acute, highly contagious, and devastating swine disease that is characterized by acute enteritis and lethal watery diarrhea, followed by dehydration frequently leading to high mortality in piglets [10,11]. PED was first observed among English fattening pigs in 1971 [10] but has increasingly become a problem in several Asian countries, including Korea [12], China [8,9,13], Japan [14], and Thailand [15]. In China, PEDV was first isolated in 1973 [9]. Almost two decades later, its prevalence has become a problem of the swine industry in China, but until 2010, the prevalence of PEDV infection was relatively low with only sporadic outbreaks. However, in late 2010, a remarkable increase in PED outbreaks occurred in the pig-producing provinces [9,16]. PED that occurred in several porcine farms and caused severe economic loss between January 2012 and March 2012 in Gansu Province, China was investigated in this study. The affected pigs exhibited watery diarrhea, dehydration, and thin-walled intestines. The disease progressed to death within four days. Pigs of all ages were affected and exhibited diarrhea and loss of appetite with different degrees of severity, which were determined to be age dependent. Among the suckling piglets, 100% became ill. Pigs >7 days of age experienced mild diarrhea and anorexia, which completely resolved within a few days. To identify the PEDV strain(s) responsible for the recent outbreak in Gansu, where located in west China, the east by Shanxi province, the south of Sichuan province, the west of Xinjiang province, and the north of Inner Mongolia province, we sequenced the full-length S gene of the isolates obtained from the diarrhea samples collected from pigs in two affected pig farms. One farm named Yongjing Tai Chi Breed Co., Ltd (Yongjing, China), and another named Hoggery of Science and Technology Breed Park of Jiugang Hongfeng Company (Jiayuguan, China).
2. Results and Discussion
2.1. Sequence Analysis of the S Gene
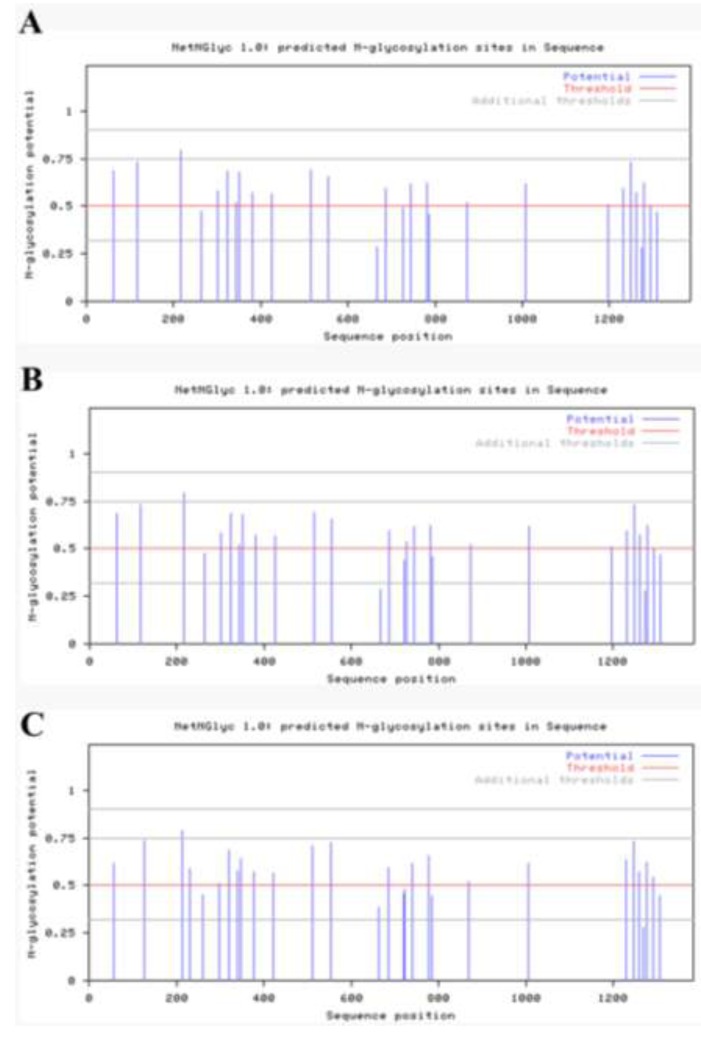
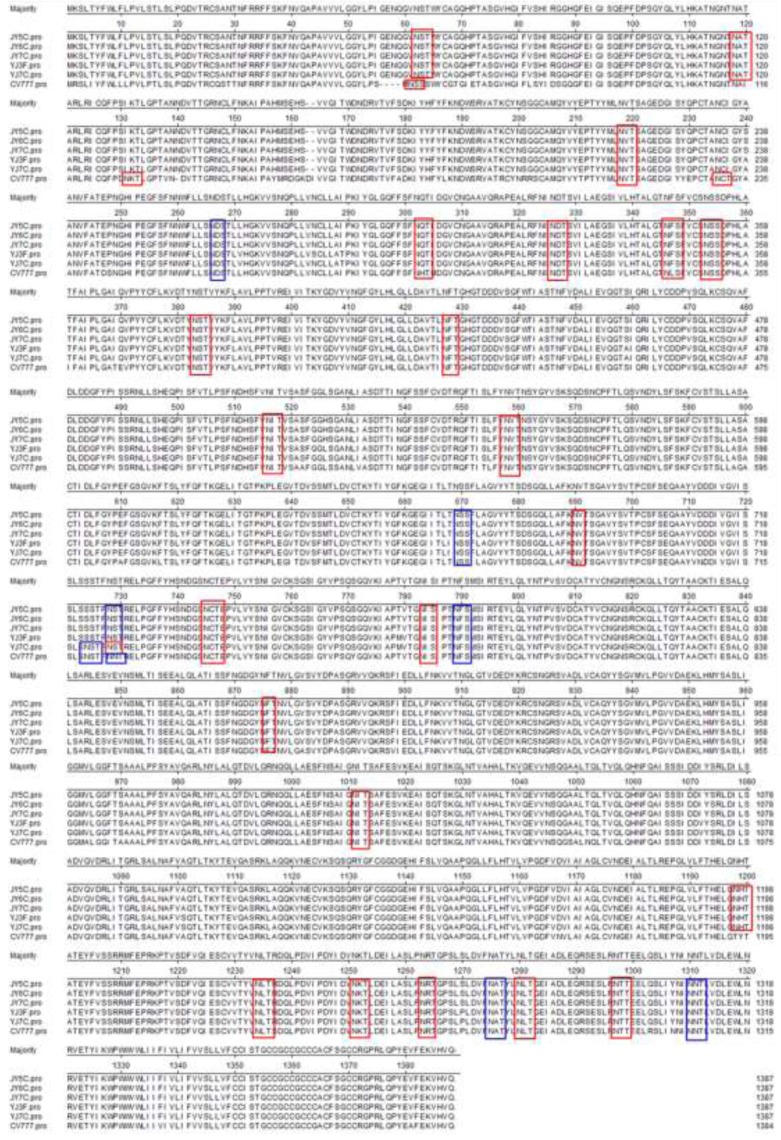
The nucleotide sequences of the S region were 4,161 bp long for JY5C, JY6C, JY7C, YJ3F, and YJ7C. Their S proteins were 1,386 aa long with a predicted Mr of 151.7 KDa. The S protein of JY5C, JY6C, JY7C, and YJ3F contained 28 Asn-Xaa-Ser/Thr sequons and 22 asparagines predicted to be N-glycosylated (Figure 1A,B). The S protein of the YJ7C strain contained 29 Asn-Xaa-Ser/Thr sequons and 23 asparagines predicted to be N-glycosylated (Figure 1C). However, the S protein of the CV777 vaccine strain contained 29 Asn-Xaa-Ser/Thr sequons and 22 asparagines predicted to be N-glycosylated (Figure 1D). For JY5C, JY6C, JY7C, and YJ3F, four (from N to V at 57, from N to I at 127, from T to I at 232, and from N to S at 719) of the changes in the predicted amino acid sequence destroyed N-linked glycosylation sites, whereas another three changes (from S to N at 58, from I to T at 116, and from T to N at 1193) created three new glycosylation sites compared with the vaccine strain CV777 (Figure 2). For the YJ7C strain, three amino acid changes (from N to V at 57, from N to I at 127, and from T to I at 232) destroyed N-linked glycosylation sites and another three changes (from S to N at 58, from I to T at 116, and from T to N at 1193) created three new glycosylation sites compared with the vaccine strain CV777 (Figure 2). The changes in the N-linked glycosylation sites between the Gansu PEDV strains from our study and the vaccine strain may influence their pathogenicity and antigenicity and should be researched in the future.
