Abstract
The fungal fruiting body or mushroom is a multicellular structure essential for sexual reproduction. It is composed of dikaryotic cells that contain one haploid nucleus from each mating partner sharing the same cytoplasm without undergoing nuclear fusion. In the mushroom, the pileus bears the hymenium, a layer of cells that includes the specialized basidia in which nuclear fusion, meiosis, and sporulation occur. Coprinopsis cinerea is a well-known model fungus used to study developmental processes associated with the formation of the fruiting body. Here we describe that knocking down the expression of Atr1 and Chk1, two kinases shown to be involved in the response to DNA damage in a number of eukaryotic organisms, dramatically impairs the ability to develop fruiting bodies in C. cinerea, as well as other developmental decisions such as sclerotia formation. These developmental defects correlated with the impairment in silenced strains to sustain an appropriated dikaryotic cell cycle. Dikaryotic cells in which chk1 or atr1 genes were silenced displayed a higher level of asynchronous mitosis and as a consequence aberrant cells carrying an unbalanced dose of nuclei. Since fruiting body initiation is dependent on the balanced mating-type regulator doses present in the dikaryon, we believe that the observed developmental defects were a consequence of the impaired cell cycle in the dikaryon. Our results suggest a connection between the DNA damage response cascade, cell cycle regulation, and developmental processes in this fungus.
Keywords: Coprinopsis cinerea, mushroom development, DNA damage response (DDR), dikaryon, cell cycle
IN fungal cells, mating—the process equivalent to fertilization—brings together two haploid nuclei in the same cytoplasm. It is generally thought that this process is essentially followed by nuclear fusion, resulting in a diploid nucleus that either enters meiosis immediately (as occurs in the fission yeast Schizosaccharomyces pombe) or is maintained and proliferates in the diploid state (as happens in the budding yeast Saccharomyces cerevisiae). However, in a large number of fungi, mating does not result in diploid nuclei. Instead, they form dikaryons, cells that contain one haploid nucleus from each mating partner sharing the same cytoplasm for a period of time without undergoing nuclear fusion or meiosis. These dikaryons continue to propagate, and eventually nuclear fusion will take place, which will be followed by meiosis, closing the sexual cycle (Brown and Casselton 2001).
Dikaryon cell division, also called conjugate division, represents a big challenge to the cell since it has to ensure that each daughter dikaryon inherits a balance of each parental genome. For that, a complex cell cycle is required that involves distinct mechanisms to maintain heterokaryosis after cell division. For example, for a large number of Basidiomycota (e.g., the mushroom Coprinopsis cinerea), conjugate division includes the formation of a structure known as the clamp connection or clamp cell, as well as the sorting of one of the nuclei to this structure (Casselton 1978; Brown and Casselton 2001). In these fungi, prior to nuclear division, a specialized projection is formed on the side of the apical cell and one nucleus migrates into this clamp while the other remains in the main cell. Nuclear divisions are synchronic and septa are laid down across the mitotic spindle planes. One septum is formed between the clamp cell and the apical cell, trapping a single nucleus in the clamp. The second septum is formed within the hyphal cell, just below the clamp and separating one nucleus in the novel subapical cell from two distinct haploid nuclei in the apical cell. Subsequently the clamp cell fuses with the subapical cell and its nucleus migrates into this cell (Supporting Information, Figure S1). This way, synchronous nuclear division occurs in two distinct subcellular compartments (Tanabe and Kamada 1994, 1996). Clamp connections therefore guarantee a correct nuclear distribution that keeps the presence of two distinct nuclei per cell. C. cinerea strains unable to produce clamps such as clp1-1 mutants showed impaired dikaryon maintenance (Inada et al. 2001). A similar structure, called the crozier cell, applies for many fungi of the phylum Ascomycota (Buller 1958). Nevertheless, the clamp connection is not essential for stable and accurate dikaryon formation since it is absent in the dikaryons of some species (Salo 1989). In these species that do not form clamp cells, the distinct sorting of each daughter nucleus most likely will depend on different spindle lengths or spindle elongation rates (Salo 1989).
Regardless of the manner in which the distinct nuclei are sorted, an important feature during nuclear division in dikaryotic cells is that even when nuclei are located in different cell compartments—one in the main hypha and the other in the clamp cell—the two nuclei have to divide synchronously (Tanabe and Kamada 1994). It seems likely that some mechanism has to be in charge to ensure that in the case that replication in one of the nuclei is delayed with respect to the other one, both nuclei will enter mitosis at the same time. However, the molecular mechanisms determining the accuracy of conjugate division are poorly understood.
The DNA damage response (DDR) pathway detects damaged as well as unreplicated DNA and coordinates its repair with the cell cycle progression (Abraham 2001; Nyberg et al. 2002). Central to this cascade are the phosphatidylinositol 3-kinase–related kinases (PIKKs) ATM and ATR, which share a significant sequence homology and canalize all signaling through this cascade (Cimprich and Cortez 2008). Downstream of these kinases, there is a group of serine–threonine kinases that belongs to two different families: Chk1 and Chk2/Rad53. They are phosphorylated in an ATR/ATM-dependent manner and their activation produces, among other effects, a cell cycle delay enabling the DNA repair before mitosis is complete (Sanchez et al. 1997; Sancar et al. 2004). These kinases are broadly conserved regulators (Abraham 2001; Bartek and Lukas 2003). Recent work performed in the phytopathogenic basidiomycete Ustilago maydis (Mielnichuk et al. 2009; de Sena-Tomas et al. 2011; Perez-Martin and de Sena-Tomas 2011) suggested a role for the kinases Atr1 and Chk1 in the regulation of the dikaryotic cell cycle. U. maydis cells defective in these kinases showed impaired dikaryon proliferation. Unfortunately, the fact that the U. maydis dikaryon is present only inside the plant precludes this fungus from a better characterization of the defects associated with these mutations with respect to dikaryon maintenance.
The fungus C. cinerea is a well-known model organism to study developmental processes associated with the formation of the fruiting body or mushroom (Kües 2000; Kamada 2002; Stajich et al. 2010). Fruiting body formation is part of the sexual cycle in C. cinerea, and it is initiated after mating of two compatible sterile monokaryotic mycelia to give rise to the fertile dikaryon carrying two distinct haploid nuclei in the hyphal segments. Since the fruiting body is composed of dikaryotic cells, this fungus has been used for many years as a model fungus for dikaryosis studies. Pioneering studies showed the importance of actin and tubulin cytoskeletons in nuclear migration as well as nuclear positioning and movements within the established dikaryon (Tanabe and Kamada 1994, 1996; Iwasa et al. 1998). Also a number of mutants affecting the cell division process, mainly the clamp formation, resulted in the description of genes involved in this process such as pcc1, num1, and clp1 (Murata et al. 1998; Inada et al. 2001; Makino and Kamada 2004).
In this work we show that Atr1 and Chk1, proteins involved in the DDR in C. cinerea, are also essential for a correct mitosis of the dikaryotic cell and that its downregulation prevents the mature fruiting body formation.
