Abstract
Constitutive transport of cellular materials is essential for cell survival. Although multiple small GTPase Rab proteins are required for the process, few regulators of Rabs are known. Here we report that EAT-17, a novel GTPase-activating protein (GAP), regulates RAB-6.2 function in grinder formation in Caenorhabditis elegans. We identified EAT-17 as a novel RabGAP that interacts with RAB-6.2, a protein that presumably regulates vesicle trafficking between Golgi, the endoplasmic reticulum, and plasma membrane to form a functional grinder. EAT-17 has a canonical GAP domain that is critical for its function. RNA interference against 25 confirmed and/or predicted RABs in C. elegans shows that RNAi against rab-6.2 produces a phenotype identical to eat-17. A directed yeast two-hybrid screen using EAT-17 as bait and each of the 25 RAB proteins as prey identifies RAB-6.2 as the interacting partner of EAT-17, confirming that RAB-6.2 is a specific substrate of EAT-17. Additionally, deletion mutants of rab-6.2 show grinder defects identical to those of eat-17 loss-of-function mutants, and both RAB-6.2 and EAT-17 are expressed in the terminal bulb of the pharynx where the grinder is located. Collectively, these results suggest that EAT-17 is a specific GTPase-activating protein for RAB-6.2. Based on the conserved function of Rab6 in vesicular transport, we propose that EAT-17 regulates the turnover rate of RAB-6.2 activity in cargo trafficking for grinder formation.
Keywords: RAB, RABGAP, Golgi-endosome trafficking, grinder formation
CELLS constitutively transport newly synthesized proteins, lipids, and other molecules to their periphery through vesicle trafficking. As the process is conserved from yeast to humans, it is essential for the function and survival of cells. Rab GTPases are small GTP binding proteins that regulate the transport of vesicles between different compartments of the cell (Zerial and McBride 2001; Jordens et al. 2005; Grosshans et al. 2006). Rab6’s are localized to Golgi membranes to mark and target both anterograde cargos from Golgi to post-Golgi compartments (such as the plasma membrane) and retrograde cargos from early/recycling endosomes to Golgi and the endoplasmic reticulum (Jasmin et al. 1992; Martinez et al. 1994, 1997; Girod et al. 1999; Opdam et al. 2000; Del Nery et al. 2006). For fast turnover, Rabs require guanine nucleotide exchange factors (Rab GEFs) for activation and GTPase-activating proteins (Rab GAPs) to turn off activity (Grosshans et al. 2006). While the role of Rab6 in membrane trafficking is well established, very few GAPs or GEFs for Rab6 have been identified; thus their physiological importance is largely unknown. The physiological roles of RabGAPs were addressed only recently in flies (Houalla et al. 2010; Uytterhoeven et al. 2011) and worms (Chotard et al. 2010). In those cases, removing the function of the RabGAPs produced phenotypes almost identical to those of removing the relevant Rabs, showing the fundamental roles of RabGAPs in modulating Rab functions. Currently, the only identified GAP for Rab6 is GAPCenA in humans, which is associated with centrosomes and regulates Golgi dynamics in dividing cells (Cuif et al. 1999). Nonetheless, misregulation of GAPs for other small G proteins, such as Ras, directly relates to diseases such as cancer, implicating the essential roles of GAPs in controlling the kinetics of G protein activity (Tanabe et al. 2006; Durkin et al. 2007; Pamonsinlapatham et al. 2009).
The Caenorhabditis elegans grinder is a complex structure required for proper grinding of food (bacteria) before it is passed to the intestine. When food is abundant, worms feed at an average rate of 200 pumps per minute (ppm) based on counts of pharyngeal pumping and grinding motions (Avery and You 2012; Raizen et al. 2012). This high frequency of grinding throughout the worm’s life suggests transport of grinder components to build a functional grinder is essential for worms' rapid growth and normal development. Considering the location of this structure in the C. elegans pharynx, grinder components are likely produced by terminal bulb muscles pm6 and pm7 and trafficked to their apical surfaces (Albertson and Thomson 1976). The exact components of the grinder and molecular mechanisms regulating their transport, however, remain unknown.
Here we report that C. elegans gene eat-17 encodes a Rab GTPase-activating protein (Rab GAP) specific for RAB-6.2, a Rab6 homolog in C. elegans. eat-17 loss-of-function and rab-6.2 deletion mutants show similar phenotypes, namely disorganized, poorly formed grinders. RNAi against rab-6.2 produces similar defects. A directed yeast two-hybrid screen using EAT-17 as bait identified RAB-6.2 as its interacting partner. Furthermore, transgenes with mutations in the conserved catalytic arginine residue required for the GAP activity of EAT-17 failed to rescue grinder defects when injected into eat-17 mutants. Based on these results and the conserved function of Rab6 in vesicle transport, we propose that RAB-6.2 and EAT-17 cooperate in the transport of grinder components to the apical surfaces of the terminal bulb muscles to form a functional grinder.
Materials and Methods
Worm culture
Worms were cultured and handled as described (Sulston and Hodgkin 1988) with the following modifications. First, they were routinely grown on NGMSR plates (Avery 1993). NGMSR differs from NGM in containing 200 pg/ml streptomycin sulfate, 10 pg/ml nystatin, and 2% agar instead of 1.7%. (Streptomycin and nystatin reduce bacterial and fungal contamination. The higher agar concentration delays the burrowing of the worms into the agar.) In addition, all worms were maintained at 20° on Escherichia coli HB101 unless indicated otherwise. The wild-type strains are Bristol N2 and Hawaiian CB4856. eat-17(ad707) and rab-6.2(ok2254) were used throughout this study after being outcrossed twice to N2. Other strains used in this study are DA2120 adEx2120[eat-17p::GFP rol-6(sd)], DA773 unc-93(e1500sd); lin-15(n309) eat-17(ad707) sup-10(n183), DA1814 ser-1(ok345), RB758 hda-4(ok518), RB787 T27A8.2(ok570), DA707 eat-17(ad707), NL2099 rrf-3(pk1426), DA1983 eat-17(ad707); adEx1983[F01G12T24D11rol-6(d)], DA1982 rrf-3(pk1426); eat-17(ad707), DA2035 adEx2035[rab-6.2p::GFP rol-6(d)], DA2033 eat-5(ad1402); adEx2033[RAB-6.2::GFP unc-122::RFP eat-5(+)], YJ89 rab-6.2(ok2254), MT993 lin-10(e1439); him-5(e1467ts).
SNP mapping
CB4856 males were crossed to DA773 hermaphrodites. unc-93; lin-15eat-17sup-10/CB4856(+) worms were isolated in the F2 generation by their ability to produce 25% Muv non-Unc self-progeny and the near absence of non-Muv non-Unc self-progeny. Sup non-Muv recombinants were isolated in the F3 generation. These were scored for the eat-17 feeding defect, and their progeny were analyzed by PCR and subsequent restriction digestion for SNPs located between lin-15 and sup-10. SNPs examined and primers used for their amplification are shown in Supporting Information, Table S1.
