Abstract
The pob3-Q308K mutation alters the small subunit of the Saccharomyces cerevisiae histone/nucleosome chaperone Facilitates Chromatin Transactions (FACT), causing defects in both transcription and DNA replication. We describe histone mutations that suppress some of these defects, providing new insight into the mechanism of FACT activity in vivo. FACT is primarily known for its ability to promote reorganization of nucleosomes into a more open form, but neither the pob3-Q308K mutation nor the compensating histone mutations affect this activity. Instead, purified mutant FACT complexes fail to release from nucleosomes efficiently, and the histone mutations correct this flaw. We confirm that pob3-T252E also suppresses pob3-Q308K and show that combining two suppressor mutations can be detrimental, further demonstrating the importance of balance between association and dissociation for efficient FACT:nucleosome interactions. To explain our results, we propose that histone H4 can adopt multiple conformations, most of which are incompatible with nucleosome assembly. FACT guides H4 to adopt appropriate conformations, and this activity can be enhanced or diminished by mutations in Pob3 or histones. FACT can therefore destabilize nucleosomes by favoring the reorganized state, but it can also promote assembly by tethering histones and DNA together and maintaining them in conformations that promote canonical nucleosome formation.
Keywords: histone mutations, FACT suppressor, nucleosome reorganization, histone rearrangement
Facilitates Chromatin Transactions (FACT) is a broadly conserved histone chaperone that is important for overcoming chromatin barriers during transcription and DNA replication and also for assembling and maintaining nucleosomes (Singer and Johnston 2004; Reinberg and Sims 2006; Winkler and Luger 2011; Formosa 2012). FACT is composed of a large Spt16 subunit and a small Pob3/SSRP1 subunit, which is found in two versions: SSRP1 with an HMGB-family DNA-binding domain at the C terminus (found in most eukaryotes) and Pob3, which lacks this domain (found in yeast and fungi). In organisms like Saccharomyces cerevisiae that have the Pob3 version, FACT activity is supported in vitro and in vivo by the separate HMGB-family protein Nhp6 (Singer and Johnston 2004; Stillman 2010; Formosa 2012). FACT was initially shown to be an H2A-H2B chaperone (Orphanides et al. 1999; Belotserkovskaya et al. 2003), but it also binds to H3-H4, intact nucleosomes, the N-terminal tails of some histones, and DNA (DNA binding is through the HMGB domain of SSRP1 or the separate Nhp6 protein in yeast) (Orphanides et al. 1999; Formosa et al. 2001; Stuwe et al. 2008; Winkler et al. 2011; Kemble et al. 2013). Structures have been determined for several individual domains of FACT (Allain et al. 1999; VanDemark et al. 2006; Stuwe et al. 2008; VanDemark et al. 2008; Hondele et al. 2013; Kemble et al. 2013) and each of these domains contributes to the interactions with nucleosomal components, suggesting that FACT can bind to the different components simultaneously through independent contacts. FACT also binds directly to several transcription and replication factors (Wittmeyer et al. 1999; Squazzo et al. 2002; Simic et al. 2003; Takahata et al. 2009a; Formosa 2012). FACT is therefore capable of interacting simultaneously with each of the separate components of nucleosomes, with intact nucleosomes, and with many factors that act on chromatin. FACT can coordinate the actions of chromatin factors, it can destabilize nucleosomes, and it can tether the components of disrupted nucleosomes together to promote their reassembly.
Purified FACT is able to destabilize nucleosomes in vitro, causing increased accessibility of the DNA to binding factors and endonucleases (Biswas et al. 2005; Xin et al. 2009). FACT binding also leads to enhanced displacement of H2A-H2B dimers from nucleosomes (Belotserkovskaya et al. 2003), but the increased accessibility of the DNA can occur without histone loss (Xin et al. 2009). FACT therefore appears to induce or stabilize an alternative, reorganized nucleosome structure in which the normal components of a nucleosome remain associated with one another, but in a way that is less compact or stable than in the canonical form (Formosa 2012). Destabilization of nucleosomes is presumably the mechanism through which FACT decreases the barrier to RNA polymerase progression posed by chromatin (Orphanides et al. 1998; Hsieh et al. 2013), enhances binding of TATA Binding Protein to sites within nucleosomal DNA (Biswas et al. 2005), and promotes displacement of nucleosomes from the promoters of inducible genes prior to the initiation of transcription (Schwabish and Struhl 2004; Ransom et al. 2009; Takahata et al. 2009b; Xin et al. 2009). However, FACT is also important for preventing dispersal of existing nucleosomes during transcription and for promoting rapid reestablishment of chromatin-based repression (Jamai et al. 2009; Hainer et al. 2012). FACT therefore promotes nucleosome disassembly in some contexts and chromatin stability in others. These opposing outcomes can be explained by a single activity, in which FACT drives both directions of a reversible transition between canonical nucleosomes and less stably associated reorganized forms (Formosa 2012).
The spt16-11 mutation was previously used to identify histone mutations that could relieve the effects of diminished FACT activity (McCullough et al. 2011). Purified Spt16-11 protein was unable to promote normal levels of nucleosome reorganization in vitro, and the histone mutations that suppressed some of the phenotypes caused by spt16-11 in vivo destabilized the (H2A-H2B):(H3-H4) interface, thereby favoring nucleosome reorganization (McCullough et al. 2011). These results strongly supported a physiological role for destabilization of nucleosomes by FACT.
The middle domains of Spt16 and Pob3 adopt similar double PH (dPH) structural motifs (VanDemark et al. 2006; Kemble et al. 2013), an architecture also found in the H3-H4 chaperone Rtt106 (Liu et al. 2010; Zunder et al. 2012). While this suggests common mechanisms for binding H3-H4, the details of the structures indicate that each chaperone has distinct properties (Hondele et al. 2013; Kemble et al. 2013). Two commonly studied alleles of FACT, spt16-11 and pob3-Q308K, map to the dPH domains of each protein (VanDemark et al. 2006; Hondele et al. 2013; Kemble et al. 2013), initially suggesting that they disrupt FACT activity in similar ways. Surprisingly, while some H2A-H2B mutations that suppressed spt16-11 were mildly beneficial in pob3-Q308K strains, most were neutral and some were strongly detrimental (McCullough et al. 2011). These two FACT mutations therefore affect structurally similar domains of the same complex, but cause very different defects in FACT activity.
Here, we describe histone mutations that suppress some of the defects caused by pob3-Q308K. Neither Pob3-Q308K protein nor its suppressors significantly affected the reorganization of nucleosomes. Instead, Pob3-Q308K caused inefficient dissociation of FACT from nucleosomes, and suppressing mutations removed this barrier. Our results suggest that the pathway leading to dissociation of FACT from nucleosomes includes multiple steps. We propose that while some features of FACT promote nucleosome destabilization by inducing or stabilizing the reorganized state, other domains configure the histones to allow them to fit together in canonical nucleosomes, a process that might include many intermediates. The results reveal the importance of balance between the destabilization and assembly activities of FACT, providing new insight into the complexity of nucleosome assembly and the multiple roles chaperones play in this process.
Materials and Methods
Yeast strains are listed in Table 1. Viability comparisons involved preparing 10-fold serial dilutions of saturated cultures, then testing 5–10 µl as indicated in each figure. The screen for suppressor mutations was performed as described previously (McCullough et al. 2011) and outlined in Results. To integrate mutations into the genome, a selectable marker integrated near the normal target histone gene was amplified using a primer with the desired mutation and the product used to transform a normal strain (see Toulmay and Schneiter 2006; McCullough et al. 2011). The markers were then removed by integrating the URA3 gene downstream of each marker (see Figure 3 below and Storici et al. 2001) and selecting/screening for coincident loss of URA3 (Boeke et al. 1987) and the marker after transforming with normal DNA flanking the site of marker integration. Candidates were sequenced to insure expression of mutant proteins from an otherwise fully native context.
