Abstract
Dynamic regulation of chromosome structure and organization is critical for fundamental cellular processes such as gene expression and chromosome segregation. Condensins are conserved chromosome-associated proteins that regulate a variety of chromosome dynamics, including axial shortening, lateral compaction, and homolog pairing. However, how the in vivo activities of condensins are regulated and how functional interactors target condensins to chromatin are not well understood. To better understand how Drosophila melanogaster condensin is regulated, we performed a yeast two-hybrid screen and identified the chromo-barrel domain protein Mrg15 to interact with the Cap-H2 condensin subunit. Genetic interactions demonstrate that Mrg15 function is required for Cap-H2-mediated unpairing of polytene chromosomes in ovarian nurse cells and salivary gland cells. In diploid tissues, transvection assays demonstrate that Mrg15 inhibits transvection at Ubx and cooperates with Cap-H2 to antagonize transvection at yellow. In cultured cells, we show that levels of chromatin-bound Cap-H2 protein are partially dependent on Mrg15 and that Cap-H2-mediated homolog unpairing is suppressed by RNA interference depletion of Mrg15. Thus, maintenance of interphase chromosome compaction and homolog pairing status requires both Mrg15 and Cap-H2. We propose a model where the Mrg15 and Cap-H2 protein–protein interaction may serve to recruit Cap-H2 to chromatin and facilitates compaction of interphase chromatin.
Keywords: condensin, homolog pairing, Mrg15, chromosome structure, transvection
CHROMOSOME structure is highly dynamic in proliferating cells as chromatin states must accommodate repeated rounds of replication, condensation, segregation, and decondensation. Although dramatic changes in chromosome morphology are usually associated with condensation of chromosomes in mitosis, dynamic three-dimensional (3D) spatial organization of interphase chromosomes is also thought to be important for gene regulation (Belmont 2006; Jackson 2010; Rajapakse and Groudine 2011). In some cell types, interphase chromosomes can maintain a Rabl conformation, while others arrange interphase chromosomes into discrete subnuclear compartments known as chromosome territories (Cremer and Cremer 2006; Lieberman-Aiden et al. 2009; Rajapakse et al. 2009). Recent evidence suggests that compaction of interphase chromosomes is sufficient to drive chromosome territories in Drosophila polyploid cells and that this is achieved through the activities of condensin II (Hartl et al. 2008b; Bauer et al. 2012). Condensin II activity is similarly required for axial compaction of mitotic chromosomes in a variety of systems (Shintomi and Hirano 2011; Green et al. 2012), and the regulation of mitotic condensin activity has been extensively studied (Bazile et al. 2010; Cuylen and Haering 2011). How condensin activity is regulated in interphase cells to modulate global chromosome organization remains unclear.
A characteristic of interphase chromosome organization is that there are extensive interactions between different chromosomes even though they are organized into globular territories (Sanyal et al. 2011). These trans-interactions can occur between homologous or nonhomologous sequences to form local functional compartments or nuclear bodies (Sutherland and Bickmore 2009; Dundr 2012). In Drosophila, allelic interactions are favored due to extensive pairing of homologs in somatic cells, and the mechanisms through which this pairing is regulated have only recently been revealed (Bosco 2012). In addition to its chromosome compaction activity, condensin II also has been shown to be a potent antagonist of homologous chromosome pairing in somatic cells and in male meiosis (Hartl et al. 2008a,b; Bateman et al. 2012; Bauer et al. 2012; Joyce et al. 2012). That condensin II, and in particular the Cap-H2 condensin subunit, is important for functional trans-interactions is evidenced by Drosophila mutants that enhance transvection (Hartl et al. 2008a). Transvection is a specific type of pairing-sensitive process in interphase cells, which was first described by Ed Lewis in the 1950s (Lewis 1954). Transvection occurs when a regulatory site on one allele activates or represses the transcriptional state of its homologous allele (Kennison and Southworth 2002). This process is thought to be dependent on the proximity of the two homologous chromosomes in 3D space and therefore can be affected by chromosomal movements altering the homologs’ proximity to each other. Trans-activation is presumed to occur by the productive interactions of enhancers and promoters on two different homologous chromosomes (Wu and Morris 1999; Kennison and Southworth 2002). Trans-repression has also been observed in Drosophila. In the case of the Drosophila bwD mutation, a 2-Mb insertion of heterochromatic repeats functions to physically move the normally euchromatic bw+ allele to a heterochromatic environment via trans-chromosomal interactions between the two bw allelic chromosomal regions (Henikoff and Dreesen 1989). Both trans-activation and trans-repression are similar in that both require homolog pairing to mediate regulation of gene expression of one allele by a second allele. Similarly, both trans-activation and trans-repression can also be hindered by chromosomal rearrangements that are thought to inhibit long/contiguous stretches of DNA homology along homologs. In mammalian systems, interchromosomal interactions are associated with gene regulation (Spilianakis et al. 2005; Lomvardas et al. 2006; Takizawa et al. 2008) and in some cases may explain sporadic reoccurring chromosomal translocations (Roix et al. 2003; Soutoglou et al. 2007). The underlying molecular mechanisms of these and other examples of chromosomal structural reorganization and movements in interphase cells are not well understood. In the Drosophila system, it has been proposed that the condensin II subunit, Cap-H2, provides a strong antipairing activity that normally antagonizes transvection (Hartl et al. 2008a). This condensin antipairing activity has also been shown in cultured Drosophila cells (Bateman et al. 2012; Joyce et al. 2012; Buster et al. 2013). A recent study showed that high levels of homolog pairing is maintained in interphase by active destruction of the Cap-H2 protein through the SCFSlimb ubiquitin E3-ligase (Buster et al. 2013). Because RNA interference (RNAi) depletion or mutations of Cap-H2 lead to increased homolog pairing, it has been proposed that low levels of Cap-H2 protein in interphase nuclei must be important for modulating pairing status (Hartl et al. 2008a; Bateman et al. 2012; Bauer et al. 2012; Joyce et al. 2012; Buster et al. 2013). However, how Cap-H2 is activated in interphase cells to oppose homolog pairing has not been studied. Moreover, whether condensins play any antipairing function in systems other than Drosophila is not known. It has been proposed that the axial compaction activity provided by condensin II is sufficient for its antipairing activity by sequestering sequences into interchromosomal globules and thus indirectly antagonizing trans-interactions between homologs and heterologous chromosomes (Hartl et al. 2008a; Bauer et al. 2012).
Condensin protein complexes were originally identified as having mitotic chromosome condensation activity in vitro (Hirano et al. 1997). Subsequent work has shown that condensins also play diverse roles in interphase chromosomes (Wood et al. 2010; Zaidi et al. 2010). Both condensin I and II contain two structural maintenance of chromosomes subunits, SMC2 and SMC4, that are highly conserved and contain ATPase domains (Hirano and Hirano 2006; Hirano 2006). Mammalian condensin I contains Cap-H, Cap-D2, and Cap-G while condensin II contains Cap-H2, Cap-D3, and Cap-G2 (Ono et al. 2003; Yeong et al. 2003). Interestingly, a Drosophila Cap-G2-encoding gene has not been identified. Condensin I and II do not completely overlap in function as it has been shown that condensin II contributes to axial shortening of chromosomes whereas condensin I promotes lateral compaction (Shintomi and Hirano 2011; Green et al. 2012). Similarly, Drosophila condensin II has recently been shown to drive axial shortening and unpairing of interphase polyploid chromosomes (Bauer et al. 2012). In cultured Drosophila cells, this antipairing activity has been shown to be dependent on condensin II-specific subunits but not condensin I-specific subunits (Joyce et al. 2012; Buster et al. 2013).
To better understand how Cap-H2 may be targeted to chromatin and its activity regulated, we wanted to take a nongenetic approach to uncover as-yet-unidentified Cap-H2-interacting proteins. Such novel interacting proteins may serve to modulate in vivo condensin activities and/or recruit condensin activity to local regions of the genome. We first performed a yeast two-hybrid screen to identify candidates that physically interacted with the Drosophila Cap-H2 protein. We show that the Drosophila homolog of the human Mortality Factor 4 (Morf4), Mrg15, was identified as physically and genetically interacting with Cap-H2.
Materials and Methods Yeast two-hybrid complementary DNA expression library screening
Total RNA was extracted from ovaries of the Drosophila melanogaster strain cn bw sp (Bloomington Stock Center #4455) using Trizol reagent (Invitrogen). Poly-A+ RNA was enriched using the Poly-ATtract mRNA Isolation System (Promega). Subsequent complementary DNA (cDNA) library construction and screening was performed using BD Matchmaker Library Construction and Screening Kits (BD Biosciences-Clontech). Briefly, poly(A) RNA was used to synthesize first-stranded cDNA with CDS III oligo(dT) primer and the BD SMART III primer, and the synthesized first-strand cDNA molecules were flanked at 5′ and 3′ ends by BD SMART III and CDS III anchors, respectively. The cDNA was amplified by PCR with primers of BD SMART III and CDS III anchors to generate a double-stranded cDNA library. The cDNA library was transformed into yeast strain AH109 together with linear vector pGADT7-Rec and Cap-H2 bait construct in vector pGBKT7 to screen the library according to the kit manual. The transformed yeast cells were selected on standard defined medium lacking adenine, histidine, leucine, and tryptophan. The number of total possible transformants was calculated by plating dilution aliquots of each transformation reaction on double-dropout plates lacking leucine and tryptophan, which merely selected for the presence of the bait (pGBKT7) and prey (pGADT7-Rec) plasmids; in this case, growth was not dependent on two-hybrid-interacting proteins. Individual colonies that supported growth on quadruple dropout plates were collected and restreaked on selective media, and putative cDNA inserts were PCR-amplified with primers of BD SMART III and CDS III anchors, as above. These PCR products were transformed back into AH109 together with linear vector pGADT7-Rec and Cap-H2 bait construct in vector pGBKT7, as above. PCR products that gave high levels of transformants on quadruple dropout media were analyzed further by PCR amplification of three different clones to confirm single insert size and by DNA sequencing. Clones with more than one insert were retransformed into yeast as above, and clones that could not be sequenced were discarded. Sequences mapping to Drosophila open reading frames were identified by BLAST searches.
S2 cell transfection, protein extraction, and immunoprecipitations
Mrg15 and Cap-H2 cDNAs were cloned in pMT/V5-His-TOPO (Invitrogen), which has metal-inducible Drosophila methallothionein (MT) promoter to drive gene expression. To make a C-terminal-tagged pMT > Cap-H2-EGFP, first EGFP was cloned into pMT using EcoRI to NotI restriction sites. Cap-H2 cDNA was PCR-amplified using primers CGGGGTACCatggagcgggttttgcc and CCGGAATTCcttcaggcgggctgtcg, where the lowercase nucleotides are Cap-H2 sequences, digested with KpnI and EcoRI and cloned in frame to EGFP. Drosophila S2 cells were transfected using an Amaxa Nucleofector 2b (Lonza) with 2 μg total plasmid DNA. For immunoprecipitations, 0.25 mg of mouse monoclonal anti-V5 antibody (Invitrogen) was bound to equilibrated ProteinG-coupled Sepharose (Sigma-Aldrich) by gently rocking overnight at 4° in 0.2 M sodium borate. For GFP immunoprecipitations, GFP-binding protein (GFPBP) (Rothbauer et al. 2008) was fused to the Fc domain of human IgG (pIg-Tail) (R&D Systems), tagged with His6 in pET28a (EMD Biosciences), expressed in Escherichia coli, and purified on Talon resin (Clontech) according to the manufacturer’s instructions. GFPBP was bound to ProteinA-coupled Sepharose, cross-linked to the resin using dimethylpimelimidate, and rocked for 1 h at 22°, and the coupling reaction was quenched in 0.2 M ethanolamine (pH 8.0) and rocked for 2 h at 22°. Antibody or GFPBP-coated beads were washed 3× with 1.5 ml of cell lysis buffer (CLB; 100 mM Tris, pH 7.2, 125 mM NaCl, 1 mM DTT, 0.1% Triton X-100, 0.1 mM PMSF). Transfected S2 cells were induced to express EGFP, V5, Mrg15-V5, or Cap-H2-EGFP with 0.35 mM CuSO4. After 24 h, cells were lysed in 0.5 ml of CLB, clarified by centrifugation, and then diluted to 2–5 mg/ml in CLB. Antibody-coated beads or GFPBP crosslinked beads were mixed with lysate for 40 min at 4°, washed 3× with CLB, and then boiled in Laemmli sample buffer. Cap-H2-EGFP was detected on immunoblots with mouse monoclonal anti-GFP (JL-8 Clontech). In all cases, 25 μl of ProteinG beads with either a saturating amount of cross-linked GFPBP or preconjugated with 0.25 mg of anti-V5 was used. For immunoblots, anti-V5 was used at 1:500 dilution.
