Abstract
Over the past few years, mass spectrometry has emerged as a technology to complement and potentially replace standard immunoassays in routine clinical core laboratories. Application of mass spectrometry to protein and peptide measurement can provide advantages including high sensitivity, the ability to multiplex analytes, and high specificity at the amino acid sequence level. In our previous study, we demonstrated excellent reproducibility of mass spectrometry-selective reaction monitoring (MS-SRM) assays when applying standardized standard operating procedures (SOPs) to measure synthetic peptides in a complex sample, as lack of reproducibility has been a frequent criticism leveled at the use of mass spectrometers in the clinical laboratory compared to immunoassays. Furthermore, an important caveat of SRM-based assays for proteins is that many low-abundance analytes require some type of enrichment before detection with MS. This adds a level of complexity to the procedure and the potential for irreproducibility increases, especially across different laboratories with different operators. The purpose of this study was to test the interlaboratory reproducibility of SRM assays with various upfront enrichment strategies and different types of clinical samples (representing real-world body fluids commonly encountered in routine clinical laboratories). Three different, previously published enrichment strategies for low-abundance analytes and a no-enrichment strategy for high-abundance analytes were tested across four different laboratories using different liquid chromatography-SRM (LC-SRM) platforms and previously developed SOPs. The results demonstrated that these assays were indeed reproducible with coefficients of variation of less than 30% for the measurement of important clinical proteins across all four laboratories in real world samples.
Keywords: SRM assay, mass spectrometry, proteomics, clinical assay
INTRODUCTION
The list of laboratory-developed tests and FDA-approved assays for proteins and peptides numbers over 200 and is growing at a rapid rate1. Many or most of these assays are offered as immunoassays running on automated clinical analyzers. Currently, a critical and unmet need exists for alternatives to immunoassays, primarily because of both the cost and time expended in development and validation of well-performing antibodies as well as their lack of specificity. This, along with the lack of standardization and certified reference materials, results in poor interplatform analyzer agreement2. Over the past few years, mass spectrometry has emerged as a technology to complement and potentially replace standard immunoassays in routine clinical core laboratories3. Application of mass spectrometry to protein and peptide measurement can provide advantages, including the ability to multiplex analytes and specificity at the amino acid sequence level4. The latter is particularly important, as it is increasingly evident that closely related protein isoforms and variants play a major role in diseases.5 Most antibody-based tests cannot provide the requisite specificity for accurate quantification of specific protein isoforms and fragments6 As the era of personalized medicine and high-throughput analysis emerges, panels of biomarkers will become the standard of care, and technologies/platforms capable of handling such throughput will become favorable.
Traditionally, a frequent criticism leveled at the use of mass spectrometers in the clinical laboratory is the perceived lack of interlaboratory reproducibility of instrumentation platforms and protocols, poor overall analytical sensitivity of MS compared to immunoassays, as well as their cost vis a vis immunoassays7. In fact, some studies, including a recent one from this group, have demonstrated excellent reproducibility of mass spectrometry-selective reaction monitoring (MS-SRM) assays when applying standardized SOPs and proteins including synthetic isotopically labeled peptides8,9 Moreover, when the throughput and capacity for multiplexing are calculated into the cost/benefit analyses, mass spectrometry-based SRM assays present an attractive alternative to standard immunoassays, especially when the latter cannot quantify the relevant protein analyte variants accurately.
An important caveat of SRM-based assays for proteins is that many low-abundance analytes require some type of enrichment before detection with MS.10–13 This adds a level of complexity to the procedure and the potential for irreproducibility increases, especially across different laboratories with different operators.14 While multidimensional sample fractionation may provide some increase in analytical sensitivity through reduced complexity, fractionation is not enrichment, and thus low -abundance analytes are still low abundance after fractionation. Consequently, efforts are underway to instill upfront sample preparation methods that concentrate and enrich the target analyte ahead of time prior to MS.
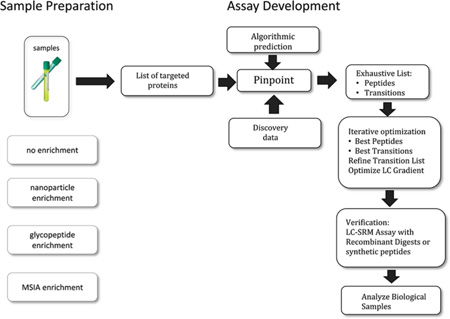
The purpose of this study was to test the interlaboratory reproducibility of SRM assays with various upfront enrichment strategies and different types of bona fide clinical samples (representing real-world body fluids commonly encountered in routine clinical laboratories). Three different, previously published enrichment strategies were tested across four different laboratories using the previously developed SRM platform and SOPs.8,15–18 We made the decision to have each laboratory perform one type of sample preparation and then distribute the samples to the other three laboratories for evaluation on the SRM platform. This was an intermediate step between running SRM assays with synthetic standards9 and completing the entire workflow (from sample preparation to MS) in each individual laboratory. Our intent was to demonstrate robustness and reproducibility of SRM peptide assays across different sample types including serum, urine, and seminal plasma in multiple laboratories. Even without completing the entire workflow in each laboratory, this was a significant challenge since protein sample preparation techniques are complex and can result in a significant degree of variability even before introduction to the mass spectrometer.15 Before SRM assays can be routinely accepted in clinical laboratories, robust results with low variability using a variety of different sample types and preparation techniques must be demonstrated. Ultimately, workflows from sample collection to introduction to the mass spectrometer will be performed in each individual laboratory. In our study, enrichment using glycopeptide capture, immunocapture, and nanoparticle as well as no enrichment were performed on the various sample types. Each laboratory prepared one type of enriched clinical sample and shipped it to the other laboratories. All four laboratories ran the SRM assay for all four types of samples. A total of six proteins, eight peptides, and two different mass spectrometers were examined. Three different bona fide clinical sample types including serum, urine, and seminal plasma were processed and tested. First, glycopeptides enriched from serum samples16 from prostate cancer patients were examined. Next, mass spectrometric immunoassay (MSIA)-SRM15 was applied to enrich prostate specific antigen (PSA) from serum samples. Then a novel enrichment strategy using nanoparticles was applied to urine samples to enrich human growth hormone17, and finally, four medium-to-high abundance proteins were analyzed in trypsin-digested seminal plasma without any additional upfront enrichment or purification.18 Figure 1 shows the four protocols used in this study. The results demonstrated that these SRM assays were sensitive enough to detect analytes at clinical relevant levels and were reproducibly analyzed in multiple laboratories with coefficients of variation of less than 30% across all laboratories. These data support the use of SRM assays with upfront enrichment strategies for measuring important clinical analytes in real samples.
Figure 1.
Protocol used in the study. Four different types of sample preparation (three enrichment protocols and one nonenrichment protocol) were used.
MATERIALS AND METHODS
Platform
The platform used in the four laboratories was described in detail in our previous study.9 All SRM assays were developed on a TSQ Quantum Ultra triple quadrupole mass spectrometer, Surveyor MS pump, Micro Autosampler and an IonMax Source equipped with a low flow metal needle (Thermo Fisher Scientific), flow rate 160–200 µL/min. Reverse phase separations were carried out on a 1 mm ×150 mm Hypersil Gold 3 µmC18 particle.
Clinical Samples, Enrichment, and Trypsin Digestion Procedures
N-Linked Glycosylated Peptides Enrichment from Human Serum
Serum samples and clinical information were obtained with informed consent and performed with the approval of the Institutional Review Board of Johns Hopkins. The samples were then prepared to enrich N-linked glycopeptides by Johns Hopkins University and distributed to the other three laboratories participating in this study. NT-Linked glycopeptides isolated from three samples were used in this multiple laboratory study:
Sample N (Normal) - glycopeptides extracted from 10 µL of pooled healthy male serum.
Sample C (Cancer) - glycopeptides extracted from 10 µL of pooled prostate cancer male serum. The PSA concentration in cancer serum was 44.5 ng/mL as measured by the clinically used PSA assay.
Sample P (PSA) - glycopeptides extracted from 10 µL of pooled healthy female serum spiked with recombinant PSA. The final PSA concentration in serum samples was 51.4 ng/mL as measured by the clinically used PSA assay16.