Deletion mutants of ser-1(ok345), hda-4(ok518), and T27A8.2(ok570) were mapped by scoring F4 progeny for deletion detected by PCR. Primers used for detecting the deletions are shown in Table S2.
Analysis of eat-17 gene structure and identification of splice variants
To determine the intron and exon structure of eat-17, primers were designed based on Genefinder predictions shown in WormBase to amplify partial and full-length transcripts by RT–PCR. First-strand poly(A)+ cDNA derived from mixed stage N2 hermaphrodites was used as template in all reactions. Trizol (Invitrogen) was used to isolate RNA and the First-Strand cDNA Synthesis kit (Roche) was used to generate cDNA. Primers used to amplify PCR fragments are shown in Table S3. All PCR reactions yielded a single product, and all exons except the first appeared to be correct. To assess the number and abundance of different splice variants, full-length eat-17 cDNAs were cloned into the pGEM-T Easy vector (Promega) and sequenced.
SL1 trans-splicing
To identify the first exon of eat-17, a forward primer recognizing the 22 nucleotide-SL1-splice leader sequence (5′-ggtttaattacccaagtttgag -3′) and two nested reverse primers recognizing sequences in exon 2 [1050: 5′-tgttcagctgctccatcttg-3′(outside primer)/1051: 5′-cgacttcatttacgcatactg-3′(inside primer)] were used. Products were purified (Qiaquick Gel Extraction kit), cloned into pGEM-T Easy vectors and sequenced. The resulting sequences were BLASTed (blastn) against C. elegans ESTs and genomic DNA using WormBase.
Cosmid rescue
Cosmids used in this study were obtained from Alan Coulson (Wellcome Trust, UK). T24D11 and F01G12 were isolated using the Qiagen Plasmid Mini kit and then co-injected into eat-17 mutants (50 ng/μl). Plasmids pPD118.20 myo-3::GFP (from Andrew Fire, Stanford University) and pRAK3 rol-6(d) were used as co-injection markers at a concentration of 10 ng/μl. Worms were fed Comamonas before injection to allow good growth (Avery and Shtonda 2003). Transgenic worms were then fed DA837 to enhance the feeding defect.
Growth rates of transgenic and nontransgenic progeny were compared to assess rescue. Eggs from transgenic mothers were placed individually onto plates seeded with DA837 and then checked every 12 hr to stage the worms. Growth rate is defined as the inverse of the time required for worms to reach adulthood and produce progeny. Worms were scored as adults if they had laid at least one egg.
Generating the eat-17 rescue construct
Rescuing fragments containing the full-length eat-17 cDNA were generated by overlap extension PCR (Ho et al. 1989). In first round PCR reactions, 5′ and 3′ eat-17 cDNA fragments were amplified using primers: 5′-ttgtcaccgccgatggcagccactgcagcgctac–3′ and 5′-tagggatgttgaagagtaattggacctagtggctatccgacagtt–3′. The resulting PCR fragments were purified and subjected to a second round of PCR using the overlap extension method to fuse the eat-17 promoter and unc-54 3′-UTR to each end. The primers used to amplify the promoter region are: 5′-taggttacggtagttggtacg–3′ and 5′-gtagcgctgcagtggctgccatcggcggtgacaa–3′. The primers used to amplify 500 bp unc-54 3′ UTR are 5′-aactgtcggatagccactaggtccaattactcttcaacatcccta–3′ and 5′-tttggtatattgggaatgtattctg–3′. The resulting PCR products were purified and injected into eat-17 worms (25 ng/μl). The final injected product contained 5.6 kb of sequence located directly upstream of the eat-17 transcriptional start site. A 2.5-kb full-length eat-17 cDNA construct was fused to this “promoter region” and the heterologous unc-54 3′-UTR was added to promote stability. This construct lacked intron elements, including the large 4-kb first intron. let-858::GFP plasmid DNA (18 ng/μl) was used as a co-injection marker.
Two transgenic lines were isolated and examined. F2 transgenic animals were identified using the SZX12GFP dissecting scope (Olympus). Because many transgenic embryos die, only animals that reached adulthood were scored for the grinder phenotype. For both transgenic lines, 100% of the adult transgenic animals were rescued for defects in grinder formation. Grinder morphology was observed using a Zeiss Axio A2 Imager at either ×630 or ×1000 magnification. Images were acquired using Zeiss Axiovision software.
Assay for EAT-17 GAP activity
To determine the functional importance of the GAP activity of EAT-17, rescuing fragments containing the catalytically inactive R116/119K and R116/119A mutations were generated using the overlap extension method as describe above. Primers used to generate the R116/119K mutation are shown in Table S4. The resulting PCR products were gel purified and injected into eat-17 worms at a concentration of 25 ng/μl. let-858::GFP plasmid DNA was used as a co-injection marker at a concentration of 100 ng/μl. Wild-type, R116/119K, and R116/119A injections were performed in parallel. To score for rescue of grinder defects, the F1 gravid adult progeny of injected mothers were picked to 4% agar pads containing 10 mM sodium azide (Sigma, St. Louis). Defects in grinder formation were recorded and then the presence or absence of GFP expression was determined. Worms were observed using a Zeiss Axio A2 Imager with a ×100 objective lens. Images were acquired using Zeiss Axiovision software.
RNAi
RNAi was performed as described with minor modifications (Kamath and Ahringer 2003). cDNA sequences of eat-17 and the 25 C. elegans Rabs were amplified by PCR using primers shown in Table S5. The HiScribe RNAi Transcription kit (New England Biolabs) was used to generate dsRNAs.
GFP fusions of EAT-17 and RAB-6.2
eat-17p::GFP:
A 5.6-kb eat-17 promoter was amplified from N2 genomic DNA using the following primers: 5′-taggttacggtagttggtacg-3′ and 5′-gaaaagttcttctcctttactcatcggcggtgacaattgg-3′. In addition to the 5′ regulatory sequence, this DNA contains the first six codons of eat-17. GFP was amplified from pPD95.75 plasmid DNA (gift from A. Fire, Stanford University) using primers: 5′-ccaattgtcaccgccgatgagtaaaggagaagaacttttc-3′ and 5′-tttggtatattgggaatggtattctg-3′. DNAs were fused together by overlap extension PCR using the following nested primers: 5′-acggtagtgttttatcagtagtg-3′ and 5′-caaacccaaaccttcttccgatc-3′.