S2 cell RNAi and chromatin fractionation
Double-stranded RNA (dsRNA) was made to a noncoding region of the pBlueScript SK plasmid (control) and to Mrg15 sequences as follows: control pBlueScript SK DNA was PCR-amplified using primers 5′-TAATACGACTCACTATAGGG ATGGATAAGTTGTCGATCG-3′ and 5′-TAATACGACTCACTATAGGG ACCAGGTTCACATGCTTGCG-3′. Mrg15 DNA was amplified from Oregon R genomic DNA using primers 5′-TAATACGACTCACTATAGGG ATCTCGTGCCTCGACGC-3′ and 5-TAATACGACTCACTATAGGG CACTGGTCGATTTCACGG-3′. The underlined nucleotides denote the T7 RNA polymerase promoter sequence, and dsRNA was made using the Ribomax Large Scale RNA Production System (Promega) according to manufacturer instructions. S2 cells were transfected by Amaxa nucleofection (Lonza) according to manufacturer instructions with 2 μg pMT-Cap-H2-V5 and treated every other day for a total of 6 days with 10 μg of dsRNA directed against pBlueScript SK as a control or against Mrg15 messenger RNA (mRNA) sequence. Induction of Cap-H2-V5 was done on day 5 using 1 mM final concentration of CuSO4. Cells were harvested and 20 μg of total cell lysate protein was analyzed by Western blot using mouse anti-V5 (Invitrogen) at 1:500, rabbit anti-Mrg15 at 1:500 (Kusch et al. 2004), and mouse anti-Lamin Dm0 at 1:1000 (ADL84.12-c, Developmental Studies Hybridoma Bank); each antibody detects only one major band (Supporting Information, Figure S5). Cells treated with dsRNA targeting Mrg15 exhibit >75% depletion of endogenous Mrg15 protein, relative to nuclear Lamin (Figure S5). This was consistent through four biological replicates. Cells were prepared into cytoplasmic, nuclear, and chromatin fractions as previously described (Wysocka et al. 2001). Total protein recovered from each fraction was quantified by Bradford’s assay, 20 μg of protein from each fraction was immunoblotted for Cap-H2-V5 or Cap-H2-EGFP and Lamin, and specific bands were quantitated by densitometry after background subtraction using ImageJ. The levels of chromatin-bound epitope-tagged Cap-H2-V5 or Cap-H2-EGFP for control RNAi and Mrg15 RNAi were calculated with the following formula: [(chromatin fraction Cap-H2/chromatin fraction Lamin)/whole-cell lysate Cap-H2/whole-cell lysate Lamin) for Mrg15 dsRNA-treated cells]/[(chromatin fraction Cap-H2/chromatin fraction Lamin)/whole-cell lysate Cap-H2/whole-cell lysate Lamin) for control dsRNA-treated cells]. Four biological replicates were done, and an average fold enrichment was calculated. P-value was calculated by two-tailed t-test assuming unequal variance in Microsoft Excel.
Chromosome length measurements in Kc167 cells
dsRNA depletion of Mrg15 and Cap-H2 was done as described above for S2 cells. Primers for Cap-H2 RNAi were as follows (underlined DNA are T7 RNA polymerase sequences): 5′-TAATACGACTCACTATAGGG ACCGGAGAAAAACGAGCGCAGGCC and 5′-TAATACGACTCACTATAGGG GGGATCCACTCTCGTGC. Image z-stacks were acquired with a Nikon A1RSi confocal microscope with Plan Apo 100× oil immersion objective lens. Three-dimensional distances between pairwise probe sets were made using the 3D distance measurement tool in Nikon elements 4.0 software package. Pairwise distance measurements were made by marking the centroid of each respective FISH focus. Statistical analyses were performed using a Student’s t-test in Microsoft Excel. BAC clones (CHORI BACPAC Resources) for FISH were as follows: X1, BACR30C13 and BACR18F10; X2, BACR20K01 and BACR35A18; X3, BACR11C13 and BACR07F15; 2L(1) BACR30M19 and BACR29P12; 2L(2), BACR15P08 and BACR14I17. Table S4 reports the number of nuclei sampled and other details for each experimental treatment.
Salivary gland suppression
We used two Drosophila stocks in which we could observe LacI-GFP spot localization: one line, we refer to as the “spots” line, is homozygous for 256× LacO arrays at cytological location 60F, hs83 > LacI-GFP on the second chromosome and Cap-H2EY09979 (Bloomington Stock Center #17627), Hsp70 > Gal4 in which both the LacI-GFP and Gal4 are under heat-shock control. The second line, which we refer to as “donut,” is identical to the “spots” line except it does not contain Cap-H2EY09979 and thus Cap-H2 cannot be overexpressed. The UAS > Mrg15-RNAi line is Vienna Drosophila RNAi Center (VDRC) #v43802. The Mrg15j6A3 (Bloomington Stock Center #10290) is a P-element insertion. Crosses were done on yeast/molasses/cornmeal media and kept at 25°. After 5–6 days, the cross larvae were heat-shocked at 32° overnight. Third instar larvae were dissected in PBS with 0.1% Triton-X (PBT), and glands were fixed for 10 min in PBS, 4% formaldehyde. Glands were rinsed three times with PBT and stained 10 min with DAPI (0.1 μg/μl final) in 1 ml of PBS with 0.005% Triton-X followed by two 10-min washes in PBT. Initial quantification of the percentage of polytene was done visually by DAPI staining using a Nikon Eclipse E800 40× objective (Figure S1, Table S2). Further imaging was done using a Zeiss LSM 510 Meta Confocal at 63× with 1-μm slices, and measurements of the number of GFP spots and distance were done using the LSM image browser software (release 4.0). At least three different glands (biological replicates) were imaged for the GFP spot counting. In cases where the number of GFP spots and distance between spots was measured, the same nuclei were used for both calculations. All Student t-tests were done using two-tailed and two samples with equal variance as the parameters in Microsoft Excel.
Quantitative PCR for Mrg15 mRNA levels was done by extracting RNA with Qiagen RNeasy Mini Kit from heat-shocked larvae resulting from “spots” crossed to UAS > Mrg15-RNAi line (VDRC #43802) or “spots” crossed to Oregon-R (control). Primers for Mrg15 were 5′-GAAAATAAAATGGGAGAAGTAAAACC-3′ and 5′-GGTTTTGTTTTCAGAACCTTGG-3′. First-strand cDNA was made with Verso cDNA synthesis kit (Thermo Scientific #AB-1453/B). mRNA levels were amplified using iQ SYBR Green Supermix (Bio-Rad) and Stratagene Mx3005P (Agilent Technologies) along with MxPro qPCR software. Primers for RpL32 were 5′-CCTTCCAGCTTCAAGATGACC-3′and 5′-ACTCTGTTGTCGATACCCTTGG-3.′ Levels of Mrg15 mRNA for each sample were calculated relative to RpL32 as follows: (1 + Mrg15 efficiency)(ΔCT Mrg15 (control – treated))/(1 + RpL32 efficiency)(ΔCt Rpl32 (control – treated)), as recommended by the manufacturer (Bio-Rad).
Nurse-cell FISH and spot quantitation
Strains used:
P-element hypomorph for Smc4 (gluon) Smc4k08819/CyO (Bloomington Stock Center #10831; in the text we refer to this allele as Smc4k08819) and the Smc488-82/CyO have been described (Cobbe et al. 2006); the Cap-H2 mutation Cap-H2Z3-0019 and Df(3R)Exel6195/TM6 B deficiency deleting the Cap-H2 locus was described (Hartl et al. 2008a); the deficiency deleting Mrg15 was Df(3R)BSC741 (Bloomington Stock Center #26839), which has breakpoints at 88E8 and 88F1.
FISH probe preparation:
BAC clones (CHORI BACPAC Resources) were used for the detection of the 34D region (RP98-16P12, RP98-48E2, and RP98-30I21), the Ubx region (RP98-24L18 and RP98-28H1), and the CapH 2 region (RP98-29B06). DNA was labeled and tissues were prepared for hybridizations as previously described (Hartl et al. 2008a; Bauer et al. 2012).
Microscopy and spot analysis:
Stage 6/7 egg chambers were imaged on a Zeiss LSM 510 Meta Confocal with an objective of 40× with z-slices of 1 μm. Spots were manually counted for each probe.
Nurse-cell RNAi depletion of Mrg15 and polytene unpairing
The maternal α-tubulin67C driver was used to express female germline-specific Gal4VP16 (Matα4-GAL-VP16; Bloomington Stock Center 7062 and 7063) (Hacker and Perrimon 1998). Virgin Matα4-GAL-VP16 females were crossed to males from a TRiP line capable of expressing an RNA-hairpin targeting the Mrg15 transcript in the germline (Bloomington Stock Center 35241: y[1] sc[*] v[1]; P[y[+t7.7]v[+t1.8]=TRiP.GL00128] attP2/TM3, Sb[1]). One- to 3-day-old Sb+ F1 females were collected and fattened on yeast paste for 2 days at 25°. Ovaries were dissected as described above for FISH and processed for DNA FISH and anti-Mrg15 immunofluorescence. As controls, F1 progeny from Matα4-GAL-VP16 females crossed to Oregon-R or a different TRiP line targeting the Set2 gene (Bloomington Stock Center 33076): y[1] sc[*] v[1]; P[y[+t7.7] v[+t1.8]=TRiP.HMS00583]attP2). Endogenous Mrg15 protein germline-specific depletion was confirmed by whole-mount immunofluorescence of egg chambers as follows: Mrg15-TRiP and control ovaries were incubated with rabbit polyclonal anti-Mrg15 serum at 1:100 and goat anti-rabbit Alexa Fluor 488 (Invitrogen) at 1:200. Image z-stacks (0.5 μm z-step size) were acquired with a Nikon A1RSi confocal microscope with Plan Apo 60× oil immersion objective lens, and relative amounts of protein were measured by drawing a line across a two-dimensional confocal z-slice of an entire stage 5/6 egg chamber. Gray-scale pixel intensity values across two follicle cells and two nurse cells (∼50–60 μm total distance) for DAPI and anti-Mrg15 were acquired with the National Institutes of Health ImageJ analysis tool “Plot Profile.” Relative protein levels in RNA-hairpin-expressing and control egg chambers were normalized to the somatic follicle cell Mrg15 immunofluorescence signal (the Matα4-GAL-VP16 driver is germline specific, and therefore protein levels in somatic follicle cells are unaffected in both control and TRiP lines).
Transvection assays
These assays were done as previously described (Hartl et al. 2008a), and stocks were a gift from Chao-Ting Wu. Scoring of Ubx phenotypes and yellow pigmentation transvection phenotypes was done blind where multiple individuals were not aware of the genotypes. Briefly, Ubx crosses were done at 25° and brooded twice. The UbxCbx-1 Ubx1/TM6B Tu, Hu, e was crossed to Oregon-R or Mrg15j6A3/TM6B Tu, Hu, e. The rearrangement of UbxCbx-1 Ubx1 (BTD in Figure S3) was #800.7 BTD24/MRS, Sb T(2;3) 50C1/C4; 81F. Wings were chosen from females with no balancers with genotypes: UbxCbx-1 Ubx1/+ and UbxCbx-1 Ubx1/Mrg15j6A3. Class A wings had loss of posterior tissue, typical of the UbxCbx-1 Ubx1/+ background but clearly not wild type; class B had a large loss of posterior tissue and blistering of the wing. See Figure 7, B and C, for representative class A and B wing images. For transvection at the yellow gene, methods and alleles were as previously described (Geyer et al. 1990; Morris et al. 1999; Hartl et al. 2008a). The following crosses were done: First, y82f29/y82f29; Cap-H2Z3-0019/TM6B, Hu females were crossed to w1118/Y; Mrg15j6A3/TM3 Sb males and F1 y82f29/Y; Mrg15j6A3/TM6B, Hu males were collected. These y82f29/Y; Mrg15j6A3/TM6B, Hu males were crossed to females from three different lines: (1) y1#8/y1#8; Cap-H2Z3-0019/TM6B, (2) y1#8/y1#8; Df(3R)Exel6159/TM6B, and (3) y1#8/y1#8. For additional controls, y1#8/y1#8; Cap-H2Z3-0019/TM6B and y1#8/y1#8; Df(3R)Exel6159/TM6B were crossed to y82f29/Y males. For each genotype, between 10 and 15 independent crosses were established, each with two females and two males. Each cross was brooded three times for 3 days, after which parental flies were then discarded. Adult F1 female progeny were collected daily (over a period of 15 days) and held in another vial for 3 days at 25°; then the abdominal stripe pigmentation of flies from each genotype that eclosed on the same day and arose from the same brood was scored relative to each fly. To prevent biasing the scorer, genotypes were concealed until after scoring completion, and females were scored blind by two individuals, as previously described (Hartl et al. 2008a). All female progeny with relevant genotypes were scored and placed into four body-pigmentation classes. Note that only abdominal stripes were considered in pigmentation scoring and not interstripe abdominal cuticle or thoracic cuticle. This pigmentation scale was developed independently of others and should not be compared to those previously described. Class 1 had no/little detectable pigmentation equal to y1#8/y1#8 and y82f29/y82f29 homozygotes (Morris et al. 1999). Class 2 had moderate pigmentation; class 3 had moderate-to-darkly pigmented posterior-most stripes only; class 4 had darkly pigmented posterior stripes and some dark pigmentation of more anterior stripes (for examples, see Figure 8 and details in its legend). Subtle variation within each class was noted. Transvection experiments at the yellow locus were done with an Mrg15j6A3 line (a P-w+ insertion) that was crossed to a w1118 female, and w+ females were backcrossed to w1118 males for seven generations, selecting for w+ female progeny, and thus allowing female meiotic recombination to occur on all chromosomes.
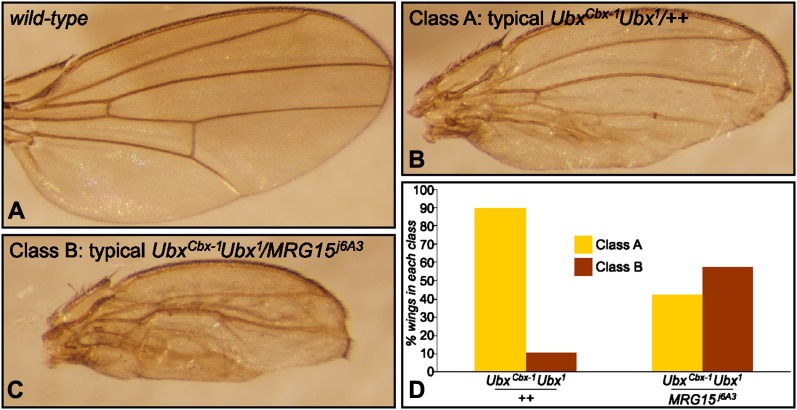
Figure 7.