Additionally, 300 fmol of heavy peptides (DKSVILLGR) with 13C- and 15N-labeled aspartic acid was spiked in each sample.16 Recombinant PSA spiked in sample P has glycosylation, so we expect the enrichment of N-linked glycopeptide from the control sample to be similar to the enrichment of endogenous PSA.
Formerly N-linked glycosylated peptides were isolated from serum samples using the N-linked glycopeptide capture procedure.19,20 Briefly, proteins from 20 µL of serum were first denatured in 90 µL of 8M urea, 0.4 M NH4HCO3, and 0.1% SDS for 2 h at 60 °C. The peptides were then reduced by adding 10 µL of 120 mM Tris(2-carboxyethyl) phosphine at room temperature for 30 min and alkylated by mixing with 10 µL of 160 mM iodoacetamide at room temperature for 30 min in the dark. The protein solution was diluted by 190 µL of Arg-C digestion buffer (100 mM Tris-HCl, 20 mM CaCl2, 10 mM DTT, I mM EDTA, 40 mM methylamine, adjust pH to 7.6). Ten micrograms of Arg-C was added to digest protein at 37 °C overnight with gentle shaking. The digested peptides were cleaned with C18 columns and oxidized by adding 45 µL of 100 mM sodium periodate in 50% acetonitrile at 4 °C for 1 h in the dark. After removal of the oxidant using C18 columns, the sample was conjugated to hydrazide resin at room temperature for 4 h in 80% acetonitrile. Non-glycosylted peptides were then removed by washing the resin three times with 800 µL of 1.5 M NaCl, H2O, and 100 mM NH4HCO3. Then, N-linked glycopeptides were released from the resin by addition 2.5 mU of PNGase F in 100 mM of NH4HCO3 and incubated at 37 °C overnight. After the final clean up by MCX columns, the peptides from each sample were dried and resuspended in 20 µL of 0.4% acetic acid solution. The 10 µL of glycopeptide mixture from 10 µL of original serum were used in each liquid chromography-mass spectrometry (LC-MS) analysis.
Nanoparticle-based Pituitary Human Growth Hormone Enrichment from Human Urine
Recombinant human growth hormone (Humatrope Somatropin, Eli Lilly, Inc.) was first used to optimize the SRM procedures in the laboratory at George Mason University. Following optimization, pituitary human growth hormore (pit-hGH, NIBCS, United Kingdom, Growth Hormone, Human, Pituitary NIBSC code: 80/505) was spiked into human urine at 100 ng/mL in order to produce “real world” samples with human-derived hGH. Serial dilutions were made with urine as the diluent to obtain the following concentrations of pit-hGH which span the known upper physiologic range of hGH in clinical samples17 50 ng/mL, 25 ng/mL, 10 ng/mL, 5 ng/mL, 2.5 ng/mL, 1.25 ng/mL, and 0.5 ng/mL). This was used to study the limit of detection (LOD)/limit of quantitation (LOQ) in-house. One millimeter of each of these samples was used for nanoparticle enrichment (the enriched sample was later brought up to 50 µL), while 50 µL of urine sample (with no pit-hGH spiked in) was used without nanoparticle enrichment as the blank sample. For interlaboratory comparison, one clinical urine sample (4 mL total volume) with hGH spiked in at 10 ng/mL (median range of known physiologic concentration) was subjected to nanoparticle enrichment and aliquots from the resultant eluate were tested across laboratories.
Briefly, 1 mL of each urine sample was centrifuged at 3000 rpm for 10 min at 4 °C. The urine supernatant was removed and incubated with 500 µL of nanoparticles (with Remazol Brilliant Blue as affinity bait) for 20 min at room temperature. Samples were centrifuged at 13 000 rpm for 40 min at room temperature to separate nanoparticles and the urine. The supernatant was removed, and the nanoparticles washed with 1 mL of water by vortexing for 10 s at room temperature. Samples were centrifuged at 13 000 rpm for 40 min at room temperature, the supernatant was removed, and the nanoparticles were washed a second time with water. After another centrifugation step, and removal of the water, proteins were eluted from the nanoparticles by briefly vortexing the nanoparticles at room temperature with 500 µL of 10% ammonium hydroxide/70% acetonitrile, followed by sonication for 2 min. Samples were centrifuged at 13 000 rpm at 25 °C for 15 min, and the eluate was collected. A second elution was performed and the two elutions were pooled. The pooled elutions, as well as the 50 µL samples of pit-hGH spiked urine not enriched with nanoparticles, were dried to completion in the speed-vac. Samples were then reduced at room temperature for 1 h in 1 M urea/50 mM ammonium bicarbonate/10 mM DTT. Iodoacetamide was added at a final concentration of 50 mM, and samples were placed in the dark at room temperature for 20 min. 500 ng of trypsin were added, and samples were placed at 37 °C overnight for digestion. One microliter of concentrated trifluoroacetic acid (TFA) was added to stop the digestions. Samples were vacuum-dried and reconstituted in 50 µL of 200 µg/mL glucagon.
MSIA Enrichment of Prostate Specific Antigen from Human Serum
All samples used in this study were acquired under an institutional review board-approved protocol, and informed consent was obtained from all study participants. One pooled serum sample was tested, for which samples were collected at Mount Sinai Hospital in Toronto, prepared at BRIMS, and then distributed to all four laboratories. Samples were frozen at −80 °C until use. Antibodies for PSA were obtained from Medix Biochemica, Kauniainen, Finland. MSIA-Tips were prepared by Intrinsic Bioprobes, Tempe, AZ, as previously described. Before analysis, samples were thawed in an ice bath, and 1 mL of serum was diluted with 750 µL of HBS-EP buffer to a total analytical volume of 1.75 mL. PSA was then extracted with the aid of a Platemate 2 × 3 96 pipetting robot (Thermo Scientific) by repeatedly (1500 repetitions) drawing and expelling (back into the analytical volume) 150 µL aliquots of the analytical volume through the antibody-prelinked MSIA-Tip. After extraction, the pipettes were rinsed with HBS-EP and H2O (in this order; each rinse consisted of 15 repetitions of 150 µL), after which PSA was immediately eluted for detection.
Bound proteins were eluted from the tips into a 96-well plate (AB-1300, Abgene) by pipetting 100 µL of 30% acetonitrile/0.5% formic acid up and down for a total of 100 cycles. Samples were lyophilized to dryness and then resuspended in 25 µL of 8 M urea/2.5% n-propanol/200 mM Tris/10 mM DTT, pH 8.5. Next the samples were reduced by incubation at 37°C for 1 h. The samples were then cooled to room temperature and iodoacetic acid (40 mM final) was added for the alkylation step. The reduced and alkylated samples were diluted with 130 µL of 50 mM Tris/5 mM CaCl2 and 1 µL of trypsin was added to each sample in 40 µL of 25 mM acetic acid. Samples were allowed to digest for 18 h at 37 °C. After digestion, samples were acidified to 1% TFA and subjected to C18 purification following the vendor’s protocol (Pierce). Samples were lyophilized and resuspended in 30 µL of 5% (v/v) acetonitrile/0.2% (v/v) formic acid/15 ng/µL glucagon/PSA heavy peptides.
Samples and Protocols for Measurement of Seminal Plasma Proteins
Seminal plasma samples were obtained with informed consent and Mount Sinai Hospital IRB approval from normal fertile men about to undergo a vasectomy (“Normal” sample, total protein concentration 45.8 mg/mL) and previously fertile men who had undergone a vasectomy (obstructive asoospermia (OA) Post-Vasectomy sample, total protein concentration 44 mg/mL). Samples were prepared at Mount Sinai and distributed to the other three laboratories participating in the interlaboratory reproducibility study. Seminal fluid was allowed to liquefy at room temperature for 1 h, after collection. Seminal fluid was aliquoted in 1 mL portions and centrifuged at 13 000 rpm for 15 min at room temperature three times, to separate plasma from cells and cellular debris. The supernatant seminal plasma was then frozen at −80°C until use. Seven microliters of seminal plasma were diluted 2-fold with 50 mM ammonium bicarbonate and subjected to trypsin digestion without prior purification or removal of high-abundance proteins. Proteins were denatured with 0.1% Rapigest (Waters) at 60 °C, and the disulfide bonds were reduced with 50 mM dithiothreitol. Following reduction, the samples were alkylated with 100 mM iodoacetamide. Samples were then trypsin-digested overnight at 37 °C. Rapigest was cleaved with 1% trifluoroacetic acid, and all samples were centrifuged at 4000 rpm for 20 min and transferred to 96-well plates.