Materials and Methods
Strains and growth conditions
C. cinerea strains are derived from the AmutBmut genetic background (A43mut B43mut pab1-1) (Swamy 1984). Strains were grown on YMG/T (0.4% yeast extract, 1% malt extract, 0.4% glucose, 100 mg/liter tryptophan, and 1% agar) and MM (0.1% KH2PO4, 0.225% Na2HPO4, 0.029% Na2SO4, 0.05% diammonium tartrate, 0.025% MgSO4 × 7H2O, 4 × 10−5% thiamine, 0.005% adenine sulfate, 0.2% asparagine, 1% glucose, and 1% agar) media at 37° (Rao and Niederpruem 1969; Granado et al. 1997). To check the sensitivity of the strains to different genotoxic stresses, a cylindrical piece of mycelium with a diameter of 4 mm was inoculated on YMG/T plates supplemented with 2.5 mM hydroxyurea or 0.015% methylmethane sulfonate. Plates were grown until the plate was fully covered with mycelium (∼6 days for control strains). Induction of fruiting body, oidia, and sclerotia formation was done as described in Granado et al. (1997; Kertesz-Chaloupkova et al. 1998; Kües et al. 1998).
RNAi procedures
To perform the silencing constructs of atr1 and chk1 genes, we constructed a recipient plasmid carrying the gpdII promoter, an efficient constitutive promoter from Agaricus bisporus previously used in C. cinerea (Burns et al. 2005); as a fungal transcriptional terminator we used the 3′-UTR region from the hygromycin-resistant gene from the pNEB-Hyg plasmid, which is used in U. maydis for transformation (Castillo-Lluva et al. 2004); and as a selectable marker we used the pab1 gene from C. cinerea, which encodes PABA synthase, necessary for para-aminobenzoic acid production (Bottoli et al. 1999). pBS(+)KS plasmid was used as backbone. First, we inserted the terminator as a SacI/NotI fragment from the pNEB-Hyg plasmid, in the respective sites of pBS(+)KS. The resulting plasmid (pBS-Hygter) was digested with ScaI and EcoRI and a ScaI/EcoRI fragment from the pYSK7 (Kilaru et al. 2006) containing the pab1 gene was inserted in the respective sites, resulting in the pHygter-pab1 plasmid. Finally, we excised the fragment containing the PgpdII promoter from pYSK7 as a EcoRI/BamHI fragment and it was inserted into the respective sites of pHygter-pab1. The resulting plasmid, pGH-pab, was used as a recipient for cloning the antisense sequences. The atr1 antisense fragment was obtained after PCR amplification with primers atr1c-1 (5′ GCGATATTCCAGACCTCATTGTCTGTCGCA 3′) and atr1c-2 (5′ TACGGATCCACTGAGAATCTTCTTGTCATGCGT 3′), which spanned from nucleotide +3271 to nucleotide +4488 (considering adenine in ATG as +1). In the chk1 silencing, the antisense construct was obtained after PCR amplification with chk1c-1 (5′ TCGCAAACGCAATCGGGCCCTCGATATGTGCCG 3′) and chk1c-2 (5′ ATGGGATCCTCATCTCTCGCCATGCCGCAGAAG 3′), which spanned from nucleotide +1760 to nucleotide +2034.
DNA transformation and regeneration of protoplasts were done as previously reported (Granado et al. 1997). From each transformation, ∼50 colonies growing on minimal media were isolated and the presence of inserted RNAi plasmid was analyzed by diagnostic PCR. Genomic DNA extraction was done as reported in Zolan and Pukkila (1986).
From all the transformants, 18 colonies from the atr1 silencing, 14 colonies from the chk1 silencing, and 8 colonies from the RNAi control were analyzed by RT-PCR for expression levels of corresponding genes (atr1 and chk1). RNA was extracted as previously reported (Chomczynski and Sacchi 1987). After the extraction, the RNA was cleaned with the High Pure RNA Isolation Kit from Roche. Subsequently, cDNA was synthesized with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and a RT-PCR was performed with the Power SYBR Green PCR Master Mix on a 7300 Real-Time PCR System from Applied Biosystems. As a housekeeping gene, benA, which codes for β-tubulin, was used (Walti et al. 2006). Data were analyzed through a mathematical equation described in Pfaffl et al. (2004). The efficiencies of the oligonucleotide pairs used were as follows: benART-1 (5′ CTTGCTTCGAGCCTGGTAAC 3′)/benART-2 (5′ TCACCACGGTAGAGGAGAGC 3′), 94.3%; atr1RT-5 (5′ CAGCTGAACTGGAAGCACAG 3′)/atr1RT-6 (5′ GGGAACATCGGGAGAATCTT 3′), 96%; chk1RT-3 (5′ ATTCTGCCGCTATCTGGATG 3′)/chk1RT-4 (5′ GGTTCACGTTCAGCATACCC 3′), 102.2%.
Microscopic observations
Samples were visualized in a Nikon Eclipse 90i microscope equipped with a Hamamatsu ORCA-ER CCD camera. Pictures were taken using the appropriate filter set, Nikon Plan Apo VC 100× NA 1.40 and Plan Apo VC 60× NA 1.40 lenses with Nikon Immersion Oil type A, nd = 1.5151. The software used with the microscope was Metamorph 6.1 and the pictures were further processed with Adobe Photoshop CS5. To perform septa and nuclei staining, C. cinerea was grown for 2 days at 37° on a thin agar layer over a glass microscope slide with a small window in the middle of the size of the cover slip. In conditions where aphidicolin (AC) was added, the agar layer carried the desired concentration of AC dissolved in DMSO. An equal volume of DMSO was added to agar pads on control slides. After this time, 70% EtOH was added to dry out the hyphae and then the hyphae were stained with 140 nM Hoechst 33258 and 273 nM Calcofluor on a 0.5% DMSO solution, for 20 min before observing (Virag et al. 2007).
Results
Silencing the genes encoding the DNA damage response regulators Atr1 and Chk1 in C. cinerea
Using the BLAST program and the sequence of DDR kinases from different fungal species as well as humans as queries, we found Atr1 (CC1G_08126.3), Atm1 (CC1G_00839.3), Chk1 (CC1G_02812.3), and Chk2 (CC1G_09319.3) homologs in the C. cinerea genome database (http://www.broadinstitute.org/annotation/genome/coprinus_cinereus/MultiHome.html) (Figure S2).