Table 1. Strains used in this study.
| Name | Bkgd | Genotype |
|---|---|---|
| Figure 1 | ||
| 8264-17-3 pTF237 | W303 | MATa ade2 can1 his3 leu2 trp1 ura3 pob3-Q308K hht1-hhf1-Δ(::HIS3) hht2-hhf2-Δ(::KanMX3) hta1-htb1-Δ(::NatMX) hta2-htb2-Δ(::HphMX) pTF237 (YCp URA3 HHT2-HHF2, HTA1-HTB1) |
| DY10003 pTF237 | W303 | MATa ade2 can1 his3 leu2 met15 trp1 ura3 spt16-11 hht1-hhf1-Δ(::HIS3) hht2-hhf2-Δ(::KanMX) hta1-htb1-Δ(::NatMX) hta2-htb2-Δ(::HphMX) pTF237 (YCp URA3 HHT2-HHF2, HTA1-HTB1) |
| DY9999 pTF237 | W303 | MATa ade2 can1 his3 leu2 trp1 ura3 hht1-hhf1-Δ(::HIS3) hht2-hhf2-Δ(::KanMX) hta1-htb1-Δ(::NatMX) hta2-htb2-Δ(::HphMX) pTF237 (YCp URA3 HHT2-HHF2, HTA1-HTB1) |
| Figure 3 | ||
| 8127-7-4 | A364a | MATa ura3-Δ0 leu2-Δ0 trp1-Δ2 his3 lys2-128∂ |
| 9408-5-2 | A364a | MATa ura3 leu2 trp1 his3 lys2-128∂ hhf1-R23S hhf2-R23S |
| 9347-6-3 | A364a | MATa ura3 leu2 trp1 his3 lys2-128∂ hhf1-N25D hhf2-N25D |
| 9273-N | A364a | MATa ura3-Δ0 leu2-Δ0 trp1-Δ2 his3 lys2-128∂ pob3-Q308K(+34, NatMX) |
| 9377-1-4 | A364a | MATa ura3 leu2 trp1 his3 lys2-128∂ pob3-Q308K(+34, NatMX) hhf1-R23S hhf2-R23S |
| 9347-4-1 | A364a | MATa ura3 leu2 trp1 his3 lys2-128∂ pob3-Q308K(+34, NatMX) hhf1-N25D hhf2-N25D |
| Figure 7 | ||
| 9202 | A364a | MATa/MATα ura3-Δ0/ura3-Δ0 leu2-Δ0/leu2-Δ0 trp1-Δ2/trp1-Δ2 his3/+ +/his7 lys2-128∂/lys2-128∂ POB3(+34, LEU2)/POB3 |
| 8700 | A364a | MATa/MATα ura3-Δ0/ura3-Δ0 leu2-Δ0/leu2-Δ0 trp1-Δ2/trp1-Δ2 his3/+ +/his7 lys2-128∂/lys2-128∂ pob3-Q308K(+34, LEU2)/POB3 |
| 9191 | A364a | MATa/MATα ura3-Δ0/ura3 leu2-Δ0/leu2 trp1-Δ2/trp1 his3/+ +/his7 lys2-128∂/lys2-128∂ pob3-Δ(::LEU2)/POB3 |
| 9193 | A364a | MATa/MATα ura3-Δ0/ura3-Δ0 leu2-Δ0/leu2-Δ0 trp1-Δ2/trp1-Δ2 his3/+ +/his7 lys2-128∂/lys2-128∂ pob3-Q308K(+34, LEU2)/pob3-Q308K |
| 9192 | A364a | ura3-Δ0/ura3 leu2-Δ0/leu2 trp1-Δ2/trp1 his3/+ +/his7 lys2-128∂/lys2-128∂ pob3-Δ(::LEU2)/pob3-Q308K |
| 8668 | A364a | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/+ +/his7 lys2-128∂/lys2-128∂ hpc2-Δ(::KanMX)/hpc2-Δ(::TRP1) |
| 8679 | A364a | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/+ +/his7 lys2-128∂/lys2-128∂ hpc2-Δ(::KanMX)/hpc2-Δ(::TRP1) pob3-Q308K(+34, LEU2)/POB3 |
| 8703 | A364a | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/+ +/his7 lys2-128∂/lys2-128∂ pob3-Δ(::TRP1)/+ hpc2-Δ(::KanMX)/hpc2-Δ(::TRP1) |
| Figure 8 | ||
| 9202-1-1 | A364a | MATa ura3-Δ0 leu2-Δ0 trp1-Δ2 his3 lys2-128∂ POB3(+34, LEU2) |
| 8324-2-2 | A364a | MATa ura3-Δ0 leu2-Δ0 trp1-Δ2 his3 lys2-128∂ pob3-Q308K(+34, LEU2) |
| 9327-7-3 | A364a | MATa ura3-Δ0 leu2-Δ0 trp1-Δ2 his3 lys2-128∂ pob3-T252E(+34, LEU2) |
| 9301-QK | A364a | MATa ura3-Δ0 leu2-Δ0 trp1-Δ2 his3 lys2-128∂ pob3-T252E, Q308K(+34, LEU2) |
| 9408-5-2 | A364a | MATa ura3 leu2 trp1 his3 lys2-128∂ hhf1-R23S hhf2-R23S |
| 9377-1-4 | A364a | MATa ura3 leu2 trp1 his3 lys2-128∂ pob3-Q308K(+34, NatMX) hhf1-R23S hhf2-R23S |
| 9411-3-1 | A364a | MATa ura3 leu2 trp1 his3 lys2-128∂ hhf1-R23S hhf2-R23S pob3-T252E(+34, LEU2) |
| 9413-2-1 | A364a | MATa ura3 leu2 trp1 his3 lys2-128∂ hhf1-R23S hhf2-R23S pob3-T252E, Q308K(+34, LEU2) |
| 9418-6-1 | A364a | MATα ura3 leu2 trp1 his3 lys2-128∂ POB3(+34, LEU2) hhf1-N25D hhf2-N25D |
| 9347-3-2 | A364a | MATα ura3 leu2 trp1 his3 lys2-128∂ pob3-Q308K(+34, NatMX) hhf1-N25D hhf2-N25D |
| 9412-5-2 | A364a | MATα ura3 leu2 trp1 his3 lys2-128∂ hhf1-N25D hhf2-N25D pob3-T252E(+34, LEU2) |
| 9417-1-1 | A364a | MATα ura3 leu2 trp1 his3 lys2-128∂ hhf1-N25D hhf2-N25D pob3-T252E, Q308K(+34, LEU2) |
Strains were constructed using standard procedures and are isogenic with either the W303 or the A364a genetic backgrounds (bkgd), as indicated. Strains with markers integrated near an ORF are listed as (+number, marker) with +number indicating the number of base pairs downstream of the ORF that sequence associated with the marker starts, and marker indicating the marker used. Strains are listed by the order of appearance in the figure noted.
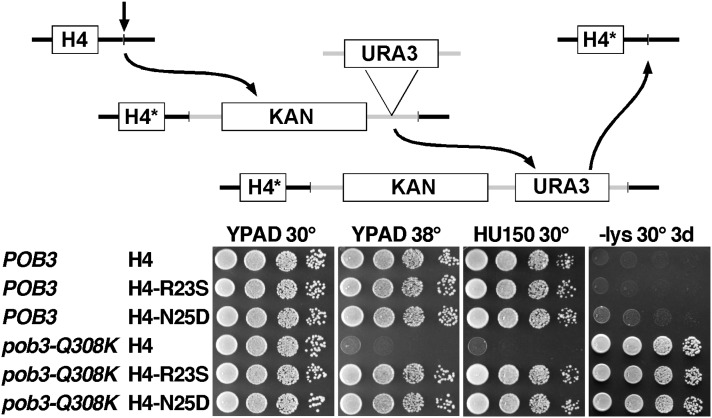
Figure 3.
Effect of mutations after integration into the genome. The H4-R23S and H4-N25D mutations were integrated at both HHF1 and HHF2, and then the markers were deleted using the scheme outlined in the top panel (with KAN as the KanMX marker and H4* denoting the desired allele of H4). Dilutions of strains lacking the markers (Table 1) were tested as in Figure 1 with −Lys indicating synthetic medium lacking lysine to reveal the Spt− phenotype.