The resulting product was purified and injected into N2 adults with pRAK3 rol-6(d) as a co-injection marker. The established transgenic lines were observed using a Zeiss Axio A2 Imager at either ×630 or ×1000 magnification. Images were acquired using Zeiss Axiovision software.
rab-6.2p::GFP:
A 2.8-kb rab-6.2 promoter fragment was amplified from N2 genomic DNA using the following primers: 5′-aatcgcacagcaggcctcc-3′ and 5′-gtgaaaagttcttctcctttactcggattaccaaagtccgacat-3′. In addition to the 5′ regulatory sequence, this DNA fragment contains the first seven codons of rab-6.2. GFP was amplified from plasmid pPD95.75 using the following primers: 5′-atgtcggactttggtaatccgagtaaaggagaagaacttttcac-3′ and 5′-tttggtatattgggaatggtattctg-3′. Products were fused by overlap extension PCR using the following primers: 5′-tttgccgaacggaagagcc-3′ and 5′-caaacccaaaccttcttccgatc-3′. The final product was purified and injected into N2 adults with rol-6(d) as a co-injection marker. GFP expression was observed as described above.
Yeast two-hybrid assay
To generate first-strand cDNA, RNA was isolated from a well-fed, mixed stage N2 population using Trizol. Poly(A)+ first-strand cDNA was prepared using the First-Strand cDNA Synthesis kit for RT–PCR (Roche).
Inserts used for preys were amplified by PCR using the Expand Long Template PCR kit (Roche). Primers to amplify the preys are shown in Table S6. PCR products were cloned into the pGEM-T Easy vector and sequenced. Constructs with wild-type sequence were subcloned into the pACT2 vector (gift from R. Lin, University of Texas Southwestern Medical Center at Dallas), in frame with the GAL4 activation domain.
Six truncated versions of eat-17 were used as baits after amplification, sequencing and cloning into the pVJL11 vector (gift from M. Cobb, University of Texas Southwestern Medical Center at Dallas), in frame with the LexA DNA binding domain. Primers are shown in Table S7. Mutant versions of the baits were generated using an in vitro mutagenesis kit (Invitrogen). Primers containing mutations are shown in Table S4.
To test for interactions, combinations of baits and preys were transformed into the yeast strain L40 (Hama et al. 1999). X-gal filter assays and quantitative ortho-nitrophenyl-β-D-galactopyranoside (ONPG) assays were performed as described (Yeast Protocols Handbook, Clontech). An average of two colonies was tested for each. For growth assays, colonies were streaked onto plates lacking histidine and supplemented with 5 mM 3-amino-1,2,4-triazole (3-AT). Miller units are calculated from OD420/t × V × OD600.
Chitin staining
Chitin was stained as described (Ruvkun and Finney 2005). Fluorescein-conjugated chitin-binding probe (P5211S, NEB) was used at a 1:100 dilution.
Grinder function assay
Worms were fed mCherry-expressing HB101 as described (You et al. 2008). Images were acquired using Zeiss Axiovision software.
Results
eat-17 mutants are Eat due to defects in grinder formation
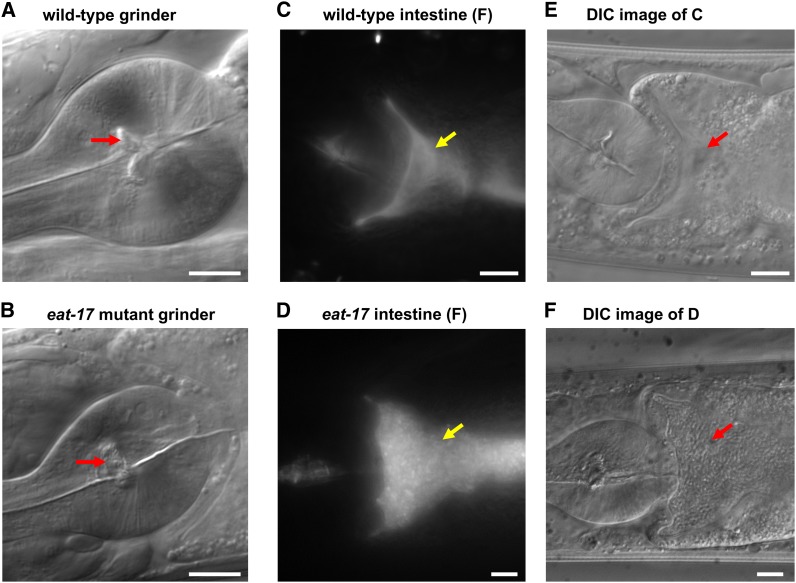
eat-17 mutants were isolated from a genetic screen for defects in feeding behavior. They are defective in trapping bacteria and have slightly asynchronous terminal bulb contractions during feeding (Avery 1993). The most striking phenotype, however, is a defect in their grinders; wild-type grinders consist of highly refractile cuticle ridges that can be observed by DIC optics (Figure 1A, arrow). These ridges are completely disorganized in eat-17 mutants (Figure 1B). In wild-type worms, the grinder is composed of three separate plates that are arranged in approximate triradial symmetry (Albertson and Thomson 1976; von Lieven 2003). These plates are secreted from the apical surfaces of pharyngeal muscle cells pm6 and pm7 in the terminal bulb (Albertson and Thomson 1976). During feeding, the plates rotate against one another, grinding any material that comes between them. Due to defects in the grinder, eat-17 mutants cannot grind bacteria efficiently. As a result, unground bacteria pass into the intestine (Figure 1, C–F) contributing to the Eat phenotype.
Figure 1.
eat-17 mutants show defects in grinder formation. (A) DIC image of wild-type pharynx. The C. elegans grinder is composed of three plates arranged in approximate triradial symmetry. Two fully formed plates are visible in lateral sections (arrow). (B) DIC image of eat-17(ad707) mutant pharynx. Most of the material comprising the grinder plates is missing in eat-17 mutants. (C) The intestine of wild-type worms fed mCherry-expressing E. coli shows smooth fluorescence due to fluorescence released from ground bacteria. (D) The intestine of eat-17 mutants show many unground bacteria indicated by an arrow. (E and F) Corresponding DIC images of wild type (E) and eat-17 mutant (F), respectively. In F, the arrow indicates DIC image of unground bacteria. For all images, anterior is shown to the left. Bars, 10 μm.