Figure 1.
(A) S proteins of the JY5C, JY6C, JY7C and JY3F strains contained the same Asn-Xaa-Ser/Thr sequons and asparagines predicted to be N-glycosylated; (B) N-glycosylated prediction of the S protein of YJ7C strain; (C) N-glycosylated prediction of the S protein of CV777 strain.
Figure 2.
Amino acid alignment of Asn-Xaa-Ser/Thr sequons and asparagines predicted to be N-glycosylated of the JY5C, JY6C, JY7C, YJ3F and YJ7C strains’ S protein. Both blue boxes and red boxes stand for the Asn-Xaa-Ser/Thr sequons, but only red boxes stand for asparagines predicted to be N-glycosylated.
2.2. Nucleotide and Deduced Amino Acid Sequence Homology
The nucleotide and deduced amino acid sequence homology results are described in Table 1. The nucleotide sequence of the five strains from our study (JY5C, JY6C, JY7C, YJ3F, and YJ7C) had 99.3% to 100% nucleotide sequence identity to one another, and their deduced amino acids had 99.0% to 100% identity to one another. The S genes’ nucleotide and deduced amino acid identities of five strains from our study (JY5C, JY6C, JY7C, YJ3F, and YJ7C) with the other PEDV reference strains are described in Table 2. The PEDV strain that had the highest DNA sequence identity to our PEDV strains was CH8 (one Chinese PEDV strain), which had 98.4% identity and the deduced amino acids had more than 98.0% identity. However, the nucleotide sequence of our PEDV strains had only 93.8% to 93.9% identity to the CV777 vaccine strain, and their deduced amino acids had 93.6% to 93.7% identity to the CV777 vaccine strain. The nucleotide sequence of our PEDV strains had lower identity (93.3% to 95.7%) to the previous domestic strains (DX, LZC, LJB-03, JS-2004-2, and CHS) and their deduced amino acids had 92.6% to 95.7% identity to the previous domestic strains (DX, LZC, LJB-03, JS-2004-2, and CHS). The nucleotide sequence of our PEDV strains also had low identity (93.2% to 94.6% and 93.7% to 93.8%) to the Japanese strains (MK, NK, and KH) and the European strain (Br1-87), respectively, and their deduced amino acids had 93.1% to 94.2% and 93.4% to 93.6% identities to the Japanese strains (MK, NK, and KH) and the Europe strain (Br1-87), respectively. The nucleotide sequence of our PEDV strains had higher identity (94.7% to 97.1%) to seven South Korean strains (KNU-0801, KNU-0802, KNU-0901, KNU-0902, KNU-0903, KNU-0904, and KNU-0905), and their deduced amino acids had 94.1% to 96.8% identity.
Table 1.
Comparison of the nucleotide and deduced amino acid sequences of S genes of PEDV (porcine epidemic diarrhea virus) reference strains and PEDV outbreak in Gansu, China.