Soluble and total histone concentrations were determined using a method based on that described by Feser et al. (2010). A total of 2 × 108 cells growing in rich medium were harvested at an absorbance of 0.8 at 600 nm by centrifugation at 1000 × g for 3 min. Cells were washed twice with harvest buffer (100 mM Tris, pH 9.4, 10 mM dithiothreitol) at 0° and then resuspended in harvest buffer and allowed to incubate at 0° for 15 min. The cells were collected again, washed once with spheroplasting buffer (20 mM HEPES, pH 7.4, 1.2 M sorbitol, and protease inhibitors; Roche, EDTA-free) at 0°, suspended in 2 ml of spheroplasting buffer, and treated with Zymolyase 100T (ICN) at a final concentration of 42 µg/ml at 30° for 45 min with gentle inversion every 15 min. A total of 80–90% spheroplasting was confirmed by examining hypotonic lysis microscopically, and then the spheroplasts were centrifuged at 150 × g for 2 min and washed twice with spheroplasting wash buffer (20 mM Tris-HCl, pH 7.4, 20 mM KCl, 1 M sorbitol, 100 mM spermine, 250 mM spermidine, protease inhibitors). Spheroplasts were lysed after the second wash by suspending the pellet with 1.5 ml of lysis buffer (20 mM Tris-HCl, pH 7.4, 20 mM KCl, 0.4 M sorbitol, 100 mM spermine, 250 mM spermidine, 1% Triton X-100, and protease inhibitors). A 75-µl aliquot was used to determine total protein by the Bradford assay, and 125 µl of 5× SDS sample buffer was added to a 500-µl aliquot to produce the total protein sample. Another 500-µl aliquot was centrifuged at 15,000 × g for 15 min at 4° to remove chromatin, and the supernatant containing soluble proteins was processed for SDS–PAGE as above. A total of 30 µg of protein from the total and soluble protein fractions was resolved by SDS–PAGE using a 4–20% gradient gel, proteins were blotted to nitrocellulose, and then histones and Pgk1 (as an internal loading control) were detected and quantitated using appropriate primary (Pgk1 and H2B from Active Motif and H3 from GenScript) and secondary (Li-Cor) antibodies and a Li-Cor Odyssey infrared scanner. Purified histone standards were included on the gels to determine absolute as well as relative histone levels and to establish linearity of the assay over the detection range.
Nucleosomes were prepared by the salt dialysis method (Luger et al. 1999) using yeast histones expressed in bacteria and a 181-bp DNA fragment derived from the sea urchin 5S rDNA repeat as described previously (Xin et al. 2009). The mutations corresponding to H4-R23S and H4-N25D were introduced using the QuikChange strategy (Stratagene). Oregon Green or tetramethyl rhodamine dyes (Molecular Probes) were attached to unique cysteine residues introduced into the yeast histones (H2B-T119C or H3-D77C), and DNA was labeled by incorporation of Cy5 into one of the PCR primers. Spt16-Pob3 or Pob3-Q308K complexes and Nhp6 were purified as previously described (Xin et al. 2009). Complexes were formed by mixing Spt16-Pob3, Nhp6, and nucleosomes (Xin et al. 2009) and then incubating for 10 min at 30°. A 4% native polyacrylamide gel electrophoresis was performed as previously described (Xin et al. 2009). Complexes were disrupted by adding sheared, unlabeled salmon testes DNA (Sigma) to a final concentration of 80–170 ng/µl. Fluorescent components were detected with a Typhoon scanner (GE).
To test the stability of nucleosomes, aliquots were incubated for 30 min at 30° after addition of increasing amounts of NaCl, and then the fraction of signal migrating in the position of nucleosomes after native PAGE was determined. Nucleosomes were also heated to 65° for 1 hr either alone or with 3.5 µg/µl of unlabeled, sonicated salmon testes DNA (Sigma). Nucleosome reorganization was measured by determining the initial rate of DraI digestion, as described previously (Xin et al. 2009). The apparent affinity of FACT for nucleosomes was determined by titration and EMSA (Ruone et al. 2003; Rhoades et al. 2004).
Results
pob3-Q308K defects can be suppressed by mutating H3 or H4
The pob3-Q308K mutation causes the Spt− phenotype, temperature sensitivity (Ts−), and sensitivity to the replication toxin hydroxyurea (HU) (Formosa et al. 2001; VanDemark et al. 2006). The Spt− phenotype here results from inappropriate initiation of transcription within the lys2-128∂ allele (Simchen et al. 1984). Pob3-Q308K protein is stable at 37° (VanDemark et al. 2008), so the temperature sensitivity must result from unmet demand for FACT activity at elevated temperatures. HU inhibits ribonucleotide reductase (RNR), presumably causing a shortage of dNTPs and thereby stressing replication progression (Aparicio 2013; Yekezare et al. 2013). Strains with the pob3-Q308K mutation are able to induce transcription of RNR genes normally (Biswas et al. 2008), so the HU sensitivity (HUs) caused by this allele is interpreted as a defect in replication fork progression or checkpoint signaling.
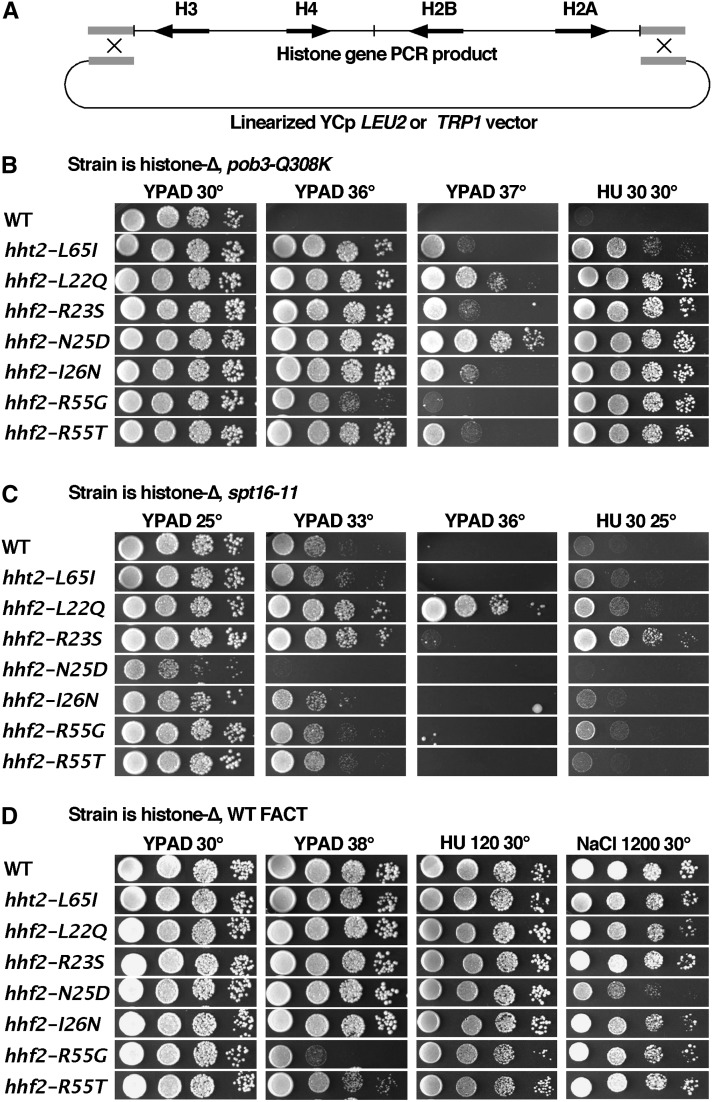
The nucleosome content of a cell must be doubled along with the DNA, so we focused on potential roles for FACT in nucleosome assembly by screening for histone gene mutations that suppress the HUs caused by pob3-Q308K. To perform the screen, a strain lacking all eight genomic histone genes and carrying the pob3-Q308K mutation was constructed (8264-17-3 pTF237; Table 1, based on strains kindly provided by M. Smith and D. Stillman). Viability of the strain was maintained by the low copy URA3 plasmid carrying HTA1-HTB1 and HHT2-HHF2 (Ahn et al. 2005). This strain was tranformed with linearized pRS414 or pRS415 (YCp TRP1 or LEU2; Sikorski and Hieter 1989) along with a PCR product with homology to the ends of the vector and containing all four histone genes (Figure 1A). Recombination between the vector and the PCR product formed a plasmid with mutagenized histone genes in vivo. After selecting for loss of the original wild-type (WT) histone plasmid by growth on medium containing 5-FOA (Boeke et al. 1987), clones carrying suppressor mutations were identified through their ability to grow in the presence of a high concentration of HU (Figure 1B). Candidate plasmids were recovered and used to transform a fresh parental strain to insure linkage of the suppression phenotype with the plasmids. Sixteen candidates that passed this test were sequenced, with 7 of these yielding single mutations in the four histone genes. The remaining 9 plasmids had multiple mutations, but this included at least one of the unique mutations in all cases so these were not studied further. The H4-N25D and H4-R55G mutations were each isolated five times in this screen, H4-I26N was isolated twice, and the other four alleles shown in Figure 1 were each isolated once, indicating that the screen was not saturated. Only one mutation affected H3 (hht2-L65I), and it produced the weakest suppression of the HUs (Figure 1B). The primary target for suppression of pob3-Q308K is therefore H4. All of the suppressors also partially suppressed the Ts− phenotype associated with pob3-Q308K, although to different extents (Figure 1B). The HUs and Ts− phenotypes are therefore partially separable, indicating that they arise from overlapping but distinct causes.