eat-17 encodes a GTPase activating protein with coiled-coil domains at the C terminus
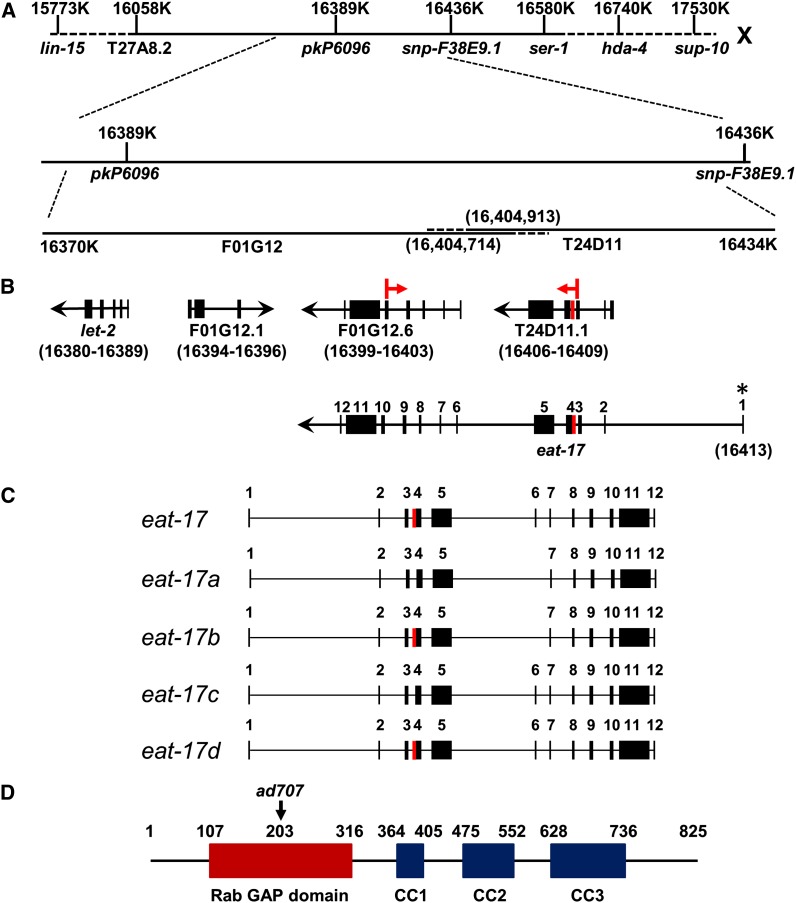
Previous work placed eat-17 on chromosome X, between lin-15 and sup-10 (Avery 1993). We further mapped eat-17 to a 47-kb region between two SNP markers, pkP6096 and snp-F38E9.2 (Figure 2A), using a multipoint SNP mapping strategy (Wicks et al. 2001). Three genes were predicted in this interval (Figure 2B): F01G12.1 encodes a putative copper transporter, T24D11.1 (currently designated as tbc-4) is predicted to encode a Rab GAP containing a TBC (Tre2/Bub/Cdc16 homology) domain—a canonical GAP domain—and F01G12.6 is predicted to encode a protein with several coiled-coil domains. Interestingly, both T24D11.1 and F01G12.6 share homology with the human protein Evi5 (Figure S1), a Rab11 GAP, whose mutation is associated with neuroblastoma (Liao et al. 1997; Dabbeekeh et al. 2007). Because T24D11.1 and F01G12.6 are transcribed in the same direction (Figure 2B) and because we were unable to retrieve cDNAs coding individual genes through RT–PCR, we hypothesized that these two predicted genes comprised a single gene. RT–PCR using primers within the third predicted exon of T24D11.1 and the fifth predicted exon of F01G12.6 identified a single transcript (indicated with arrows in Figure 2B), confirming T24D11.1 and F01G12.6 are parts of the same gene. Furthermore, by injecting two cosmids, T24D11 and F01G12, which completely cover this region, we rescued the growth, feeding, and grinder defects of eat-17 mutants. Injecting either alone did not rescue the phenotype (data not shown).
Figure 2.
eat-17 gene structure. (A) Region in C. elegans genome where eat-17 maps. eat-17 was mapped to 47 kb between two SNP markers pkP6096 and snp-F38E9.2 using a multipoint SNP mapping strategy. (B) Genes predicted to be located in the interval and the gene structure of eat-17. Red arrows indicate the exons targeted to amplify and validate eat-17 transcripts. (C) Exon usages of four possible splice forms deduced from SL1 and RT–PCR analysis. The red box indicates exon 4B. The two different splice forms eat-17a and eat-17b encode two proteins differing by three amino acids. (D) Predicted protein structure of EAT-17 and the location of the mutation in the ad707 allele. An N-terminal Rab GAP domain is followed by three C-terminal predicted coiled-coil domains. The arrow indicates the position of the mutated amino acid in ad707.
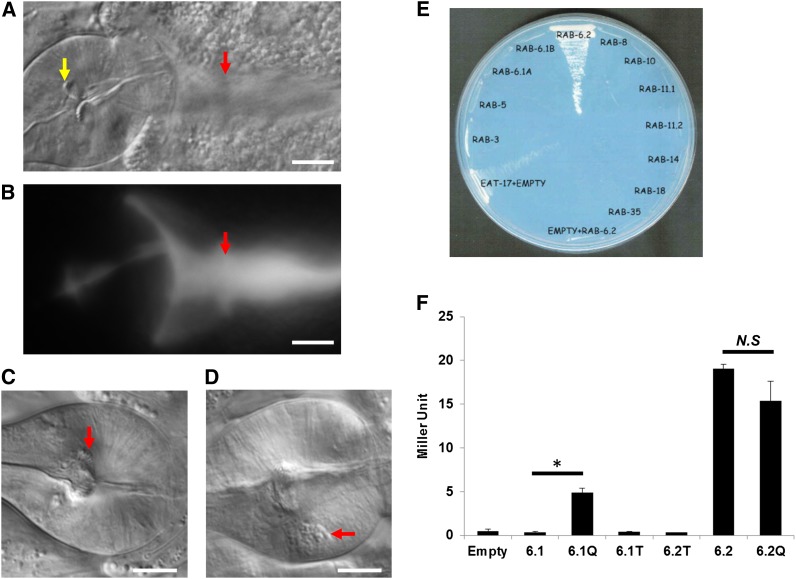
To determine the gene structure of eat-17, we first tried to isolate full-length transcripts with the intron/exon structure predicted in WormBase (www.wormbase.org) but failed. Next we identified individual intron/exon boundaries and found that the first exon was incorrectly predicted. To identify the 5′ end of the transcript, we used SL1 trans-splicing analysis. In C. elegans, ∼60% of all transcripts are either SL1 or SL2 trans-spliced at their 5′ ends (Blumenthal 1995; Conrad et al. 1995). The results show that the first exon of eat-17 is located ∼4 kb upstream of exon 2 (indicated with * in Figure 2B). We also learned that this transcript contains ∼40 bp of 5′-UTR. Using a primer targeting the newly identified first exon and one targeting the last predicted exon of eat-17, we were able to amplify a single 2.5-kb fragment. To confirm that we had isolated the full-length transcript, we generated a rescuing transgene in which this 2.5-kb sequence was attached to an ∼5.6-kb region of sequence upstream of the predicted eat-17 transcriptional start site (eat-17 promoter) at its 5′ end and to a 500-bp unc-54 3′-UTR. When this construct was injected into eat-17 mutants, we found that 100% of surviving F2 transgenic progeny were rescued for defects in their grinders (Figure 3A).
Figure 3.