| Virus strian | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 JY5C | 100.0 | 100.0 | 99.3 | 99.3 | 97.6 | 95.1 | 93.4 | 93.4 | 95.1 | 95.2 | 94.4 | 98.4 | 97.7 | 94.6 | 95.3 | 97.5 | 95.0 | 93.3 | 94.9 | 95.7 | 93.4 | 95.1 | 97.1 | 95.9 | 95.0 | 95.8 | 94.7 | 96.5 | 93.8 | 93.2 | 93.6 | 93.2 | 94.5 | 94.3 | 93.8 | 93.7 | |
| 2 JY6C | 100.0 | 100.0 | 99.3 | 99.3 | 97.6 | 95.1 | 93.4 | 93.4 | 95.1 | 95.2 | 94.4 | 98.4 | 97.7 | 94.6 | 95.3 | 97.5 | 95.0 | 93.3 | 94.9 | 95.7 | 93.4 | 95.1 | 97.1 | 95.9 | 95.0 | 95.8 | 94.7 | 96.5 | 93.8 | 93.2 | 93.6 | 93.2 | 94.5 | 94.3 | 93.8 | 93.7 | |
| 3 JY7C | 100.0 | 100.0 | 99.3 | 99.3 | 97.6 | 95.1 | 93.4 | 93.4 | 95.1 | 95.2 | 94.4 | 98.4 | 97.7 | 94.6 | 95.3 | 97.5 | 95.0 | 93.3 | 94.9 | 95.7 | 93.4 | 95.1 | 97.1 | 95.9 | 95.0 | 95.8 | 94.7 | 96.5 | 93.8 | 93.2 | 93.6 | 93.2 | 94.5 | 94.3 | 93.8 | 93.7 | |
| 4 YJ3F | 99.0 | 99.0 | 99.0 | 100.0 | 97.6 | 95.1 | 93.5 | 93.5 | 95.1 | 95.2 | 94.4 | 98.4 | 97.7 | 94.7 | 95.3 | 97.5 | 95.0 | 93.3 | 94.9 | 95.6 | 93.5 | 95.2 | 97.0 | 95.8 | 94.9 | 95.7 | 94.7 | 96.7 | 93.8 | 93.3 | 93.7 | 93.2 | 94.6 | 94.2 | 93.8 | 93.8 | |
| 5 YJ7C | 99.0 | 99.0 | 99.0 | 99.9 | 97.6 | 95.1 | 93.6 | 93.5 | 95.1 | 95.2 | 94.4 | 98.4 | 97.7 | 94.7 | 95.4 | 97.6 | 95.0 | 93.4 | 94.9 | 95.6 | 93.6 | 95.2 | 97.0 | 95.9 | 95.0 | 95.8 | 94.8 | 96.7 | 93.9 | 93.4 | 93.7 | 93.2 | 94.6 | 94.3 | 93.9 | 93.8 | |
| 6 CH1 | 97.9 | 97.9 | 97.9 | 97.6 | 97.6 | 94.5 | 93.4 | 93.3 | 94.5 | 94.6 | 95.8 | 98.4 | 98.5 | 94.2 | 94.9 | 97.1 | 94.6 | 92.9 | 94.5 | 94.9 | 93.0 | 94.8 | 96.5 | 95.4 | 94.8 | 95.3 | 94.5 | 96.2 | 93.5 | 93.2 | 93.3 | 93.1 | 94.3 | 94.1 | 93.4 | 93.4 | |
| 7 CH2 | 95.1 | 95.1 | 95.1 | 94.9 | 94.9 | 94.6 | 95.8 | 95.8 | 99.5 | 99.6 | 97.6 | 95.3 | 94.0 | 96.8 | 95.9 | 94.4 | 97.3 | 95.2 | 97.2 | 98.4 | 95.6 | 93.1 | 93.9 | 93.8 | 92.9 | 93.8 | 92.8 | 93.6 | 95.8 | 95.6 | 92.6 | 95.2 | 93.0 | 92.7 | 95.7 | 95.6 | |
| 8 CH3 | 92.8 | 92.8 | 92.8 | 92.8 | 93.0 | 92.9 | 95.3 | 99.7 | 95.8 | 96.0 | 95.8 | 93.7 | 93.1 | 97.6 | 96.6 | 94.4 | 96.0 | 96.2 | 95.9 | 96.3 | 96.6 | 94.0 | 93.4 | 93.5 | 93.0 | 93.4 | 94.2 | 93.6 | 96.8 | 99.5 | 93.8 | 97.6 | 94.2 | 94.2 | 96.8 | 96.7 | |
| 9 CH4 | 92.8 | 92.8 | 92.8 | 92.8 | 93.0 | 92.9 | 95.3 | 99.4 | 95.8 | 95.9 | 95.7 | 93.6 | 93.0 | 97.5 | 96.6 | 94.3 | 95.9 | 96.2 | 95.8 | 96.2 | 96.6 | 94.0 | 93.3 | 93.4 | 92.9 | 93.3 | 94.1 | 93.5 | 96.8 | 99.4 | 93.7 | 97.5 | 94.2 | 94.1 | 96.8 | 96.7 | |
| 10 CH5 | 94.6 | 94.6 | 94.6 | 94.5 | 94.5 | 94.1 | 99.1 | 94.9 | 94.9 | 99.6 | 97.6 | 95.3 | 94.0 | 96.7 | 95.9 | 94.3 | 97.3 | 95.2 | 97.2 | 98.4 | 95.6 | 93.1 | 93.9 | 93.8 | 92.9 | 93.8 | 92.8 | 93.6 | 95.8 | 95.6 | 92.6 | 95.2 | 93.0 | 92.7 | 95.7 | 95.6 | |
| 11 CH6 | 95.1 | 95.1 | 95.1 | 94.9 | 94.9 | 94.6 | 99.6 | 95.4 | 95.4 | 99.1 | 97.7 | 95.4 | 94.1 | 96.9 | 96.1 | 94.5 | 97.4 | 95.4 | 97.3 | 98.5 | 95.7 | 93.3 | 94.0 | 93.9 | 93.0 | 93.9 | 93.0 | 93.8 | 96.0 | 95.7 | 92.8 | 95.3 | 93.2 | 92.9 | 95.9 | 95.8 | |
| 12 CH7 | 94.6 | 94.6 | 94.6 | 94.3 | 94.3 | 95.6 | 97.5 | 95.3 | 95.3 | 97.0 | 97.5 | 94.7 | 95.1 | 96.5 | 95.6 | 94.0 | 97.2 | 94.9 | 97.0 | 97.8 | 95.3 | 92.8 | 93.5 | 93.5 | 92.9 | 93.5 | 92.6 | 93.4 | 95.4 | 95.5 | 92.3 | 95.1 | 92.7 | 92.4 | 95.4 | 95.3 | |