Figure 1.
Suppression of pob3-Q308K by H3-H4 mutations. 8264-17-3 pTF237 (Table 1) was transformed with linear vector and a histone gene fragment derived from pQQ18 (Ahn et al. 2005) by PCR using Pfu polymerase under standard conditions. (B) After recovery of candidate plasmids with only the mutations shown, retransformation of 8264-17-3 pTF237, and loss of pTF237, viability was compared under the conditions indicated (see Materials and Methods). (C and D) The same plasmids were tested as the sole source of histones in strains with the spt16-11 mutation (DY10004) or normal FACT (DY9999). YPAD is rich medium; HU and NaCl indicate the concentrations of hydroxyurea or NaCl added to YPAD (mM).
Suppressors of pob3-Q308K were less beneficial in an spt16-11 strain (Figure 1C). Effects ranged from mild enhancement of growth defects to moderate suppression, but no strong suppression was observed. Similar tests in a strain with the pob3-L78R mutation (Schlesinger and Formosa 2000) also showed some neutral and some detrimental combinations, but no suppression (not shown). Different FACT mutations therefore reveal multiple functions of FACT, as each allele responded differently to these and other histone mutations (VanDemark et al. 2008; McCullough et al. 2011).
The suppressor mutations caused moderate or no phenotypes in a strain with normal FACT (Figure 1D). H4-R55G/T caused growth defects at 38°, and H4-N25D caused some sensitivity to high levels of NaCl, but in general the suppressor mutations were tolerated well. The strong benefits conferred to strains with the pob3-Q308K mutation therefore appear to reveal a mechanism of suppression that mainly affects the specific defect in FACT activity caused by this allele.
Suppressors of pob3-Q308K cluster near the H4 N-terminal tail
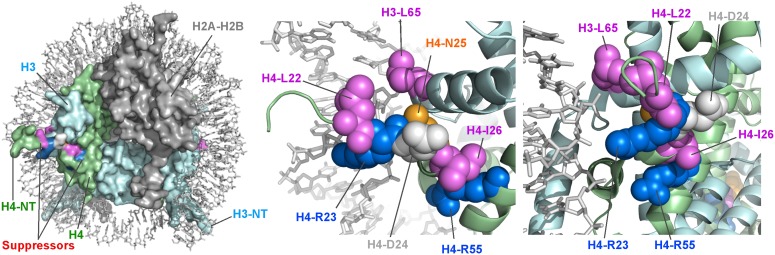
The residues altered in the pob3-Q308K suppressors are near one another in the nucleosome structure, clustering at the junction between the unstructured basic tail of H4 and the structured globular domain (Figure 2). Four of the five residues in the sequence from 22 to 26 (H4-L22Q, -R23S, -N25D, and -I26N) were identified, and the remaining two sites identified (H3-L65I and H4-R55G/T) are near to one of these residues in the nucleosome structure. The suppressors therefore reveal a small region of the nucleosome that must play an important functional role in a FACT activity disturbed by the pob3-Q308K mutation.
Figure 2.
Locations of the residues involved in pob3-Q308K suppression in a nucleosome. Residues that gave rise to suppression when mutated are mapped on the yeast nucleosome structure (Protein Data Bank, I1D3, White et al. 2001, images rendered in Pymol). (Left) Intact nucleosome with the histones shown as surfaces and the DNA as sticks. The suppressor mutations cluster in the region where the N-terminal tail (NT) of H4 (green) meets the globular core of the nucleosome. The first 17 residues of the H4 NT are unstructured. (Middle) A closer view in the same orientation as the left panel, with histones shown as cartoons with the mutated residues as spheres. Basic residues are rendered in blue (H4-R23 and H4-R55), hydrophobic in magenta (H3-L65, H4-L22, and H4-I26), and polar in orange (H4-N25). H4-D24 (gray) was not identified in the screen and mutation to alanine did not produce suppression (not shown). (Right) The same as the middle panel, except rotated ∼90° about the vertical (dyad) axis.
The region defined by this analysis partially overlaps the binding site for the BAH domain of Sir3 (Armache et al. 2011), an interaction that is important for maintaining fertility of yeast cells through transcriptional repression of the silent mating type information cassette HML (Johnson et al. 1990). The H4-N25D mutation caused low mating efficiency in MATa strains (not shown) but no similar defect was noted for the other suppressor mutations. Suppression of pob3-Q308K therefore does not appear to be directly related to fertility or Sir3 binding, but rather to a region of nucleosomes important for FACT activity that coincidentally overlaps a site important for Sir3 function.
Integration of suppressor mutations into the genome
To insure that suppression is not affected by the known interactions between FACT mutations and histone gene copy number (Formosa et al. 2002), we integrated two of the suppressor mutations into the genome to analyze their effects under more native expression conditions. We chose hhf2-R23S (which suppressed spt16-11 moderately) and hhf2-N25D (which caused synthetic defects with spt16-11) as the different effects with the other subunit of FACT suggests these mutations are likely to reveal any mechanistic differences among the suppressors. We integrated the mutations at both sources of H4 (HHF1 and HHF2) using the procedure outlined in Figure 3 (also see McCullough et al. 2011), a method that leaves selectable markers integrated downstream of the altered genes. Subsequent analysis showed that the presence of marker genes alone caused noticeable effects, especially in strains with FACT mutations. We therefore removed the markers, leaving the desired mutations expressed from otherwise native genomic contexts (see Materials and Methods).
After integration, the H4-R23S and H4-N25D mutations strongly suppressed the Ts− and HUs caused by pob3-Q308K, they had the same opposing effects on spt16-11 noted above, and they caused no significant phenotypes on their own (Figure 3 and data not shown). Genomic expression of these mutations as the sole source of H4 therefore largely recapitulated the results obtained with a single plasmid-borne HHF1-HHF2 locus. The Spt− phenotype (inappropriate growth on −Lys plates in these lys2-128∂ strains) can be caused by altered histone gene expression, so we tested it only after integration of the H4 mutations. Unlike the results with the Ts− and HUs phenotypes, these histone mutations had little effect on the Spt− phenotype caused by pob3-Q308K (Figure 3). Two interpretations of this observation are considered. (1) FACT has at least two independent activities that are impacted by the pob3-Q308K mutation, with one defect leading to the Spt− phenotype and the other causing Ts− and HUs. The histone mutations correct only the latter defect. (2) FACT has a single activity that is impaired in pob3-Q308K strains but different cellular functions require different levels of that activity. The histone mutations only partially restore FACT function, and this is sufficient to reverse the Ts− and HUs phenotypes, but not to reverse the Spt− phenotype. In one potential scenario, the Ts− and HU phenotypes could reveal a need for FACT activity at ∼400 replication forks during DNA replication (Yekezare et al. 2013), which is an essential cell cycle event but one that occurs only periodically. In contrast, the Spt− phenotype might reveal the need to maintain a repressive chromatin state simultaneously in all regions of the genome during all cell cycle phases, and might therefore have more stringent requirements for FACT function. We favor the latter interpretation and note that suppression of the Ts− and HUs but not the Spt− phenotype is typical for all extragenic suppressors of FACT mutations that we have isolated (McCullough et al. 2011 and our unpublished results).