Analysis
Development of SRM Assays, General Platform
SRM assays were developed and run as previously published9. Briefly, assays were developed on either a TSQ Quantum (Johns Hopkins and George Mason University laboratories) or TSQ Vantage Ultra (Mount Sinai and BRIMS laboratories) triple quadrupole mass spectrometers, Surveyor or Accela MS pump, Micro or Accela Autosampler and an IonMax Source equipped with a low flow metal needle (Thermo Fisher Scientific), flow rate 160–200 µL/min. Reverse-phase separations were carried out on a 1 mm × 150 mm Hypersil Gold 3 µL C18 particle. Solvent A was LC-MS grade water with 0.2% (v/v) formic acid, and solvent B was LC-MS grade acetonitrile with 0.2% (v/v) formic acid (Optima grade reagents, Thermo Fisher Scientific). The HPLC was plumbed using 1/32 red peek tubing. The instrument divert valve was switched to waste before and after the peptides eluted in order to keep the source free of excess salts and debris. Pinpoint software (version 1.1, Thermo Fisher Scientific) was used for developing SRM assays. The algorithm facilitates selection of proteotypic peptides based on the identification data and prediction of optimal fragment ions for SRM assay design, iterative method development, automatic peptide identity confirmation and quantitative data processing. For verification, ratios of transitions monitored should be within 15% of the reference ratios (internal standard peptide); otherwise, the sample is reported as not containing the peptide (signal below 1e2) or having interferences. Peptide sequences were checked for uniqueness for each protein of interest against the Human IPI database (version 3.87, Sept 2011). Pinpoint software is available for download at www.thermo.com/pinpoint. Peptide sequences and transitions for all assays are given in Supplemental Table 1. Polytyrosine-1,3,6 calibrant was obtained from CS Bio Company, product number CS0272S. Glucagon was obtained from Sigma Chemical Co, product number G2044-25MG. The mass spectrometers were operated in unit resolution with Q1 and Q3 set to 0.7 fwhm. The instrument operating software was Xcalibur 2.0.7 and TSQ 2.2.0. Nonscheduled methods were run with a cycle time of 0.8 s. Scheduled methods were run with a cycle time of 1 s. Cycle times were optimized to ensure a minimum of 12 scans across each peak. All samples were analyzed in quadruplicate.
Light and Heavy Labeled Peptides
Light and isotopically heavy labeled versions (incorporating 13C- and 15N-labeled arginine or lysine) of each target peptide were synthesized (Thermo Fisher Scientific). Heavy peptides had identical sequences to the light peptides, but the C-terminal lysine or arginine was fully labeled (>98.5%) with 13C and 15N. The high purity (>97%) of these peptides enabled confident characterization of their ionization, elution, and fragmentation and therefore, facilitated optimization of the SRM assay.
Development of SRM Assay: PSA, Glycosylated Peptide, in Serum
The enrichment protocol for this assay specifically enriches for glycopeptides16. We targeted the formerly N-linked glycosylated peptide from PSA, DKSVILLGR. The enrichment protocol captures the glycosylated peptides with subsequent cleavage of the glycan residue resulting in the peptide with sequence DKSVILLGR. Using the synthetic heavy labeled peptide, we specifically monitored all b and y fragments and chose the two most intense ions as described previously9. Next, collision energy optimization was performed for each of the two transitions. We then used the same transitions for endogenous peptide (light peptide) measurement, completing the development of the final assay for PSA measurement.
Development of SRM Assay: PSA, Tryptic Peptides, in Serum
We developed a technique for the rapid creation of a sensitive SRM assay starting with the recombinant form of any targeted protein. In this case, we started with recombinant proteins (Lee Biosolutions) for each of the two different isoforms of PSA, isoforms 3 and 4.24,25 In the first step, we identified transitions for all b and y fragments from all possible tryptic peptides arising from digestion of the recombinants. A total of 11 peptides and 501 transitions (including transitions for both for +2 and +3 precursors if an internal histidine was present) were predicted for both isoforms. This complete set was divided into two sets in order to allow optimal scheduling, and MS data were acquired from tryptic digests of the recombinant proteins. After automated and iterative refinement, the five optimal peptides and eight optimal transitions for each recombinant protein isoform were chosen.