EAT-17 interacts with RAB-6.2. (A) eat-17 rescue. Expressing a wild-type eat-17 cDNA construct in mutant worms rescues defects in grinder formation. Grinder plates are of normal size (yellow arrow) and there are no unground bacteria in the intestine (red arrow). (B) Fluorescence image of A. Smooth fluorescence in the intestine indicates the bacteria were ground (red arrow). (C) rab-6.2(ok2254) mutants show a grinder defect. Grinder is formed but the size is small. (D) eat-17(ad707) grown at 15° containslarge refractile body in the terminal bulb (arrow). (E) Yeast two-hybrid screen using EAT-17 as bait identified RAB-6.2 as the only interacting Rab protein. (F) A constitutively GTP bound form of RAB-6.1 enhances binding of EAT-17 to RAB-6.1. On the other hand, a constitutively GDP bound form of RAB-6.2 (6.2T) abolishes binding of EAT-17 to RAB-6.2. The EAT-17 construct used was a truncated protein (amino acids 1–460) containing only first coiled coil domain. The truncated protein binds to RAB-6.2 very efficiently, suggesting the first coiled-coil domain is sufficient for the binding. Abbreviations are as follows: 6.1, RAB-6.1 full-length wild type 6.1Q, RAB-6.1 with Q70L mutation; 6.1T, RAB-6.1 with T27N mutation; 6.2T, RAB-6.2 with T27N mutation; 6.2, RAB-6.2 full-length wild type 6.2Q, RAB-6.2 with Q69L mutation. *P < 0.05; NS, not statistically significant by Student t-test. Miller units calculated as OD420/t × V × OD600. Bars, 10 μm.
To determine the exact splicing pattern(s) of eat-17, we cloned the amplified PCR fragments into vectors and sequenced the inserts (see Materials and Methods). We named the possible splice products EAT-17 a, b, c, and d (Figure 2C). EAT-17a has intron/exon boundaries consistent with WormBase/GeneFinder predictions. EAT-17b uses an alternative splice acceptor site at the exon 3/4 boundary (exon 4A, labeled in red, Figure 2, B and C). For both of these, predicted exon 5 of T24D11.1 was truncated by 180 nucleotides and spliced directly to predicted exon 2 of F01G12.6 in WormBase. Neither splice form contained the predicted first exon of F01G12.6. The proteins encoded by EAT-17a and EAT-17b differ by only 3 amino acids. When we designed a set of primers to amplify sequences between exon 1 and exon 6 (corresponding to the predicted first exon of F01G12.6), we could isolate transcripts that included exon 6. We named the predicted transcripts that include exon 6 EAT-17c and EAT-17d (Figure 2C). GeneFinder predictions suggested a splicing pattern that would create a stop codon at the junction between exon 5 and exon 6. This transcript would encode a truncated protein lacking most of the C terminus. It seems most likely that the a and b splice forms encoding full-length proteins are the functional ones.
Analysis of the predicted protein sequence revealed two conserved domains in EAT-17: a Rab GTPase activating (Rab GAP) domain and three coiled-coil domains (CC1–3, Figure 2D). The Rab GAP domain implicates EAT-17 in vesicle trafficking, a role consistent with the grinder defects in eat-17 mutants. To identify the specific mutation in eat-17(ad707), we sequenced the predicted eat-17 coding region (based on our RT–PCR results) and found a C-to-T base-pair transition at nucleotide 607 in exon 5 of the ad707 mutant allele, creating a stop codon at amino acid 203 (Figure 2D). This mutation would generate a protein with a truncated RabGAP domain, likely rendering it nonfunctional.
To determine whether EAT-17 contains GAP activity, we mutated the catalytic arginine residue (R116) to either lysine (K) or alanine (A) to abolish the GAP activity and expressed these mutated constructs in eat-17 mutants (see Materials and Methods). A total of 21% of F1’s carrying a wild-type copy of eat-17 were rescued for defects in grinder formation (Figure 3, A and B), while only 4.1 and 1.6% of mutants expressing either the R116K or R116A mutations were rescued (Table 1). Collectively, these data show that the catalytic activity of EAT-17 is critical for its function and that EAT-17 is a GAP protein.
Table 1. GAP activity is required for full EAT-17 function.
| No. rescued | Total no. examined | % rescue | |
|---|---|---|---|
| Wild-type eat-17 construct | 16 | 76 | 21 |
| R116K | 2 | 49 | 4.1 |
| R116A | 1 | 64 | 1.6 |
eat-17 mutants were injected with DNA constructs encoding either wild-type or one of two mutant versions (R116K or R116A) of the EAT-17 protein (see Materials and Methods). F1 transgenic progeny were blindly assayed for rescue of grinder defects. GFP was used to identify transgenic progeny afterward.
EAT-17 and RAB-6.2 interact to regulate grinder formation
To identify the substrates of the GAP activity of EAT-17, we performed RNA interference against 25 of the 29 predicted Rab genes in the C. elegans genome (Audhya et al. 2007) and looked for a grinder phenotype similar to that of eat-17 mutants. RNAi was performed in worms mutant for rrf-3, a hypersensitive background for RNAi (Simmer et al. 2002). Among the 25 Rab genes tested, RNAi against rab-6.2 alone produced a phenotype similar to eat-17 (Table 2), whereas rrf-3 mutants show no obvious defects in grinder formation (data not shown). When we examined rab-6.2 deletion mutants, the defects in the size and organization of the grinder plates were identical to those of eat-17 mutants (Figures 1B and 3C), confirming our RNAi results.
Table 2. RNAi of rab-6.2 phenocopies eat-17 mutants.
| dsRNA injected | Phenotype |
|---|---|
| None | Wild type |
| rab-1 | Embryonic lethal |
| rab-2 | Slight growth delay |
| rab-3 | Wild type |
| rab-5 | Embryonic lethal |
| rab-6.1 | Wild type |
| rab-6.2 | Growth delay with abnormal grinder |
| rab-7 | Wild type |
| rab-8 | Loopy movement, exaggerated body bends |
| rab-10 | NDa |
| rab-11.1 | Embryonic lethal |
| rab-11.2 | Arrested as L1s |
| rab-14 | Some show growth delay, Dpyb |
| rab-18 | Growth delay, infrequent pumping but normal grinder |
| rab-19 | Wild type |
| rab-21 | Sick, starved, and strong Dpy |
| rab-27 | Wild type |
| rab-28 | Wild type |
| rab-30 | Wild type |
| rab-33 | Slightly Uncc, otherwise wild type |
| rab-35 | Wild type |
| rab-37 | Loopy movement, otherwise wild type |
| rab-39 | Wild type |
| 4R79.2 | Loopy movement, Unc, otherwise wild type |
| K02E10.1 | ND |
| F11A5.4 | Wild type |
| F11A5.3 | Wild type |
| C56E6.2 | Loopy movement, Unc |
Among 25 Rabs tested, RNAi of rab-6.2 shows an identical grinder phenotype to eat-17. Full-length dsRNAs for each Rab listed were injected into rrf-3 RNAi hypersensitive mutants. Worms were grown at 15° to enhance the phenotype.