| 13 CH8 | 98.1 | 98.1 | 98.1 | 98.0 | 98.0 | 98.6 | 95.0 | 92.9 | 92.9 | 94.6 | 95.0 | 94.6 | 97.5 | 94.6 | 95.3 | 97.4 | 94.9 | 93.3 | 94.8 | 95.6 | 93.4 | 95.2 | 96.9 | 95.8 | 95.0 | 95.7 | 94.8 | 96.5 | 93.8 | 93.5 | 93.5 | 93.4 | 94.7 | 94.5 | 93.8 | 93.7 | |
| 14 CHGD01 | 98.1 | 98.1 | 98.1 | 97.9 | 97.9 | 98.2 | 93.9 | 92.7 | 92.7 | 93.5 | 93.9 | 94.6 | 97.4 | 94.3 | 95.0 | 97.1 | 94.4 | 92.9 | 94.4 | 94.7 | 92.9 | 94.6 | 96.5 | 95.3 | 94.5 | 95.3 | 94.4 | 96.3 | 93.4 | 92.9 | 93.4 | 92.9 | 94.2 | 93.9 | 93.4 | 93.4 | |
| 15 CH-FJND-1-2011 | 93.8 | 93.8 | 93.8 | 94.0 | 94.1 | 93.6 | 96.0 | 97.3 | 97.2 | 95.6 | 96.1 | 95.7 | 93.8 | 93.7 | 98.9 | 96.7 | 96.8 | 95.9 | 96.7 | 97.3 | 96.0 | 93.2 | 94.2 | 94.2 | 93.2 | 94.1 | 93.3 | 94.1 | 96.4 | 97.3 | 93.1 | 96.1 | 93.4 | 93.3 | 96.4 | 96.3 | |
| 16 CH-FJND-2-2011 | 95.1 | 95.1 | 95.1 | 95.2 | 95.4 | 94.9 | 95.0 | 96.1 | 96.0 | 94.6 | 95.1 | 94.7 | 94.8 | 94.9 | 98.6 | 97.5 | 96.1 | 95.2 | 96.0 | 96.5 | 95.3 | 93.6 | 94.9 | 94.5 | 93.6 | 94.5 | 93.7 | 94.7 | 95.7 | 96.4 | 93.4 | 95.3 | 93.8 | 93.5 | 95.8 | 95.7 | |
| 17 CH-FJND-3-2011 | 97.0 | 97.0 | 97.0 | 97.1 | 97.3 | 96.8 | 93.8 | 93.8 | 93.8 | 93.3 | 93.8 | 93.4 | 96.7 | 96.6 | 96.3 | 97.5 | 94.5 | 93.6 | 94.5 | 94.9 | 93.7 | 95.1 | 96.9 | 95.9 | 95.0 | 95.8 | 95.1 | 96.5 | 94.1 | 94.2 | 93.9 | 93.7 | 94.9 | 94.8 | 94.1 | 94.1 | |
| 18 DX | 95.3 | 95.3 | 95.3 | 95.2 | 95.2 | 94.8 | 97.2 | 95.4 | 95.4 | 96.9 | 97.2 | 97.0 | 95.0 | 94.6 | 96.5 | 95.5 | 94.4 | 95.5 | 99.1 | 98.1 | 95.9 | 93.2 | 94.2 | 94.3 | 93.4 | 94.2 | 93.1 | 94.0 | 96.0 | 95.7 | 93.0 | 95.3 | 93.2 | 93.0 | 96.0 | 95.9 | |
| 19 LZC | 92.8 | 92.8 | 92.8 | 92.6 | 92.8 | 92.3 | 94.8 | 94.9 | 94.9 | 94.4 | 95.0 | 94.6 | 92.3 | 92.6 | 94.9 | 94.2 | 92.7 | 94.8 | 95.4 | 95.9 | 95.8 | 93.4 | 93.1 | 93.2 | 93.0 | 93.2 | 93.5 | 93.5 | 99.4 | 96.0 | 93.8 | 95.5 | 93.7 | 93.3 | 99.5 | 99.4 | |
| 20 LJB-03 | 94.9 | 94.9 | 94.9 | 94.8 | 94.8 | 94.4 | 96.9 | 95.2 | 95.2 | 96.6 | 96.9 | 96.7 | 94.6 | 94.3 | 96.0 | 95.0 | 93.8 | 98.6 | 94.5 | 98.0 | 95.8 | 93.2 | 94.2 | 94.1 | 93.3 | 94.1 | 93.1 | 93.9 | 96.0 | 95.6 | 92.9 | 95.2 | 93.1 | 92.9 | 96.0 | 95.9 | |
| 21 JS-2004-2 | 95.7 | 95.7 | 95.7 | 95.5 | 95.5 | 94.9 | 98.0 | 95.8 | 95.8 | 97.6 | 98.0 | 97.6 | 95.3 | 94.7 | 96.7 | 95.7 | 94.4 | 98.0 | 95.4 | 97.8 | 96.2 | 93.6 | 94.7 | 94.6 | 93.7 | 94.6 | 93.3 | 94.4 | 96.4 | 96.1 | 93.1 | 95.7 | 93.4 | 93.2 | 96.4 | 96.3 | |
| 22 CHS | 93.3 | 93.3 | 93.3 | 93.3 | 93.5 | 93.0 | 95.4 | 96.1 | 96.1 | 95.2 | 95.5 | 95.2 | 93.1 | 93.0 | 95.6 | 94.6 | 93.3 | 95.7 | 95.2 | 95.4 | 96.1 | 94.0 | 93.9 | 93.9 | 93.4 | 93.8 | 93.7 | 93.7 | 96.3 | 96.3 | 93.5 | 95.8 | 94.0 | 93.4 | 96.4 | 96.3 | |
| 23 KUN-0901 | 94.7 | 94.7 | 94.7 | 94.7 | 94.8 | 94.4 | 92.9 | 93.4 | 93.3 | 92.5 | 93.1 | 92.5 | 94.7 | 94.4 | 92.7 | 93.5 | 94.5 | 93.0 | 92.8 | 92.7 | 93.3 | 93.6 | 95.4 | 95.5 | 95.7 | 95.4 | 97.9 | 95.8 | 93.9 | 93.7 | 96.0 | 93.4 | 96.7 | 94.7 | 93.9 | 93.9 | |
| 24 KUN-0902 | 96.4 | 96.4 | 96.4 | 96.1 | 96.3 | 95.9 | 93.1 | 92.4 | 92.4 | 92.9 | 93.1 | 92.8 | 96.0 | 95.9 | 93.0 | 94.2 | 96.0 | 93.6 | 92.3 | 93.2 | 94.1 | 93.5 | 94.5 | 97.8 | 95.3 | 97.6 | 94.9 | 96.5 | 93.6 | 93.1 | 93.8 | 92.9 | 94.6 | 94.3 | 93.6 | 93.6 | |
| 25 KUN-0903 | 95.0 | 95.0 | 95.0 | 94.8 | 95.0 | 94.4 | 93.1 | 92.6 | 92.5 | 92.8 | 93.3 | 93.1 | 94.7 | 94.4 | 93.1 | 94.1 | 94.8 | 93.6 | 92.3 | 93.1 | 94.1 | 93.5 | 95.1 | 96.5 | 96.8 | 99.8 | 95.1 | 95.8 | 93.7 | 93.2 | 94.1 | 92.9 | 95.1 | 94.6 | 93.8 | 93.7 | |