The second step was collision energy optimization. Traditionally, this step is done by manually infusing the synthetic form of the peptide, a costly, time-consuming, and labor-intensive process. We optimized this step by creating multiple virtual transitions for each transition. For example, one of the transitions identified in Step 1 was 379.250 (Q1) → 571.392 (Q3) for the peptide SVILLGR y51+ precursor +2). We create 12 virtual transitions at 12 different collision energies by modifying the product mz values by 0.02 amu.
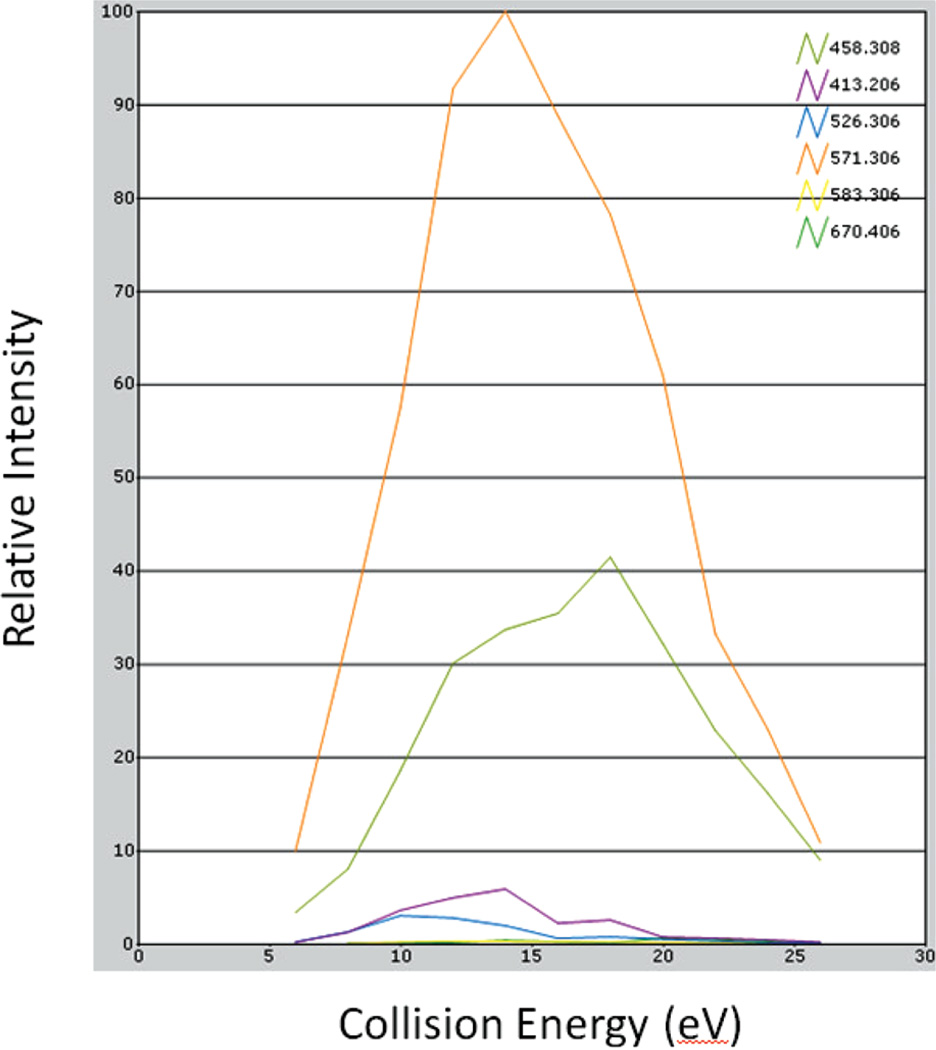
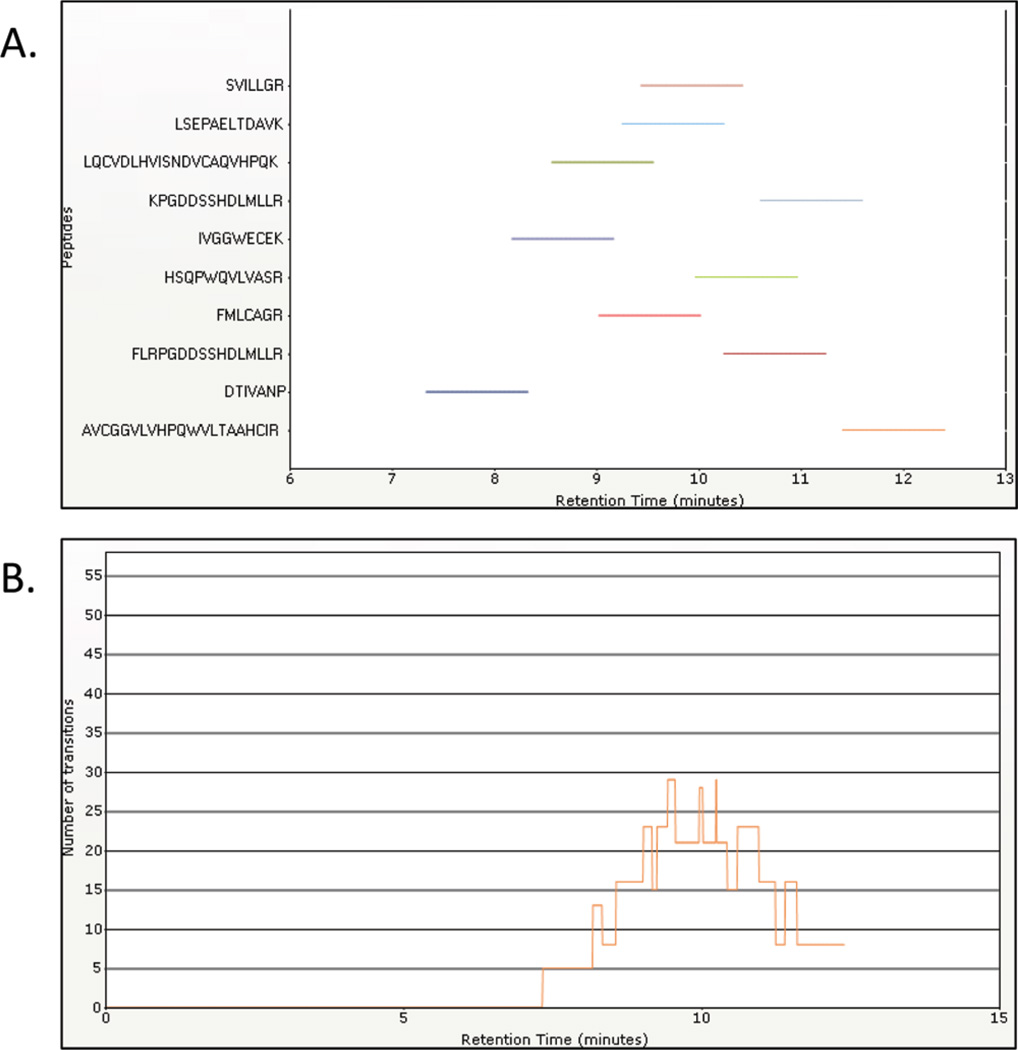
Since the Q3 resolution was set to 1 amu, all the virtual transitions monitored the same y51+ transition because they differed by less than 0.03 amu (the difference between 571.306 and 571.326). This is done because the instrument method is not able to take the exact same transition (Q1/Q3 values) more than once. However, the channel intensity was set at different collision energies. The sum of the area under the peak for each of the virtual transitions is equivalent to the signal of the corresponding collision energy for that transition. If the summed areas are plotted versus all the collision energies, the traditional breakdown curve (Figure 2) is obtained. In Figure 2, the orange curve shows the breakdown of y51+ at different collision energy values. This curve allows the identification of the optimal collision energy value for each transition of the peptide SVILLGR. The collision energy values for each of the eight transitions for the other nine peptides were optimized in the same chromatographic run in this manner. Thus, using a recombinant protein and this method allowed for optimization of collision energy values for 10 different peptides in a single LC run. This methodology has also been used by other research groups26. The observed retention times permitted the generation of a scheduled method. Figure 3 shows the benefit of scheduling: (a) shows the various start and stop times for the various peptides that were monitored, and (b) shows the number of transitions that were being acquired at any given time. For the 10 targeted peptides, there are a total of 73 transitions. However, with scheduling a maximum of only 29 transitions are monitored at any given time, thus providing a 2.5 fold increase in dwell times. The assay was then further optimized by selecting the best four transitions for each peptide.
Figure 2.
Breakdown curve plotted for six transitions of peptide SVILLGR. The response of a transition at a particular collision energy value is obtained by calculating the area under the peak of the SRM at that CE.
Figure 3.
Scheduled SRM assay for 10 peptides from PSA. (A) The start and stop times for the various peptides that were monitored, and (B) the number of transitions that were being acquired at any given time during the acquisition.
In the next step, the same transitions were monitored by spiking the recombinant protein into a background of serum matrix. Transition ratios that exhibited greater than 15% variation compared to the recombinant only experiment were removed. Last, we selected transitions from two peptides from each of the protein isoforms for the final assay.
Development of SRM Assay: hGH in Urine
In order to develop a sensitive and specific assay for hGH, we used the same protocol as described above for PSA. Starting with recombinant hGH (Humatrope Somatropin, Eli Lilly, Inc.), we evaluated all 12 possible tryptic peptides and their associated 325 transitions (including b and y fragments, and +3 precursors if an internal histidine was present). This allowed the selection of the best seven peptides (the other five were not observed) and the top eight transitions for each. Subsequently, on-column collision energy optimization was performed (as described above) for each of the transitions and the top four transitions for each peptide were selected. The assay was performed in a background of urine matrix and less-than-optimal transitions were removed.
Development of SRM Assay: Proteins in Seminal Plasma
SRM assays were developed using LTQ-Orbitrap discovery data as described previously27,28 and then optimized using synthetic peptides18. Since the targeted proteins were highly abundant in prevasectomy seminal plasma, no additional upfront enrichment was used to generate SRM assay and analyze the targeted peptides in the trypsin-digested seminal plasma.