ND, Not determined.
Dpy, dumpy phenotype, worms are shorter than wild type.
Unc, uncoordinated movement phenotype, worms move abnormally.
To examine whether EAT-17 and RAB-6.2 directly bind to each other and whether the binding is specific, we performed directed yeast two-hybrid assays with EAT-17 and each of the 25 RAB proteins. Among all the RABs, only RAB-6.2 interacted with EAT-17 (Figure 3E, data not shown). To examine if the guanylate nucleotide binding status of RAB-6.2 is critical, we introduce two types of mutations into RAB-6.2: (1) glutamine (Q69) to leucine (L) to produce a constitutively GTP bound form and (2) threonine (T27) to asparagine (N) to produce a constitutively GDP bound form (Martinez et al. 1994, 1997). The interaction between EAT-17 and RAB-6.2 was completely abolished when the T27N mutation was introduced (Figure 3E, 6.2T), but it was not further enhanced with the Q69L mutation (Figure 3C, 6.2Q), as compared to wild-type RAB-6.2 (Figure 3F, 6.2). This suggests that the reaction reached maximum binding with wild-type RAB-6.2 and that we could not enhance binding any further under the conditions in which we performed the assay.
RAB-6.1, the only other Rab6 in the C. elegans genome, shares 81% amino acid sequence identity with RAB-6.2. Because of the high similarity between the two Rab6’s, we tested whether EAT-17 could bind to RAB-6.1 as well. Although we did not detect any binding between wild-type RAB-6.1 and EAT-17 in our assay, RAB-6.1(Q70L) (equivalent to Q69L in RAB-6.2) showed enhanced binding to EAT-17, suggesting that EAT-17 binds to RAB-6’s through the conserved guanylate nucleotide binding site. Taken together, these data strongly support the hypothesis that RAB-6.2 is a specific substrate of EAT-17 and that EAT-17 promotes hydrolysis of GTP by binding the active (GTP bound) form of RAB-6.
Our studies identified EAT-17 as a GAP protein with three coiled-coil domains. Coiled-coil domains are generally involved in protein–protein interactions, and GAPCenA, a mammalian RabGAP, interacts with Rab6 through a coiled-coil domain (Cuif et al. 1999). When we tested several truncated EAT-17 constructs, EAT-17 (aa 1–316), EAT-17 (aa 1–364), and EAT-17 (aa 1–405), they did not interact with RAB-6.2, showing that the GAP domain alone is not sufficient for this interaction (data not shown). A construct of EAT-17 containing the first coiled-coil domain [EAT-17 (aa 1–460)], however, shows significant interaction with RAB-6.2 (Figure 3E). Collectively, our results show that the GAP domain of EAT-17 mediates its catalytic activity and the first coiled-coil domain mediates its interaction with RAB-6.2. Presumably, the coiled-coil domains are important to provide EAT-17 with specificity toward RAB-6.2. Altogether, these data suggest a conserved structural mechanism of interaction between Rab6’s and their RabGAP proteins through the coiled-coil domain (Cuif et al. 1999).
EAT-17 and RAB-6.2 are co-expressed in terminal bulb muscle, the site of grinder secretion
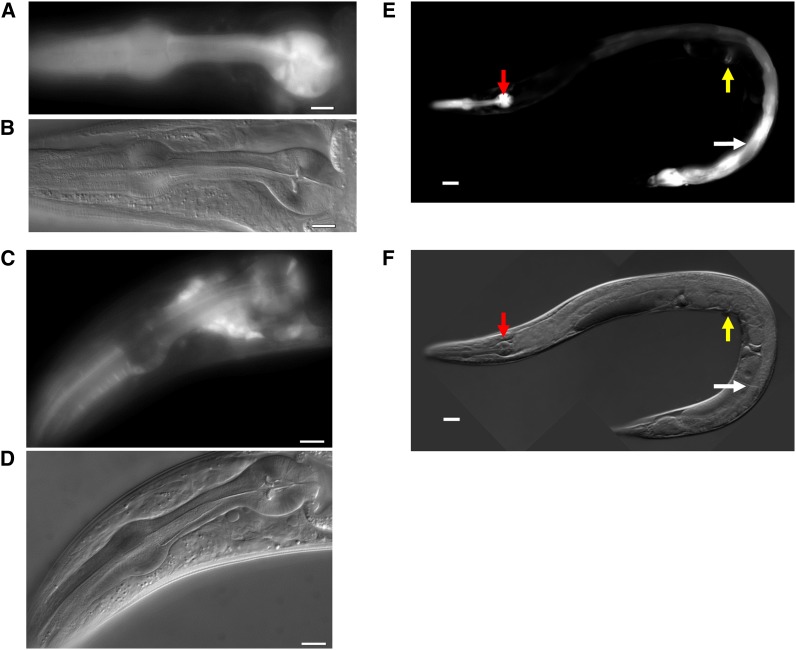
To determine whether EAT-17 and RAB-6.2 function together in the terminal bulb, we examined the expression patterns of transgenic lines that carry the promoters of each gene fused with GFP (see Materials and Methods). Both EAT-17 and RAB-6.2 are strongly expressed in the terminal bulb muscles that secrete the components of the grinder (Figure 4, A–D, see Materials and Methods). Our construct also shows EAT-17 expression outside of the pharynx, such as in intestine and vulva (Figure 4, E and F).
Figure 4.
EAT-17 is expressed in pharyngeal muscle, intestine, and vulva. (A and B) Corresponding GFP and DIC images of EAT-17::GFP. (C and D) Corresponding GFP and DIC images of RAB-6.2::GFP. Both EAT-17 and RAB-6.2 are highly expressed in the pharyngeal muscle. RAB-6-2 is also highly expressed in neurons. (E and F) Corresponding GFP and DIC images of EAT-17::GFP in the pharynx (red arrows), intestine (white arrows), and vulva (yellow arrows). For Figure 4, A–D, bars, 10 μm. For Figure 4, E and F, bars, 30 μm.
This tissue expression pattern of EAT-17 overlaps largely with that previously reported for RAB-6.2 (Zhang et al. 2012), supporting the inference that EAT-17 is a specific GAP to regulate RAB-6.2 activity in those tissues.