| 26 KUN-0904 | 94.1 | 94.1 | 94.1 | 94.0 | 94.2 | 93.9 | 92.5 | 92.0 | 92.0 | 92.1 | 92.8 | 92.4 | 93.9 | 93.8 | 92.3 | 93.3 | 93.9 | 92.8 | 91.9 | 92.5 | 93.1 | 92.8 | 95.3 | 94.0 | 96.5 | 96.7 | 95.1 | 96.2 | 93.5 | 92.7 | 93.9 | 92.3 | 95.0 | 94.2 | 93.5 | 93.5 | |
| 27 KUN-0905 | 94.7 | 94.7 | 94.7 | 94.6 | 94.7 | 94.2 | 93.1 | 92.4 | 92.3 | 92.8 | 93.2 | 93.0 | 94.4 | 94.2 | 92.9 | 93.8 | 94.6 | 93.4 | 92.2 | 92.9 | 94.0 | 93.3 | 95.0 | 96.2 | 99.6 | 96.4 | 95.0 | 95.7 | 93.6 | 93.1 | 94.0 | 92.9 | 94.9 | 94.5 | 93.7 | 93.6 | |
| 28 KUN-0801 | 94.1 | 94.1 | 94.1 | 94.1 | 94.2 | 94.2 | 92.3 | 93.6 | 93.5 | 91.9 | 92.5 | 92.2 | 94.2 | 94.2 | 92.8 | 93.6 | 94.5 | 92.8 | 92.7 | 92.4 | 93.0 | 93.3 | 97.9 | 94.1 | 94.7 | 94.5 | 94.4 | 95.4 | 94.1 | 93.9 | 97.8 | 93.6 | 97.9 | 95.0 | 94.1 | 94.0 | |
| 29 KUN-0802 | 96.6 | 96.6 | 96.6 | 96.6 | 96.8 | 96.2 | 93.5 | 92.8 | 92.8 | 93.1 | 93.6 | 93.2 | 96.3 | 96.5 | 93.6 | 94.7 | 96.2 | 93.8 | 92.7 | 93.4 | 94.2 | 93.4 | 95.4 | 96.4 | 95.5 | 94.9 | 95.2 | 95.5 | 94.0 | 93.3 | 94.2 | 93.0 | 95.1 | 94.6 | 94.1 | 94.0 | |
| 30 Parent DR13 | 93.7 | 93.7 | 93.7 | 93.6 | 93.7 | 93.3 | 95.7 | 95.9 | 95.9 | 95.4 | 96.0 | 95.4 | 93.3 | 93.5 | 95.7 | 95.0 | 93.5 | 95.7 | 98.9 | 95.4 | 96.2 | 96.0 | 93.6 | 93.1 | 93.1 | 92.7 | 93.0 | 93.6 | 93.5 | 96.6 | 94.3 | 96.0 | 94.2 | 93.9 | 99.9 | 99.8 | |
| 31 Attenuated DR13 | 92.8 | 92.8 | 92.8 | 92.8 | 93.0 | 92.8 | 95.2 | 99.2 | 99.1 | 94.8 | 95.3 | 95.2 | 92.8 | 92.6 | 97.0 | 95.9 | 93.7 | 95.3 | 94.8 | 95.0 | 95.7 | 95.8 | 93.1 | 92.3 | 92.5 | 92.0 | 92.3 | 93.3 | 92.8 | 95.7 | 93.5 | 97.3 | 94.0 | 93.9 | 96.5 | 96.4 | |
| 32 Chinju99 | 92.5 | 92.5 | 92.5 | 92.5 | 92.6 | 92.3 | 90.9 | 92.0 | 92.0 | 90.5 | 91.2 | 90.9 | 92.3 | 92.5 | 91.6 | 92.2 | 92.8 | 91.5 | 92.0 | 91.2 | 91.8 | 92.0 | 95.4 | 92.5 | 92.8 | 92.7 | 92.6 | 97.3 | 93.6 | 92.8 | 91.9 | 93.3 | 96.3 | 93.8 | 94.4 | 94.4 | |
| 33 MK | 93.4 | 93.4 | 93.4 | 93.1 | 93.3 | 93.5 | 94.9 | 96.6 | 96.5 | 94.5 | 94.9 | 95.1 | 93.3 | 93.3 | 95.4 | 94.7 | 93.5 | 95.2 | 94.8 | 94.9 | 95.5 | 95.5 | 93.3 | 93.1 | 92.9 | 92.2 | 92.7 | 93.8 | 93.4 | 95.6 | 96.3 | 92.5 | 93.8 | 93.7 | 96.1 | 96.0 | |
| 34 NK | 93.9 | 93.9 | 93.9 | 94.1 | 94.2 | 93.9 | 92.8 | 93.7 | 93.6 | 92.3 | 93.0 | 92.6 | 94.0 | 93.8 | 93.0 | 93.7 | 94.2 | 93.1 | 92.9 | 92.7 | 93.5 | 93.8 | 96.3 | 93.7 | 94.4 | 94.2 | 94.2 | 97.5 | 94.7 | 93.8 | 93.5 | 95.5 | 94.1 | 95.2 | 94.2 | 94.2 | |
| 35 KH | 93.9 | 93.9 | 93.9 | 93.7 | 93.9 | 93.8 | 92.7 | 93.1 | 93.0 | 92.3 | 92.8 | 92.3 | 94.0 | 93.7 | 92.3 | 93.2 | 93.8 | 92.7 | 92.3 | 92.3 | 92.9 | 93.1 | 94.2 | 93.4 | 94.1 | 93.4 | 93.9 | 94.6 | 94.1 | 93.3 | 92.9 | 92.8 | 93.7 | 94.9 | 93.8 | 93.8 | |
| 36 CV777 | 93.7 | 93.7 | 93.7 | 93.6 | 93.7 | 93.3 | 95.7 | 95.9 | 95.9 | 95.3 | 95.9 | 95.4 | 93.3 | 93.6 | 95.9 | 95.2 | 93.6 | 95.7 | 99.1 | 95.4 | 96.2 | 96.2 | 93.7 | 93.2 | 93.2 | 92.8 | 93.1 | 93.6 | 93.6 | 99.7 | 95.7 | 93.0 | 95.7 | 93.8 | 93.2 | 99.9 | |
| 37 Br1-87 | 93.6 | 93.6 | 93.6 | 93.4 | 93.6 | 93.1 | 95.4 | 95.6 | 95.6 | 95.1 | 95.6 | 95.2 | 93.1 | 93.4 | 95.6 | 94.9 | 93.5 | 95.4 | 98.8 | 95.2 | 96.0 | 95.9 | 93.6 | 93.1 | 93.1 | 92.7 | 93.0 | 93.5 | 93.5 | 99.4 | 95.4 | 93.0 | 95.4 | 93.8 | 93.1 | 99.7 |
Nucleotide identity (%) in upper triangle; Deduced amino acid identity (%) in lower triangle.
Table 2.