Effects of mutations on histone levels
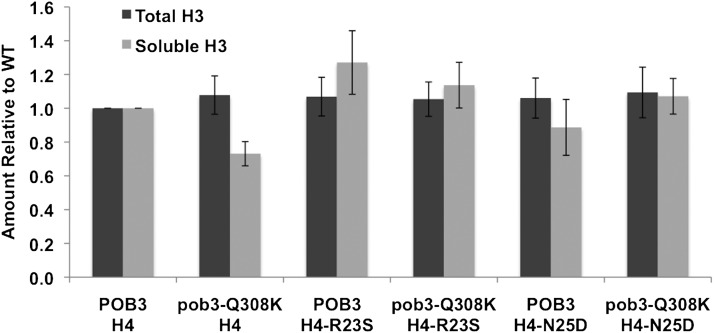
FACT mutants are sensitive to the levels of histone expression (Formosa et al. 2002), and they also affect histone stability. Normal FACT reduces the turnover of chromatin caused by transcription (Jamai et al. 2009; Hainer et al. 2012), and it acts as a chaperone for any excess histones synthesized during S phase that are destined for degradation (Morillo-Huesca et al. 2010). FACT therefore minimizes the level of free histones in a cell by reducing the displacement of histones from existing chromatin and by promoting the degradation of excess histones. We therefore considered the possibility that a FACT defect could be suppressed by a histone mutation that reduces the size of the free histone pool, reducing the need for an activity of FACT. We adapted a cell lysis and centrifugation assay to separate histones associated with chromatin from soluble histones likely to be associated with chaperones but not with chromatin (Feser et al. 2010) and then used quantitative Western blots to determine the amount of H3 in each fraction (see Materials and Methods). The results showed a slight decrease in the amount of soluble H3 in a pob3-Q308K strain, but no other significant changes that would explain the suppression (Figure 4). We conclude that suppression of pob3-Q308K is not related to changes in the total amounts or the soluble pools of histones.
Figure 4.
Effect of mutations on total and soluble histone H3 levels. Histone levels were measured by Western blotting (Materials and Methods) using whole cell extracts (total) or after removing chromatin by centrifugation to measure the free pool of histones (soluble). Signals were normalized in each case to the level observed with strains carrying wild-type POB3 and H4; the absolute level of soluble histone was ∼0.6% of the total level. Error bars indicate the standard deviation from three independent measurements.
Suppressing mutations do not affect nucleosome stability or reorganization in vitro
Histone mutations that suppressed spt16-11 also promoted formation of the reorganized nucleosome state in vitro, reducing the need for FACT to be fully active (McCullough et al. 2011). To test for similar effects with the pob3-Q308K suppressors, we purified FACT with the Pob3-Q308K mutant protein and constructed nucleosomes with the H4-R23S and H4-N25D mutations.
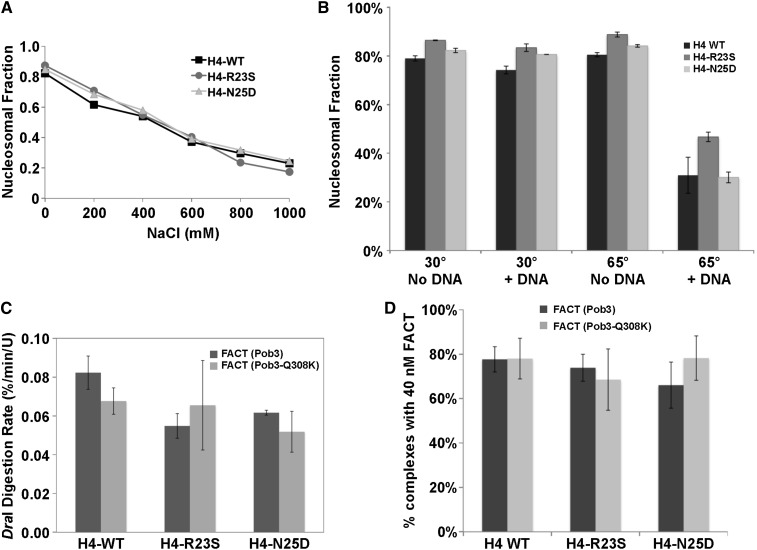
We first measured nucleosome integrity after a challenge with NaCl (Figure 5A). The dissociation profiles were essentially identical for WT and mutant, indicating similar overall stability. As a second test, we heated nucleosomes to 65° for 1 hr, roughly the maximal tolerated temperature for these nucleosomes. Again, the mutants displayed normal stability (Figure 5B). As a variation on this assay, we have noted that performing the 65° incubation in the presence of a high concentration of sheared genomic DNA significantly increases the level of H2A-H2B dimer displacement. Presumably, incubation at 65° causes reversible breathing of the nucleosome, creating the possibility of capture by DNA. Dimers are not evicted at this temperature if excess DNA is not present, so an intermediate must exist at 65° in which dimers are tenuously held and can be captured by a competitor but are not yet fully free. This provides a stringent way to challenge the ability to retain H2A-H2B dimers in nucleosomes. As shown in Figure 5B, the suppressing mutations did not reduce H2A-H2B dimer retention, in fact H4-R23S may have slightly increased it. The suppressing H4 mutations therefore form nucleosomes in vitro with normal ability to resist disruption by increased salt concentrations or elevated temperatures.
Figure 5.
Mutant histones form nucleosomes in vitro with normal stability and normal reorganization. (A) Nucleosomes with the H4 sequence indicated were incubated for 30 min at different final concentrations of NaCl and then subjected to native PAGE. The fraction of the total DNA migrating in the nucleosomal position was calculated. (B) Nucleosomes were incubated at 30° or 65° for 1 hr either in the absence or presence of sheared genomic DNA (no DNA or +DNA) and then tested for integrity as in A. (C) The initial rate of DraI digestion was measured using normal and mutant nucleosomes with normal and mutant FACT. (D) The fraction of nucleosomes incorporated into complexes with FACT was determined by native PAGE after incubating with 40 nM Spt16-Pob3 with 3 µM Nhp6. Error bars indicate the standard deviation among samples tested in triplicate.
Nucleosome reorganization is observed in vitro as an increased rate of digestion by the restriction endonuclease DraI at its recognition site in the 5S rDNA sequence, which is normally protected by histone binding (Xin et al. 2009). Purified FACT containing Spt16-11 protein had a significant defect in this activity (McCullough et al. 2011), but FACT with the Pob3-Q308K mutation was comparable to WT (Figure 5C). Nucleosomes with the H4-R23S or H4-N25D mutations were slightly less prone to reorganization than normal, and the rates were similar using WT FACT or FACT with Pob3-Q308K. The pob3-Q308K mutation therefore does not appear to cause a defect in promoting nucleosome reorganization, and its suppression does not involve altering nucleosomes to favor the reorganized state.
These endonuclease digestion assays were performed with saturating levels of FACT, leaving open the possibility that the Pob3-Q308K protein has decreased affinity for nucleosomes but high concentrations compensate for the defect. To test this, we measured the apparent affinity of FACT for nucleosomes using an EMSA binding test with variable concentrations of FACT (Ruone et al. 2003). FACT with Pob3 or Pob3-Q308K displayed half-maximal binding in the expected 10–20 nM range with normal and mutant nucleosomes (not shown) and ∼75% complex formation at 40 nM FACT (Figure 5D). Therefore, neither Pob3-Q308K nor the H4 mutants appear to affect the bulk affinity of FACT for nucleosomes.
Suppression involves the release of FACT from nucleosomes
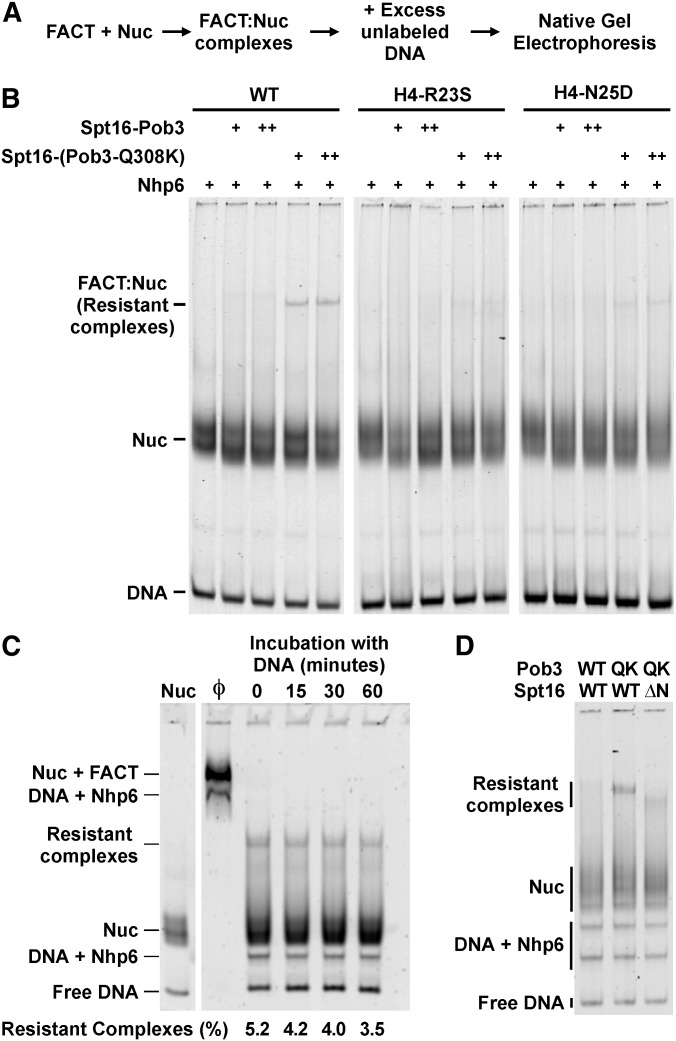
FACT forms a 1:1 complex with nucleosomes only in the presence of a 10-fold molar excess of the DNA-binding protein Nhp6, so the complexes can be disrupted rapidly by the addition of sufficient amounts of DNA to bind the Nhp6 (Xin et al. 2009). As shown in Figure 6B, FACT–nucleosome complexes were quantitatively disrupted by the addition of excess DNA, but ∼5% of the complexes formed using FACT with Pob3-Q308K remained stable after this treatment. Importantly, complexes resistant to disruption were absent or significantly less abundant when nucleosomes with the H4-R23S or H4-N25D proteins were used. This correlates with the suppression of phenotypes observed in vivo and suggests that the formation of unusually stable FACT–nucleosome complexes in vitro is related to the physiological defects caused by pob3-Q308K and the mechanism of suppression by the histone mutations.