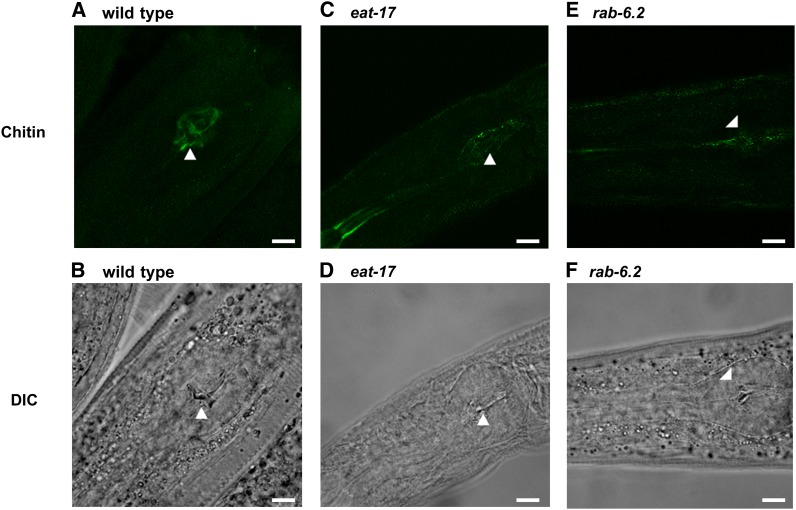
Chitin deposition is superficially intact in eat-17 and rab-6.2 mutants
Chitin provides mechanical support to exoskeletons in many invertebrate animals. In yeast, proper chitin deposition is essential and strictly regulated by controlling the trafficking of its synthase, chs3p (Schorr et al. 2001). Because chitin is one of the components of the grinder and its synthase is regulated by trafficking, we stained the grinders of eat-17 and rab-6.2 mutants for chitin to examine if chitin deposition is misregulated in these mutants. Despite the global disorganization of the grinder, the overall chitin deposition was indistinguishable among eat-17, rab-6.2, and wild-type (Figure 5, A–F). However, when grown at a low temperature, eat-17 mutants occasionally have large refractory bodies in the terminal bulbs that secrete the components of the grinder (Figure 3D). Interestingly, at least in appearance, those bodies share similar refractile characteristics with that of grinder. These results may suggest that the materials trafficked by RAB-6.2 from terminal bulb could be the components of the grinder and that the refractile bodies in eat-17 could be these components, accumulating in the muscle because of the trafficking defect.
Figure 5.
Chitin deposition in the grinders of eat-17 and rab-6.2 mutants is normal. (A and B) Chitin staining of grinder from wild type and corresponding DIC image. (C and D) From eat-17 mutants and corresponding DIC image. (E and F) From rab-6.2 mutants and corresponding DIC image. Arrowheads indicate the position of the grinder. Bars, 10 μm.
Discussion
In this study we mapped and characterized eat-17, a new GTPase-activating protein gene in C. elegans. Using two different approaches, RNA interference and yeast two hybrid, we showed that EAT-17 interacts with RAB-6.2, a conserved small G protein regulating membrane trafficking from Golgi (Del Nery et al. 2006; Girod et al. 1999). The fact that both approaches identify only RAB-6.2 as a substrate of EAT-17 among 25 tested Rabs strongly suggests EAT-17 is a specific GAP for RAB-6.2. Both eat-17 and rab-6.2 mutants show identical defects in their grinders, the part of the feeding organ that crushes food, and which, therefore, is essential for rapid growth and normal development. A new grinder replaces the old one during every molt, implying that the grinder components needed to be transported during every molt to build a new one. Our observations that a mutation in rab-6.2, which functions in cargo trafficking, and a mutation in its GAP protein EAT-17 show their most striking phenotypes in the grinder, suggest that precise regulation of trafficking of grinder components is critical to build a functional grinder.
Although we do not know the identities of the cargos in RAB-6.2 targeted vesicles, we found that eat-17 mutants grown at 15° have refractory bodies in the terminal bulbs. The bodies appear similar (i.e., highly refractile) to the grinder. This suggests that those bodies may be grinder components accumulating in the muscle because of slow trafficking at low temperature. In fact, Chotard et al. (2010) showed that the mutant of tbc-2, which encodes another RabGAP in C. elegans, accumulates enormous RAB-7-positive late endosome in the intestine containing refractile materials. These results strongly suggest that the refractile bodies can be a common phenotype in endosomal trafficking mutants and that EAT-17 regulates endosomal trafficking.
Small G proteins are substrates of GAPs for their fast turnover. The importance of GAPs is demonstrated in many examples such as the oncogene Evi5, which is homologous to EAT-17 (Liao et al. 1997; Dabbeekeh et al. 2007). Only one other GAP for Rab6 has been identified so far, GAPCenA, which also interacts with Rab6 through a coiled-coil domain. The requirement of the coiled-coil domain for interaction appears to be unique for the Rab6 GAPs. Studies have shown that a class of Rab6 effector, Golgins, interacts with Rab6’s through their coiled-coil domains to direct vesicle capture and sorting within the Golgi. It is possible that Rab6’s bind to their GAPs and effectors through a similar mechanism mediated by coiled-coil domains (Sinka et al. 2008).
Our discovery of EAT-17 and its specificity toward RAB-6.2 strongly suggest that there are specific GAPs yet to be discovered for RAB-6.1, the only other Rab6 in C. elegans, sharing 81% amino acid sequence identity with RAB-6.2. rab-6.1 and rab-6.2 are partially redundant; rab-6.1(RNAi) worms are superficially normal, and rab-6.2(RNAi) worms have defects only in the grinder. Knockout of both genes, however, produces gross defects in molting and growth. The fact that EAT-17 does not interact with RAB-6.1 to regulate grinder formation and the similarity of the eat-17 phenotype to that of the rab-6.2 single mutant or rab-6.2(RNAi) is consistent with the proposed EAT-17 specificity toward RAB-6.2.
Recently Zhang et al. (2012) identified LIN-10 as an RAB-6.2 effector to regulate glutamate receptor recycling in C. elegans neurons. However, we did not see any grinder defect in lin-10 mutants (data not shown). In addition, we could not detect EAT-17 expression in neurons in which LIN-10 interacts with RAB-6.2. Although this does not exclude the possibility that LIN-10 functions as an effector for RAB-6.2 and EAT-17 pathway in other places, LIN-10 does not obviously function in grinder formation. This suggests LIN-10 affects RAB-6.2 function probably specifically in neurons, supporting the notion that differential expression of specific GEFs and GAPs in different places and at different times provides spatial and temporal specificity to the G proteins (Segev and Kahn 2012).
That mutations in a Rab and one of its GAPs cause an identical phenotype demonstrates how critical the precise kinetics of the Rab provided by their GAPs are. Furthermore, this implies that we could identify Rab interactors such as their GAPs by looking for mutants exhibiting phenotypes similar to those of Rab mutants. Among 20 TBC GAP domain-containing proteins, only two have been characterized (Mukhopadhyay et al. 2007; Chotard et al. 2010). Our finding provides an insight that may be useful for discovering more GAPs for these Rabs. In addition, using the Eat phenotype caused by the grinder defect in eat-17 mutants, we could identify other interactors such as RAB-6.2 effectors through genetic screens to isolate suppressors or enhancers of eat-17 mutants.