RESULTS
SRM Assays for N-Linked Glycosylated Peptide of PSA Isolation from Serum
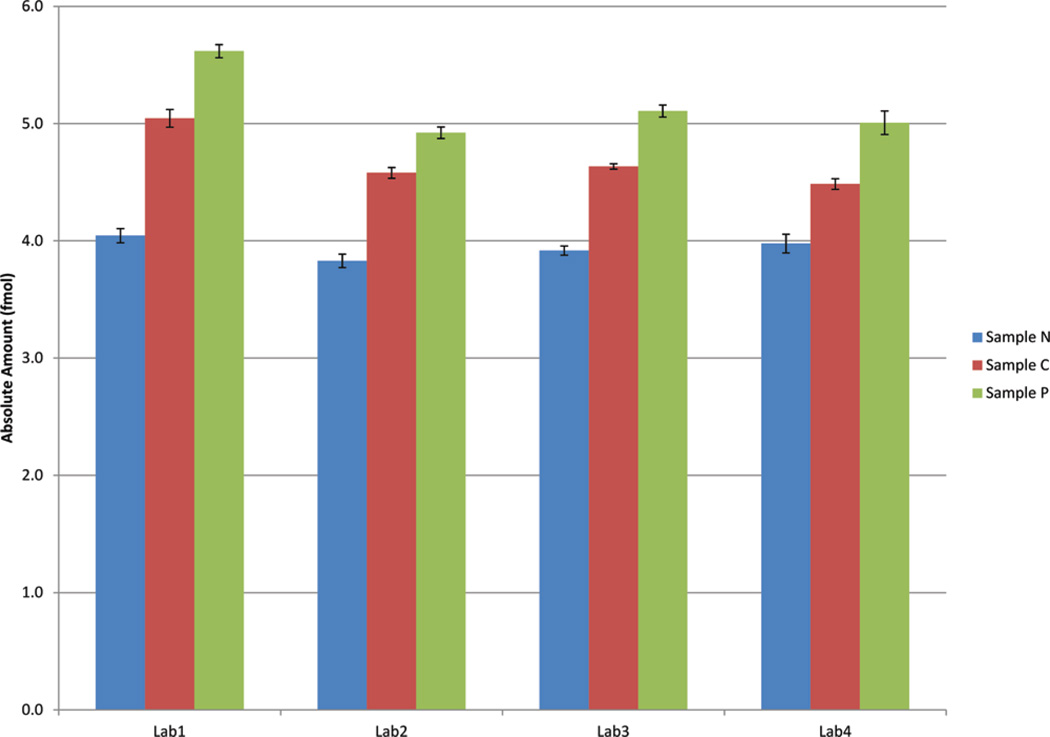
Transitions for peptide DKSVILLGR and C-13 and N-15 labeled DKSVILLGR were monitored for the three samples N, C, and P as described in the Materials and Methods. Each sample was analyzed in each of the four laboratories in quadruplicate. Relative amounts of the glycopeptide16 in the cancer serum and control serum spiked with PSA were calculated using the heavy-isotope-labeled peptide as internal standard. All the laboratories detected PSA in prostate cancer serum (sample C) and control serum spiked with PSA protein (sample P). The PSA in healthy men were below detectable range. Table 1 and Figure 4 shows relative amounts in samples N, C, and P. Sample C (from prostate cancer patients) had higher levels of PSA than level from sample N (from healthy men). Sample P had higher PSA levels than sample C, also consistent with expectations, as a much higher level of recombinant PSA was spiked in sample P (51.4 ng/mL PSA) comparing to sample C (44.5 ng/mL PSA). The interlab CVs for sample C and sample P were calculated at 28% and 21%, respectively.
Table 1.
Absolute Amounts (fmol), and Intra- and InterLab CVs for the Various Assays and Proteins Calculated from Data Acquired at Four Laboratories
| Lab 1 |
Lab 2 |
Lab 3 |
Lab 4 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| amount (fmol) |
CV% | amount (fmol) |
CV% | amount (fmol) |
CV% | amount (fmol) |
CV% | interlab CV% |
||
| PSA isoform 3 - clinical serum | SVILLGR | 217.5 | 8 | 262.7 | 13 | 199.1 | 20 | 204.5 | 11 | 13 |
| PSA isoform 4 - clinical serum | DTIVANP | 140.5 | 4 | 165.2 | 7 | 129.7 | 16 | 106.0 | 26 | 18 |
| hGH - clinical urine | FPTIPLSR | 3.8 | 19 | 4.1 | 10 | 2.9 | 10 | 2.0 | 7 | 30 |
| LDHC - normal seminal plasma | EELFLSIPC[Carboxyamidomethyl] VLGR | 206.9 | 10 | 201.7 | 7 | 233.0 | 16 | 245.6 | 22 | 9 |
| LDHC - OA seminal plasma | EELFLSIPC[Carboxyamidomethyl] VLGR | below LOD | below LOD | below LOD | below LOD | N/A | ||||
| PTGDS - normal seminal plasma | AQGFTEDTIVFLPQTDK | 2257.2 | 8 | 2943.0 | 10 | 3156.0 | 16 | 2878.5 | 22 | 14 |
| PTGDS - OA seminal plasma | AQGFTEDTIVFLPQTDK | below LOD | below LOD | below LOD | below LOD | N/A | ||||
| SPAG11B - normal seminal plasma | IC[Carboxyamidomethyl]VDFLGPR | 28.3 | 14 | 30.2 | 9 | 29.3 | 15 | 26.8 | 12 | 5 |
| SPAG11B - OA seminal plasma | IC[Carboxyamidomethyl]VDFLGPR | below LOD | below LOD | below LOD | below LOD | N/A | ||||
| FAM12B - normal seminal plasma | NAYVWVQ NPLK | 213.0 | 15 | 249.6 | 14 | 212.4 | 22 | 185.2 | 11 | 12 |
| FAM12B - OA seminal plasma | NAYVWVQ NPLK | below LOD | below LOD | below LOD | below LOD | N/A | ||||
| PSA - N sample | DKSVILLGR | below LOD | below LOD | below LOD | below LOD | N/A | ||||
| PSA - C sample | DKSVILLGR | 1.0 | 3 | 0.8 | 2 | 0.7 | 1 | 0.5 | 2 | 28 |
| PSA - P sample | DKSVILLGR | 1.6 | 2 | 1.1 | 2 | 1.2 | 2 | 1.0 | 4 | 21 |
Figure 4.
Amounts of DKSVILLGR (representing PSA) calculated by the four laboratories in samples N, C, and P.
Absolute Amounts in Serum Sample for MSIA Enrichment of Prostate Specific Antigen
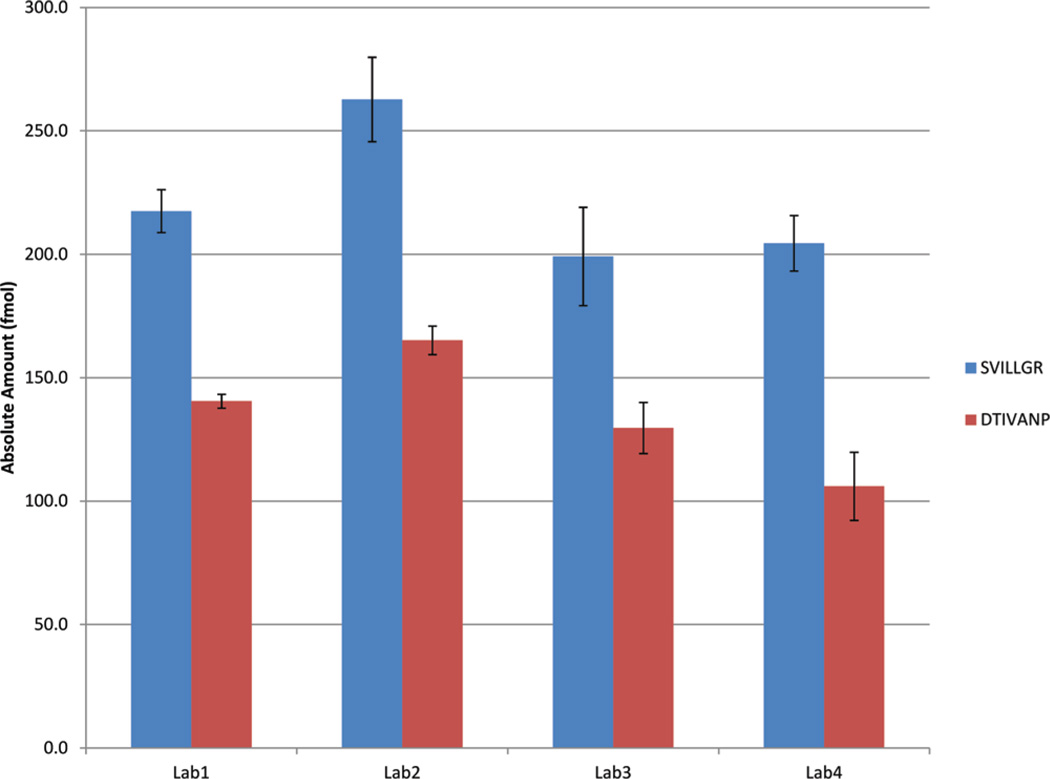
We monitored two peptides specific to the two PSA isoforms, SVILLGR corresponding to isoform 3 and DTIVANP corresponding to isoform 4. One MSIA-enriched serum sample was analyzed in each of the four laboratories in quadruplicate, and the absolute amounts of these peptides were calculated using the known amount of the internal standard (SVILLGR and DTIVANP). Figure 5 and Table 1 show the calculated amounts from each of the four laboratories, with isoform 3 peptide unambiguously reported as more abundant compared to isoform 4. The interlab CVs were calculated at 13% and 18% for isoforms 3 and 4, respectively.