Based on our previous results in U. maydis (de Sena-Tomas et al. 2011), we played with the hypothesis that Atr1 and Chk1 might have some role during the dikaryotic cell cycle in C. cinerea. To address this hypothesis we tried to impair the function of these regulators during the dikaryotic cell cycle, using dsRNA-mediated gene silencing (Walti et al. 2006; Heneghan et al. 2007). Antisense constructs were designed from different chk1 and atr1 exons (RNAi plasmids, see Materials and Methods for a description). As a receptor strain for these silencing plasmids we used the self-compatible homokaryon AmutBmut. This particular strain carries specific mutations in both mating-type loci (A43mut and B43mut) that enable the formation of fruiting bodies without prior mating to another strain (Swamy 1984; Boulianne et al. 2000). It shows typical characteristics of the dikaryon such as the formation of fused clamp cells at hyphal septa and it has been used to screen for mutations affecting dikaryotic phenotypes, such as clamp connection formation (Inada et al. 2001), fruiting body development (Chiu and Moore 1990; Granado et al. 1997; Muraguchi et al. 2008), and meiosis (Cummings et al. 1999).
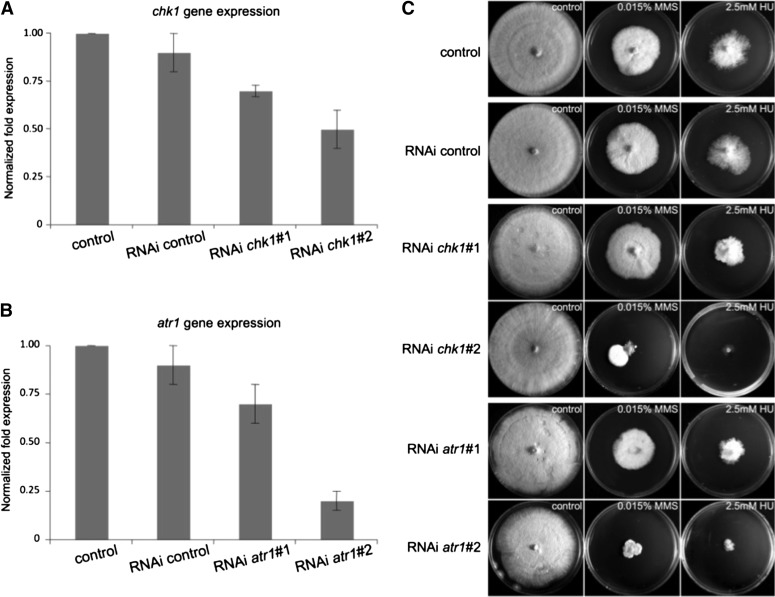
We transformed AmutBmut with chk1 and atr1 RNAi plasmids as well as the empty plasmid used for the silencing as a control (RNAi control). Two independent silenced clones for each gene—showing a different silencing degree—as well as one clone carrying the control construct were chosen for further analysis. Strain RNAi chk1#1 showed ∼70% of expression of chk1 relative to the control strain while strain RNAi chk1#2 showed 50% of mRNA levels (Figure 1A). We were unable to find clones showing a much higher level of silencing for this gene. In the atr1 clones, strains RNAi atr1#1 and RNAi atr1#2 showed 70% and 20% mRNA levels in comparison to the control strain, respectively (Figure 1B). Silenced strains were affected to various degrees in their ability to grow. Control strains (AmutBmut and RNAi control) took ∼6 days at 37° to fully cover the YMG/T plates, while RNAi chk1#1 and RNAi chk1#2 needed 7 days to grow to the same extent. RNAi atr1#1 and RNAi atr1#2 needed 8 days and 10 days, respectively (Figure S3). The gene-silencing process was stable in all strains with the exception of strain RNAi chk1#2, in which colony sectors with cells showing low levels of silencing raised frequently when plated in conditions in which gene silencing impaired growth (see below).
Figure 1.
Silencing chk1 and atr1 expression in Coprinopsis cinerea resulted in sensitivity to DNA-damage agents. (A and B) Relative gene expression values measured by RT-qPCR of chk1 and atr1 in control and silenced strains, respectively. The data show the average and SE of three biological replicates (P < 0.01). Expression rates are normalized with respect to transcript abundance of benA (control), a housekeeping gene. (C) Sensitivity of control and silenced strains to the DNA-damage agents, hydroxyurea (HU) and methylmethane sulfonate (MMS). Plates were incubated for 8 days at 37°.
The kinases Atr1 and Chk1 are required for the DNA damage response in C. cinerea
Atr1 and Chk1 are the main players of the DDR pathway in eukaryotic organisms and its lack of function produces a high sensitivity to genotoxic agents in the cells, in particular to agents that cause DNA replication stress such as methylmethane sulfonate (MMS), which induces DNA alkylation (Lundin et al. 2005), and hydroxyurea (HU), which inhibits ribonucleotide reductase and therefore affects replication by depletion of dNTPs (Koc et al. 2004). Therefore, the sensitivity of the selected silenced clones against these genotoxic agents was tested. Strains were grown on rich media supplemented with HU or MMS. As expected, when the atr1 or chk1 gene expression was silenced, the cells were more sensitive to these genotoxic stresses (Figure 1C). We found a good grade of correlation with the level of silencing in these strains and the level of sensitivity to HU or MMS. It is worth noting that for chk1, a dramatic difference with respect HU and MMS sensitivity was observed between RNAi chk1#1 (70% of wild-type expression) and RNAi chk1#2 (50% of wild-type expression). Moreover, plating RNAi chk1#2 cells in the presence of genotoxic stress often resulted in the appearance of fast-growing sectors (Figure S4A). Further analysis of these fast-growing cells revealed that they expressed the chk1 gene at levels as high as 90% of wild-type expression (Figure S4B) while retaining the RNAi transgene and regaining the ability to survive in the presence of genotoxic stress (Figure S4C). One of these silencing revertants, named RNAi chk1#2 Rev, was used as an additional control in the characterization of the roles of these kinases (see below).