Supplementary Material
Acknowledgments
Several strains used in these studies were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). We thank Melanie Cobb and Rueyling Lin for DNA constructs and reagents and David Raizen, Jim McKay, Scott Cameron, and Pamela Marshall for invaluable discussions. This work was supported by NIH grant HL46154 (L.A.) and 09SDG2150070 from the American Heart Association (Y.-J.Y.).
Footnotes
Communicating editor: D. I. Greenstein
Literature Cited
- Albertson D. G., Thomson J. N., 1976. The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275: 299–325. [DOI] [PubMed] [Google Scholar]
- Audhya A., Desai A., Oegema K., 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133: 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., Shtonda B. B., 2003. Food transport in the C. elegans pharynx. J. Exp. Biol. 206: 2441–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery, L. and You, Y.J. C. elegans feeding (May 21, 2012), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.150.1, http://www.wormbook.org. [Google Scholar]
- Blumenthal T., 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11: 132–136. [DOI] [PubMed] [Google Scholar]
- Chotard L., Mishra A. K., Sylvain M. A., Tuck S., Lambright D. G., et al. , 2010. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell 21: 2285–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R., Lea K., Blumenthal T., 1995. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA 1: 164–170. [PMC free article] [PubMed] [Google Scholar]
- Cuif M. H., Possmayer F., Zander H., Bordes N., Jollivet F., et al. , 1999. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 18: 1772–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbeekeh J. T., Faitar S. L., Dufresne C. P., Cowell J. K., 2007. The EVI5 TBC domain provides the GTPase-activating protein motif for RAB11. Oncogene 26: 2804–2808. [DOI] [PubMed] [Google Scholar]
- Del Nery E., Miserey-Lenkei S., Falguieres T., Nizak C., Johannes L., et al. , 2006. Rab6A and Rab6A’ GTPases play non-overlapping roles in membrane trafficking. Traffic 7: 394–407. [DOI] [PubMed] [Google Scholar]
- Durkin M. E., Yuan B. Z., Zhou X., Zimonjic D. B., Lowy D. R., et al. , 2007. DLC-1:a Rho GTPase-activating protein and tumour suppressor. J. Cell. Mol. Med. 11: 1185–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A., Storrie B., Simpson J. C., Johannes L., Goud B., et al. , 1999. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat. Cell Biol. 1: 423–430. [DOI] [PubMed] [Google Scholar]
- Grosshans B. L., Ortiz D., Novick P., 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 103: 11821–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H., Tall G. G., Horazdovsky B. F., 1999. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J. Biol. Chem. 274: 15284–15291. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R., 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59. [DOI] [PubMed] [Google Scholar]
- Houalla T., Shi L., van Meyel D. J., Rao Y., 2010. Rab-mediated vesicular transport is required for neuronal positioning in the developing Drosophila visual system. Mol. Brain 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmin B. J., Goud B., Camus G., Cartaud J., 1992. The low molecular weight guanosine triphosphate-binding protein Rab6p associates with distinct post-Golgi vesicles in Torpedo marmorata electrocytes. Neuroscience 49: 849–855. [DOI] [PubMed] [Google Scholar]
- Jordens I., Marsman M., Kuijl C., Neefjes J., 2005. Rab proteins, connecting transport and vesicle fusion. Traffic 6: 1070–1077. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Liao X., Du Y., Morse H. C., 3rd, Jenkins N. A., Copeland N. G., 1997. Proviral integrations at the Evi5 locus disrupt a novel 90 kDa protein with homology to the Tre2 oncogene and cell-cycle regulatory proteins. Oncogene 14: 1023–1029. [DOI] [PubMed] [Google Scholar]
- Martinez O., Schmidt A., Salamero J., Hoflack B., Roa M., et al. , 1994. The small GTP-binding protein rab6 functions in intra-Golgi transport. J. Cell Biol. 127: 1575–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O., Antony C., Pehau-Arnaudet G., Berger E. G., Salamero J., et al. , 1997. GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 94: 1828–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Pan X., Lambright D. G., Tissenbaum H. A., 2007. An endocytic pathway as a target of tubby for regulation of fat storage. EMBO Rep. 8: 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdam F. J., Echard A., Croes H. J., van den Hurk J. A., van de Vorstenbosch R. A., et al. , 2000. The small GTPase Rab6B, a novel Rab6 subfamily member, is cell-type specifically expressed and localised to the Golgi apparatus. J. Cell Sci. 113(Pt 15): 2725–2735. [DOI] [PubMed] [Google Scholar]
- Pamonsinlapatham P., Hadj-Slimane R., Lepelletier Y., Allain B., Toccafondi M., et al. , 2009. P120-Ras GTPase activating protein (RasGAP): a multi-interacting protein in downstream signaling. Biochimie 91: 320–328. [DOI] [PubMed] [Google Scholar]
- Raizen D., Song B. M., Trojanowski N., You Y. J., 2012. Wormbook, Methods for measuring pharyngeal behaviors. Available at http://www.ncbi.nlm.nih.gov/pubmed/23255345. [DOI] [PMC free article] [PubMed]
- Ruvkun, G., and M. Finney, 2005 Antibody staining of formaldehyde-fixed animals. Available at http://www.wormatlas.org/EMmethods/Antibodystaining.htm in Wormatlas, edited by D. Hall. WormBook.
- Schorr M., Then A., Tahirovic S., Hug N., Mayinger P., 2001. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr. Biol. 11: 1421–1426. [DOI] [PubMed] [Google Scholar]
- Segev N., Kahn R. A., 2012. Kinetic and cell-based analyses of GTPase regulators. Cell. Logist. 2: 138–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., et al. , 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Sinka R., Gillingham A. K., Kondylis V., Munro S., 2008. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J. Cell Biol. 183: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Hodgkin J. G., 1988. Methods, pp. 587–606 in The Nematode Caenorhabditis elegans, edited by Wood W. B. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Tanabe K., Kon S., Natsume W., Torii T., Watanabe T., et al. , 2006. Involvement of a novel ADP-ribosylation factor GTPase-activating protein, SMAP, in membrane trafficking: implications in cancer cell biology. Cancer Sci. 97: 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uytterhoeven V., Kuenen S., Kasprowicz J., Miskiewicz K., Verstreken P., 2011. Loss of skywalker reveals synaptic endosomes as sorting stations for synaptic vesicle proteins. Cell 145: 117–132. [DOI] [PubMed] [Google Scholar]
- von Lieven A. F., 2003. Functional morphology and evolutionary origin of the three-part pharynx in nematodes. Zoology (Jena) 106: 183–201. [DOI] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H., 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- You Y. J., Kim J., Raizen D. M., Avery L., 2008. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 7: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., McBride H., 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2: 107–117. [DOI] [PubMed] [Google Scholar]
- Zhang D., Isack N. R., Glodowski D. R., Liu J., Chen C. C., et al. , 2012. RAB-6.2 and the retromer regulate glutamate receptor recycling through a retrograde pathway. J. Cell Biol. 196: 85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.