Figure 5.
Amounts of the two isoforms of PSA calculated by the four laboratories in the MSIA-enriched clinical sample. SVILLGR and DTIVANP uniquely measure isoform 3 and isoform 4, respectively.
Absolute Amounts in Urine Samples for Nanoparticle Capture of Pituitary Human Growth Hormone
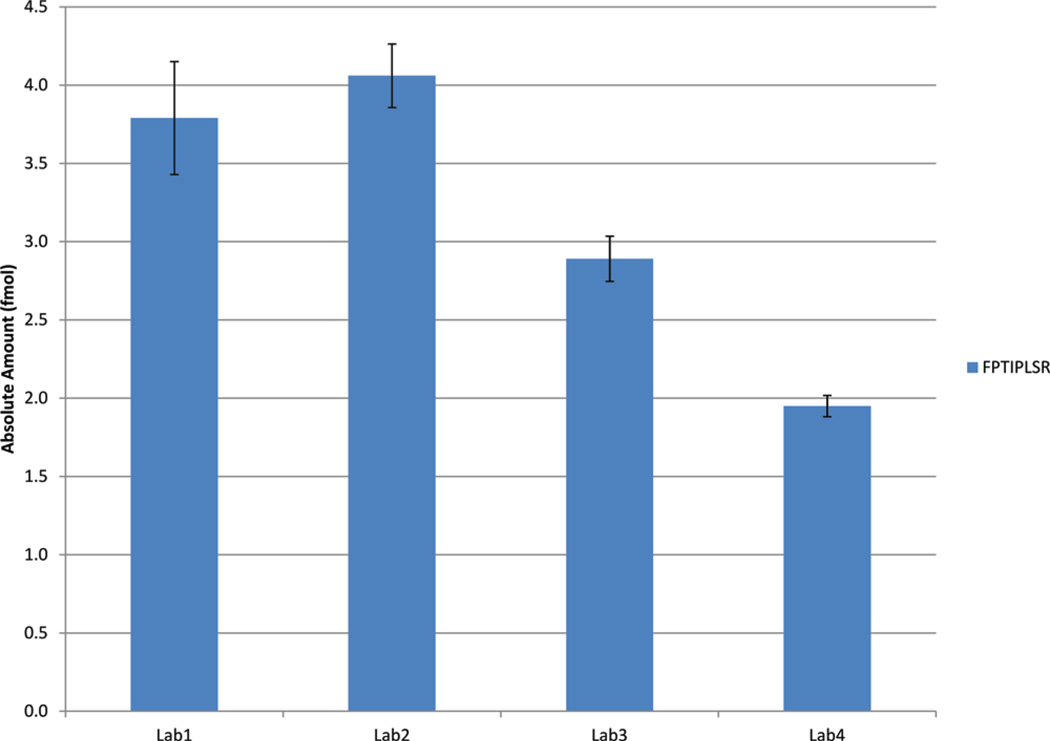
In order to evaluate the interlaboratory performance of SRM-based measurements of hGH in a real clinical matrix such as urine, we established the LLOD of measurement of hGH within one (Petricoin/Liotta) laboratory by exploring a urine sample study set with a concentration range (0.5–50 ng/mL) of hGH that spans known concentrations of the analyte in clinical samples.17,21–23 Using this study set, urine samples were processed for SRM analysis either with or without nanoparticle capture and elution of hGH. As shown in Figure 6, nanoparticle capture greatly enhances the analytical sensitivity of a resultant SRM assay, where the LLOD was at least 500 pg/mL with nanoparticle enrichment and only 5 ng/mL without nanoparticle enrichment. Upon the basis of this result, a preparation of a single sample containing 4 mL of urine with 10 ng/mL of hGH was prepared and subjected to nanoparticle processing, and the eluate was distributed among the participating laboratories such that each laboratory analyzed an equivalent of a 1 mL urine sample. Transitions for peptide FPTIPLSR and heavy labeled FPTIPLSR were monitored in one clinical urine sample in quadruplicate at each of the four laboratories. The absolute amount of this peptide was calculated using the known amount of the internal standard (300 fmol = 10 ng/mL). Figure 7 and Table 1 show the calculated amounts and CVs from each of the four laboratories. The calculated interlab CV was 30% for a 1 mL urine sample containing 10 ng/mL.
Figure 6.
SRM peak AUC values of peptide 46S2+ from pituitary hGH spiked into urine with and without nanoparticle enrichment. A concentration curve was obtained by serial dilution into a normal urine sample and subjected to nanoparticle capture, or untreated. A magnified view of the lower concentration range is shown in the inset graph.
Figure 7.
Amounts of FPTIPLSR (peptide representing hGH) calculated by the four laboratories in the nanoparticle-enriched clinical urine sample.
Absolute Amounts in Samples for Measurement of Seminal Plasma Proteins
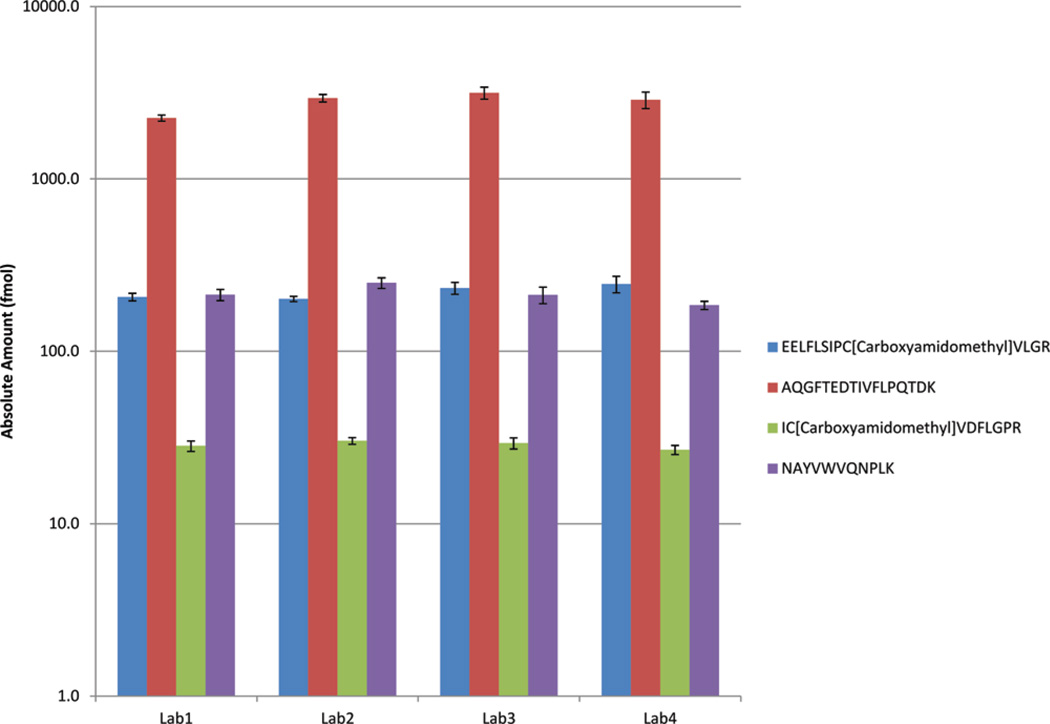
Transitions for four peptides corresponding to four plasma proteins (LDHC, PTGDS, SPAG11B, and FAM12B) were monitored in one prevasectomy seminal plasma sample and one postvasectomy seminal plasma sample. The peptides and the transitions that were monitored are given in Supplementary Table 1, Supporting Information; all samples were analyzed in quadruplicate at all four laboratories. Absolute amounts of the four peptides were calculated using the known amount of the corresponding internal standard peptides. None of the laboratories detected the peptides in the postvasectomy sample (Table 1). This was an expected result since these proteins are primarily produced by the testis and will not be found in postvasectomy seminal plasma due to ligation of vas deferens in vasectomized men. The absolute amount calculated in the prevasectomy samples is shown in Figure 8 and Table 1, demonstrating consistent trends for the four peptides across the four laboratories. The interlab CVs were calculated at 9%, 14%, 5%, and 12% respectively, for the four peptides.