Mrg15 antagonizes transvection at Ubx. (A) Wild-type wing from Oregon-R. (B) Wing of class A typical of UbxCbx-1 Ubx1/++. (C) Wing of class B typical of UbxCbx-1 Ubx1/Mrg15j6A3. (D) Quantification of phenotypic classes for UbxCbx-1 Ubx1/++ and UbxCbx-1 Ubx1/Mrg15j6A3. n > 50 for both genotypes. P < 10−3 for either class using the χ-square test. See Figure S3 for additional supporting data.
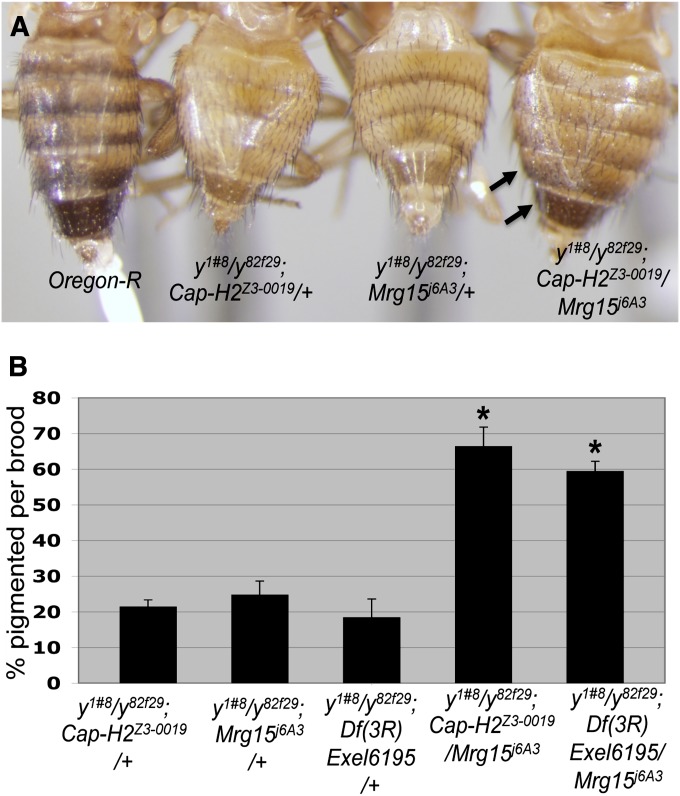
Figure 8.
Mrg15 cooperates with Cap-H2 to antagonize transvection at yellow. (A) Similarly aged females with the following genotypes from left to right: Oregon-R, y1#8/y82f29; Cap-H2Z3-0019/+, y1#8/y82f29;Mrg15j6A3/+ and y1#8/y82f29; Cap-H2Z3-0019/Mrg15j6A3. Arrows point to darkly pigmented posterior stripes. (B) Percentage of all female progeny exhibiting darkly pigmented posterior stripes per brood for the following genotypes left to right: y1#8/y82f29; Cap-H2Z3-0019/+ (n = 53), y1#8/y82f29;Mrg15j6A3/+ (n = 77), y1#8/y82f29;Df(3R)Exel6195/+ (n = 45), y1#8/y82f29; Cap-H2Z3-0019/Mrg15j6A3 (n = 96), and y1#8/y82f29;Df(3R)Exel6195/Mrg15j6A3 (n = 91). Asterisks denote P < 0.001, two-tailed t-test assuming unequal variance.
Results
Cap-H2 and Mrg15 proteins interact
Cap-H2 is required for polytene chromosome unpairing as well as unpairing of homologous sequences in cultured Drosophila cells (Hartl et al. 2008a; Bauer et al. 2012; Joyce et al. 2012; Buster et al. 2013). Although this activity is dependent on other condensin genes, such as SMC4 and Cap-D3, it is not known what other factors may function to recruit and/or activate this condensin activity on chromatin. To identify proteins that interact with Cap-H2 protein, a yeast two-hybrid screen was done using Cap-H2 cDNA fused to the GAL4 DNA-binding domain (GAL4BD) as bait protein. Because the primary phenotypes observed in Cap-H2 mutants are in the ovary, we reasoned that potential regulators of Cap-H2 would be most abundant in the ovarian transcriptome. Therefore, a cDNA library from Drosophila ovarian mRNA was constructed as GAL4-activation domain fusions and was used as prey protein. Of ∼105 total putative cDNA clones that were plated, 124 were able to grow under selection that required a bait–prey interaction. Clone inserts were PCR-amplified using universal vector primers and recloned into the prey vector by homologous recombination in yeast. Of these, 84 clones retested as positive and inserts were sequenced (Table S1). We found that a disproportionate number of clones encoded known or predicted ribosomal proteins (21 clones) and translation regulatory factors (7 clones). In addition, yolk protein 1 (CG2985) was represented by 7 independent clones. We speculate that these interactions may be nonspecific, and they are overrepresented because of the vast abundance of ovarian transcripts encoding protein synthesis, chorion, and yolk proteins.
Two additional genes were represented by three or more Cap-H2-interacting clones: Rack1 (CG7111) was recovered in seven clones while Mrg15 (CG6363) was recovered in three clones (Table S1). In an in vitro protein translation system, Rack1 protein exhibited only weak protein–protein interactions with Cap-H2, and three different Rack1 mutants (Rack11.8, Rack1EY00128, Rack1EE) failed to modify nurse cell polytene phenotypes caused by Cap-H2 and Smc4 mutations (data not shown). Therefore, we did not pursue Rack1 as an interactor of condensin function. An interaction between Cap-H2 and Mrg15 proteins previously was identified by a large-scale protein interaction screen (Giot et al. 2003). Here, we report further analysis of the Cap–H2 interaction with Mrg15.
The Mrg15 gene is found in all metazoans. The Mrg15 homolog in the budding yeast is Eaf3, and the human homolog is Morf4 (Bertram et al. 1999; Bertram and Pereira-Smith 2001; Chen et al. 2010). The yeast Eaf3 protein can bind histone H3 monomethyl, dimethyl and trimethyl lysine-36 as well as H3 trimethyl lysine-4 (Joshi and Struhl 2005). The human Morf4 chromodomain binds to H3 dimethyl and trimethyl lysine-36 (Zhang et al. 2006). The Drosophila Mrg15 protein is enriched at sites of active gene expression (Filion et al. 2010), and it has been reported to bind H3 trimethyl lysine-36 and monomethyl and dimethyl lysine-20 on histone H4 (Moore et al. 2010). The MRG domain is less well characterized, but it has been reported to contain HLH and leucine-zipper motifs, suggesting that the MRG domain may mediate interactions with other proteins (Bertram and Pereira-Smith 2001; Chen et al. 2010). A previously described Mrg15 protein-binding consensus sequence (FxLP) has been determined through MRG domain structure–function studies (Xie et al. 2012). Interestingly, a FKLP sequence exists within all four reported Drosophila Cap-H2 protein isoforms.
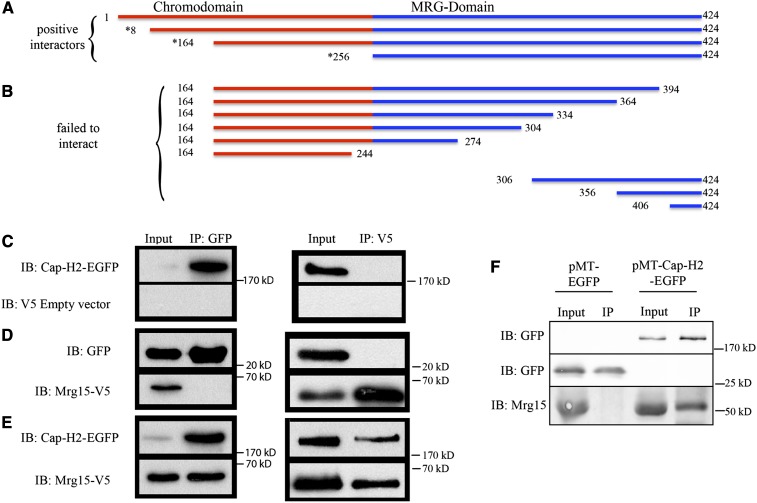
The protein domain illustration in Figure 1A shows the full-length 424-amino-acid protein and portions of Mrg15 that were recovered in three independent clones from the yeast two-hybrid screen. All three of the interacting Mrg15 cDNA inserts contain the C-terminal MRG domain while one clone is lacking the entire N-terminal chromodomain, suggesting that the MRG domain is sufficient for the interaction with Cap-H2. C-terminal deletions were constructed and tested, and these did not exhibit any interaction with the GAL4BD-Cap-H2 bait (Figure 1B and data not shown), further demonstrating that the MRG domain is necessary for a two-hybrid interaction with Cap-H2. These data suggest that amino acids 256–424 are necessary for the Mrg15 protein interaction with Cap-H2.
Figure 1.
Mrg15 protein interacts with Cap-H2. (A) Linear protein map showing Mrg15 (not to scale). The red line shows the chromo-barrel domain and the blue line shows the MRG domain. Numbers represent amino acid positions relative to the full-length 424-amino-acid predicted protein (GenBank accession AAF55161). Three Mrg15 clones were recovered from a yeast two-hybrid screen (asterisk) and the full-length proteins were found to interact. (B) Linear depictions of the constructs that failed to interact with Cap-H2 in the yeast two-hybrid assay. (C–E) Immunoprecipitations (IP) were done with GFP-binding protein (IP:GFP) or anti-V5 (IP:V5), and 10% of IP supernatant from S2 cell extracts were immunoblotted (IB) with anti-GFP (IB:GFP) or anti-V5 (IB:V5). (C) S2 cells cotransfected with pMT-Cap-H2-EGFP and pMT(empty vector). (D) Cells cotransfected with pMT-EGFP and pMT-Mrg15-V5. (E) Cells cotransfected with pMT-Cap-H2-EGFP and pMT-Mrg15-V5. (F, top) Cells transfected with pMT-Cap-H2-EGFP (right) produce a protein band at ∼175 kDa that is immunoprecipitated by GFP-binding protein and detected by anti-GFP (IB:GFP). Cells transfected with pMT-EGFP (left) do not produce a protein band at ∼175 kDa on immunoblots probed with anti-GFP. Middle panel: Cells transfected with pMT-EGFP (left) produce a GFP band at ∼27 kDa, while cells transfected with pMT-Cap-H2-EGFP (right) do not produce an ∼27-kDa protein band. (Bottom) Cells transfected with pMT-EGFP (left) have endogenous Mrg15 protein that does not IP with GFP-binding protein. GFP-binding protein can IP endogenous Mrg15 from cells transfected with pMT-Cap-H2-EGFP (right).
The interaction between Mrg15 and Cap-H2 was tested in cell culture using S2 cells. Both Mrg15 and Cap-H2 cDNA clones were inserted into pMT expression vectors under the control of the metallothionein promoter. pMT-Mrg15-V5 contains a V5-epitope C-terminal tag while the pMT-Cap-H2-EGFP contains a C-terminal EGFP or GFP. Cotransfection of pMT-Cap-H2-EGFP and an empty vector control (pMT with no insert) resulted in a Cap-H2-EGFP band to immunoprecipitate (IP) with recombinant GFPBP (Rothbauer et al. 2008) while EGFP was detected on an immunoblot (IB) with anti-GFP antibody (Figure 1C). The same IP reactions were immunoblotted with anti-V5 antibodies and did not yield detectable Cap-H2-EGFP bands (Figure 1C, bottom). When pMT-EGFP was cotransfected with pMT-Mrg15-V5, GFPBP was able to IP EGFP but not Mrg15-V5 (Figure 1D). In these same extracts, anti-V5 antibodies were able to IP Mrg15-V5 but not EGFP alone (Figure 1D, right). These data show that in S2 cell extracts anti-V5 antibodies cannot IP EGFP and GFPBP cannot IP overexpressed Mrg15-V5 protein. Next we co-transfected pMT-Cap-H2-EGFP with pMT-Mrg15-V5. In these extracts expressing Cap-H2-EGFP and Mrg15-V5, GFPBP was able to IP both Cap-H2-EGFP and Mrg15-V5 (Figure 1E, top). In the reciprocal experiment, anti-V5 antibodies were able to IP both Mrg15-V5 and Cap-H2-EGFP (Figure 1E, bottom).
Because proteins are likely overexpressed from the pMT-plasmid, we tested whether endogenous Mrg15 protein could IP with overexpressed Cap-H2-EGFP. First, we transfected S2 cells with either pMT-EGFP alone or pMT-Cap-H2-EGFP alone. GFPBP immunoprecipitations did not detect any protein bands at ∼175 kDa in the pMT-EGFP only cells (Figure 1F, top left), whereas GFPBP could IP an ∼175-kDa protein band from cells transfected with pMT-Cap-H2-EGFP (Figure 1F, top right). In cells expressing pMT-EGFP alone, anti-GFP immunoblots (IB:GFP) specifically recognize EGFP at ∼27 kDa from a GFPBP IP and do not detect a polypeptide at the predicted ∼175-kDa size for Cap-H2-EGFP (Figure 1F, left). This again demonstrates that anti-GFP immunoblots specifically recognize GFP-tagged proteins only. In cells transfected with pMT-EGFP alone, GFPBP was not able to IP endogenous Mrg15 detected at ∼50 kDa, even though 10% of total input extract used has abundant levels of Mrg15 protein (Figure 1F, bottom left). In cells transfected with pMT-Cap-H2-EGFP, GFPBP was able to IP both Cap-H2-EGFP and endogenous Mrg15 (Figure 1F, bottom right).