The S genes’ nucleotide and deduced amino acid identities of five strains from our study (JY5C, JY6C, JY7C, YJ3F, and YJ7C) with the other PEDV reference strains.
| The other strains | Nucleotide identities | Deduced amino acid identities |
|---|---|---|
| CH8 (one Chinese PEDV strain) | 98.40% | 98.0%-98.1% |
| CV777 vaccine strain | 93.8%–93.9% | 93.6%–93.7% |
| Previous domestic strains (DX, LZC, LJB-03, JS-2004-2, and CHS) | 93.3%–95.7% | 92.6%–95.7% |
| Japanese strains (MK, NK, and KH) | 93.2%–94.6% | 93.1%–94.2% |
| European strain (Br1-87) | 93.7%–93.8% | 93.4%–93.6% |
| South Korean strains (KNU-0801, KNU-0802, KNU-0901, KNU-0902, KNU-0903, KNU-0904, and KNU-0905) | 94.7%–97.1% | 94.1%–96.8% |
2.3. Phylogenetic Analysis of the S Gene
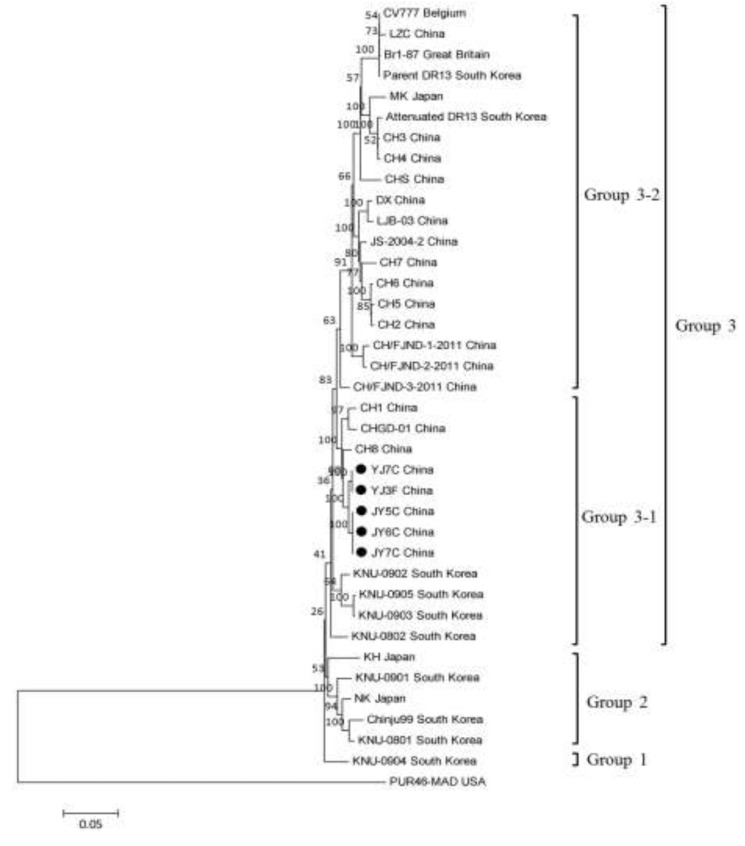
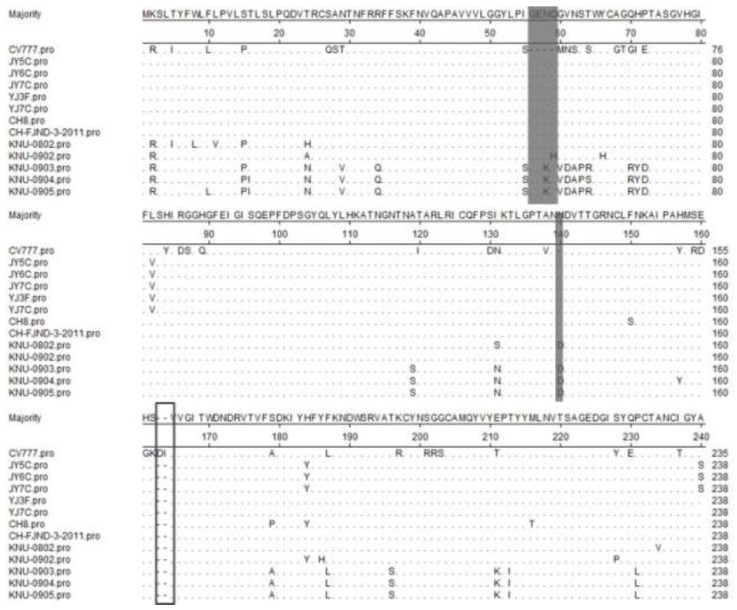
Phylogenetic analysis of the nucleotide sequences of the S gene revealed three major clusters, and the third group has two subgroups (3-1 and 3-2). All PEDVs isolated from China in our study belonged to subgroup 3-1 (Figure 3). Group 1 comprised one strain from South Korea (KNU-0904). Group 2 comprised three strains from South Korea (KNU-0801, KNU-0901, and Chinju 99) and two strains from Japan (NK and KH). Subgroup 3-1 comprised five strains from our study (JY5C, JY6C, JY7C, YJ3F, and YJ7C), three strains (CH1, CH8, CHGD-01) that were identified from China in 2011 and four strains from South Korea KNU-0802, KNU-0902, KNU-0903, and KNU-0905). Group 3-2 comprised 15 strains from China (vaccine strain CV777, CH2, CH3, CH4, CH5, CH6, CH7, CHS, LZC, DX, LJB-03, JS-2004-2, CH/FJND-1-2011, CH/FJND-2-2011, and CH/FJND-3-2011), two strains from South Korea (parent DR13 and attenuated DR13), one strain from Great Britain (Br1-87), and one strain from Japan (MK). The five variant strains from our study (JY5C, JY6C, JY7C, YJ3F, and YJ7C), two strains (CH8 and CH/FJND-3-2011) that were identified from China, and five PEDV isolates from South Korea (KNU-0802, KNU-0902, KNU-0903, and KNU-0905) shared a 4-aa insertion (at positions 56–59 of the S protein), 1-aa insertion (at position 140 of the S protein), and 2-aa deletions (at positions 163–164 of the S protein), compared with CV777 (Figure 4). Our results indicate that the North Chinese PEDV strains from our study had a close relationship with the South Korean strains and mapped phylogenetically to the same branch. However, they differed genetically from the European strains (including the vaccine strain CV777) and the early domestic strains. Similar to the result by Li et al. [9], the appearance of strains from China similar to those from South Korea and their function in the recent PED outbreak in South China, should be further investigated.
Figure 3.
Phylogenetic trees of PEDV strains generated by the neighbor-joining method with nucleotide sequences of the full-length spike genes. Bootstrapping with 1,000 replicates was performed to determine the percentage reliability for each internal node. PUR46-MAD is an out group control. Horizontal branch lengths are proportional to genetic distances between PEDV strains. Black circles indicate PEDV isolates from the 2012 outbreak in Gansu Province, China. Scale bar indicates nucleotide substitutions per site.
Figure 4.
Alignment of amino terminal 1–238 amino acid of S proteins of Gansu PEDV strains and reference strains. Ellipses represent the consensus amino acids. Boxes indicate deleted amino acids compared with CV777. Shadows indicate the inserted amino acids compared with CV777.