Downregulation of Atr1 and Chk1 kinases resulted in an increase in abnormal mitosis in the dikaryon
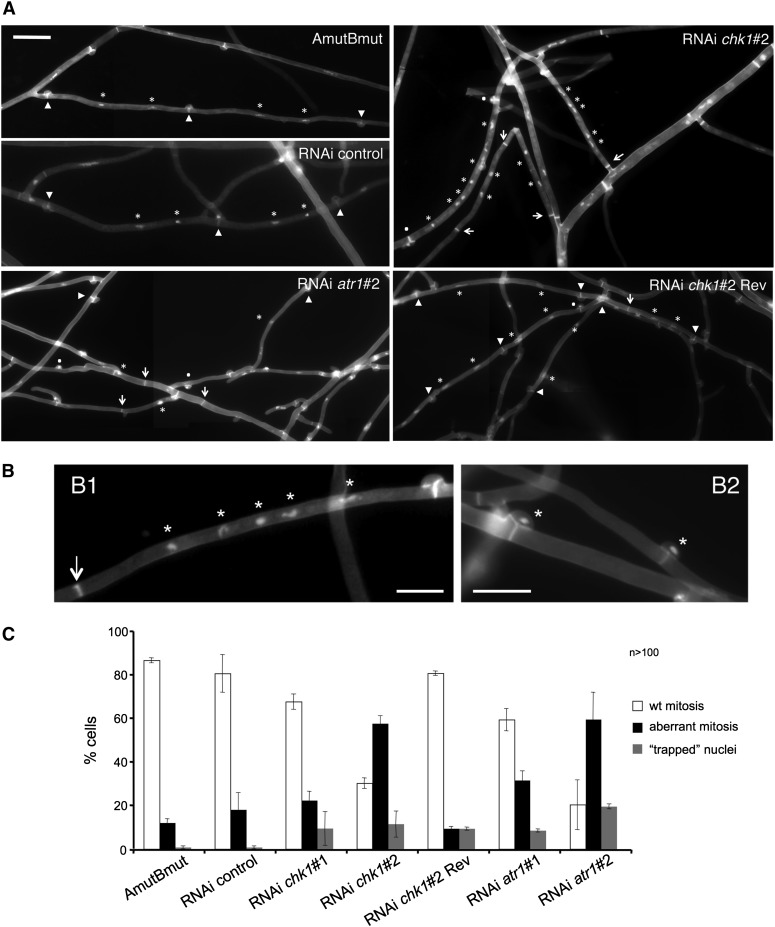
We analyzed the consequence of knocking down atr1 or chk1 for the dikaryotic cell cycle in C. cinerea. For this, we performed microscopic analysis of the atr1 and chk1 silenced strains as well as the controls (AmutBmut, RNAi control, and a chk1-revertant obtained from a fast-growing sector of RNAi chk1#2). The different strains were grown on a thin agar layer upon a microscope glass slide for 2 days. Then, they were costained with Hoechst and Calcofluor White, to visualize nuclei and septa, respectively. Hyphae from control strains showed cell compartments flanked by septa associated to clamp connections and carrying two nuclei each (Figure 2A). In contrast, chk1 and atr1 silenced strains cells showed a range of abnormalities such as more than two nuclei per cell compartment, septa without an associated clamp connection, or clamp connections with “trapped” nuclei (Figure 2, A and B). We quantified the abnormalities observed in the chk1 and atr1 silenced strains. For this, the number of nuclei per cellular compartment as well as the presence of normal or aberrant clamps in each of these cells was counted. Figure 2C shows the resulting graphic, where “wt mitosis” gathers all the cells where the presence of nuclei and clamps was normal, “aberrant mitosis” groups those cells where either the nuclear distribution or the clamp formation was aberrant, and “trapped nuclei” shows the nuclei that were observed “locked” inside the clamps. In the control strains the number of aberrant phenotypes was very low and similar to previously reported results (Polak et al. 1997). In contrast, in the silenced strains the number of cells resulting from aberrant mitosis increased dramatically (Figure 2C). As we noted with the sensitivity to genotoxic stress, we observed a nice correlation between the degree of silencing and the severity of the phenotype.
Figure 2.
Downregulation of atr1 and chk1 resulted in an increase in abnormal mitosis in the dikaryon. (A) Cell images from the indicated strains. Septa with associated clamp connections are marked with arrowheads, while septa without clamp connections are marked with arrows. Nuclei are marked with asterisks. Dots mark the presence of nuclei trapped by a clamp connection. Bar: 20 μm. (B) Examples of cells resulting from abnormal mitosis. In B1, a cell compartment is flanked by a septum with no associated clamp connection (arrow) and five nuclei (asterisk). In B2, nuclei are trapped inside the clamp connection (asterisk). Bar: 10 μm. (C) Quantification of the number of aberrant mitoses (cell compartments with clamp-free septa and/or more than two nuclei) as well as clamps with trapped nuclei inside in hyphae from the indicated strains. The graph shows the results from three independent experiments, counting more than 100 cell compartments each. Means and standard deviations are shown.
Effect of atr1 and chk1 downregulation on synchronous mitosis in the dikaryon
An important feature during nuclear division in C. cinerea dikaryotic cells is that the two nuclei have to enter mitosis synchronously (Tanabe and Kamada 1994, 1996). The importance of this synchrony was uncovered upon the study of the consequences in the dikaryotic cell cycle of several α- and β-tubulin mutants (Tanabe and Kamada 1994). The cell cycle abnormalities that we observed in conditions of Atr1 and Chk1 deficiency are compatible with a cell cycle in which this synchronous entry of nuclei into mitosis was affected, and as a consequence the connections between clamp formation and nuclear division were impaired, resulting in a range of abnormal cell divisions. We decided to quantify how frequently asynchronous divisions occurred in mutant strains vs. control strains. For that we analyzed apical cells and sorted the samples between synchronous mitosis and nonsynchronous mitosis (Figure 3A). We observed a significant proportion of asynchronous mitosis in mutant strains, in contrast to a low percentage of this class in control strains (Figure 3B).
Figure 3.
Effect of atr1 and chk1 downregulation on synchronous mitosis in the dikaryon. (A) Examples of asynchronous mitosis. In A1 a hyphal tip cell is undergoing synchronous mitosis with one nucleus dividing at the clamp cell and the other one in the main hypha. In A2 and A3 nuclei are dividing at different times. Asterisks mark the dividing nuclei. Bar: 10 μm. (B) Quantification of the presence of asynchronous mitosis. The graph shows the result from two independent experiments, counting >50 mitotic cells each. Means and standard deviations are shown. Only apical cells that possessed two nuclei were analyzed. (C) Growing the cells in the presence of sublethal amounts of aphidicolin (AC), a replication inhibitor, increased the frequency of aberrant mitosis in chk1- and atr1-silenced strains. A total of 50 cells per experiment (two independent experiments) were counted per mutant. The lethal dose (i.e., absence of growth) of AC for wild-type cells in the experimental conditions was 100 μM. Means and standard deviations are shown.
These results suggest the idea that the Atr1/Chk1 pathway could be the surveillance system that avoids the entry into mitosis until both nuclei have finished their respective S phases. Following this idea, in the absence of Atr1 or Chk1 this coupling is lost and then different replication rates in the distinct nuclei will lead to an asynchronous entry into mitosis, which may affect the ability to produce or to resolve an appropriated clamp connection, and will result in an abnormal division. To test this prediction we reasoned that conditions affecting DNA replication would result in an increase in abnormal mitosis in those cells defective in the DDR pathway while they would not affect wild-type cells retaining this checkpoint activity. We incubated the different strains on a thin agar layer upon a microscope glass slide in the presence of two different sublethal concentrations of aphidicolin, a specific inhibitor of α-DNA polymerase (Ikegami et al. 1978), and quantified the proportion of cells in which abnormal mitosis (both aberrant mitosis and trapped nuclei as defined above) took place. We observed that in cells showing downregulation of atr1 or chk1 expression even the lower amount of aphidicolin severely affected the ability to produce correct mitosis (Figure 3C). Interestingly, in control cells these conditions decreased the proportion of cells resulting from abnormal mitosis (see Discussion).