Figure 6.
Pob3-Q308K releases from normal nucleosomes inefficiently, and this is reversed by the suppressor histone mutations. (A) The outline for the experiments shown in B–D. (B) Samples of normal and mutant nucleosomes were mixed with 3 µM Nhp6 and 50 nM (+) or 200 nM (++) Spt16-Pob3 or Spt16-Pob3-Q308K, incubated for 10 min at 30°, challenged with competitor DNA, and then subjected to native PAGE. The migration of FACT–nucleosome complexes (FACT:Nuc), nucleosomes (Nuc), and free DNA were detected by scanning for Cy5 (DNA). (C) WT nucleosomes were treated as in B using Spt16-Pob3-Q308K and then separated by native PAGE without (Ø) or after incubating at 30° with competitor DNA for the time shown. The DNA signal in the resistant complexes as a fraction of the total signal in the lane is given. Sequestration of the Nhp6 alters migration of the complexes, so the resistant complexes do not comigrate with the Nuc + FACT complexes. The “DNA + Nhp6” complexes in lane 2 (Ø) contain saturating levels of Nhp6, whereas those in lanes 3–6 (and in D) contain one or two molecules of Nhp6 due to incomplete sequestration of the Nhp6 (Ruone et al. 2003; Rhoades et al. 2004). (D) As in B, except Spt16-Pob3 contained normal subunits (WT), Pob-Q308K (QK), or Spt16 lacking the first 468 amino acids (ΔN; Vandemark et al. 2008).
To further analyze the dissociation of FACT from nucleosomes, we attempted to measure the rate of disruption of the complexes. About 95% of the FACT–nucleosome complexes were disrupted by addition of excess unlabeled DNA as quickly as the samples could be tested, but the resistant fraction seen with Pob3-Q308K remained intact for at least 1 hr at 30° (Figure 6C). This suggests that the resistant complexes represent a subset of the population that cannot dissociate. One explanation for this result is that Pob3-Q308K binds too tightly to a conformation of the histones that is a necessary intermediate along the pathway to forming a nucleosome, and FACT cannot dissociate until this intermediate is resolved (see Discussion).
Another explanation for the presence of complexes after sequestering Nhp6 is that the mutant Pob3-Q308K protein allows FACT to bind to nucleosomes at a slow rate without Nhp6, leading to a steady state in which a low level of complexes is always present. To test this, we attempted to form complexes in the absence of Nhp6 or by adding the unlabeled genomic DNA to the Nhp6 and nucleosomes before adding Spt16-Pob3 or Spt16-Pob3-Q308K, but no complexes were observed under these conditions (not shown). The defect caused by Pob3-Q308K therefore occurs during dissociation of the complexes, not during their formation. The persistent complexes contain DNA, H2A-H2B, and H3-H4 as demonstrated by using nucleosomes with each of these components labeled (Figure 6 shows the DNA, the histone labels are not shown). They also contain Spt16-Pob3 as demonstrated by Western blotting of the native gels (not shown) and by observing the expected increase in migration of the complexes after deleting the N-terminal domain of Spt16 (Figure 6D). This deletion significantly alters the mass of the Spt16-Pob3 dimer but does not affect the affinity of FACT for nucleosomes (VanDemark et al. 2008), so this confirms the presence of Spt16-Pob3 in the resistant complexes.
These results support a model in which Pob3-Q308K can block the release of FACT from a subset of complexes with nucleosomes. This model makes several predictions, which we test below.
The pob3-Q308K mutation has partially dominant effects
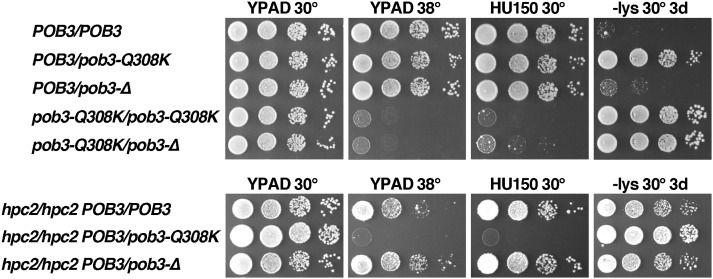
Persistent binding to nucleosomes is a gain of function for FACT, suggesting that pob3-Q308K should be genetically dominant. As shown in Figure 7, a POB3/pob3-Q308K diploid displays an Spt− phenotype almost as strong as a pob3-Q308K/pob3-Q308K strain. This is not due to haploinsufficiency, as a strain with POB3 over a deletion of the gene (pob3-Δ) did not display this phenotype. Notably, the POB3/pob3-Q308K strain was Ts+ and HU resistant, so the mutation is recessive for these phenotypes. This again supports the interpretation that optimal FACT function is more important for preventing inappropriate transcription globally than for supporting the response to elevated temperatures or a replication toxin.
Figure 7.
The pob3-Q308K effects are partially dominant, consistent with a gain of function. Diploid strains (Table 1) were constructed and tested as described in Figure 1. The top panel shows the five viable combinations of normal (POB3), mutated (pob3-Q308K), and deleted (pob3-Δ) versions of the POB3 locus. The bottom panel shows the three viable combinations that also lack HPC2.
To probe this further, we tested the dominance of pob3-Q308K in a background sensitized for FACT function. Many FACT mutations, including pob3-Q308K, are lethal in cells lacking the HIR/HPC complex, a histone chaperone complex that is also involved in regulating the expression of some of the histone genes as well as having a direct role in replication-independent histone deposition (Formosa et al. 2002). This synthetic lethality indicates that FACT and the HIR/HPC complex have partially overlapping or codependent functions, probably through a common ability to chaperone histones during assembly and disassembly of nucleosomes (Formosa et al. 2002; Groth et al. 2007). Diploids homozygous for deletion of a component of this complex (Hpc2) were therefore constructed to increase the reliance of the cells on FACT function. Indeed, in the absence of Hpc2, pob3-Q308K was dominant for Ts−, HUs, and the Spt− phenotypes (Figure 7). This shows that pob3-Q308K is at least partially dominant for all defects, but the replication defects can be masked by the actions of the Hir/Hpc complex, whereas maintaining fully repressive chromatin genome-wide to prevent the Spt− phenotype cannot. The genetic dominance of pob3-Q308K supports the model that this mutation inhibits release of FACT from nucleosomes.
Balanced interaction between Pob3 and H3-H4 is important for FACT function
Both pob3-Q308K and pob3-Q308R were isolated multiple times in our original screen for HU-sensitive alleles of POB3 (VanDemark et al. 2006), but mutating Q308 to alanine or even deleting this residue caused no phenotypes (VanDemark et al. 2006), suggesting that it is the presence of a basic residue at this site rather than the loss of the glutamine that is problematic. The structures of the Pob3-M and Rtt106 proteins show that Pob3-Q308K might expand a basic surface implicated in H3-H4 binding (Liu et al. 2010; Zunder et al. 2012). Supporting this, the amount of H3 that coprecipitated from a whole cell lysate with Myc-tagged Pob3 protein increased when the Q308K mutation was introduced (Zunder et al. 2012). Notably, the increased yield of H3 and the other phenotypes caused by pob3-Q308K were reversed by introducing a negatively charged residue into the surface adjacent to Pob3-Q308 (T252E; Zunder et al. 2012), strongly suggesting that the total charge of this surface is an important determinant of the strength of the interaction between Pob3 and histones.