2.4. Antigenicity Analysis of the S Gene
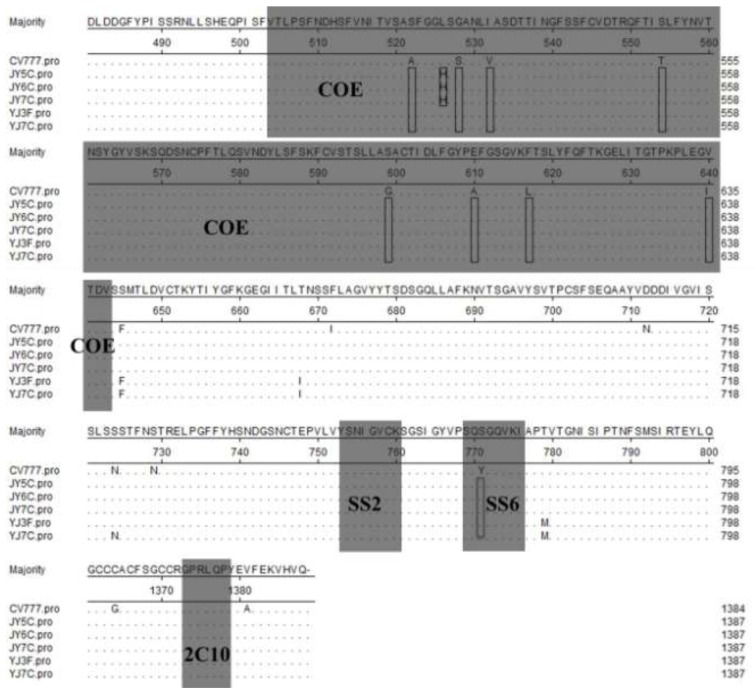
The PEDV S protein is a type I glycoprotein. Its neutralizing epitopes contain COE (499–638 aa), SS2 (748–755 aa), SS6 (764–771 aa), and 2C10 (1,368–1,374 aa) [3,4,5], and regions of the alignment sequences (Figure 5) correspond to these regions are COE (504–643 aa), SS2 (753–760 aa), SS6 (769–776 aa), and 2C10 (1,373–1,379 aa). In our study, compared with the vaccine strain CV777, eight mutations (A→S at 517, S→G at 523, V→I at 527, T→S at 549, G→S at 594, A→E at 605, L→F at 612, and I→V at 635) were found in all the five PEDV strains from our study (JY5C, JY6C, JY7C, YJ3F, and YJ7C) and one mutation (L→H at 521) was found in JY5C, JY6C, and JY7C in the neutralizing epitope COE. Compared with the vaccine strain CV777, one mutation (Y→S at 766) was also found in all the five PEDV strains from our study (JY5C, JY6C, JY7C, YJ3F, and YJ7C) in the neutralizing epitope SS6. However, compared with the vaccine strain CV777, no mutations were found in the five PEDV strains from our study (JY5C, JY6C, JY7C, YJ3F, and YJ7C) in the neutralizing epitopes SS2 and 2C10 (Figure 5). Similar to other coronavirus S proteins, the PEDV S protein is a glycoprotein peplomer (surface antigen) on the viral surface, with a pivotal function in stimulating induction of neutralizing antibodies in the natural host. Thus, the S glycoprotein is a primary target for the development of effective vaccines against PEDV. Further research is needed to determine whether the amino acid changes in the Gansu strains from our study result in any antigenicity changes.
Figure 5.
Alignment of amino acid sequences of S proteins of Gansu PEDV strains and reference strains. Ellipses represent the consensus amino acids. Boxes indicate substitution amino acids compared with CV777. Shadows indicate the neutralizing epitopes (COE, SS2, SS6, and 2C10 motif).
2.5. Discussion
RT-PCR amplification and sequencing analysis of the full-length PEDV S genes were used to investigate the isolates from diarrhea samples from local pig farms with severe diarrhea in piglets. The variant strains were detected in this study, implying an identical distribution profile for PEDV on pig farms in Gansu, China. The sequence insertions and deletions in the S gene and mutations in the antigenic regions found in variant strains possibly provided stronger pathogenicity and antigenicity to the new PEDV variants that influenced the effectiveness of the CV777-based vaccine, ultimately causing the 2012 outbreak of severe diarrhea in the porcine farms of Gansu Province, China. Future studies should investigate the biological functions of these particular insertions, deletions, and mutations.
3. Experimental
3.1. Sample Collection
A total of 17 samples (fecal and intestinal) were collected from seven dead piglets showing signs of watery diarrhea and dehydration from two farms in Gansu Province, China (Table 3). These samples were collected individually and placed in separate sterile specimen containers. Samples were homogenized with PBS. The suspensions were vortexed and centrifuged for 10 min at 10,000 × g. The supernatants were stored at −80 °C before use.
Table 3.
Current farm status in China.
| Farm | No. of sows | Vaccination a | Illness rate (%/y) | Mortality rate (%) |
|---|---|---|---|---|
| (Yongjing Tai Chi Breed Co., Ltd.) YJ | 400 | Yes | 80 | 60 |
| (Hoggery of Science and Technology Breed Park of Jiugang Hongfeng Company) JY | 2000 | Yes | 80 | 60 |
a Sows were vaccinated with divalent inactivated transmissible gastroenteritis (TGE) and porcine epidemic diarrhea (PED) vaccine before delivery.
3.2. RNA Extraction and Reverse Transcription
All samples were evaluated by reverse transcription PCR (RT-PCR) using PEDV special primers (Table 4). In brief, viral RNA was extracted from the supernatants of the homogenized samples with TRIzol LS (Invitrogen Co., Carlsbad, CA, USA) according to the manufacturer’s instructions. RT was carried out using random hexamer primers (TaKaRa Bio Inc., Otsu, Japan), and the cDNA was immediately amplified with primers, which were designed based on the sequences of PEDV reference strains (Table 4) under the following conditions: denaturation at 95 °C for 5 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 90 s. The RT-PCR products were analyzed by 1.5% agarose gel electrophoresis and visualized by ultraviolet illumination after ethidium bromide staining.
Table 4.
Amplification primers for the S gene of PEDV in Gansu, China in 2012 a.
| Primers | Nucleotide sequence, 5'→3' | Primer location b | Length (bp) c |
|---|---|---|---|
| PEDVS1-P1 | CCATTAGTGATGTTGTGTTAG | 20, 535–20, 555 | 1031 |
| PEDVS1-P2 | GCACAGCAGCTCCATT | 21, 565–21, 550 | |
| PEDVS2-P1 | CCACATACCAGAAGGTTTTAG | 21, 372–21, 392 | 1146 |
| PEDVS2-P2 | CCAGTAATCAACTCACCCTT | 22, 517–22, 498 | |
| PEDVS3-P1 | CCCTGAGTTTGGTAGTGG | 22, 446–22463 | 1154 |
| PEDVS3-P2 | CATCCGTCTGTAGAGCAAG | 23, 599–23, 581 | |
| PEDVS4-P1 | CTCATCGGTGGTATGGTGCT | 23, 497–23, 516 | 1355 |
| PEDVS4-P2 | AGCAGACTTTGAGACATCTTTGAC | 24, 851–24, 828 |
a PEDV, porcine epidemic diarrhea virus; P1, forward; P2, reverse. b Numbers correspond to the nucleotide positions within the CV777 genome. c Length of PCR products.