Atr1 and Chk1 are necessary for mature fruiting body formation in C. cinerea
Fruiting body (mushroom) formation is the most complex developmental process in the life cycle of C. cinerea that starts from a mesh of free undifferentiated hyphae and ends up as a compact structure with differentiated tissues inside. The production of fruiting bodies depends on environmental conditions that include temperature, humidity, light, and nutrients (Kües and Liu 2000). Several studies indicated the importance in C. cinerea of the correctness of the dikaryotic cell cycle for appropriated developmental decisions: nuclear division and septation have to be coordinated in such a way that each daughter cell inherits a balance of each parental genome. Interferences in this process often resulted in an impairment in the formation of fruiting bodies (Tanabe and Kamada 1994; Muraguchi et al. 2008). We wondered about the possible consequences of the impaired dikaryosis observed in Atr1-Chk1-defective cells on the ability of C. cinerea to produce fruiting bodies. For this, we evaluated in the silenced strains the formation of mushrooms under nutrient-controlled conditions.
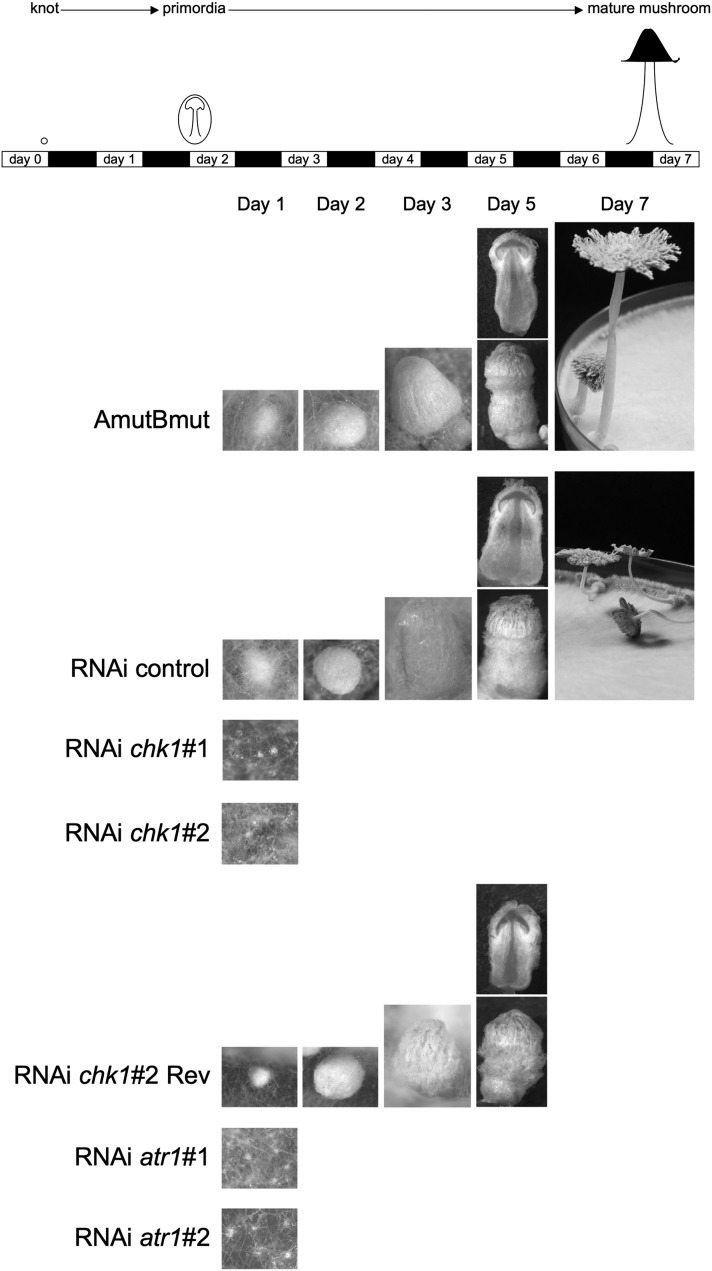
To induce fruiting, each strain was grown on 15 YMG/T plates until the mycelium filled up all the plate surface and just before the hyphae reached the border, the growth conditions of these plates were changed from 37° and 24 hr dark to 25°, >90% humidity and a 12-hr light/12-hr dark regime. This experiment was done three times with similar results. In the control strains, all the different developmental stages that had been previously described (Boulianne et al. 2000) were observed: loosely aggregated hyphal knots became primordia with differentiated tissues and finally gave rise to mature fruiting bodies. In contrast, in the silenced strains no fully developed mushrooms were observed (Figure 4). Both atr1- and chk1-silenced strains could form only hyphal knots. Interestingly, for the RNAi chk1#2 Rev strain we observed that primordia aborted, when in the dikaryotic basidia premeiotic S-phase replication should happen and karyogamy should be induced (Raudaskoski and Lu 1980; Kües et al. 2002a), indicating that even a small reduction in chk1 expression levels affected the ability to produce a full life cycle in this fungus (see Discussion).
Figure 4.
Fruiting defects in chk1 and atr1 knockdown strains. The top panel shows the approximate time course of fruiting and the times when photographs (bottom panels) were taken. Note that hyphal knots in knockdown mutants never developed into fruiting body primordia. Only the reverted strain RNAi chk1#2 Rev was able to develop into fully differentiated fruiting body primordia, but development stopped at the prekaryogamy stage.
Downregulation of the DDR response differentially affects other development choices
C. cinerea dikaryotic cells have a wide-ranging developmental potential. This potential ranges from the above-mentioned fruiting body (that produces the meiotic basidiospores) to production of asexual spores (oidia) and mitotic submerged spores (chlamydospores) as well as a plethora of multicellular structures such as sclerotia, mycelial cords, pseudorhizas, and rockeries (Kües 2000). This variety of developmental choices is a response to changing environmental conditions. For instance, oidia formation is dependent on light/darkness incubation (Kertesz-Chaloupkova et al. 1998) while sclerotia arise when the fungus has been kept for a long period in the dark (Moore 1981).