Taken together with the yeast two-hybrid interaction these co-immunoprecipitation experiments suggest that Cap-H2 and Mrg15 proteins interact. This observation is consistent with a previously reported large-scale Drosophila yeast two-hybrid screen demonstrating a Cap–H2–Mrg15 protein interaction (Giot et al. 2003).
Mrg15 is required for Cap-H2-mediated salivary gland polytene dispersal
To ask if Mrg15 and Cap-H2 interact in vivo, a functional genetic test was done using salivary gland cells. Wild-type larval salivary gland nuclei are polyploid and contain 1000–2000 copies of each chromosome arranged in register to form a structure called polytene chromosomes where chromatids and homologs are paired in a homology-dependent manner. We previously have shown that Cap-H2 overexpression in the larval salivary gland was sufficient to drive unpairing of polytene chromosomes, and this unpairing activity was completely suppressed by Cap-D3 loss-of-function mutations (Hartl et al. 2008a). We tested whether Mrg15 function was also necessary for this Cap-H2-mediated disassembly of salivary gland chromosomes. The LacO/LacI-GFP system (Vazquez et al. 2002) allows visualization of the pairing status of a single locus (the exogenous LacO array) in the genome. In salivary gland cells expressing wild-type levels of Cap-H2, there is one GFP spot or band because polytene chromosomes have all homologous sequences paired (Hartl et al. 2008a), and transgenic lines carrying UAS > Mrg15-RNAi similarly exhibit one large LacI-GFP spot or polytene band (Figure 2, A–C). In larvae containing Hs > Gal4, UAS > Cap-H2 chromosomes become nonpolytene, and homologs and sister chromatids unpair and result in multiple GFP spots after heat-shock induction (Figure 2, D–F). We previously demonstrated that this Cap-H2 overexpression phenotype could be suppressed by mutations in Cap-D3, suggesting that this polytene unpairing phenotype could be modulated by altering genetic factors (Hartl et al. 2008a). Here, we extend this observation by demonstrating that in vivo RNAi to Cap-D3, Cap-H2, or Smc4 also suppresses the salivary gland Cap-H2 overexpression phenotype (Figure S1, Table S2). To test whether suppression could also be achieved by RNAi, we used UAS > RNAi to other condensins and condensin-interacting genes: RNAi targeting of Cap-G, Cap-H (barren), Smc3, Smc5, and Polo kinase. These transgenic lines gave minimal to partial rescue of the polytene dispersal phenotype (Figure S1), although at present it is unclear how RNAi depletion of Smc3 or Smc5 results in rescue. UAS > RNAi transgenes targeting Cap-D2, Smc1, trithorax (trx), enhancer of zest [E(z)], and Tip60 had little or no significant suppression of the polytene dispersal phenotype (Figure S1). Together, these data support a model where Cap-H2 most likely functions in the context of the condensin complex to disperse polytene chromosomes since knockdown of other condensin subunits produces partial or complete suppression, consistent with previous observations suggesting that this is a condensin complex function (Hartl et al. 2008a; Bauer et al. 2012; Joyce et al. 2012; Buster et al. 2013). More importantly, this demonstrates that ds RNA expression in the salivary gland cells can be an effective assay for Cap-H2-interacting genes. We note that not all UAS-driven hairpin transgenes target efficient depletion of mRNA, and therefore for those genes where little or no effects were observed, we cannot exclude the possibility that these genes were not sufficiently depleted.
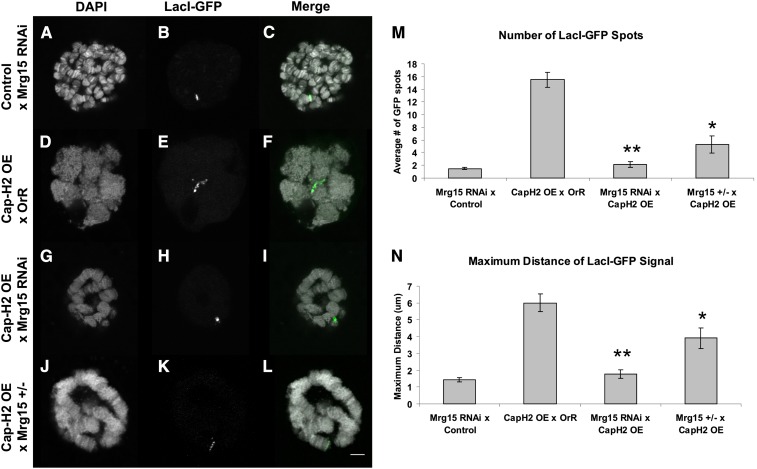
Figure 2.
Mrg15-RNAi suppresses Cap-H2 overexpression phenotypes in salivary glands. (A–C) Single salivary gland nucleus of a control UAS > Mrg15-RNAi/+, Hs > LacI-GFP/+, LacO (60F)/+; Hs > Gal4/+. (D–F) Cap-H2 overexpression (OE) line crossed to Oregon-R resulting in larvae as follows: Hs > LacI-GFP/+, LacO (60F)/+; Hs > Gal4/+, and UAS > Cap-H2EY09979/+. (G–I) Cap-H2 overexpression line crossed to a Mrg15-RNAi line resulting in larvae as follows: UAS > Mrg15-RNAi/+, Hs > LacI-GFP/+, LacO (60F)/+; Hs > Gal4/+, and UAS > Cap-H2EY09979/+. (J–L) Cap-H2 overexpression in an Mrg15j6A3 heterozygous background resulted in larvae as follows: Hs > LacI-GFP/+, LacO (60F)/+; Hs > Gal4/+, UAS > Cap-H2EY09979/+, and Mrg15j6A3/+. The left column (A, D, G, and J) is the DAPI channel. The middle column (B, E, H, and K) is the GFP channel. The right column (C, F, I, and L) is a merge of the two channels with DAPI in white and GFP in green. The white scale bar in L indicates 5 μm. (M) The average number of GFP spots per nucleus and standard error are plotted. One asterisk indicates a P-value of 1.9 × 10−4, and two asterisks indicate a P-value of 3.9 × 10−9 using a two-tailed, equal variance Student t-test. (N) The maximum distance between two GFP signals was calculated, and the average distance with standard error was plotted. Using the Student t-test, one asterisk indicates P = 0.02 and two asterisks indicate P = 1.1 × 10−6. For each genotype, n = 10 nuclei.
We used this Hs > Gal4/UAS>RNAi system to test whether Mrg15 interacts with Cap-H2. When UAS > Mrg15 RNAi alone is crossed to the heat-shock-driven Gal4, the polytene structure is maintained after heat shock, demonstrating that Mrg15 depletion alone does not impact polytene structure (Figure 2, A–C). This was apparent by qualitative DAPI polytene visualization as well as by LacI-GFP tethering to LacO arrays (Figure 2, A–C). By contrast, Cap-H2 overexpression produced a nonpolytene phenotype as indicated by an average of 15 GFP spots (Figure 2, D–E and M). The introduction of UAS > Mrg15 RNAi into the Cap-H2 overexpression line suppressed the nonpolytene phenotype as indicated by one GFP signal (Figure 2, G–I). Using this LacI-GFP system, the number of distinct spots and the maximum distance between the two farthest spots was calculated (Figure 2, M and N). In larvae with Mrg15 RNAi Cap-H2 overexpression, both the number of GFP spots and the maximum distance between spots were reduced significantly compared to Cap-H2 overexpression alone (P < 10−8 and <10−5, respectively). To confirm that this Mrg15 RNAi line effectively depleted Mrg15, we measured Mrg15 mRNA levels by quantitative RT-PCR. We found that the Mrg15 RNAi larvae had Mrg15 mRNA levels that were 50 ± 6% (P = 0.032) that of control larvae not containing the Mrg15 RNAi transgene (relative to RpL32 mRNA levels; see Materials and Methods). This is consistent with a previous report showing that this very same RNAi line can deplete Mrg15 mRNA by 10–36% in whole larvae (Zhang et al. 2010). It is also noteworthy that an Mrg15 overlapping gene, l(3)neo43 (CG14865), shares no sequence homology to the 332-bp predicted RNA hairpin produced by the Mrg15-RNAi line.
Given that a 50% RNAi depletion of Mrg15 mRNA was sufficient to suppress the Cap-H2 overexpression phenotype (Figure 2, G–I), this predicted that reducing the Mrg15 gene dosage to half could also suppress the salivary polytene phenotype. An Mrg15 mutant, Mrg15j6A3, was also crossed into the LacO/LacI-GFP Hs > Gal4, UAS > Cap-H2 overexpression line. Using qualitative polytene DAPI staining, we observed an intermediate degree of suppression (Figure 2, J–L); however, the number of spots was significantly reduced (P < 10−4) as was the maximum distance between GFP foci (P = 0.02). This suggests that the Cap-H2 overexpression phenotype is very sensitive to Mrg15 dosage, and 50% reduction by RNAi or by a mutation in one allele can lead to suppression. Additionally, because both the RNAi transgene and the loss-of-function mutant independently produce similar suppression of the Cap-H2 gain-of-function phenotype, it is unlikely that this suppression is caused by genetic background effects. Therefore, this suggests that Mrg15 function is required in vivo for Cap-H2-mediated salivary gland polytene unpairing.
Mrg15 mutants enhance condensin II partial loss of function in ovarian nurse cells
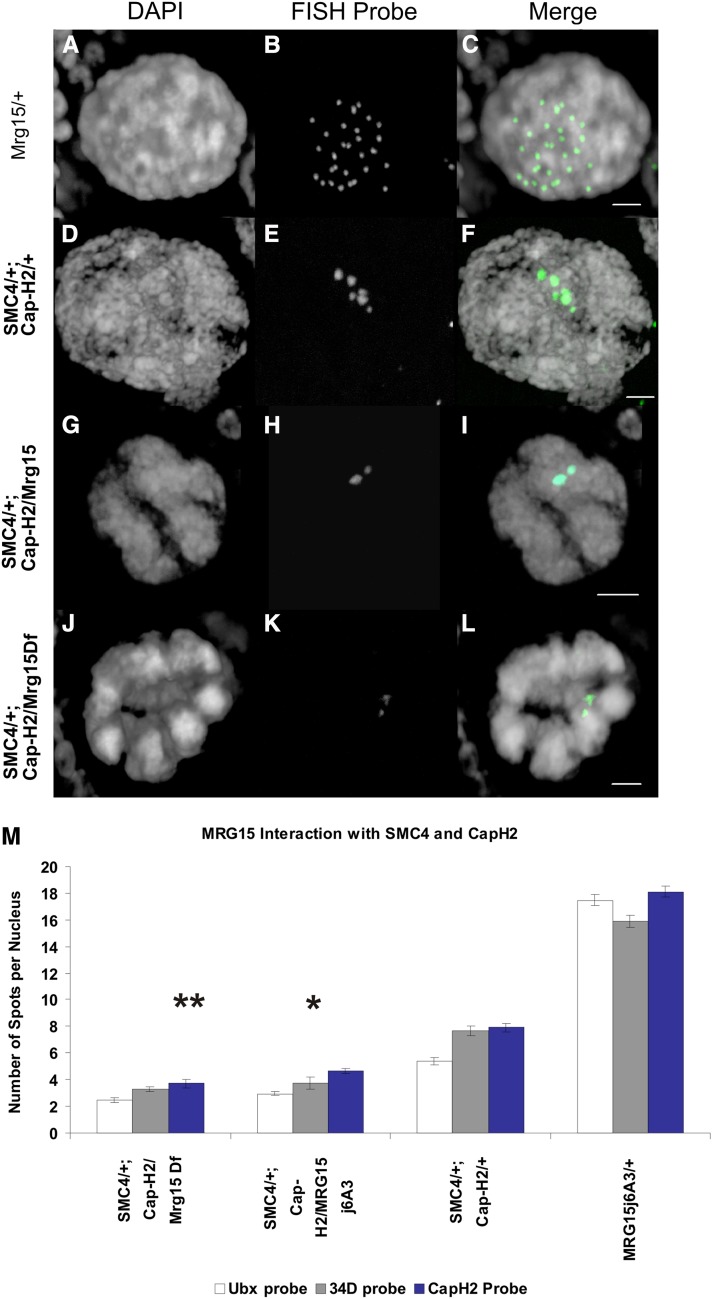
In wild-type egg chambers, polyploid nurse cells undergo a transition from the polytene structure to nonpolytene during stage 5 (Dej 1999; Dej and Spradling 1999). This developmentally regulated chromosome reorganization is dependent on Cap-H2, Smc4, and Cap-D3, and thus we used this polytene-to-nonpolytene transition to test whether Mrg15 was also required. We used FISH with probes spanning ∼350 kb to each of three different genomic locations at the Ubx, Cap-H2, and 34D loci. Confocal 3D images of stage 6/7 egg chamber nurse cells were acquired, and the number of discrete FISH signals in each nurse cell nucleus for each probe is a direct measure of chromatid and homolog pairing within the polytene structure (Dej 1999; Hartl et al. 2008a; Bauer et al. 2012). In Oregon-R stage 6/7 egg chambers, we detected 15–17 spots per wild-type nurse cell, depending on the probe used (Figure S2). Homozygous mutant Cap-H2 nurse cells show complete polytene structure, have only one large detectable FISH spot, and therefore were not useful for testing enhancers of nurse cell polytene structure (Hartl et al. 2008a). To assay for a genetic interaction, we desired a sensitized background that is not completely polytene. Double-heterozygous mutations in the condensin II complex (SMC4k08819/+; Cap-H2Z3-0019/+) give an intermediate phenotype in nurse cells (Hartl et al. 2008a) and therefore were ideal for use in our assay. Furthermore, this double-heterozygous phenotype can be enhanced by an additional heterozygous mutation in the Cap-D3 gene and suppressed by a heterozygous mutation in the Slimb E3-ubiquitin ligase (Hartl et al. 2008a; Buster et al. 2013). Therefore, the SMC4k08819/+; Cap-H2Z3-0019/+double-heterozygous line is ideal for testing genetic interactors. We used this sensitized genetic background to test for interactions between Cap-H2 and Mrg15 in ovarian nurse cells.