3.3. Sequence Analysis
Five of 17 pig samples were positive for PEDV by RT-PCR. Sequencing analysis of the full-length S gene was performed for the five samples designated as JY5C, JY6C, JY7C, YJ3F, and YJ7C. In brief, bands of the corresponding size of the gene were excised, and the synthesized DNA was purified using a QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions and sequenced by BGI Company (Peking, China). The five PEDV S gene sequences were aligned with the sequences of 32 previously published PEDV S genes (Table 5) using the DNASTAR, DNAMAN, and MegAlign version 5.0 (DNAStar Inc., Madison, WI, USA) software packages [17]. To investigate their molecular and epidemiological characteristics and to determine their profile of genetic diversity, phylogenetic trees were constructed using molecular evolutionary genetics analysis MegAlign version 5.0 [17] with the neighbor-joining (NJ) method to calculate distance. Bootstrap values were estimated for 1,000 replicates. SignalP 4.1 software [18] was used to predicte the N-glycosylated sites.
Table 5.
Isolates and reference strains used in studies of PEDV outbreak in Gansu, China.
| Virus strain | GenBank accession No. | Country and year of isolation |
|---|---|---|
| JY5C | KF177254 | China 2012 |
| JY6C | KF177255 | China 2012 |
| JY7C | KF177256 | China 2012 |
| YJ3F | KF177257 | China 2012 |
| YJ7C | KF177258 | China 2012 |
| CH1 | JQ239429 | China 2011 |
| CH2 | JQ239430 | China 2011 |
| CH3 | JQ239431 | China 2011 |
| CH4 | JQ239432 | China 2011 |
| CH5 | JQ239433 | China 2011 |
| CH6 | JQ239434 | China 2011 |
| CH7 | JQ239435 | China 2011 |
| CH8 | JQ239436 | China 2011 |
| CHGD01 | JN980698 | China 2011 |
| CH-FJND-1-2011 | JN543367.1 | China 2011 |
| CH-FJND-2-2011 | JN315706.1 | China 2011 |
| CH-FJND-3-2011 | JN381492.1 | China 2011 |
| DX | EU031893 | China 2007 |
| LZC | EF185992 | China 2006 |
| LJB-03 | DQ985739 | China 2006 |
| JS-2004-2 | AY653204 | China 2004 |
| CHS | JN547228.1 | China 1986 |
| KUN-0901 | GU180144 | South Korea 2009 |
| KUN-0902 | GU180145 | South Korea 2009 |
| KUN-0903 | GU180146 | South Korea 2009 |
| KUN-0904 | GU180147 | South Korea 2009 |
| KUN-0905 | GU180148 | South Korea 2009 |
| KUN-0801 | GU180142 | South Korea 2008 |
| KUN-0802 | GU180143 | South Korea 2008 |
| Parent DR13 | DQ862099 | South Korea 2006 |
| Attenuated DR13 | DQ462404.2 | South Korea 2006 |
| Chinju99 | AY167585 | South Korea 1999 |
| MK | AB548624.1 | Japan 1996 |
| NK | AB548623.1 | Japan |
| KH | AB548622.1 | Japan |
| CV777 | AF353511.1 | Belgium 1988 |
| Br1-87 | Z25483 | Great Britain 1993 |
| PUR46-MAD | M94101 | USA 1992 |
4. Conclusions
There were few positives (5) by RT-PCR when 17 piglets with water diarrhea and dehydration were sampled. There may be two reasons for this phenomenon. Firstly, inability to amplify virus from all piglets might impact our results. Secondly, the piglets maybe have coinfections that might skew our results.
Our study of the full-length S gene revealed a more comprehensive distribution profile that reflects the current PEDV status in pig farms in Gansu Province, China, including the presence of strains similar to those from South Korea. These data indicate that the new variant PEDV strains in Gansu Province, which were first found in 2011, may have originated from South China. Thus, certain actions must be taken to prevent the continued transmission of this virus, including the development of novel vaccines for prevention.
Acknowledgments
This work was supported by the National Key Technologies R&D Program (No. 2013BAD12B04) and the Special Fund for Agro-scientific Research in the Public Interest (No. 201303042).
Conflict of Interest
The authors declare no conflict of interest.
References and Notes
- 1.Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song D., Park B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W., Laude H, Yang M.S., Jang Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- 4.Cruz D.J., Kim C.J., Shin H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun D., Feng L., Shi H., Chen J., Cui X., Chen H., Liu S., Tong Y., Wang Y., Tong G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X., Yang J., Yu F., Ge J., Lin T., Song T. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) samples from field cases in Fujian, China. Virus Genes. 2012;36:366–364. doi: 10.1007/s11262-012-0794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z.L., Zhu L., Ma J.Y., Zhou Q.F., Song Y.H., Sun B.L., Chen R.A., Xie Q.M., Bee Y.Z. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field strains in south China. Virus Genes. 2012;45:181–185. doi: 10.1007/s11262-012-0735-8. [DOI] [PubMed] [Google Scholar]
- 9.Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- 11.Debouck P., Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980;41:219–223. [PubMed] [Google Scholar]
- 12.Park S.J., Kim H.K., Song D.S., Moon H.J., Park B.K. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field isolates in Korea. Arch. Virol. 2011;156:577–585. doi: 10.1007/s00705-010-0892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J.F., Sun D.B., Wang C.B., Shi H.Y., Cui X.C., Liu S.W., Qiu H. J., Feng L. Molecular characterization and phylogenetic analysis of membrane protein genes of porcine epidemic diarrhea virus isolates in China. Virus Genes. 2008;36:355–364. doi: 10.1007/s11262-007-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sueyoshi M., Tsuda T., Yamazaki K., Yoshida K., Nakazawa M., Sato K., Minami T., Iwashita K., Watanabe M., Suzuki Y., et al. An immunohistochemical investigation of porcine epidemic diarrhoea. J. Comp. Pathol. 1995;113:59–67. doi: 10.1016/S0021-9975(05)80069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puranaveja S., Poolperm P., Lertwatcharasarakul P., Kesdaengsakonwut S., Boonsoongnern A., Urairong K., Kitikoon P., Choojai P., Kedkovid R., Teankum K., et al. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg. Infect. Dis. 2009;15:1112–1115. doi: 10.3201/eid1507.081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Y., Zhang J., Deng X., Ye Y., Liao M., Fan H. Complete genome sequence of a highly prevalent isolate of porcine epidemic diarrhea virus in South china. J. Virol. 2012;86 doi: 10.1128/JVI.01455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 18.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]