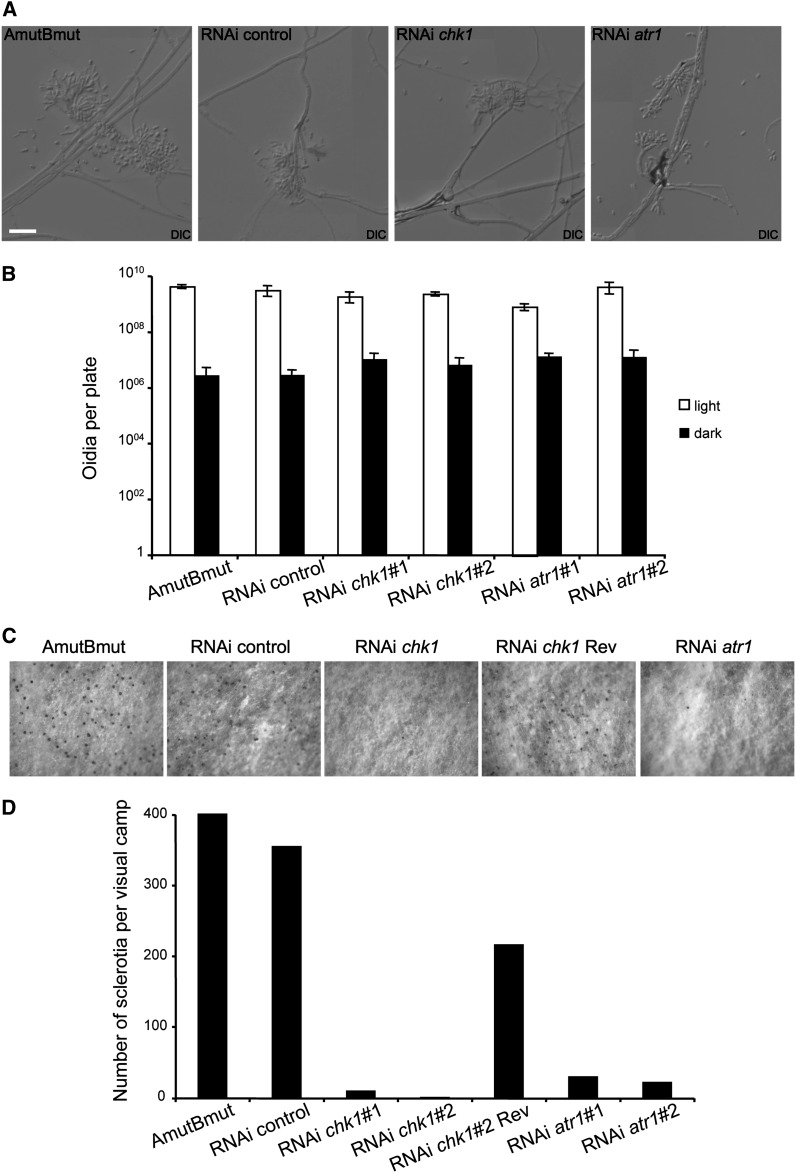
We wondered whether downregulation of DDR response in C. cinerea also affected other developmental responses, in addition to fruiting body formation. We analyzed the ability to produce oidia and sclerotia. Surprisingly, we found a distinct response in silenced strains with respect to these distinct developmental choices. We observed no differences between control and silenced strains in oidia production (Figure 5, A and B). However, we found that formation of the globose dormant sclerotia from hyphal knots as an alternative pathway to fruiting (Moore 1981; Kües et al. 2002a) was also dramatically impaired in strains carrying the silenced genes (Figure 5, C and D).
Figure 5.
Silencing of atr1 and chk1 affects other developmental decisions. (A) Images of oidiophores and oidia in the indicated strains. The ability to form oidiophores after light induction was not affected in silenced strains. Bar: 20 μm. (B) Quantification of the ability to produce oidia in the silenced strains. A similar number of oidia were produced by silenced and control strains, both in light and in dark conditions. (C and D) Sclerotia formation was severely inhibited in silenced strains. Sclerotia can be observed as black dots in the surface of the aerial mycelium. Note the scarce number of sclerotia per mycelial camp in silenced strains.
In summary, alteration of expression levels of the DDR kinases Atr1 and Chk1 resulted in an inability to carry out some developmental choices such as fruiting body formation or sclerotia formation, while other processes such as oidia formation were unaffected.
Discussion
The main conclusion of this study is that the Atr1 and Chk1 kinases, required for the response to DNA damage in a number of eukaryotic organisms, also have roles in the control of the dikaryotic cell cycle as well as in the developmental choices of C. cinerea.
During the dikaryotic cell cycle, nuclear division and septation have to be coordinated in such a way that each daughter cell inherits a balance of each parental genome. Although in some fungi the formation of clamp connections is not a requisite for this intricate process, in C. cinerea clamp connections are necessary for appropriated cell division (Inada et al. 2001). We found that the downregulation of Chk1 and Atr1 levels seem to result in a high frequency of abnormal mitosis. The outcome of these abnormal mitoses is the presence of an excess of nuclei per hyphal cell, nuclei trapped in the clamp cell, or septa with no associated clamp structure. How Atr1 and Chk1 deficiency is connected at a molecular level with these abnormalities is not fully addressed in our study. We envisioned several possible explanations. One is a putative connection between the Atr1/Chk1 regulatory cascade and the formation of the clamp structure. In S. cerevisiae it has been reported that DDR checkpoint kinase Rad53 is able to control morphogenetic events during bud formation via its interaction with septins (Enserink et al. 2006; Smolka et al. 2006). Although no study has addressed so far whether septins are involved in clamp formation in C. cinerea, most likely it will be the case, as it was reported recently in dikaryons from Cryptoccocus neoformans, another basidiomycete (Kozubowski and Heitman 2010). Clamp formation is coupled somehow to nuclear position as well as cell cycle stage although the details of such a coupling are not well understood (Tanabe and Kamada 1994). It could be possible that Atr1 and Chk1 are elements involved in this connection. When the clamp formation is impaired or disconnected from nuclear position and/or division, then abnormal mitosis is likely to arise.
We also entertained a second explanation more related with the “canonical” role of the Atr1-Chk1 cascade during the cell cycle (to delay the G2/M transition in the presence of unreplicated or damaged DNA). Since during nuclear division in dikaryotic cells, the two nuclei have to divide synchronously (Tanabe and Kamada 1994), it could well be that the Atr1/Chk1 pathway acts as a surveillance system that avoids the entry into mitosis until both nuclei have finished their respective S phases. In the absence of Atr1 or Chk1, this coupling is lost and then different rates of replication in the distinct nuclei will result in a differential entry into mitosis, which may affect the ability to produce or to resolve an appropriated clamp connection and would result in an abnormal division. Our results support this view. We found a much higher frequency of asynchronous mitosis in the silenced strains than in control strains. In addition, incubation of the silenced strains in the presence of a DNA replication inhibitor resulted in a higher percentage of cells that originated from abnormal divisions. Surprisingly, in these same conditions we found that for control strains the mitosis seemed more accurate since the percentage of cells resulting from abnormal mitosis decreased. We explain these results assuming that in normal conditions there is a certain degree of leakiness in the Atr1-Chk1 axis during cell division and that having the Atr1-Chk1 axis chronically activated at a low level (as may happen in the presence of a sublethal concentration of aphidicolin) may correct this leakiness. We observed that a higher level of “mitotic accuracy” seems not to affect the ability to grow or to produce fruiting bodies in control strains (Table S1). We can speculate about the advantages for the fungus to have a small population of cells that escape from dikaryosis. Aside from the described consequences on cell fitness (Clark and Anderson 2004), one envisioned advantage of dikaryotic vs. diploid status is its reversibility [diploids raise haploid cells only after meiosis while dikaryons can produce haploid cells just by nuclear sorting (Kües et al. 2002a)]. It is possible that in nature for a dikaryotic mycelium to have a small population of haploid cells (as a result of this low level of abnormal mitosis) may increase the chances of the fungus to cope with changes on the external conditions, increasing the well-known adaptability of fungi to environmental cues (Kües 2000; Kües et al. 2002a).