If Mrg15 is working with Cap-H2 to unpair polytenes, then a mutation in Mrg15 should further limit condensin function and therefore enhance the SMC4k08819/+; Cap-H2Z3-0019/+ intermediate phenotype. The heterozygous Mrg15j6A3 alone does not affect nurse cell polytene unpairing (Figure 3, A–C) since these nurse cells exhibit the same number of FISH spots as wild-type Oregon-R nurse cells (Figure S2). In the sensitized SMC4k08819/+; Cap-H2Z3-0019/+ condensin mutant line, nurse cells exhibit five to eight spots per nucleus (Figure 3, D–F). By contrast, when the Mrg15j6A3 mutant was crossed into the double-heterozygous condensin mutant, there was a decrease in the number of spots, indicating a more polytene-like structure (Figure 3, G–I). For each of the three FISH probes, there was a significant difference between the double heterozygote (Smc4k08819/+; Cap-H2Z3-0019/+) and the Smc4k08819/+; Cap-H2Z3-0019/Mrg15 j6A3 (P < 10−11 for Ubx probe, P < 10−7 for 34D probe, P < 10−12 for Cap-H2 probe). A deficiency [Df(3R)BSC741] that deletes the entire Mrg15 locus crossed into the sensitized background also decreased the number of FISH spots (Figure 3, J–L). The Mrg15 deficiency [Df(3R)BSC741] alone did not change the number of FISH spots, as compared to wild-type cells, and exhibited no other obvious nurse cell phenotypes (Figure S2). To ensure that this was not a condensin allele-specific interaction, we used a different Smc4 allele combined with a small deficiency that deletes the Cap-H2 locus. This condensin Smc488-82/+; Df(3R)Exel6195/+ double heterozygote exhibited seven to nine FISH spots, similar to the Smc4k08819/+; Cap-H2Z3-0019/+ background (Figure S2). By contrast, the Smc488-82/+; Df(3R)Exel6195/Mrg15 j6A3 exhibited an average of four to five FISH spots (P < 10−12 for Ubx, P < 10−11 for 34D, P < 10−5 for Cap-H2; Figure S2). This suggests that Mrg15 genetically interacts with Smc4 and Cap-H2 in nurse cells. This interaction is not specific to Smc4 or Cap-H2 mutant alleles and therefore is also unlikely to be due to nonspecific genetic background effects in the condensin mutant lines.
Figure 3.
Mrg15 mutants enhance condensin II partial loss of function in ovarian nurse cells. Single ovarian nurse cell nuclei from stage 6/7 egg chambers with the following genotypes: Mrg15j6A3/+ (A–C); SMC4k08819/+; Cap-H2Z3-0019/+ (D–F); SMC4k08819/+; Cap-H2Z3-0019/Mrg15j6A3 (G–I); and SMC4k08819/+; Cap-H2Z3-0019/Df(3R)BSC741 (J–L). The left column (A, D, G, and J) shows the DAPI channel staining for DNA. The middle column (B, E, H, and K) is the Ubx FISH probe. The right column (C, F, I, and L) is a merge of the DAPI in white and the FISH probe in green. Bars, 5 μm. (M) The bars in the graph represent the average number of spots per nucleus for each of three FISH probes: quantitation of FISH spots for Ubx is shown in white, 34D is shown in gray, and Cap-H2 FISH is shown in blue. A single asterisk indicates a significant difference compared to double heterozygote (SMC4k08819/+; Cap-H2Z3-0019/+) with P = 1.58 × 10−12 (Ubx), P = 3.72 × 10−8 (34D), and P = 3.92 × 10−13 (Cap-H2). Two asterisks indicate a significant difference from the double heterozygote (SMC4k08819/+; Cap-H2Z3-0019/+) with P = 2.63 × 10−15 (Ubx), P = 6.69 × 10−18 (34D), and P = 1.82 × 10−13 (Cap-H2). The genotypes are listed on the x-axis. For each of the three probes, the number (n) of nuclei scored was as follows: n ≥ 69 nurse cells for Mrg15j6A3/+. n ≥ 59 for SMC4k08819/+; Cap-H2Z3-0019. n ≥ 19 for SMC4k08819/+; Mrg15j6A3/Cap-H2Z3-0019. See Figure S2 and Table S3 for additional supporting data.
RNAi depletion of Mrg15 in nurse cells leads to polytene unpairing defects
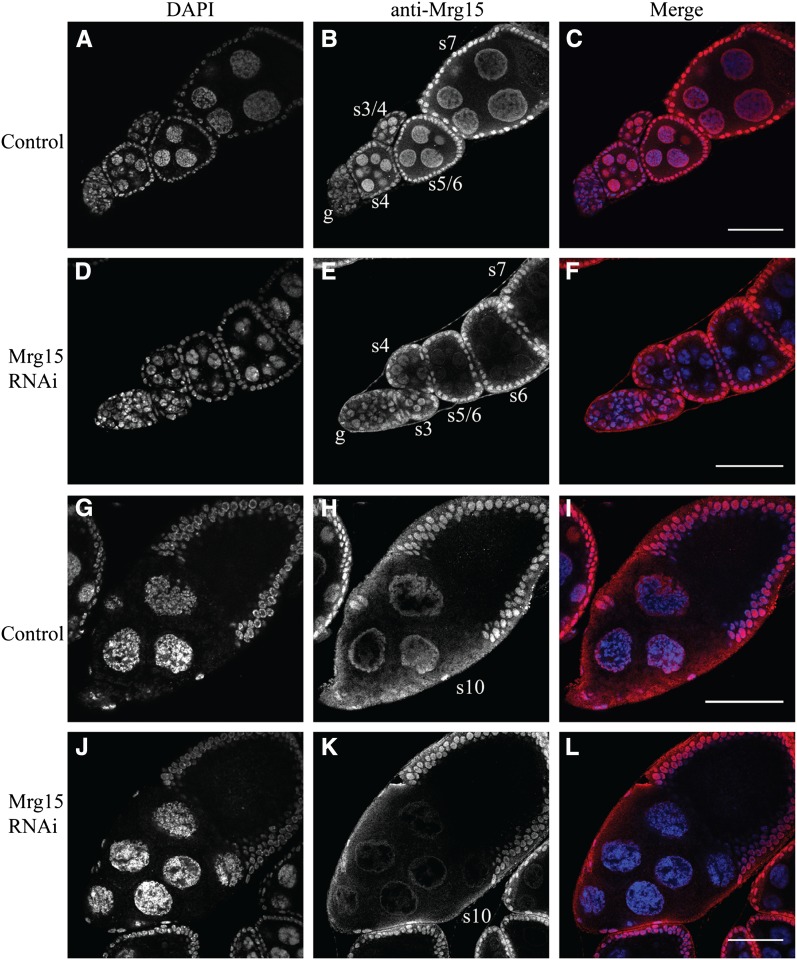
To further test the function of Mrg15 in nurse cell polytene unpairing, we used a TRiP RNAi line capable of expressing an RNA hairpin in the female germline. We used this Mrg15-TRiP in combination with a germline-specific Matα4-GAL-VP16 driver (Hacker and Perrimon 1998; Januschke et al. 2002). First, we confirmed that endogenous Mrg15 protein was expressed in somatic and germline cells of control ovaries from Matα4-GAL-VP16 transgenic females (Figure 4, A–C) (Kusch et al. 2004). Second, we validated RNAi depletion of Mrg15 in ovaries from germline-specific RNAi (Figure 4, D–F). The first detectable Mrg15 depletion was found in stage 4 nurse cells, and by stage 5/6 protein levels were barely detectable (Figure 4, A and F). Note that stage 5/6 and stage 7 nurse cell DNA has blob-like chromosomes (compare Figure 4A to Figure 4D and Figure 4G to Figure 4J), typical of polytene chromosomes that have failed to unpair (Dej 1999). This suggested that RNAi depletion of endogenous Mrg15 in the ovary could be accomplished.
Figure 4.
Mrg15 is efficiently depleted by RNAi in the ovary. Egg chambers stained with DAPI and anti-Mrg15 in Matα4-GAL-VP16 control (A–C) and Matα4-GAL-VP16, UAS > TRiP:Mrg15 RNAi (D–F). Germarium (g) and stages 3–7 are shown. Stage 10 egg chamber from Matα4-GAL-VP16 control (G–I) and Matα4-GAL-VP16, UAS > TRiP:Mrg15 RNAi (J–L). Note the decreases in Mrg15 protein levels in nurse cells but not in follicle cells in stage 4–10 egg chambers. Bar, 50 μm.
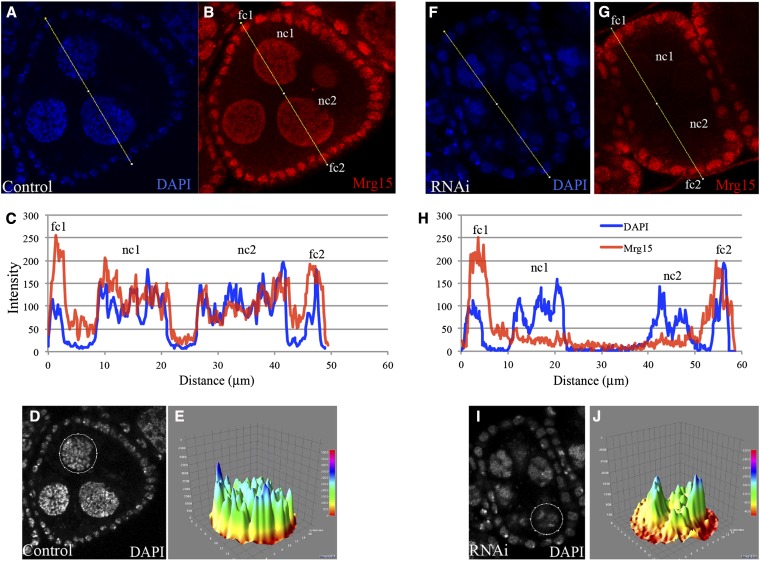
Because the Gal4 driver is germline specific, the somatic follicle cells that are not affected by the RNAi serve to normalize the Mrg15 protein within control and RNAi-treated egg chambers (Figure 5). We used this to further asses Mrg15 protein levels. A z-slice confocal image of a stage 5/6 egg chamber was taken from Figure 4, and pixel intensities were acquired along a single 50- to 60-μm line that crossed exactly two follicle cells (fc1, fc2) and two nurse cells (nc1, nc2) (Figure 5, A and B and F and G). Intensities along these lines for both DAPI and anti-Mrg15 signal were plotted, and we observed that follicle-cell Mrg15 levels were approximately equal in control and RNAi tissues (Figure 5, C and H). Nuclear-localized Mrg15 protein was also apparent in follicle cells of later stage egg chambers of both control and Mrg15 RNAi ovaries (Figure 4). However, nurse cells from control egg chambers exhibit high levels of Mrg15 while RNAi-expressing nurse cells have vastly reduced Mrg15 staining (Figure 5, A–C and F–H). The position at which nurse cell nuclei (nc1 and nc2) are found within each egg chamber, relative to the Mrg15-positive follicle cells, is noted by the DAPI signal (Figure 5, C and H). Nurse cell DNA in the control egg chamber had the expected unpaired polyploid nuclei with DNA distributed throughout the nucleus (Figure 5, D and E). Nurse cell DNA in the Mrg15 RNAi-depleted egg chamber had nondispersed DNA with blob-like features (Figure 5, I and J).
Figure 5.
Mrg15 RNAi-depleted nurse cells have altered chromosome organization. Confocal slice of a stage 5/6 egg chamber from control Matα4-GAL-VP16 stained with DAPI and Mrg15 (A and B). Pixel intensity for DAPI (blue) and Mrg15 (red) was determined along a 50-μm line through the egg chamber. Two follicle cells (fc1, fc2) and two nurse cells (nc1, nc2) were included in the quantitation (C). Single DAPI-stained nucleus from control (D) egg chamber and topographical map of signal intensity showing DNA distribution (E). Confocal slice of a stage 5/6 egg chamber from Matα4-GAL-VP16, TRiP:Mrg15 stained with DAPI and Mrg15 (F and G). Pixel intensity for DAPI (blue) and Mrg15 (red) was determined along a 58-μm line through the egg chamber. Two follicle cells (fc1, fc2) and two nurse cells (nc1, nc2) were included in the quantitation (H). Note that the signal intensity for the follicle-cell nuclei in the TRiP:Mrg15 sample is comparable to that of the follicle-cell nuclei in the control shown in C. Signal for the Mrg15 protein is vastly diminished in the nurse cells (nc1, nc2). Single DAPI-stained nucleus from the control (I) egg chamber and topographical map of signal intensity showing DNA distribution (J). Note the uneven blob-like distribution of DAPI signal in nurse cells from RNAi egg chambers.