Figure 8.
Amounts of four peptides (corresponding to four plasma proteins LDHC, PTGDS, SPAG11B, and FAM12B) calculated in the four laboratories in one seminal plasma sample.
DISCUSSION
Immunoassays are currently the gold standard for the measurement of protein analytes in clinical samples. However, immunoassays are subject to variability and may give different results in different laboratories.29,30 Mass spectrometry-based assays are poised to revolutionize the world of clinical diagnostics for proteins.4,31,32 As opposed to traditional protein immunoassays, mass spectrometry-based assays deliver required sequence specificity to discriminate and measure protein isoforms and are cost-effective because they can be multiplexed. Over the past 10 years, the steady trickle of evidence that protein isoforms (including sequence variants, post-translationally modified forms and truncations) are important and key to diseases and pathologies has recently increased to a flood.5,32–35 The accurate and quantitative measurement of protein isoforms will be a crucial part of the new age of “personalized medicine”36–40 In particular, SRM-based mass spectrometry assays have recently come to the forefront as a potentially preferred method for clinical assays because they have the key attributes of speed, throughput, excellent quantification, and multiplexing capabilities. ‘ A potential caveat is the measurement of low-abundance analytes because enrichment or depletion techniques may add an unacceptable degree of irreproducibility to the measurements.43–46
This study was designed to test several different, previously published15–18 sample preparation techniques, focused especially on analyte concentration methodology, coupled to a standardized platform for SRM-MS assays9 across different laboratories. We demonstrated the rapid development of sensitive and specific SRM assays and related SOPs for proteins derived from several sample types using four different sample preparation methods, three employing enrichment and one with no enrichment. Preparing the samples in each laboratory and distributing to the other three laboratories for analysis on the triple quads was an intermediate step to completing the entire workflow in each laboratory. However, the reproducible results obtained in this study demonstrate that various sample preparation and assay workflows for quantitative peptide measurement can be reproducible at multiple sites. Looking at Table 1, we can also observe a strong relationship between CV and absolute amount (same as peak area). This is expected, as the lower the amount of the analyte, the higher will be the stochastic variation resulting in a higher CV. The highest interlab CV observed in our study was 30% for the hGH peptide, which also had the lowest peak area.
In conclusion, this study helps to confirm the feasibility of using SRM-based assays for low-abundance protein analyte measurement in clinical specimens.
Supplementary Material
ABBREVIATIONS
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- SPE
solid phase extraction
- ESI
electrospray ionization
- ACN
acetonitrile
- m/z
mass to charge ratio
- SRM
selective reaction monitoring
- apo
apolipoprotein
- LOD
limit of detection
- LOQ
limit of quantitation
- MSIA
mass spectrometric immunoassay
- SOP
standard operating procedure
- PTH
parathyroid hormone
- PSA
prostate specific antigen
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- TFA
trifluoroacetic acid
- OA
obstructive asoospermia
- HPLC
high performance liquid chromatography
- hGH
human growth hormone
- CV
coefficient of variation
Footnotes
ASSOCIATED CONTENT
Supporting Information
Table of the peptides and the transitions that were monitored. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin. Chem. 2010;56(2):177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. Immunol. Methods. 2009;347:3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agger SA, Marney LC, Hoofnagle AN. Simultaneous quantification of apolipoprotein A-I and apolipoprotein B by liquid-chromatography- multiple-reaction-monitoring mass spectrometry. Clin. Chem. 2010;56(12):1804–1813. doi: 10.1373/clinchem.2010.152264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Q, Schoenhoff FS, Savage WJ, Zhang P, Van Eyk JE. Multiplex assays for biomarker research and clinical application: translational science coming of age. Proteomics Clin. Appl. 2010;4(3):271–284. doi: 10.1002/prca.200900217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borges CR, Render DS, Jarvis JW, Schaab MR, Oran PE, Nelson RW. Full-length characterization of proteins in human populations. Clin. Chem. 2010;6:202–211. doi: 10.1373/clinchem.2009.134858. [DOI] [PubMed] [Google Scholar]
- 6.Miller WL, Phelps MA, Wood CM, Schellenberger U, Van Le A, Perichon R, Jaffe AS. Comparison of mass spectrometry and clinical assay measurements of circulating fragments of B-type natriuretic peptide in patients with chronic heart failure. Circ: Heart Failure. 2011;4(3):355–360. doi: 10.1161/CIRCHEARTFAILURE.110.960260. [DOI] [PubMed] [Google Scholar]
- 7.Vogeser M, Seger C. Pitfalls associated with the use of liquid chromatography-tandem mass spectrometry in the clinical laboratory. Clin. Chem. 2010;56(8):1234–1244. doi: 10.1373/clinchem.2009.138602. [DOI] [PubMed] [Google Scholar]
- 8.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DG, Paulovich AG, Regnier FE, Tempst P, Carr S. A Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 2009;27(7):633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prakash A, Rezai T, Krastins B, Sarracino D, Athanas M, Russo P, Ross MM, Zhang H, Tian Y, Kulasingam V, Drabovich AP, Smith G, Batruch I, Liotta L, Petricoin E, Diamandis EP, Chan DW, Lopez MF. A platform for establishing inter-laboratory reproducibility of selected reaction monitoring-based mass spectrometry peptide assays. J. Proteome Res. 2010;9:6678–6688. doi: 10.1021/pr100821m. [DOI] [PubMed] [Google Scholar]
- 10.Nelson RW, Krone JR, Bieber AL, Williams P. Mass spectrometric immunoassay. Anal. Chem. 1995;67(7):1153–1158. doi: 10.1021/ac00103a003. [DOI] [PubMed] [Google Scholar]
- 11.Ackermann BL, Berna MJ. Coupling immunoaffinity techniques with MS for quantitative analysis of low-abundance protein biomarkers. Expert Rev. Proteomics. 2007;4:175–186. doi: 10.1586/14789450.4.2.175. [DOI] [PubMed] [Google Scholar]
- 12.Makawita S, Diamandis EP. The bottleneck in the cancer biomarker pipeline and protein quantification through mass spectrometry-based approaches: current strategies for candidate verification. Clin. Chem. 2010;56(2):212–222. doi: 10.1373/clinchem.2009.127019. [DOI] [PubMed] [Google Scholar]
- 13.Brewis IA, Brennan P. Proteomics technologies for the global identification and quantification of proteins. Adv. Protein Chem. Struct. Biol. 2010;80:1–44. doi: 10.1016/B978-0-12-381264-3.00001-1. [DOI] [PubMed] [Google Scholar]
- 14.Ly L, Wasinger VC. Protein and peptide fractionation, enrichment and depletion: tools for the complex proteome. Proteomics. 2011;11(4):513–534. doi: 10.1002/pmic.201000394. [DOI] [PubMed] [Google Scholar]