In addition to the observed cell cycle defects, a striking result we found upon downregulation of atr1 and chk1 was the inability to assume some developmental choices, such as the fruiting body initiation or the induction of sclerotia. We believe that the most obvious explanation is that the developmental defects were a consequence of the cell cycle impairment. In C. cinerea developmental decisions are controlled by environmental conditions as well as by an appropriated balance of the mating-type regulators (Tymon et al. 1992; Kües et al. 1998, 2002b). Therefore it seems likely that any condition affecting the correct dose of mating-type regulators will affect the further developmental decisions. Fruiting body initiation is dependent on the balanced mating-type regulator doses present in the dikaryon. In the same way, sclerotia production in the dikaryon is enhanced by the mating-type proteins compared to the monokaryotic situation (Kües et al. 1998). Therefore, we believe that in the silenced strains, the disruption in nuclear communication/synchronization could mimic the monokaryotic situation affecting these processes. In contrast, oidiation is a process that involves—even in the dikaryon—only a single nucleus that undergoes a pathway of specific development of uninuclear cells as defined on the monokaryon (Polak et al. 1997; Kües et al. 2002a) and therefore the loss of nuclear synchronization may be not so important for this developmental decision.
However, although we consider unlikely a direct role of the Atr1-Chk1 cascade in the developmental choices in C. cinerea, that does not necessarily exclude that this regulatory cascade may have additional roles during fruiting body formation. Since karyogamy in C. cinerea coincides with onset of fruiting body maturation (Kües et al. 2002a), it seems likely that arrest in fruiting body development of reverted strain RNAi chk1#2 Rev occurs at premeiotic S phase. It has been reported that in C. cinerea, treatment of primordial caps at premeiotic S phase with hydroxyurea arrests further fruiting body development, while treatment after karyogamy at later meiotic stages also affects sporulation but still allows fruiting body maturation (Raudaskoski and Lu 1980). In animal germ cells, the ATR/CHK1 checkpoint pathway is activated during entry into meiosis (Miles et al. 2010; Chen et al. 2012), further supporting the idea that in C. cinerea this signaling cascade might be required in the replicative premeiotic S phase. In addition, recent work indicated the presence of checkpoint arrest in several meiotic mutants in C. cinerea (Anderson et al. 2012).
We believe that future studies dedicated to analyzing the connections between developmental decisions and dikaryotic cell cycle regulation in C. cinerea will help to increase our knowledge of these important processes.
Supplementary Material
Acknowledgments
We thank Jimmy Correa-Bordes (Universidad de Badajoz) for stimulating discussions. We also thank the anonymous referees for excellent suggestions. C.d.S.-T. was supported by FPI (Formación de Personal Investigador) contract (BES-2007-16813). This work was supported by grants from the Spanish Government (BIO2008-04054 and BIO2011-27773).
Footnotes
Communicating editor: A. Houben
Literature Cited
- Abraham R. T., 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15: 2177–2196. [DOI] [PubMed] [Google Scholar]
- Anderson E., Burns C., Zolan M. E., 2012. Global gene expression in Coprinopsis cinerea meiotic mutants reflects checkpoint arrest. G3 2: 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J., Lukas J., 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3: 421–429. [DOI] [PubMed] [Google Scholar]
- Bottoli A. P., Kertesz-Chaloupkova K., Boulianne R. P., Granado J. D., Aebi M., et al. , 1999. Rapid isolation of genes from an indexed genomic library of C. cinereus in a novel pab1+ cosmid. J. Microbiol. Methods 35: 129–141. [DOI] [PubMed] [Google Scholar]
- Boulianne R. P., Liu Y., Aebi M., Lu B. C., Kues U., 2000. Fruiting body development in Coprinus cinereus: regulated expression of two galectins secreted by a non-classical pathway. Microbiology 146: 1841–1853. [DOI] [PubMed] [Google Scholar]
- Brown A. J., Casselton L. A., 2001. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 17: 393–400. [DOI] [PubMed] [Google Scholar]
- Buller A. H. R., 1958. The Formation of Hyphal Fusions in the Fusions in the Mycelium of the Higher Fungi. Hafner Publishing, New York. [Google Scholar]
- Burns C., Gregory K. E., Kirby M., Cheung M. K., Riquelme M., et al. , 2005. Efficient GFP expression in the mushrooms Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet. Biol. 42: 191–199. [DOI] [PubMed] [Google Scholar]
- Casselton L. A., 1978. Dikaryon Formation in Higher Basidiomycetes. Edward Arnold, London. [Google Scholar]
- Castillo-Lluva S., Garcia-Muse T., Perez-Martin J., 2004. A member of the Fizzy-related family of APC activators is regulated by cAMP and is required at different stages of plant infection by Ustilago maydis. J. Cell Sci. 117: 4143–4156. [DOI] [PubMed] [Google Scholar]
- Chen L., Chao S. B., Wang Z. B., Qi S. T., Zhu X. L., et al. , 2012. Checkpoint kinase 1 is essential for meiotic cell cycle regulation in mouse oocytes. Cell Cycle 11: 1948–1955. [DOI] [PubMed] [Google Scholar]
- Chiu S. W., Moore D., 1990. A mechanism for gill pattern formation in Coprinus cinereus. Mycol. Res. 94: 320–326. [Google Scholar]
- Chomczynski P., Sacchi N., 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D., 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T. A., Anderson J. B., 2004. Dikaryons of the basidiomycete fungus Schizophyllum commune: evolution in long-term culture. Genetics 167: 1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings W. J., Celerin M., Crodian J., Brunick L. K., Zolan M. E., 1999. Insertional mutagenesis in Coprinus cinereus: use of a dominant selectable marker to generate tagged, sporulation-defective mutants. Curr. Genet. 36: 371–382. [DOI] [PubMed] [Google Scholar]
- de Sena-Tomas C., Fernandez-Alvarez A., Holloman W. K., Perez-Martin J., 2011. The DNA damage response signaling cascade regulates proliferation of the phytopathogenic fungus Ustilago maydis in planta. Plant Cell 23: 1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J. M., Smolka M. B., Zhou H., Kolodner R. D., 2006. Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J. Cell Biol. 175: 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granado J. D., Kertesz-Chaloupkova K., Aebi M., Kues U., 1997. Restriction enzyme-mediated DNA integration in Coprinus cinereus. Mol. Gen. Genet. 256: 28–36. [DOI] [PubMed] [Google Scholar]
- Heneghan M. N., Costa A. M., Challen M. P., Mills P. R., Bailey A., et al. , 2007. A comparison of methods for successful triggering of gene silencing in Coprinus cinereus. Mol. Biotechnol. 35: 283–296. [DOI] [PubMed] [Google Scholar]
- Ikegami S., Taguchi T., Ohashi M., Oguro M., Nagano H., et al. , 1978. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature 275: 458–460. [DOI] [PubMed] [Google Scholar]