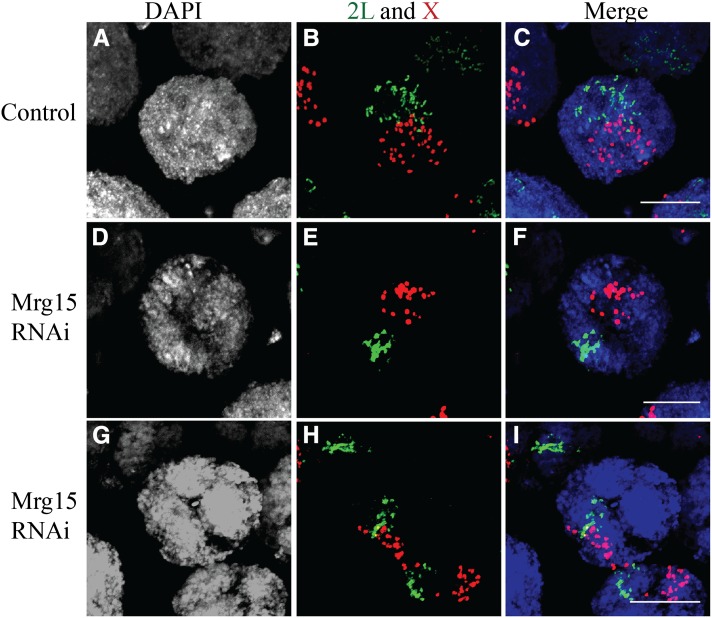
The level of polytene pairing in nurse cells expressing Mrg15-targeting dsRNA was further evaluated by DNA FISH. In control stage 10 nurse cells, chromosomes were dispersed, as visualized by probes on the X chromosome and chromosome II (Figure 6, A–C). In contrast, 100% (n > 100 nuclei) of stage 10 nurse cell nuclei from Mrg15 RNAi-expressing tissue had partially paired or completely paired polytene chromosomes (Figure 6, D–I). The failure to unpair was so severe in nurse cells earlier than stage 6 that it was not possible to count individual FISH spots. In stage 10 egg chambers where individual FISH spots could be counted, we observed that control nurse cells had an average of 42 ± 3 2L-chromosome spots and 40 ± 3 X-chromosome spots (Figure S3). Stage 10 nurse cells from Mrg15 RNAi ovaries had 14 ± 1 2L-chromosome spots and 19 ± 2 X-chromosome spots per nucleus (Figure S3). This represents a significant defect in nurse cell polytene chromosome unpairing (2L probe, P < 10−8; X probe, P < 10−6; Figure S3). We conclude that Mrg15 protein function is important for normal nurse cell polytene unpairing regardless of condensin status. That both Mrg15 and condensin gene functions are required to normally drive unpairing of polytene chromosomes in ovarian nurse cells is consistent with our observations in the salivary gland where Cap-H2 and Mrg15 are polytene-pairing antagonists (Figure 2).
Figure 6.
Mrg15 is required for nurse cell polytene unpairing. Confocal projection of a stage 10 single nurse cell nucleus from control Matα4-GAL-VP16 stained with DAPI (A) and DNA FISH to the 2L (green) and X chromosomes (red) (B) and the merged image (C). Confocal projection of a stage 10 single nurse cell nucleus from Matα4-GAL-VP16, TRiP:Mrg15 stained with DAPI (D and G) and DNA FISH (E and H) and the merged images (F and I). Note the clustering and fewer FISH spots in the TRiP:Mrg15 nuclei (E and H) compared to the control in B. Bar, 20 μm. See Figure S3 for additional images and quantitation of FISH spots.
We note that a previous report has shown that a second-site lethal mutation is also present on the Mrg15 j6A3 chromosome (Qi et al. 2006). Therefore, with experiments where the Mrg15 j6A3 chromosome was used, we cannot exclude the possibility that this undefined lethal mutation may interact with mutations in Smc4 and Cap-H2. However, because the RNAi depletion of Mrg15 in the salivary gland and in nurse cells (using two different RNAi-transgene systems) gives similar results as the Mrg15 j6A3 mutation, we suggest that it is highly unlikely that a second-site mutation on this chromosome is responsible for suppression of condensin phenotypes in the salivary gland and enhancement of condensin phenotypes in nurse cells.
Mrg15 antagonizes transvection at Ubx and yellow
Our observations indicated that Mrg15, like Cap-H2, inhibits interchromosomal interactions in polyploid cells, and we wanted to determine whether this was also true in diploid cells. Previously, we showed that in diploid cells Cap-H2 mutations enhance trans-activation (transvection) at the yellow and Ubx loci, while overexpression can suppress transvection at Ubx (Hartl et al. 2008a). This trans-activation of one mutant allele by the second allele is thought to require extensive physical interactions between the two homologous chromosomes, and chromosomal rearrangements that disrupt diploid homolog pairing also suppress transvection (Lewis 1954). The UbxCbx-1 Ubx1/++ is a genetic assay for homologous chromosome interactions (Lewis 1954). Ubx is normally expressed in nonwing tissues to repress wing-specific gene expression. However, in the UbxCbx-1 Ubx1/++ genetic background the Cbx enhancer can express Ubx in the wing and disrupt development of the ventral side of the wing. This is true despite the fact that the Ubx1 mutation, in cis with the Cbx enhancer, is a Ubx null (Figure 7, A and B). Lewis proposed that the Cbx enhancer activates, in trans, the wild-type copy of Ubx on the homologous chromosome (Lewis 1954). If Mrg15 inhibits these trans-interactions, like Cap-H2, then Mrg15 mutants are predicted to enhance transvection. We observed that a heterozygous Mrg15 mutation enhances the UbxCbx-1 Ubx1 transvection phenotype (Figure 7, C and D) similar to the Cap-H2 mutant (Hartl et al. 2008a). Transvection tests done at the same time with a Set21 loss-of-function mutation in the dSet2 histone methyl transferase did not enhance transvection at Ubx (Figure S4). We considered the possibility that Mrg15 is a general repressor of Ubx transcription and that this could explain the increase in wing phenotype. One control for this is the transposition of the wild-type allele of Ubx to a nonallelic position by a chromosomal rearrangement (BTD), which would be expected to have no effect on the ability of the Mrg15 mutation to increase Ubx expression in the wing. However, if the enhancement of transvection caused by the Mrg15 mutation is homolog-pairing dependent, then the transposed Ubx should not show a phenotype. We found that enhancement of transvection at Ubx by a mutation in Mrg15 does not occur in the translocation background (Figure S4).
Cap-H2 mutations also have been shown to be recessive enhancers of transvection at the yellow locus (Hartl et al. 2008a). We asked whether a Mrg15 mutation (alone or in combination with Cap-H2 mutations) could also modify the y1#8/y82f29-transvecting alleles. We scored all female progeny from different crosses into four classes based on darkly pigmented abdominal stripes (see Materials and Methods and Figure 8), as previously described (Hartl et al. 2008a). We observed that ∼20% of all y1#8/y82f29 ; Mrg15 j6A3/+ females were darkly pigmented (class 3 and 4), and this was not significantly different from y1#8/y82f29 ; Cap-H2Z3-0019/+, and y1#8/y82f29 ; Df(3R)Exel6195/+ females (Figure 8). Thus, these three heterozygous controls do not differ from each other in the proportion of darkly pigmented females (pigmentation class 3 and 4). By contrast, we observed that 60–65% of all y1#8/y82f29;Cap-H2Z3-0019/Mrg15 j6A3 and y1#8/y82f29;Df(3R)Exel6195/Mrg15 j6A3 females were darkly pigmented (Figure 8). This represents a significant enhancement of transvection at the yellow locus when both Cap-H2 and Mrg15 are heterozygous. Because Mrg15 is an essential gene (Kusch et al. 2004), it is not possible to test homozygous effects on transvection. Furthermore, because the Mrg15 j6A3 chromosome has previously been shown to contain a second lethal mutation (Qi et al. 2006), these studies were done in a line where Mrg15 j6A3 was allowed to recombine over seven generations in a w1118 background. These data suggest that Mrg15, like Cap-H2, normally functions to inhibit transvection at Ubx and yellow and/or interacts with Cap-H2 to regulate the expression of Ubx and yellow. However, we cannot exclude the possibility that Mrg15 could alter expression of both the Ubx and yellow genes in a manner that is independent of chromosomal interactions leading to transvection.
Cap-H2 levels on chromatin are dependent on Mrg15 levels
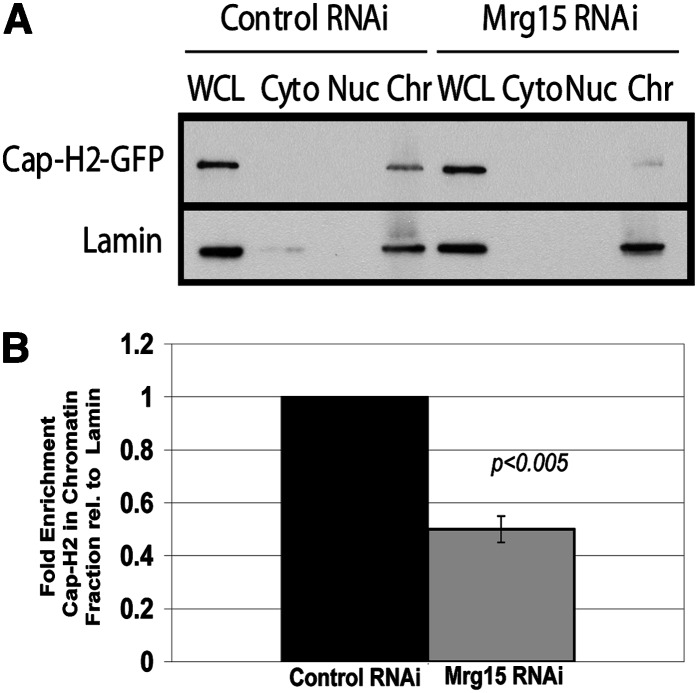
The physical interaction between Cap-H2 and Mrg15 maps to the Mrg domain that is not overlapping with the chromodomain (Figure 1). These results suggest that the MRG domain of Mrg15 could act as a region to bind to Cap-H2, while the chromodomain serves to bind Mrg15 to chromatin; therefore, Mrg15 may serve as a tether to recruit the Cap-H2 protein to chromatin. To test this possibility, we measured levels of Cap-H2 bound to chromatin in cultured Drosophila S2 cells as previously done for Cap-H2 chromatin-fractionation studies (Buster et al. 2013). We compared control RNAi chromatin-bound Cap-H2 levels in S2 cells to those depleted of Mrg15 by RNAi. To accomplish this, pMT-Cap-H2-EGFP or pMT-Cap-H2-V5 were transfected into S2 cells, and we used anti-GFP or anti-V5 antibody to determine levels of Cap-H2 in the whole-cell lysate, cytosolic, soluble nuclear, and chromatin-bound fractions in control RNAi-treated cells and in cells depleted by RNAi of Mrg15. First, whole-cell lysates show a single major band when probed with anti-Mrg15, anti-V5, or anti-lamin (Figure 9 and Figure S5). Treatment of cells with dsRNA directed to Mrg15 sequences depleted endogenous Mrg15 protein by ∼75% relative to extracts from dsRNA to a control sequence within the pBlueScript SK plasmid (Figure S5), and this level of depletion was consistent in four biological replicates. Upon cell fractionation followed by immunoblot analysis, Cap-H2 chromatin-bound levels were normalized to lamin chromatin-bound levels for each treatment (mock RNAi control and Mrg15 RNAi); the same calculation was performed for whole-cell lysate levels of Cap-H2 and lamin in each treatment [Materials and Methods and as previously described (Buster et al. 2013)]. Subsequently, the chromatin-bound Cap-H2/lamin ratio was divided by the Cap-H2/lamin whole-cell lysate ratio to calculate chromatin-bound enrichment. We then compared the chromatin-bound enrichment value between the control RNAi and Mrg15 RNAi treatments. We found that Cap-H2-EGFP (or Cap-H2-V5) bound to chromatin in Mrg15 RNAi-treated cells was reduced by >50% (P < 0.005) compared to that in cells treated with a control RNAi (Figure 9). By normalizing to whole-cell lysate lamin levels, we also determined that levels of Cap-H2 expression did not significantly differ between control RNAi and Mrg15 RNAi treatments. These data suggest that in S2 cells Mrg15 protein facilitates Cap-H2 tethering to chromatin. Although anti-Cap-H2 antibodies have been reported, we found that this reagent is not of sufficient quality to detect endogenous protein in whole-cell lysates, and thus the effects of Mrg15 on endogenous chromatin-bound Cap-H2 could not be ascertained.
Figure 9.
Chromatin-bound Cap-H2 is partially dependent on Mrg15 levels. S2 cells transfected with pMT-Cap-H2-EGFP: (A) Cells were treated with control RNAi and RNAi targeting Mrg15. Cells expressing Cap-H2-EGFP were fractionated, and extracts were immunoblotted with anti-GFP and anti-Lamin. Anti-Mrg15 immunoblots were used to confirm RNAi depletion of Mrg15 (see Figure S5). Each lane represents 20 μg of whole-cell lysates (WCL), cytosolic (Cyto), soluble nuclear (Nuc), and chromatin bound (Chr). (B) The fold enrichment of Cap-H2-EGFP in the chromatin fraction was calculated from four biological replicates as described in Materials and Methods (P < 0.005, two tailed t-test assuming unequal variance). Control RNAi is shown with the solid bar and Mrg15 RNAi is shown with the shaded bar. (See Figure S5 for supporting data.)