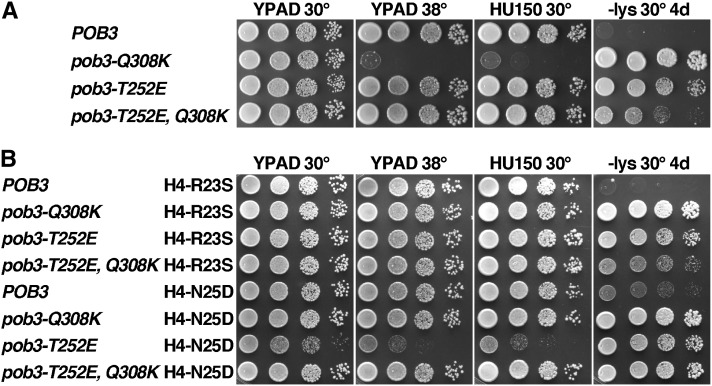
When integrated into A364a strains, pob3-T252E caused a moderate Spt− phenotype (Lys+ after 4 days, whereas pob3-Q308K strains take 2 days), but the HU sensitivity reported previously in a W303 background was not observed (Figure 8A; a mild defect was reported and we consistently observe weaker phenotypes for FACT mutations in the W303 background relative to A364a due to the different SSD1 alleles in these strains) (O’Donnell et al. 2009; and our unpublished observations). However, we did observe the reported suppression of the defects caused by pob3-Q308K, including complete reversal of the Ts− and HU phenotypes as well as strong, but not complete, reversal of the Spt− phenotype (Figure 8A). Introduction of a negative charge to the surface of Pob3 near the site of the Q308K mutation therefore significantly ameliorated the defects caused by the extra lysine residue.
Figure 8.
Effects of pob3-T252E support the importance of balanced interaction between FACT and nucleosomes. (A) and (B) Strains (Table 1) with the relevant genotypes shown were grown to saturation and tested as described in Figure 1.
If two mutations each suppress pob3-Q308K by weakening the interaction between FACT and histones, combining these mutations in the absence of pob3-Q308K might weaken the binding excessively. Consistent with this prediction, Figure 8B shows that combining pob3-T252E with H4-N25D was severely detrimental. H4-R23S did not have a similar effect, again showing that while these two histone mutations each suppress pob3-Q308K, their effects on H4 appear to be distinct as they interact differently with both spt16-11 and pob3-T252E. A strain with pob3-T252E, Q308K, and H4-N25D did not display growth defects (Figure 8B), supporting the interpretation that the detrimental effects were caused by imbalanced interaction between FACT and histones.
Overall, the results show that optimal FACT function includes the ability to release from nucleosomes efficiently, and that the pathway of release may be more complex than simple dissociation of binding between two static complementary surfaces.
Discussion
We describe a set of histone mutations that suppress some of the defects caused by the pob3-Q308K allele of the small subunit of FACT in S. cerevisiae. The spt16-11 allele of the large subunit of FACT caused a defect in the ability to promote reorganization of nucleosomes to a more open configuration, and suppression was accomplished by mutating H2A-H2B to make nucleosomes that adopt the reorganized form more readily. Here, we show that pob3-Q308K can also be suppressed by histone mutations, but in this case Pob3-Q308K protein promotes reorganization normally, and suppression does not appear to involve destabilization of nucleosomes. Instead, Pob3-Q308K protein is defective in releasing from nucleosomes, during the step when FACT should be converting the loosely assembled histones and DNA into canonical nucleosomes. The histone mutations that suppress the physiological effects of pob3-Q308K promote this release of Pob3-Q308K protein from nucleosomes, supporting the importance of this step in FACT activity in vivo. These results stress the involvement of FACT in both destabilizing and reassembling nucleosomes and suggest that reassembly involves a series of steps that are guided by FACT, as discussed below.
The clustering of the suppressor mutations suggested an interaction surface important for binding FACT. This model is especially attractive given that pob3-Q308K enhances the binding of H3-H4 in cell lysates, inhibits release of FACT from reconstituted nucleosomes in vitro, and can be suppressed by inserting an acidic residue near the site of the basic Q308K mutation (pob3-T252E; Zunder et al. 2012 and Figure 8). Together, these observations suggest a simple model in which the surface of Pob3 containing T252 and Q308 directly contacts a surface on H3-H4 containing the sites identified here by suppressor mutations. Q308K inappropriately increases the affinity of this interaction, leading to inefficient resolution of FACT:nucleosome complexes, and this is suppressed by mutations that weaken the interaction back to the normal range.
We tested this model by measuring the affinity of FACT for nucleosomes using normal or mutated components, but failed to find the predicted enhancement of binding by Pob3-Q308K or weaker binding with mutated histones (Figure 5). We considered the possibility that the important binding interaction occurs between FACT and H3-H4 outside of the context of a nucleosome while different surfaces of H3-H4 are exposed. However, the EMSA test we developed previously for measuring the interaction between free H3-H4 and FACT (Kemble et al. 2013) also gave normal values with Pob3-Q308K and mutant H4 (not shown). These EMSA-based titrations have produced results similar to those determined by fluorescence quenching or anisotropy (Ruone et al. 2003; Winkler et al. 2011; Kemble et al. 2013), but physiological effects could result from changes in affinity too small to detect this way, so we cannot rule out a simple binding model for suppression.
FACT:nucleosome complexes dissociate rapidly when Nhp6 is sequestered by addition of competitor DNA, but ∼5% of the complexes formed with Spt16-Pob3-Q308K were resistant to even long incubations in the absence of free Nhp6. This unexpected result shows that while most of the complexes can release normally, a subset is at least temporarily stuck, suggesting a pathway of dissociation with multiple steps or potential outcomes. Pob3-Q308K protein is then inefficient at performing one of these steps, leading to a persistent or perhaps permanent block to resolution. This subset of the complexes is too small to skew the affinity measured by EMSA, but is large enough to provide a physiological effect. An S. cerevisiae haploid cell contains ∼25,000 Spt16-Pob3 heterodimers and 70,000 nucleosomes (VanDemark et al. 2008), so prolonged binding of 5% of the FACT molecules to nucleosomes could block ∼2% of the chromatin in a cell. This would affect processes like transcription and replication that require efficient progression of polymerases through large numbers of nucleosomes and their rapid reassembly afterward. Because FACT:nucleosome complexes are rapidly reversible, converting 5% of the complexes to a persistently blocked form could also have a disproportionate effect on the yield of nucleosomes (and therefore histone H3 molecules) that copurify with FACT in an immunoprecipitation, as observed (Zunder et al. 2012). We did not detect an increase in the total or soluble pools of H3 in a pob3-Q308K strain (Figure 4), and we did not see an increase in affinity of Pob3-Q308K protein for nucleosomes or H3-H4, so we propose that the excess H3 that copurifies with Pob3-Q308K protein is due to persistently associated FACT:nucleosome complexes.
We propose a “structural rearrangement” model to explain the prolonged binding of Spt16-Pob3-Q308K to nucleosomes and suppression of this defect by H3-H4 mutations. In this model, the histones are not static structures with fixed binding surfaces but instead they undergo structural rearrangements between their free (chaperoned) or reorganized forms and their nucleosomal forms. There is precedent for this type of structural switch between chaperoned histones and nucleosomes, as an alpha helix of H4 is reoriented between the Asf1-bound form and the nucleosomal structure (Antczak et al. 2006; English et al. 2006). The H4 N-terminal region makes a series of sharp bends as it joins the globular core, contacting the DNA, H3, and more internal regions of itself as it does so. In the nucleosome, these bends are supported by many contacts, including interaction between H4-N25 and a surface of H3 (including H3-P66, -R69, -L70, and -E73), and several contacts involving H4-R55. Notably, H4-N25D and H4-R55G were the two most commonly identified residues in our screen, and the other residues identified also support the shape of this region of H4 in a nucleosome. We propose that this section of H4 is more extended or at least in some other configuration when it is outside of a nucleosome or in the reorganized form bound by FACT, and that the transition from this state to the canonical nucleosomal state requires passage through one or more intermediates. FACT might guide these steps, stabilize the conformation found in nucleosomes during reorganization, or prevent inappropriate off-pathway interactions from occurring. Pob3-Q308K protein could either fail to efficiently retain or form the appropriate conformation, or fail to efficiently block the formation of a dead-end product. Suppression therefore occurs by easing this structural transition between extended and compact forms of H4, either by weakening the hold of Pob3 on the histones or by altering the structure of the histones such that finding the correct conformation is less difficult or less prone to errors. The suppressors therefore might reveal a binding site for FACT or they could be hinge regions in H3-H4 whose rearrangement is promoted by or coordinated with FACT activity to allow the transition between nucleosomal and nonnucleosomal states.