- 15.Lopez MF, Rezai T, Sarracino DA, Prakash A, Krastins B, Athanas M, Singh RJ, Barnidge DR, Oran P, Borges G, Nelson RW. Selected reaction monitoring-mass spectrometric immunoassay responsive to parathyroid hormone and related variants. Clin. Chem. 2010;56:281–290. doi: 10.1373/clinchem.2009.137323. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Tian Y, Rezai T, Prakash A, Lopez MF, Chan DW, Zhang H. Simultaneous analysis of glycosylated and sialylated prostate-specific antigen revealing differential distribution of glycosylated prostate-specific antigen isoforms in prostate cancer tissues. Anal. Chem. 2011;83:240–245. doi: 10.1021/ac102319g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredolini G, Tamburro D, Gambara G, Lepene BS, Espina V, Petricoin EF, 3rd, Liotta LA, Luchini A. Nanoparticle technology: amplifying the effective sensitivity of biomarker detection to create a urine test for hGH. Drug Test Anal. 2009;(No. 9–10):447–454. doi: 10.1002/dta.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drabovich AP, Jarvi K, Diamandis EP. Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.004127. No. M110.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu AY, Zhang H, Sorensen GM, Diamond DL. Analysis of prostate cancer by proteomics using tissue specimens. Urol. 2005;173:73–78. doi: 10.1097/01.ju.0000146543.33543.a3. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Liu AY, Loriaux P, Wollscheid B, Zhou Y, Watts JD, Aebersold R. Mass spectrometric detection of tissue proteins in plasma. Mol. Cell. Proteomics. 2007;6:64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Fredolini C, Meani F, Reeder KA, Rucker S, Paanarut A, Botterell PJ, Bishop B, Longo C, Espina V, Petricoin EF, Liotta LA, Luchini A. Concentration and preservation of very low abundance biomarkers in urine, such as human growth hormone (hGH), by Cibacron Blue F3G-A loaded hydrogel particles. Nana Res. 2008;1:502–518. doi: 10.1007/s12274-008-8054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luchini A, Geho DH, Bishop B, Tran D, Xia C, Dufour RL, Jones GD, Espina V, Patanarut A, Zhou W, Ross MM, Tessitore A, Petricoin EF, Liotta LA. Smart hydrogel particles: biomarker harvesting: one-step affinity purification, size exclusion, and protection against degradation. Nana Lett. 2008;8(1):350–361. doi: 10.1021/nl072174l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russo P, Rezai T, Prakash A, Lopez MF, Tamburro D, Fredolini C, Luchini A, Liotta LA, Petricoin EF. Human growth hormone quantititation in urine by isoform ratio analysis using nanoparticle capture/multiple reaction monitoring-mass spectrometry. ASMS. 2011 [Google Scholar]
- 24.Kote-Jarai Z, Amin Al, Olama A, Leongamornlert D, Tymrakiewicz M, Saunders E, Guy M, Giles GG, Severi G, Southey M, Hopper JL, Sit KG, Harris JM, Batra J, Spurdle AB, Clements JA, Hamdy F, Neal D, Donovan J, Muir K, Pharoah PD, Chanock SJ, Brown N, Benlloch S, Castro E, Mahmud N, Brien L, Hall A, Sawyer E, Wilkinson R, Easton DF, Eeles RA. Identification of a novel prostate cancer susceptibility variant in the KLK3 gene transcript. Hum. Genet. 2011;(No. 6):687–694. doi: 10.1007/s00439-011-0981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penney KL, Schumacher FR, Kraft P, Mucci LA, Sesso HD, Ma J, Niu Y, Cheong JK, Hunter DJ, Stampfer MJ, Hsu SI. Association of KLK3 (PSA) genetic variants with prostate cancer risk and PSA levels. Carcinogenesis. 2011;32(6):853–859. doi: 10.1093/carcin/bgr050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherwood GA, Eastham A, Lee LW, Risler J, Mirzaei H, Falkner JA, Martin DB. Rapid optimization of MRM-MS instrument parameters by subtle alteration of precursor and product m/z targets. J. Proteome Res. 2009;8(7):3746–3751. doi: 10.1021/pr801122b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batruch I, Lecker I, Kagedan D, Smith GR, Mullen BJ, Grober E, Lo KG, Diamandis EP, Jarvi KA. Proteomic analysis of seminal plasma from normal volunteers and post-vasectomy patients identifies over 2000 proteins and candidate biomarkers of the urogenital system. J. Proteome Res. 2011;10(3):941–953. doi: 10.1021/pr100745u. [DOI] [PubMed] [Google Scholar]
- 28.Drabovich AP, Diamandis EP. Combinatorial peptide libraries facilitate development of multiple reaction monitoring assays for low-abundance proteins. J. Proteome Res. 2010;9(3):1236–1245. doi: 10.1021/pr900729g. [DOI] [PubMed] [Google Scholar]
- 29.Sturgeon GM, Sprague SM, Metcalfe W. Variation in parathyroid hormone immunoassay results—a critical governance issue in the management of chronic kidney disease. Nephrol. Dial. Transplant. 2011;26:3440–3445. doi: 10.1093/ndt/gfr614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sturgeon GM, Seth J. Why do immunoassays for tumour markers give differing results? A view from the UK National External Quality Assessment Schemes. Eur. J. Clin. Chem. Clin. Biochem. 1996;34:755–759. [PubMed] [Google Scholar]
- 31.Hoofnagle AN, Becker JO, Oda MN, Cavigiolio G, Mayer P, Vaisar T. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clin Chem. 2012;58(4):777–781. doi: 10.1373/clinchem.2011.173856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez MF, Kuppusamy R, Sarracino DA, Prakash A, Athanas M, Krastins B, Rezai T, Sutton JN, Peterman S, Nicolaides K. Mass spectrometric discovery and selective reaction monitoring (SRM) of putative protein biomarker candidates in first trimester trisomy 21 maternal serum. J. Proteome Res. 2011;10(1):133–142. doi: 10.1021/pr100153j. [DOI] [PubMed] [Google Scholar]
- 33.Boja ES, Rodriguez H. The path to clinical proteomics research: integration of proteomics, genomics, clinical laboratory and regulatory science. Korean J. Lab. Med. 2011;(2):61–71. doi: 10.3343/kjlm.2011.31.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Eyk JE. Overview: the maturing of proteomics in cardiovascular research. Circ. Res. 2011;108(4):490–498. doi: 10.1161/CIRCRESAHA.110.226894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wells L, Hart GW. O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 2003;546:154–158. doi: 10.1016/s0014-5793(03)00641-0. [DOI] [PubMed] [Google Scholar]
- 36.Shen F, Kirmani KZ, Xiao Z, Thirlby BH, Hickey RJ, Malkas LH. Nuclear protein isoforms: implications for cancer diagnosis and therapy. J Cell. Biochem. 2011;112(3):756–760. doi: 10.1002/jcb.23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emami N, Diamandis EP. Human tissue kallikreins: a road under construction. Clin. Chim. Acta. 2007;381(1):78–84. doi: 10.1016/j.cca.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 38.Niesler B. 5-HT(3) receptors: potential of individual isoforms for personalised therapy. Curr. Opin. Pharmacol. 2011;11(1):81–86. doi: 10.1016/j.coph.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Hein DW. N-acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opin. Drug Metab. Toxicol. 2009;5(4):353–366. doi: 10.1517/17425250902877698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danser AH, Batenburg WW, van den Meiracker AH, Danilov SM. ACE phenotyping as a first step toward personalized medicine for ACE inhibitors. Why does ACE genotyping not predict the therapeutic efficacy of ACE inhibition? Pharmacol. Ther. 2007;113(3):607–618. doi: 10.1016/j.pharmthera.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Black JL, 3rd, Kane DJ, Mrazek DA. The impact of CYP allelic variation on antidepressant metabolism: a review. Expert Opin. Drug Metab. Toxicol. 2007;3(1):21–31. doi: 10.1517/17425255.3.1.21. [DOI] [PubMed] [Google Scholar]
- 42.Reymond MA, Steinert R, Kahne T, Sagynaliev E, Allal AS, Lippert H. Expression and functional proteomics studies in colorectal cancer. Pathol. Res. Pract. 2004;200(2):119–127. doi: 10.1016/j.prp.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Calvo E, Camafeita E, Fernandez-Gutierrez B, Lopez JA. Applying selected reaction monitoring to targeted proteomics. Expert Rev. Proteomics. 2011;8(2):165–173. doi: 10.1586/epr.11.11. [DOI] [PubMed] [Google Scholar]
- 44.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods. 2010;7(1):43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 45.Pernemalm M, Lewensohn R, Lehtio J. Affinity prefractionation for MS-based plasma proteomics. Proteomics. 2009;9(6):1420–1427. doi: 10.1002/pmic.200800377. [DOI] [PubMed] [Google Scholar]
- 46.Becker CH, Bern M. Recent developments in quantitative proteomics. Mutat. Res. 2011;722(2):171–182. doi: 10.1016/j.mrgentox.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.