Mrg15 and Cap-H2 are required to maintain chromosome compaction and function as antipairing factors in cultured cells
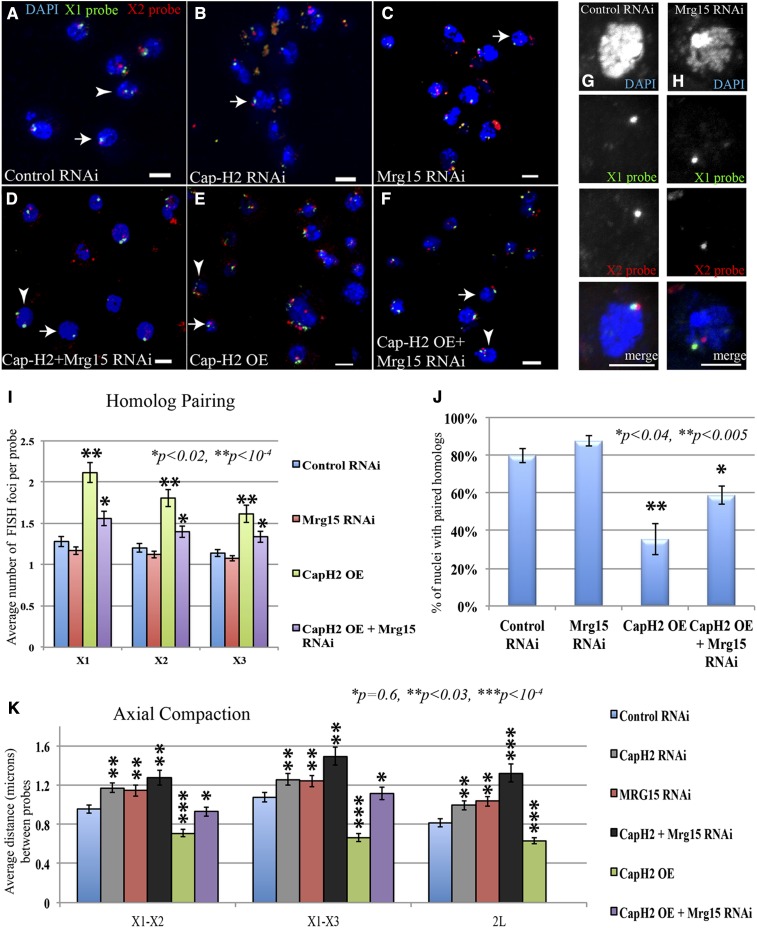
The loss of condensin II activity results in chromosome length increases in interphase polyploid cells, coincident with a failure to unpair polytene chromosomes (Bauer et al. 2012). This suggests that condensins are necessary for maintaining some level of interphase chromosome compaction. In addition, Cap-H2, Cap-D3, and Smc2 condensin subunits have recently been identified as heterochromatin antipairing factors in cultured Kc167 cells (Joyce et al. 2012). To determine whether the Cap-H2 axial compaction and antipairing functions was Mrg15 dependent, we used 3D DNA FISH in Kc cells to measure chromosome compaction and homolog pairing. Note that because homologs are normally paired at high frequency in most Drosophila cells, including Kc cells, each unique FISH probe is expected to give only one spot in most nuclei (Figure 10, A–F, arrow). In some cells two or more FISH spots can be seen for each of the probes (Figure 10, A–F, arrowhead), and multiple probes on the same chromosome allow for an indirect measure of axial compaction (Figure 10, G and H). Using three different probes on the X chromosome (all in euchromatin), we found that RNAi depletion of Mrg15 (validated by immunoblots; Figure S5) gave a slight decrease in the average number of FISH spots per nucleus, relative to a control RNAi treatment (Figure 10I). Overexpression of Cap-H2, however, resulted in a dramatic increase in the average number of FISH foci per nucleus (Figure 10I). Cap-H2 overexpression also led to a decrease in the proportion of nuclei with paired homologs from 80% of cells with paired homologs in control cells compared to 35% (P < 0.005) paired in Cap-H2-overexpressing cells (Figure 10J). This suggests that Cap-H2 overexpression is sufficient to drive unpairing of homologs, consistent with its previously reported antipairing function. By contrast, overexpression of Cap-H2 in combination with RNAi depletion of Mrg15 both rescued the average number of FISH spots per nucleus and increased the percentage of nuclei with paired homologs to 59% (P < 0.04) (Figure 10, I and J).
Figure 10.
Cap-H2 and Mrg15 are required to maintain chromosome length and control axial compaction. (A–F) Cultured Kc cell nuclei stained with DAPI (blue), X-chromosome probe 1 (X1 probe, green), and X-chromosome probe 2 (X2 probe, red). Arrows indicate nuclei with a single FISH spot for each probe showing paired homologs. Arrowheads indicate nuclei with multiple spots of unpaired homologs. (A) Cells treated with control RNAi. (B) Cells treated with RNAi targeting Cap-H2. (C) Cells treated with RNAi targeting Mrg15. (D) Cells treated with RNAi targeting both Cap-H2 and Mrg15. (E) Cells with Cap-H2 overexpression (OE) from pMT-Cap-H2. (F) Cells with Cap-H2 overexpression plus RNAi targeting Mrg15. (G) Single nucleus of control RNAi and (H) Mrg15 RNAi probed with two different probes on the X chromosome. (I) The average number of FISH spots per probe per cell is indicated as a measure of homolog pairing. Spots were counted as one spot if they were merged or their centers were <0.2 μm apart. (J) The proportion of nuclei with paired homologs is shown. (K) Pairwise distances were measured in 3D image reconstructions for three different FISH probe pairs: X1–X2 (∼2.05 Mb apart), X1–X3 (∼3.6 Mb apart) probes were on the X chromosomes, and 2L (∼2 Mb apart) probes were on the left arm of the second chromosome (see Materials and Methods and Table S4). Significance levels were calculated by t-test, assuming equal variance in Microsoft Excel (*P = 0.6, **P < 0.03, and ***P < 10−4). All P-values are calculated relative to control RNAi samples. Table S4 contains further details including number of nuclei measured, average distances, standard error, and P-values. Bars, 5 μm.
To measure axial compaction, we used pairs of probes on the same chromosome. Using three different probes on the X chromosome and two different probes on the second chromosome (all in euchromatin), we found that RNAi depletion of Cap-H2 or Mrg15 increased the 3D distance between these sequences by ∼0.2 μm (17–23%) for each pairwise probe distance, when compared to a control RNAi treatment (P < 0.03, Figure 10K). RNAi depletion of both Cap-H2 and Mrg15 resulted in an even greater increase in distance between probes, ranging from 0.31 to 0.51 μm (24–63%) longer than the control RNAi (**P < 0.03; Figure 10K). By contrast, overexpression of Cap-H2 led to axial shortening of chromosomes by 0.18–0.41 μm (20–61%), relative to control cells (***P < 10−4; Figure 10K). This Cap-H2-induced axial shortening was almost completely rescued by RNAi depletion of Mrg15, where probe distances differed by 0.03–0.04 μm compared to the control RNAi-treated cells (P = 0.6; Figure 10K).
Although our previous studies were on nonmitotic polyploid cells, here we considered the possibility that altering the Cap-H2 levels could change the number of cells in mitosis, thus skewing chromosome compaction measurements. To determine whether the fraction of cells in mitosis was altered by depletion or overexpression of Cap-H2, we stained cells with antiphospho-histone H3. We found that 0.01% of control cells were phospho-H3 positive, while 0.014% of Cap-H2 RNAi cells and 0.01% of Cap-H2 overexpression cells were phospho-H3 positive (n > 395 cells for each treatment, P > 0.2). These observations are consistent with previous findings that altering Cap-H2 levels results in changes in interphase chromosome length in Drosophila (Bauer et al. 2012). Mrg15 RNAi depletion did not lead to mitotic arrest or delay as 0.009% of cells were phospho-H3 positive, while 0.01% of control RNAi-treated cells were phospho-H3 positive (n > 600 cells, P = 0.21). In addition, nuclear size for cells after any RNAi treatment or Cap-H2 overexpression was not significantly different from control RNAi-treated cells. We conclude that both Cap-H2 and Mrg15 are required to maintain interphase chromosome compaction in cultured Kc cells, consistent with a condensin II axial shortening function (Shintomi and Hirano 2011; Bauer et al. 2012).
Discussion
A previously reported large-scale yeast two-hybrid screen found that Drosophila Cap-H2 and Mrg15 proteins interact (Giot et al. 2003). Here, we have replicated this result and show that Cap-H2 interacts with the MRG domain of Mrg15 while the chromodomain is not sufficient for an interaction with Cap-H2 (Figure 1A). Immunoprecipitation with S2 cell extracts further supports this interaction (Figure 1). In ovarian nurse cells, where condensin is normally required for polytene unpairing, two different Mrg15 heterozygous mutants enhanced condensin mutant chromosome unpairing defects (Figure 3). Similarly, Mrg15 RNAi knockdown or a decrease in its gene dosage by 50% in the salivary glands suppressed Cap-H2-induced polytene dispersal (Figure 2). Mutations in Mrg15 showed similar enhancement of transvection at the Ubx locus and interactions with Cap-H2 mutations to enhance transvection at the yellow locus (Figure 7 and Figure 8). We cannot exclude the possibility that both Mrg15 and Cap-H2 proteins cooperate to directly regulate Ubx and yellow transcription, as opposed to productive homolog pairing that leads to transvection. However, our observations reported here and previous studies in polyploid, diploid, and male meiotic chromosomes suggesting that Cap-H2 is required for chromosome unpairing (Hartl et al. 2008a,b) support a model in which Cap-H2 and Mrg15 antagonize transvection by inhibiting trans-interactions. That Cap-H2 provides a homolog antipairing activity has recently been shown for euchromatic and heterochromatic sequences in cultured Drosophila cells (Joyce et al. 2012; Buster et al. 2013) also suggests a more direct function in antagonizing transvection. Interestingly, the Mrg15 homolog in Caenorhabditis elegans (Mrg-1) has also been suggested to antagonize ectopic pairing in meiosis (Dombecki et al. 2011). Additionally, using cellular fractionation we observed that RNAi depletion of endogenous Mrg15 protein in S2 cells results in an approximately twofold decrease of chromatin-bound Cap-H2 protein (Figure 9). RNAi depletion of Mrg15 in cultured Kc cells leads to axial lengthening of interphase chromosomes (Figure 10K), consistent with a chromatin compaction function thought to be provided by condensins (Shintomi and Hirano 2011; Bauer et al. 2012). Importantly, axial compaction driven by overexpression of Cap-H2 can be suppressed by RNAi depletion of Mrg15 (Figure 10K). This suggests that interphase Cap-H2-mediated compaction in polyploid cells in vivo and in cultured cells is Mrg15 dependent.
These data raise the possibility that a protein–protein interaction between Cap-H2 and Mrg15 is important for Cap-H2 activity in vivo. Because chromatin-bound Cap-H2 levels are partially Mrg15 dependent (Figure 9), and because Cap-H2 interacts with the MRG domain, we speculate that this interaction allows the chromodomain of Mrg15 to recruit Cap-H2 to chromatin. These data do not exclude the possibility that Mrg15 also functions to regulate Cap-H2 activity in a manner that is independent of a possible chromatin-tethering role. A model in which Mrg15 allows tethering or enrichment of Cap-H2 to chromatin through direct protein–protein interactions with the MRG domain of Mrg15 is attractive because the Mrg15 chromodomain is known to bind to methylated histones (Kusch et al. 2004; Joshi and Struhl 2005; Zhang et al. 2006; Filion et al. 2010; Moore et al. 2010). We speculate that such a mechanism could be utilized in interphase cells to recruit condensin activity to specific genomic regions marked by histone methylation. This idea is similar to what has been proposed for the Cap-D3 condensin II subunit that interacts with pRB, and pRB is proposed to tether Cap-D3 to centromeric chromatin to effect compaction during mitosis (Longworth et al. 2008; Manning et al. 2010). Currently, it is not clear if histone methylation or histone methyl transferases, such as Set2, play any role in Cap-H2 tethering to chromatin (Figure S2 and Figure S3). Future studies that systematically examine histone methyl transferases will be necessary.
Mrg15 can also associate with chromatin through interactions with the Tip60 complex (Kusch et al. 2004) and potentially provide an indirect mechanism of recruiting condensins to chromatin. We tested reptin and Tip60 mutants in both the salivary glands and the ovary. RNAi to reptin led to high larval lethality, and those few larvae that were alive had very small salivary glands (data not shown). In the ovary, two different reptin mutants showed similar levels of enhancement when crossed to the Smc4k08819/+; Cap-H2Z3-0019/+ double heterozygote (Figure S2, top). The level of enhancement was equal if not greater than that of the Mrg15 mutants (Figure S2 and Table S3). This is consistent with the idea of the two proteins, reptin and Mrg15, working in the same complex. In Drosophila, Mrg15 and reptin proteins have been copurified and also show similar genetic interactions in position-effect variegation (Kusch et al. 2004; Qi et al. 2006). Interestingly, the reptin protein and its binding partner pontin, also known as RVB2/RVB1, may have DNA/RNA-binding functions and participate in a variety of chromatin-remodeling complexes (Jha and Dutta 2009), further linking condensins and Mrg15 to chromatin- and histone-modifying activities. When we performed RNAi to Tip60 in conjunction with Cap-H2 overexpression in the salivary glands, there was no significant suppression observed (Figure S1). However, when a Tip60 mutant (“Tip60G,” Tip60GG01739; Table S3) was crossed into the SMC4k08819/+; Cap-H2Z3-0019/+ double heterozygote, we did observe significant suppression of the polytene unpairing defect (Figure S2, top). This means that the ovarian nurse cells became less polytene and looked more like wild type, which is contrary to the enhancing effect of Mrg15 and reptin mutants (Figure 3 and Figure S2). A different transposon insertion at Tip60 (“Tip60E,” Tip60e02395) did not modify the condensin polytene nurse cell phenotype (Figure S2, top). Although intriguing, the exact role of Tip60 in polytene structure and Cap-H2 activity is not clear, and further studies will be required to determine whether Tip60 directly contributes to these observed phenotypes.