FACT is known as a factor that is useful for destabilizing nucleosomes and paradoxically for preventing dispersal of chromatin components during transcription. We propose that FACT has a functional role in insuring that the loosely associated components of nucleosomes in the reorganized state adopt shapes compatible with formation of canonical nucleosomes. Different alleles of FACT have defects in different stages of this reversible transition, explaining their very different interactions with histone mutations. The results described here stress the previously less appreciated role of FACT in promoting the formation of nucleosomes, not just their disassembly.
Acknowledgments
We thank Mitch Smith and David Stillman for providing strains, Robert Rawlins and Liz Kendall for valuable technical assistance, and David Stillman and members of his lab for thoughtful discussion and comments on this project and this manuscript. This project was supported by a grant from the National Institutes of Health to T.F.
Footnotes
Communicating editor: M. Hampsey
Literature Cited
- Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., et al. , 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120: 25–36. [DOI] [PubMed] [Google Scholar]
- Allain F. H., Yen Y. M., Masse J. E., Schultze P., Dieckmann T., et al. , 1999. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 18: 2563–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antczak A. J., Tsubota T., Kaufman P. D., Berger J. M., 2006. Structure of the yeast histone H3–ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct. Biol. 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O. M., 2013. Location, location, location: it’s all in the timing for replication origins. Genes Dev. 27: 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache K. J., Garlick J. D., Canzio D., Narlikar G. J., Kingston R. E., 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 334: 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., et al. , 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093. [DOI] [PubMed] [Google Scholar]
- Biswas D., Yu Y., Prall M., Formosa T., Stillman D. J., 2005. The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 25: 5812–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D., Takahata S., Xin H., Dutta-Biswas R., Yu Y., et al. , 2008. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics 178: 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R., 1987. 5-fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175. [DOI] [PubMed] [Google Scholar]
- English C. M., Adkins M. W., Carson J. J., Churchill M. E., Tyler J. K., 2006. Structural basis for the histone chaperone activity of Asf1. Cell 127: 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J., Truong D., Das C., Carson J. J., Kieft J., et al. , 2010. Elevated histone expression promotes life span extension. Mol. Cell 39: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., 2012. The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819: 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., et al. , 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20: 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Ruone S., Adams M. D., Olsen A. E., Eriksson P., et al. , 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway. Polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A., Corpet A., Cook A. J., Roche D., Bartek J., et al. , 2007. Regulation of replication fork progression through histone supply and demand. Science 318: 1928–1931. [DOI] [PubMed] [Google Scholar]
- Hainer, S. J., B. A. Charsar, S. B. Cohen, and J. A. Martens, 2012 Identification of mutant versions of the Spt16 histone chaperone that are defective for transcription-coupled nucleosome occupancy in Saccharomyces cerevisiae. G3 (Bethesda) 2: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., et al. , 2013. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499: 111–114. [DOI] [PubMed] [Google Scholar]
- Hsieh F. K., Kulaeva O. I., Patel S. S., Dyer P. N., Luger K., et al. , 2013. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 110: 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A., Puglisi A., Strubin M., 2009. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell 35: 377–383. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Kayne P. S., Kahn E. S., Grunstein M., 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87: 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble D. J., Whitby F. G., Robinson H., McCullough L. L., Formosa T., et al. , 2013. Structure of the Spt16 middle domain reveals functional features of the histone chaperone FACT. J. Biol. Chem. 288: 10188–10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Huang H., Zhou B. O., Wang S. S., Hu Y., et al. , 2010. Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing. J. Biol. Chem. 285: 4251–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Rechsteiner T. J., Richmond T. J., 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304: 3–19. [DOI] [PubMed] [Google Scholar]
- McCullough L., Rawlins R., Olsen A. E., Xin H., Stillman D. J., et al. , 2011. Insight into the mechanism of nucleosome reorganization from histone mutants that suppress defects in the FACT histone chaperone. Genetics 188: 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo-Huesca M., Maya D., Munoz-Centeno M. C., Singh R. K., Oreal V., et al. , 2010. FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1. PLoS Genet. 6: e1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A. F., Stevens J. R., Kepkay R., Barnes C. A., Johnston G. C., et al. , 2009. New mutant versions of yeast FACT subunit Spt16 affect cell integrity. Mol. Genet. Genomics 282: 487–502. [DOI] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C.-H., Luse D. S., Reinberg D., 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu W. H., Lane W. S., Hampsey M., Reinberg D., 1999. The chromatin-specific transcription elongation factor FACT comprises the human SPT16/CDC68 and SSRP1 proteins. Nature 400: 284–288. [DOI] [PubMed] [Google Scholar]
- Ransom M., Williams S. K., Dechassa M. L., Das C., Linger J., et al. , 2009. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J. Biol. Chem. 284: 23461–23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D., Sims R. J., 3rd, 2006. de FACTo nucleosome dynamics. J. Biol. Chem. 281: 23297–23301. [DOI] [PubMed] [Google Scholar]
- Rhoades A. R., Ruone S., Formosa T., 2004. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 24: 3907–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruone S., Rhoades A. R., Formosa T., 2003. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and reorganize nucleosomes. J. Biol. Chem. 278: 45288–45295. [DOI] [PubMed] [Google Scholar]
- Schlesinger M. B., Formosa T., 2000. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 155: 1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish M. A., Struhl K., 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24: 10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen G., Winston F., Styles C. A., Fink G. R., 1984. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc. Natl. Acad. Sci. USA 81: 2431–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R., Lindstrom D. L., Tran H. G., Roinick K. L., Costa P. J., et al. , 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22: 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer R. A., Johnston G. C., 2004. The FACT chromatin modulator: genetic and structure/function relationships. Biochem. Cell Biol. 82: 419–427. [DOI] [PubMed] [Google Scholar]
- Squazzo S. L., Costa P. J., Lindstrom D., Kumer K. E., Simic R., et al. , 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21: 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., 2010. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1799: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F., Lewis L. K., Resnick M. A., 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773–776. [DOI] [PubMed] [Google Scholar]
- Stuwe T., Hothorn M., Lejeune E., Rybin V., Bortfeld M., et al. , 2008. The FACT Spt16 “peptidase” domain is a histone H3–H4 binding module. Proc. Natl. Acad. Sci. USA 105: 8884–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S., Yu Y., Stillman D. J., 2009a The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 28: 3378–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S., Yu Y., Stillman D. J., 2009b FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell 34: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A., Schneiter R., 2006. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast 23: 825–831. [DOI] [PubMed] [Google Scholar]
- VanDemark A. P., Blanksma M., Ferris E., Heroux A., Hill C. P., et al. , 2006. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 22: 363–374. [DOI] [PubMed] [Google Scholar]
- VanDemark A. P., Xin H., McCullough L., Rawlins R., Bentley S., et al. , 2008. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J. Biol. Chem. 283: 5058–5068. [DOI] [PubMed] [Google Scholar]
- White C. L., Suto R. K., Luger K., 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20: 5207–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler D. D., Luger K., 2011. The histone chaperone Fact: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286: 18369–18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler D. D., Muthurajan U. M., Hieb A. R., Luger K., 2011. The histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286: 41883–41892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J., Joss L., Formosa T., 1999. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38: 8961–8971. [DOI] [PubMed] [Google Scholar]
- Xin H., Takahata S., Blanksma M., McCullough L., Stillman D. J., et al. , 2009. yFACT induces global accessibility of nucleosomal DNA without H2A–H2B displacement. Mol. Cell 35: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekezare M., Gomez-Gonzalez B., Diffley J. F., 2013. Controlling DNA replication origins in response to DNA damage: inhibit globally, activate locally. J. Cell Sci. 126: 1297–1306. [DOI] [PubMed] [Google Scholar]
- Zunder R. M., Antczak A. J., Berger J. M., Rine J., 2012. Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing. Proc. Natl. Acad. Sci. USA 109: E144–E153. [DOI] [PMC free article] [PubMed] [Google Scholar]