- Inada K., Morimoto Y., Arima T., Murata Y., Kamada T., 2001. The clp1 gene of the mushroom Coprinus cinereus is essential for A-regulated sexual development. Genetics 157: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa M., Tanabe S., Kamada T., 1998. The two nuclei in the dikaryon of the homobasidiomycete Coprinus cinereus change position after each conjugate division. Fungal Genet. Biol. 23: 110–116. [DOI] [PubMed] [Google Scholar]
- Kamada T., 2002. Molecular genetics of sexual development in the mushroom Coprinus cinereus. Bioessays 24: 449–459. [DOI] [PubMed] [Google Scholar]
- Kertesz-Chaloupkova K., Walser P. J., Granado J. D., Aebi M., Kues U., 1998. Blue light overrides repression of asexual sporulation by mating type genes in the basidiomycete Coprinus cinereus. Fungal Genet. Biol. 23: 95–109. [DOI] [PubMed] [Google Scholar]
- Kilaru S., Hoegger P. J., Majcherczyk A., Burns C., Shishido K., et al. , 2006. Expression of laccase gene lcc1 in Coprinopsis cinerea under control of various basidiomycetous promoters. Appl. Microbiol. Biotechnol. 71: 200–210. [DOI] [PubMed] [Google Scholar]
- Koc A., Wheeler L. J., Mathews C. K., Merrill G. F., 2004. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 279: 223–230. [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Heitman J., 2010. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of Cryptococcus neoformans. Mol. Microbiol. 75: 658–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües U., 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64: 316–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües U., Liu Y., 2000. Fruiting body production in Basidiomycetes. Appl. Microbiol. Biotechnol. 54: 141–152. [DOI] [PubMed] [Google Scholar]
- Kües U., Granado J. D., Hermann R., Boulianne R. P., Kertesz-Chaloupkova K., et al. , 1998. The A mating type and blue light regulate all known differentiation processes in the basidiomycete Coprinus cinereus. Mol. Gen. Genet. 260: 81–91. [DOI] [PubMed] [Google Scholar]
- Kües U., Polak E., Hollenstein M., Bottoli A. P. F., Walser P. J.et al, 2002a Vegetative development in Coprinus cinereus, pp. 133–164 in Molecular Biology of Fungal Development, edited by Osiewacz H. D. Marcel Dekker, New York. [Google Scholar]
- Kües U., Walser P. J., Klaus M. J., Aebi M., 2002b Influence of activated A and B mating-type pathways on developmental processes in the basidiomycete Coprinus cinereus. Mol. Genet. Genomics 268: 262–271. [DOI] [PubMed] [Google Scholar]
- Lundin C., North M., Erixon K., Walters K., Jenssen D., et al. , 2005. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33: 3799–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino R., Kamada T., 2004. Isolation and characterization of mutations that affect nuclear migration for dikaryosis in Coprinus cinereus. Curr. Genet. 45: 149–156. [DOI] [PubMed] [Google Scholar]
- Mielnichuk N., Sgarlata C., Perez-Martin J., 2009. A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J. Cell Sci. 122: 4130–4140. [DOI] [PubMed] [Google Scholar]
- Miles D. C., van den Bergen J. A., Sinclair A. H., Western P. S., 2010. Regulation of the female mouse germ cell cycle during entry into meiosis. Cell Cycle 9: 408–418. [DOI] [PubMed] [Google Scholar]
- Moore D., 1981. Developmental genetics of Coprinus cinereus: genetic evidence that carpophores and sclerotia share a common pathway of initiation. Curr. Genet. 3: 145–150. [DOI] [PubMed] [Google Scholar]
- Muraguchi H., Abe K., Nakagawa M., Nakamura K., Yanagi S. O., 2008. Identification and characterisation of structural maintenance of chromosome 1 (smc1) mutants of Coprinopsis cinerea. Mol. Genet. Genomics 280: 223–232. [DOI] [PubMed] [Google Scholar]
- Murata Y., Fujii M., Zolan M. E., Kamada T., 1998. Molecular analysis of pcc1, a gene that leads to A-regulated sexual morphogenesis in Coprinus cinereus. Genetics 149: 1753–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A., 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36: 617–656. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J., de Sena-Tomas C., 2011. Dikaryotic cell cycle in the phytopathogenic fungus Ustilago maydis is controlled by the DNA damage response cascade. Plant Signal. Behav. 6: 1574–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P., 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26: 509–515. [DOI] [PubMed] [Google Scholar]
- Polak E., Hermann R., Kues U., Aebi M., 1997. Asexual sporulation in coprinus cinereus: structure and development of oidiophores and oidia in an amut bmut homokaryon. Fungal Genet. Biol. 22: 112–126. [DOI] [PubMed] [Google Scholar]
- Rao P. S., Niederpruem D. J., 1969. Carbohydrate metabolism during morphogenesis of Coprinus lagopus (sensu Buller). J. Bacteriol. 100: 1222–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudaskoski M., Lu B. C., 1980. The effect of hydroxyurea on meiosis and genetic recombination in the fungus Coprinus lagopus. Can. J. Genet. Cytol. 22: 41–50. [DOI] [PubMed] [Google Scholar]
- Salo V., Niini S. S., Virtanen I., Raudaskoski M., 1989. Comparative immunocytochemistry of the cytoskeleton in filamentous fungi with dikaryotic and multinucleate hyphae. J. Cell Sci. 94: 11–24. [Google Scholar]
- Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S., 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73: 39–85. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Wong C., Thoma R. S., Richman R., Wu Z., et al. , 1997. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277: 1497–1501. [DOI] [PubMed] [Google Scholar]
- Smolka M. B., Chen S. H., Maddox P. S., Enserink J. M., Albuquerque C. P., et al. , 2006. An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J. Cell Biol. 175: 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J. E., Wilke S. K., Ahren D., Au C. H., Birren B. W., et al. , 2010. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. USA 107: 11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy S., 1984. Morphogenetic effects of mutations at the A and B incompatibility factors in Coprinus cinereus. Microbiology 130: 3219–3224. [Google Scholar]
- Tanabe S., Kamada T., 1994. The role of Astral microtubules in conjugate division in the dikaryon of Coprinus cinereus. Exp. Mycol. 18: 338–348. [Google Scholar]
- Tanabe S., Kamada T., 1996. Dynamics of the actin cytoskeleton, hyphal tip growth and the movement of the two nuclei in the dikaryon of Coprinus cinereus. Mycoscience 37: 339–344. [Google Scholar]
- Tymon A. M., Kues U., Richardson W. V., Casselton L. A., 1992. A fungal mating type protein that regulates sexual and asexual development contains a POU-related domain. EMBO J. 11: 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag A., Lee M. P., Si H., Harris S. D., 2007. Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Mol. Microbiol. 66: 1579–1596. [DOI] [PubMed] [Google Scholar]
- Walti M. A., Villalba C., Buser R. M., Grunler A., Aebi M., et al. , 2006. Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs. Eukaryot. Cell 5: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolan M. E., Pukkila P. J., 1986. Inheritance of DNA methylation in Coprinus cinereus. Mol. Cell. Biol. 6: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.