In both mammalian and Drosophila cells, the retinoblastoma (Rb) transcriptional repressor has been shown to directly interact with the Cap-D3 condensin II subunit (Longworth et al. 2008). The Rb protein also forms a complex with Mrg15, and Mrg15 blocks the Rb transcriptional repressor activity (Leung et al. 2001). Together, these observations suggest that chromatin factors, such as Mrg15 and Rb, facilitate condensin activities in vivo, perhaps by recruitment of condensins to specific genomic regions. The existence of these protein complexes also raises the interesting possibilities that, in proliferating cells, condensins interact with chromatin-remodeling complexes to coordinate cell-cycle progression and/or transcriptional regulation to changes in global chromosome compaction states. Coordination of chromosome organization, condensation, and transcriptional “bookmarking” is likely to be important for maintenance of epigenetic expression states, and local inactivation of condensin activity has been posited to serve such a bookmarking function (Xing et al. 2005, 2008). Our observations that mutations in Cap-H2 and Mrg15 result in enhancement of transvection at two different loci is consistent with the idea that condensin inactivation may be required for inheritance of active transcriptional states, possibly by regulating trans-interactions such as those observed at transvecting loci. Recently, it has been shown that trans-splicing of mRNA can occur in Drosophila and that homolog-pairing status was proposed to play a role in facilitating trans-splicing between paired alleles (McManus et al. 2010). Interestingly, in mammalian cells alternative RNA splicing is thought to be influenced by histone H3K36 methylation of intron/exon boundaries (Luco et al. 2010, 2011). It will be of great interest to determine whether Mrg15 and Cap-H2 modulation of allelic interactions use histone methylation patterns to also regulate pre-mRNA trans-splicing in Drosophila.
That interphase chromosome length may be actively regulated is a novel observation (Figure 10K) (Bauer et al. 2012). Recent reports have proposed that local and global condensin-mediated chromatin compaction is important for mammalian T-cell quiescence and erythroid maturation (Xu et al. 2006; Rawlings et al. 2011). Thus, it is intriguing that both Cap-H2 and Mrg15 are necessary for interphase chromosome length maintenance in Drosophila cells (Figure 10K), as the Cap-H2 mouse homolog has been shown to be required for chromatin compaction necessary for maintenance of T-cell quiescence (Rawlings et al. 2011). Whether Cap-H2, and its interaction with Mrg15, have a function in coordinating global interphase chromosome structure with transcriptional processes remains to be determined. It is also unclear whether histone methylation states serve as docking sites for the Mrg15–Cap–H2 complexes or whether other factors serve to recruit Cap-H2 activity to specific genomic regions. It will be of interest to reveal genome-wide condensin localization patterns and how these relate to local functions such as interphase chromatin compaction and gene activity, propensity for transvection, RNA trans-splicing, and general chromatin states.
Supplementary Material
Acknowledgments
We thank all of the Bosco lab members for helpful discussion and are grateful for the essential community resources provided by FlyBase, the University of California at Santa Cruz Genome Browser, the Bloomington Stock Center, the Vienna Drosophila RNAi Center, the Harvard TRiP collection, the University of Iowa Developmental Studies Hybridoma Bank for antibodies, and the Drosophila Genomics Resource Center for cell lines. We thank the anonymous reviewer for extensive comments making this third revision possible and M. Heck, J. L. Workman, and T. Kusch for antibodies. This work was funded by National Institutes of Health grant R01GM69462 to G.B.
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Bateman J. R., Larschan E., D’Souza R., Marshall L. S., Dempsey K. E., et al. , 2012. A genome-wide screen identifies genes that affect somatic homolog pairing in Drosophila. G3 2: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. R., Hartl T. A., Bosco G., 2012. Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet. 8: e1002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazile F., St-Pierre J., D’Amours D., 2010. Three-step model for condensin activation during mitotic chromosome condensation. Cell Cycle 9: 3243–3255. [DOI] [PubMed] [Google Scholar]
- Belmont A. S., 2006. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 18: 632–638. [DOI] [PubMed] [Google Scholar]
- Bertram M. J., Pereira-Smith O. M., 2001. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene 266: 111–121. [DOI] [PubMed] [Google Scholar]
- Bertram M. J., Berube N. G., Hang-Swanson X., Ran Q., Leung J. K., et al. , 1999. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 19: 1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G., 2012. Chromosome pairing: a hidden treasure no more. PLoS Genet. 8: e1002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buster D. W., Daniel S. G., Nguyen H. Q., Windler S. L., Skwarek L. C., et al. , 2013. SCFSlimb ubiquitin ligase suppresses condensin II-mediated nuclear reorganization by degrading Cap-H2. J. Cell Biol. 201: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Tominaga K., Pereira-Smith O. M., 2010. Emerging role of the MORF/MRG gene family in various biological processes, including aging. Ann. N. Y. Acad. Sci. 1197: 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbe N., Savvidou E., Heck M. M., 2006. Diverse mitotic and interphase functions of condensins in Drosophila. Genetics 172: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer C., 2006. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur. J. Histochem. 50: 223–272. [PubMed] [Google Scholar]
- Cuylen S., Haering C. H., 2011. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 21: 552–559. [DOI] [PubMed] [Google Scholar]
- Dej K. J., 1999. The structure and function of the nurse cell chromosomes during oogenesis in Drosophila melanogaster, Ph.D. Dissertation, The Johns Hopkins Unversity, Baltimore. [Google Scholar]
- Dej K. J., Spradling A. C., 1999. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development 126: 293–303. [DOI] [PubMed] [Google Scholar]
- Dombecki C. R., Chiang A. C., Kang H. J., Bilgir C., Stefanski N. A., et al. , 2011. The chromodomain protein MRG-1 facilitates SC-independent homologous pairing during meiosis in Caenorhabditis elegans. Dev. Cell 21: 1092–1103. [DOI] [PubMed] [Google Scholar]
- Dundr M., 2012. Nuclear bodies: multifunctional companions of the genome. Curr. Opin. Cell Biol. 24: 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., et al. , 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Green M. M., Corces V. G., 1990. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 9: 2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., et al. , 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736. [DOI] [PubMed] [Google Scholar]
- Green L. C., Kalitsis P., Chang T. M., Cipetic M., Kim J. H., et al. , 2012. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J. Cell Sci. 125: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker U., Perrimon N., 1998. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila. Genes Dev. 12: 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T. A., Smith H. F., Bosco G., 2008a Chromosome alignment and transvection are antagonized by condensin II. Science 322: 1384–1387. [DOI] [PubMed] [Google Scholar]
- Hartl T. A., Sweeney S. J., Knepler P. J., Bosco G., 2008b Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 4: e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Dreesen T. D., 1989. Trans-inactivation of the Drosophila brown gene: evidence for transcriptional repression and somatic pairing dependence. Proc. Natl. Acad. Sci. USA 86: 6704–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M., Hirano T., 2006. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol. Cell 21: 175–186. [DOI] [PubMed] [Google Scholar]
- Hirano T., 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7: 311–322. [DOI] [PubMed] [Google Scholar]
- Hirano T., Kobayashi R., Hirano M., 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89: 511–521. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., 2010. Spatial epigenetics: linking nuclear structure and function in higher eukaryotes. Essays Biochem. 48: 25–43. [DOI] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Dass S., Kaltschmidt J. A., Lopez-Schier H., et al. , 2002. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 12: 1971–1981. [DOI] [PubMed] [Google Scholar]
- Jha S., Dutta A., 2009. RVB1/RVB2: running rings around molecular biology. Mol. Cell 34: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A. A., Struhl K., 2005. Eaf3 chromodomain interaction with methylated H3–K36 links histone deacetylation to Pol II elongation. Mol. Cell 20: 971–978. [DOI] [PubMed] [Google Scholar]
- Joyce E. F., Williams B. R., Xie T., Wu C. T., 2012. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8: e1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J. A., Southworth J. W., 2002. Transvection in Drosophila. Adv. Genet. 46: 399–420. [DOI] [PubMed] [Google Scholar]
- Kusch T., Florens L., Macdonald W. H., Swanson S. K., Glaser R. L., et al. , 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087. [DOI] [PubMed] [Google Scholar]
- Leung J. K., Berube N., Venable S., Ahmed S., Timchenko N., et al. , 2001. MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem. 276: 39171–39178. [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88: 225–239. [Google Scholar]
- Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., et al. , 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S., Barnea G., Pisapia D. J., Mendelsohn M., Kirkland J., et al. , 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413. [DOI] [PubMed] [Google Scholar]
- Longworth M. S., Herr A., Ji J. Y., Dyson N. J., 2008. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 22: 1011–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco R. F., Pan Q., Tominaga K., Blencowe B. J., Pereira-Smith O. M., et al. , 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco R. F., Allo M., Schor I. E., Kornblihtt A. R., Misteli T., 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A. L., Longworth M. S., Dyson N. J., 2010. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 24: 1364–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus C. J., Duff M. O., Eipper-Mains J., Graveley B. R., 2010. Global analysis of trans-splicing in Drosophila. Proc. Natl. Acad. Sci. USA 107: 12975–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. A., Ferhatoglu Y., Jia Y., Al-Jiab R. A., Scott M. J., 2010. Structural and biochemical studies on the chromo-barrel domain of male specific lethal 3 (MSL3) reveal a binding preference for mono- or dimethyllysine 20 on histone H4. J. Biol. Chem. 285: 40879–40890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Chen J., Filandrinos S. T., Dunn R. C., Fisk R., et al. , 1999. An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics 151: 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M. P., Neuwald A. F., et al. , 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115: 109–121. [DOI] [PubMed] [Google Scholar]
- Qi D., Jin H., Lilja T., Mannervik M., 2006. Drosophila Reptin and other TIP60 complex components promote generation of silent chromatin. Genetics 174: 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I., Groudine M., 2011. On emerging nuclear order. J. Cell Biol. 192: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I., Perlman M. D., Scalzo D., Kooperberg C., Groudine M., et al. , 2009. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc. Natl. Acad. Sci. USA 106: 6679–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings J. S., Gatzka M., Thomas P. G., Ihle J. N., 2011. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 30: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roix J. J., McQueen P. G., Munson P. J., Parada L. A., Misteli T., 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat. Genet. 34: 287–291. [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M. C., et al. , 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7: 282–289. [DOI] [PubMed] [Google Scholar]
- Sanyal A., Bau D., Marti-Renom M. A., Dekker J., 2011. Chromatin globules: A common motif of higher order chromosome structure? Curr. Opin. Cell Biol. 23: 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Hirano T., 2011. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 25: 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E., Dorn J. F., Sengupta K., Jasin M., Nussenzweig A., et al. , 2007. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9: 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis C. G., Lalioti M. D., Town T., Lee G. R., Flavell R. A., 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645. [DOI] [PubMed] [Google Scholar]
- Sutherland H., Bickmore W. A., 2009. Transcription factories: Gene expression in unions? Nat. Rev. Genet. 10: 457–466. [DOI] [PubMed] [Google Scholar]
- Takizawa T., Gudla P. R., Guo L., Lockett S., Misteli T., 2008. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 22: 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J., Belmont A. S., Sedat J. W., 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Wood A. J., Severson A. F., Meyer B. J., 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. T., Morris J. R., 1999. Transvection and other homology effects. Curr. Opin. Genet. Dev. 9: 237–246. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Reilly P. T., Herr W., 2001. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 21: 3820–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Graveline R., Kumar G. S., Zhang Y., Krishnan A., et al. , 2012. Structural basis for molecular interactions involving MRG domains: implications in chromatin biology. Structure 20: 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H., Wilkerson D. C., Mayhew C. N., Lubert E. J., Skaggs H. S., et al. , 2005. Mechanism of hsp70i gene bookmarking. Science 307: 421–423. [DOI] [PubMed] [Google Scholar]
- Xing H., Vanderford N. L., Sarge K. D., 2008. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol. 10: 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Leung C. G., Lee D. C., Kennedy B. K., Crispino J. D., 2006. MTB, the murine homolog of condensin II subunit CAP-G2, represses transcription and promotes erythroid cell differentiation. Leukemia 20: 1261–1269. [DOI] [PubMed] [Google Scholar]
- Yeong F. M., Hombauer H., Wendt K. S., Hirota T., Mudrak I., et al. , 2003. Identification of a subunit of a novel Kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr. Biol. 13: 2058–2064. [DOI] [PubMed] [Google Scholar]
- Zaidi S. K., Young D. W., Montecino M., Lian J. B., Stein J. L., et al. , 2010. Architectural epigenetics: mitotic retention of mammalian transcriptional regulatory information. Mol. Cell. Biol. 30: 4758–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li Y., Yang J., Tominaga K., Pereira-Smith O. M., et al. , 2010. Conditional inactivation of MRG15 gene function limits survival during larval and adult stages of Drosophila melanogaster. Exp. Gerontol. 45: 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Du J., Sun B., Dong X., Xu G., et al. , 2006. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 34: 6621–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.